Abstract
Background
Although the prevalence of the carbapenem-resistant A. baumannii (CRAB) has increased in Taiwan, few studies have elucidated the prevalence of different carbapenemase genes in Taiwan. The first objective of this study was to identify the types and prevalence of different carbapenemase genes, and the second objective was to determine the carbapenem antimicrobial susceptibility of carbapenemase producing isolates.
Methods
In total, 269 CRAB blood isolates from four medical centres in Taiwan from 1/1/2009 to 31/12/2013 were analysed. Antimicrobial susceptibilities were determined using the Vitek 2 system. Carbapenemase genes were identified by polymerase chain reaction (PCR) and sequencing. Pulsed-field gel electrophoresis (PFGE) was used to identify the different pulsotypes.
Results
All 269 CRAB isolates had the blaOXA-51-like gene, while 237 (88.1%) had the blaOXA-23-like gene, and 11 (4.09%) had the blaOXA-24-like gene. Twenty-one CRAB isolates (7.81%) contained only the blaOXA-51-like gene. None of the isolates had the blaOXA-58-like gene or the metallo-β-lactamases (MBL)-encoding genes. In 28.69% of isolates with the blaOXA-23-like gene and 90.91% of isolates with the blaOXA-24-like gene, the minimum inhibitory concentrations (MICs) for imipenem were 64 mg/L or more. In 37.55% of isolates with the blaOXA-23-like gene and 100% of isolates with the blaOXA-24-like gene, meropenem MICs were 64 mg/L or more. PFGE analyses indicated that six highly similar genomes which harbored the blaOXA-24-like gene came from three different medical centres.
Conclusion
Our study determined the prevalence of CRAB, the types and prevalence of carbapenemase genes, carbapenem susceptibility among CRAB isolates, and documented that the blaOXA-24-like gene had greater resistance to carbapenem than the blaOXA-23-like gene. We also demonstrated inter-hospital transmission of the highly resistant bla OXA-24-like gene.
Keywords: Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex, Carbapenem-resistant Acinetobacter baumannii (CRAB), blaOXA-23-like, blaOXA-24-like, blaOXA-51-like
Background
Acinetobacter is a Gram-negative bacillus that is common in water and soil. Acinetobacter species are recognized as opportunistic pathogens of increasing relevance in healthcare-associated infections, and are particularly likely to cause opportunistic infections in intensive care units (ICUs) [1]. Analysis of healthcare-associated infection data from the Taiwan Nosocomial Infections Surveillance System (TNIS) in 2007–2012 showed that A. baumannii complex (AB complex) composed by A. baumannii, A. nosocomialis and A. pittii was among the top three common causes of infections in ICUs of medical centres and regional hospitals [2].
The Acinetobacter calcoaceticus-Acinetobacter baumannii (ACB) complex is composed of A. calcoaceticus (genospecies 1), A. baumannii (genospecies 2), A. pittii (genospecies 3) and A. nosocomialis (genospecies 13TU). A. baumannii often causes bloodstream infections, and the associated mortality rate is about 43.3% in ICUs [3]. ACB complex species have similar appearances and biochemical reactions, and cannot be distinguished by traditional biochemical reactions or by automatic or semi-automatic identification machines, such as the Vitek 2 system (BioMérieux). Therefore, ACB complex species have often mistakenly been categorized as A. baumannii. Precise molecular biological techniques can now be used to identify and accurately distinguish these species. A. calcoaceticus is less likely to cause severe disease. However, A. baumannii, A. nosocomialis, and A. pittii (AB complex) can cause healthcare-associated infections in ICUs. In recent years, the gradual increase of carbapenem-resistant forms of each of these species from hospital isolates has caused great concern. The resistance rate of Acinetobacter to carbapenems has exceeded 50% in the United States, South America, India, and China [4]. Data from the TNIS indicated the percentage of carbapenem-resistant Acinetobacter calcoaceticus-Acinetobacter baumannii (CRACB) complex increased from 49.0% in 2007 to 71.2% in 2012 among ICUs in medical centres in Taiwan, and increased from 49.8 to 63% in regional hospitals during the same period [2].
A. baumannii is the most common and important genospecies of Acinetobacter and the presence of carbapenemases is the commonest cause of resistance to carbapenems. The carbapenemases in the AB complex include carbapenem-hydrolyzing class D β-lactamases (CHDLs), which consist of OXA-51-like, OXA-23-like, OXA-24-like, and OXA-58-like genes, and class B metallo-β-lactamases, mainly VIM, IMP, and SIM [5]. Our study utilized molecular biological methods to analyze carbapenemase genes in 269 nosocomial carbapenem-resistant A. baumannii (CRAB) blood isolates from four medical centres in northern and central Taiwan. The first objective was to identify the types and prevalence of different carbapenemase genes, and the second objective was to determine the carbapenem antimicrobial susceptibility testing of carbapenemase producing isolates.
Methods
Study design
The Inclusion criteria were listed as follows: all the tested strains were isolated from infected patients from 4 tertiary medical centres in Taiwan, randomized to collect up to 100 clones of ACB complex with imipenem or meropenem MICs of at least 8 mg/L in each tertiary medical centres from January of 2009 to December of 2013. The exclusion criteria were that only the first episode was enrolled from patients with ≥2 positive blood cultures, the other blood isolates were excluded. We collected the ACB complex blood isolates using the Vitek 2 system, and further species identification of ACB complex was confirmed by analysis of the RNA polymerase β subunit (rpoB) gene. The prevalence of different carbapenemase genes among CRAB was performed by conventional PCR and sequencing. The imipenem and meropenem MICs were tested in all CRAB blood isolates. Additionally, tigecycline, ceftazidime, cefepime, amikacin, ampicillin/sulbactam, piperacillin/tazobactam and colistin MICs were tested for the blaOXA-24-like highly resistant gene only. Antimicrobial susceptibility was tested using the Vitek-2 system and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines except for the MICs of tigecycline which were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints for Enterobacteriaceae spp. (MIC > 2 mg/L was defined as resistant). Pulsed-field gel electrophoresis was used to analyse the high resistant blaOXA-24-like gene pulsotypes.
Each medical centre was expected to collect 100 ACB complex blood isolates. MacKay Memorial Hospital is a 2200-bed tertiary medical centre in northern Taiwan. Taipei Veterans General Hospital is a 3000-bed tertiary medical centre in northern Taiwan. Tri-Service General Hospital is an 1800-bed tertiary medical centre in northern Taiwan. Changhua Christian Hospital is an 1500-bed tertiary medical centre in central Taiwan. These four medical centres provide both primary and tertiary care, including cancer therapy, intensive care and organ transplantation. They are premier hospitals in Taiwan.
Collection of bacterial isolates
All the tested strains were isolated from infected patients. A total of 357 blood isolates of ACB complex, with imipenem or meropenem MICs of at least 8 mg/L, were collected from January of 2009 to December of 2013 from 4 tertiary medical centres in Taiwan. A total of 357 nonduplicate ACB complex blood isolates were collected from the 4 medical centres. ACB complex blood isolates were stored at − 70 °C in tryptic soy broth (BD, MD, USA) supplemented with 20% glycerol (v/v). Freezing at − 70 °C in a high percentage of glycerol were employed as cryoprotective agents. All patients were deidentified locally and re-coded. Then the blood isolates were transported to the research laboratory at MacKay Memorial hospital for further study. This study protocol (14MMHIS125) was reviewed and approved by the Institutional Review Boards of the MacKay Memorial Hospital, Taipei, Taiwan.
Species identification
The plates were incubated under aerobic conditions at 36 °C and evaluated for growth after 24 and 48 h. Suspected colonies were further cultivated on blood agar and identified at species level using the automated Vitek MS system (bioMerieux). Antimicrobial susceptibility was tested using Vitek-2 (card AST-N196 and/or N248). All isolates were stored at − 70 °C for analysis of genetic relatedness. Genospecies identification of ACB complex was confirmed by analysis of the RNA polymerase β subunit (rpoB) gene according to the protocol by La Scola et al. [6].
Antimicrobial susceptibility testing
Antimicrobial susceptibility was tested using Vitek-2 system. The imipenem and meropenem MICs were tested in all CRAB blood isolates. However, tigecycline, ceftazidime, cefepime, amikacin, ampicillin/sulbactam, piperacillin/tazobactam and colistin MICs were tested for the blaOXA-24-like highly resistant gene only. The MICs of tested antibiotics were interpreted according to the CLSI guidelines, but the MICs of tigecycline were interpreted according to the EUCAST breakpoints for Enterobacteriaceae spp. (MIC > 2 mg/L was defined as resistant). Staphylococcus aureus ATCC 29213, Escherichia coli ATCC25922, Pseudomonas aeruginosa ATCC 27853 were measured for quality control. All results were within quality control ranges.
Polymerase chain reaction and sequencing
The ACB complex blood isolates were screened for the following carbapenemase genes by use of conventional PCR and sequencing: blaIMP,blaVIM, blaNDM, blaSPM, blaGIM, blaSIM, blaKPC, blaGES, blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, blaOXA-143-like, blaOXA-235-like. The sequences of primers are shown in Table 1 [7–12]. Deionized distilled water was used as the negative control. Isolates whose sequences were previously confirmed was used as the positive control. Multiplex PCR were performed. A separate group of 2~ 10 primers (such as blaIMP, blaVIM, blaNDM, blaSPM, blaGIM, blaSIM, blaKPC, blaGES, blaOXA-23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, blaOXA-143-like and blaOXA-235-like) were used to test multiplex PCR each time. The PCR condition was one cycle of 95 °C for 10 min, followed by 40 cycles of 94 °C for 1 min, 52 °C for 1 min, 72 °C for 2 min and finally, one cycle of 72 °C for 10 min followed by cooling to 4 °C. A sample of amplified DNA was electrophoresed in an agarose gel and stained with ethidium bromide to assess the purity and size of the more multiplex PCR products. PCR products were visualized by agarose gel electrophoresis stained with ethidium bromide followed by UV light crosslinking. The PCR amplicons were purified for DNA sequencing using a ExoProStar™ reagent (GE Healthcare, UK) as per manufacturer instructions and incubated at 37 °C for 30 min, followed by incubation at 80 °C for 15 mins to inactivate the enzymes. The enzyme-treated PCR products were sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) following instructions provided by the manufacturer. The PCR products were purified using by gel filtration with Sephadex G-50 (GE Healthcare, UK) using spin columns in a MultiScreen™ Filtration System 96 well filter plate (Millipore) to remove unincorporated dye terminators. The final DNA sequence reading was performed in an ABI Prism 377 DNA sequencer analyzer (Applied Biosystems, Foster City, CA, USA). Sequence similarity searches were performed with the basic local alignment search tool (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) | Reference |
|---|---|---|
| OXA-51-like forward | TAA TGC TTT GAT CGG CCT TG | 6 |
| OXA-51-like reverse | TGG ATT GCA CTT CAT CTT GG | |
| OXA-23-like forward | GAT CGG ATT GGA GAA CCA GA | 6 |
| OXA-23-like reverse | ATT TCT GAC CGC ATT TCC AT | |
| OXA-24-like forward | GGT TAG TTG GCC CCC TTA AA | 6 |
| OXA-24-like reverse | AGT TGA GCG AAA AGG GGA TT | |
| OXA-58-like forward | AAG TAT TGG GGC TTG TGC TG | 6 |
| OXA-58-like reverse | CCC CTC TGC GCT CTA CAT AC | |
| OXA-143-F | TGGCACTTTCAGCAGTTCCT | 7 |
| OXA-143-R | TAATCTTGAGGGGGCCAACC | |
| OXA-235-F | TTGTTGCCTT TACTTAGTTGC | 8 |
| OXA-235-R | CAAAATTTTAAGACGGAT CG | |
| Imp-F | GGA ATA GAG TGG CTT AAY TCT C | 9 |
| Imp-R | CCA AAC YAC TAS GTT ATC T | |
| Vim-F | GAT GGT GTT TGG TCG CAT A | 9 |
| Vim-R | CGA ATG CGC AGC ACC AG | |
| Sim-F | TAC AAG GGA TTC GGC ATC G | 9 |
| Sim-R | TAA TGG CCT GTT CCC ATG TG | |
| Gim-F | TCG ACA CAC CTT GGT CTG AA | 9 |
| Gim-R | AAC TTC CAA CTT TGC CAT GC | |
| Spm-F | AAA ATC TGG GTA CGC AAA CG | 9 |
| Spm-R | ACA TTA TCC GCT GGA ACA GG | |
| NDM-F | GGTTTGGCGATCTGGTTTTC | 10 |
| NDM-R | CGGAATGGCTCATCACGATC | |
| KPC-Fm | CGTCTAGTTCTGCTGTCTTG | 10 |
| KPC-Rm | CTTGTCATCCTTGTTAGGCG | |
| GES-C | GTTTTGCAATGTGCTCAACG | 11 |
| GES-D | TGCCATAGCAATAGGCGTAG |
Pulsed-field gel electrophoresis
Eleven isolates with blaOXA-24-like genes among the 357 ACB complex blood isolates were typed by pulsed-field gel electrophoresis following digestion of intact genomic DNA with ApaI (New England Biolabs) [13]. The DNA fragments were separated on 1% (w/v) SeaKem Gold agarose gels in 0.5% Tris-borate-ethylene diamine tetra-acetic acid buffer using a contour-clamped homogeneous electric field (CHEF) Mapper apparatus (Bio-Rad, Hercules, CA, USA) at a potential of 6 V/cm pulsed from 5 s to 20 s for 19 h at 14 °C [13]. The gels were stained with ethidium bromide and photographed under ultraviolet light. The ApaI restriction profiles were initially compared by visual inspection, and isolates were considered closely related if they had differences in fewer than three bands [14]. Computer-assisted analysis using BioNumerics software (Applied Maths, Sint-Martens-Latem, Belgium) was also performed. Cluster analysis was performed by the unweighted pair group method with mathematical averaging. DNA similarity was calculated using the band-based Dice coefficient with a tolerance setting of 1.0% and an optimization setting of 1.0% for the whole profile. Isolates were considered to be in the same cluster if the similarity coefficient was greater than 87% [13].
Results
We collected a total of 357 ACB complex isolates, including 57 from Taipei Veterans General Hospital, and 100 each from the other 3 medical centres. Among these, 334 were CRACB complex truly (93.6%); of which 269 were CRAB isolates (80.5%).
All 269 of the CRAB isolates had the blaOXA-51-like gene, a gene considered intrinsic to A. baumannii (Table 2) [15]. The combination of blaOXA-23-like gene and blaOXA-51-like gene had the highest prevalence, (237 isolates, 88.1%). The combination of blaOXA-24-like gene and blaOXA-51-like gene was present in 11 isolates (4.09%), and 21 isolates (7.81%) contained solely the blaOXA-51-like gene without combination with other blaOXA-like genes. None of the isolates had a blaOXA-58-like gene or any of the MBL-encoding genes.
Table 2.
Carbapenem resistance among ACB complex blood isolates and distribution of OXA carbapenemase genes in Taiwan
| Target allele(s) | No. (%) of isolates |
|---|---|
| ACB complex by Vitek 2 method | 357 |
| CRACB complex by Vitek 2 & AST method | 334 |
| CRAB by La Scola’s protocol method | 269 |
| blaOXA-51-like+blaOXA-23-like | 237 (237/269, 88.10%) |
| blaOXA-51-like+blaOXA-24-like | 11 (11/269, 4.09%) |
| blaOXA-51-like solely | 21 (21/269, 7.81%) |
Abbreviations here and below: ACB complex Acinetobacter calcoaceticus-Acinetobacter baumannii complex, CRACB complex carbapenem-resistant Acinetobacter calcoaceticus-Acinetobacter baumannii complex, AST antimicrobial susceptibility testing, CRAB carbapenem-resistant Acinetobacter baumannii
Table 3 shows the prevalence of the different carbapenemase genes among CRAB isolates from the four institutions. The highest prevalence in all 4 hospitals was for the combination of blaOXA-23-like gene and blaOXA-51-like gene (75.93–92.77%). The prevalence foe blaOXA-51-like gene solely was 2.17–22.22%, and 0–6.52% for the combination of blaOXA-24-like gene and blaOXA-51-like gene.
Table 3.
Prevalence of CRAB/ACB complex and OXA carbapenemase genes from four hospitals in Taiwan
| 2009–2013 | MacKay Memorial Hospital | Taipei Veterans General Hospital | Tri-Service General Hospital | Changhua Christian Hospital |
|---|---|---|---|---|
| CRAB/ACB complex | 83/100 = 83% | 54/57 = 94.74% | 40/100 = 40% | 92/100 = 92% |
| blaOXA-23-like + blaOXA-51-like | 77 (77/83 = 92.77%) | 41 (41/54 = 75.93%) | 35 (35/40 = 87.50%) | 84 (84/92 = 91.30%) |
| blaOXA-24-like + blaOXA-51-like | 4 (4/83 = 4.82%) | 1 (1/54 = 1.85%) | 0 (0/40 = 0.00%) | 6 (6/92 = 6.52%) |
| blaOXA-51-like solely | 2 (2/83 = 2.41%) | 12 (12/54 = 22.22%) | 5 (5/40 = 12.50%) | 2 (2/92 = 2.17%) |
Figures 1 and 2 show the distributions of imipenem and meropenem MICs in 269 A. baumannii isolates. In isolates with the blaOXA-23-like gene, 28.69% had imipenem MICs of 64 mg/L or more, and 90.91% of isolates with the blaOXA-24-like gene had imipenem MICs of 64 mg/L or more. In isolates with the blaOXA-23-like gene, 37.55% had meropenem MICs of 64 mg/L or more, and all isolates with the blaOXA-24-like gene had meropenem MICs of 64 mg/L or more. Our study showed that the blaOXA-24-like genes were more resistant than blaOXA-23-like genes to carbapenem. Hence we performed PFGE in these blaOXA-24-like highly resistant genes.
Fig. 1.

Distribution of imipenem MICs in 269 CRAB blood isolates with the blaOXA-23-like and blaOXA-24-like genes
Fig. 2.
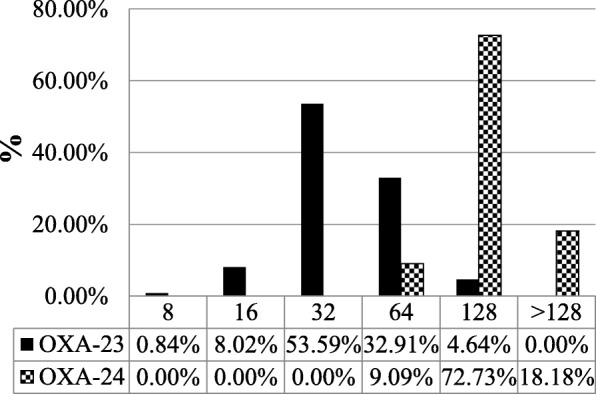
Distribution of meropenem MICs in 269 CRAB blood isolates with the blaOXA-23-like and blaOXA-24-like genes
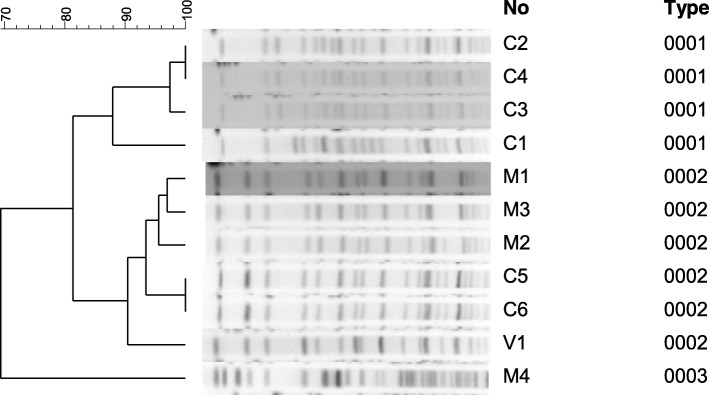
PFGE analysis indicated the presence of 11 distinct strains of CRAB among isolates that had both the blaOXA-24-like gene and the blaOXA-51-like gene (Fig. 3). This analysis also indicated the presence of three main PFGE pulsotypes (Fig. 3). Interestingly, six blaOXA-24-like genes of same cluster originated from three different medical centres. It meant that inter-hospital transmission of the highly resistant blaOXA-24-like gene may occur in Taiwan. The MICs values of 11 blaOXA-24-like +blaOXA-51-like CRAB isolates against six antimicrobial agents are shown in Table 4. Among the eleven CRAB isolates which harbored both the blaOXA-24-like genes and the blaOXA-51-like genes, the highest sensitivity was for colistin (MICs≦1 mg/L, showed 100% sensitive). Only 9% had MICs≦1 mg/L sensitive for tigecycline, while 64% had MICs = 2 mg/L and intermediate sensitivity, and 27% MICs≧2 mg/L were resistant to tigecycline. Besides, only 9% showed sensitivity to amikacin with MICs = 8 mg/L. All eleven isolates demonstrated total resistance to cefepime (MICs ≧ 64 mg/L), ceftazidime (MICs ≧ 128 mg/L), piperacillin/tazobactam (MICs ≧ 128 mg/L) and ampicillin/sulbactam (MICs ≧ 32 mg/L).
Fig. 3.
Pulsed-field gel electrophoresis results of A. baumannii isolates that had the blaOXA-24-like and blaOXA-51-like genes. There were 11 distinct genospecies and 3 PFGE dominant types. The scale indicates the percentage of overall genetic similarity. Isolates with the same letter were from the same hospital. M1, M2, M3, C5, C6, and V1 with the same gene cluster came from 3 different hospitals. Letter C: Changhua Christian Hospital. Letter M: MacKay Memorial Hospital. Letter V: Taipei Veterans General Hospital
Table 4.
The MICs of 11 blaOXA-24-like +blaOXA-51-like CRAB isolates for six antimicrobial agents
| OXA-24+ OXA-51 (n = 11) | CL | TGa | AMK | FEP | CAZ | PTZ | AMS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MICs | ≦0.5 | 1 | 1 | 2 | 4 | ≧8 | 8 | ≧128 | 64 | ≧128 | ≧128 | ≧128 | 32 | 64 | 128 |
| Percentage | 45% | 55% | 9% | 64% | 0% | 27% | 9% | 91% | 9% | 91% | 100% | 100% | 9% | 36% | 55% |
| Number of isolates | 5 | 6 | 1 | 7 | 0 | 3 | 1 | 10 | 1 | 10 | 11 | 11 | 1 | 4 | 6 |
| susceptibility | S | S | S | I | R | R | S | R | R | R | R | R | R | R | R |
Abbreviations: CL colistin, TG tigecycline, AMK amikacin, FEP cefepime, CAZ ceftazidime, PTZ piperacillin-tazobactam, AMS ampicillin/sulbactam, R resistant, S sensitive, I intermediate
aThe MICs of tigecycline were interpreted according to the European Committee on Antimicrobial Susceptibility Testing breakpoint for Enterobacteriaceae spp. (MIC > 2 mg/L as resistant)
The MICs of the other tested antibiotics were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines
Discussions
Antibiotic over-use has led to an increase in the prevalence of drug-resistant strains of Acinetobacter. In the United States, South America, India, and China, more than 50% of Acinetobacter species are resistant to carbapenems [4]. At the same time, carbapenem-resistant A. baumannii has become the most common genospecies of the CRACB complex. Our study showed that different hospitals had different CRAB/CRACB complex ratios, but a common feature was that CRAB accounted for most of our CRACB complex blood isolates. Researchers previously reported similar results for three other hospitals in Taiwan and in the US, but different results for Hong Kong, Denmark, and Norway [16–20].
In our study, all CRAB isolates had the blaOXA-51-like gene. The blaOXA-23–like gene was the second commonest carbapenemase gene (88.10%), and its prevalence among CRAB isolates was higher than in previous reports in Taiwan. For example, Kuo et al. reported that the prevalence of the blaOXA-23-like gene was 58% in a hospital in northern Taiwan [21], while Lin et al. reported a 4.2% prevalence in a hospital in northern Taiwan [22]. Chuang et al. reported a prevalence of 7.7% in Taiwan [23]. Our data thus indicate that the prevalence of the blaOXA-23like gene among CRAB isolates is increasing in Taiwan. In agreement with our findings, a report from Italy showed that the blaOXA-23-like enzyme was the most common carbapenemase (81.7%) [24]. Chusri et al. also reported that the prevalence of the blaOXA-23-like gene was 95% in their hospital in Thailand [25]. We think that carbapenem and/or cephalosporin antibiotics overuse could be responsible for the increased prevalence of the blaOXA-23like gene among CRAB isolates. Lack of strict infection control strategy is another cause. We also found that only 4.09% of CRAB isolates had the blaOXA-24–like gene, similar to two other studies in Taiwan (3% and 7.7%) [21, 23]. The findings of Hu et al. report are similar to our study [26]. They found that the prevalence of the blaOXA-23-like gene was 93.5% and the blaOXA-24-like gene was 4.6% in MacKay Memorial Hospital in Taiwan. Our study differed from Hu et al. in that firstly, we investigated the prevalence of different carbapenemase genes among CRAB in multiple tertiary centres (all > 1500 beds) in northern and central Taiwan. Secondly, we compared the MICs to imipenem and meropenem between the blaOXA-23-like gene and the blaOXA-24-like gene. Lastly, we compared the MICs of the blaOXA-24-like highly resistant gene to ceftazidime, cefepime, amikacin, ampicillin/sulbactam, colistin and piperacillin/tazobactam. Although the blaOXA-24-like gene accounts for only a small percentage of carbapenemase genes, it confers high resistance to carbapenems (Figs. 1 and 2). Since the occurrence of the blaOXA-24–like gene was so rare, we performed PFGE analysis and antimicrobial susceptibility tests for this group. We found that the blaOXA-24–like gene was almost totally resistant to amikacin, cefepime, ceftazidime, piperacillin/tazobactam and ampicillin/sulbactam. In our study, the isolates with blaOXA-24–like gene were 100% sensitive to colistin. The second recommended antibiotic is tigecycline despite the absence of Clinical and Laboratory Standards Institute (CLSI) MICs breakpoints for tigecycline.
Our study showed that the MICs to imipenem and meropenem among CRAB isolates with the blaOXA-23-like gene were much lower than that for those with the blaOXA-24-like gene. Thus, the blaOXA-24-like gene appears to confer greater resistance to carbapenem than the blaOXA-23-like gene. We also identified 3 major PFGE types of blaOXA-24-like genes in Taiwan based on ApaI digestion. Six blaOXA-24-like genes of same cluster came from 3 different hospitals (Fig. 3). This suggested that inter-hospital transmission of this resistant gene could occur in Taiwan, and may help to explain the increasing prevalence of more drug-resistant strains of CRAB in Taiwan. There are two reasons that could lead to the facilitated transmission of CRAB strains among hospitals. Firstly, in Taiwan, a patient can gain admission to any tertiary teaching hospital freely, without a doctor’s referral. Usually, the same patient had also visited several different tertiary teaching hospitals for different diseases. Secondly, after discharge, many patients carry resistant pathogens from different hospitals in which they were cared for back to the private nursing home or private respiratory care unit where they reside. The resistant strains are easily transmitted in these sites there if no strict infection control strategy. Therefore, we recommend additional infection control interventions for these patients to reduce the spread of resistant strains of CRAB.
What is the prevalence in other countries of the carbapenemase genes found in our study and do they have similar or differing antibiotic susceptibilities? What are the MICs of the blaOXA-23-like gene and the blaOXA-24-like gene to ceftazidime-avibactam, meropenem-vaborbactam or fosfomycin? Future multicenter, multi-country investigations could provide more data on the worldwide prevalence of carbapenemase genes in CRAB isolates and help the treatment of CRAB.
There were some limitations in our study. Firstly, the analysis of antibiotics consumption is not listed in our study. We could not analyse the relationship between the prevalence of the OXA genes and antibiotic drug consumption data in these 4 tertiary teaching hospitals. We were also unable to compare the relationship between the prevalence of the OXA genes and infection control strategy between these 4 tertiary teaching hospitals. Secondly, we did not perform PFGE and antimicrobial susceptibility tests on all CRAB blood isolates. Thirdly, under or over estimation of imipenem and meropenem non-sensitivity for the blaOXA-24-like gene is possible due to the small number of blaOXA-24-like genes (only 11 isolates). Large-scale studies throughout Taiwan are needed to address this limitation.
Conclusions
Our study determined the prevalence of CRAB, the types and prevalence of carbapenemase genes, antibiotic susceptibility among CRAB isolates, and documented that the blaOXA-24-like gene showed greater resistance to carbapenem than the blaOXA-23-like gene. We also found evidence of inter-hospital transmission of the highly antimicrobial-resistant blaOXA-24-like gene in Taiwan. Results of this study are reliable because all isolates were all from blood cultures and not from bacterial colonization. Our findings are helpful in that identification of the type of gene associated with carbapenemase resistance could assist in devising strategies to reduce the transmission of CRAB strains between patients and among hospitals and thus reduce mortality from CRAB infection.
Acknowledgments
We thank the Microbiology Laboratory personnel who are responsible for collection of bacterial isolates in MacKay Memorial Hospital, Taipei Veterans General Hospital, Tri-Service General Hospital, and Changhua Christian Hospital.
Funding
This study was funded by the grant MMH 103–54, MMH 104–15 and MMH 104–69 from MacKay Memorial Hospital, Taipei, Taiwan.
Availability of data and materials
Please contact author for data requests.
Abbreviations
- AB complex
Acinetobacter calcoaceticus-Acinetobacter baumannii complex
- AB
Acinetobacter baumannii
- CHDLs
Carbapenem-hydrolyzing class D β-lactamases
- CHEF
Contour-clamped homogeneous electric field
- CRAB complex
Carbapenem-resistant Acinetobacter baumannii complex
- CRAB
Carbapenem-resistant Acinetobacter baumannii
- ICUs
Intensive care units
- MBL
Metallo-β-lactamases
- MICs
Minimum inhibitory concentrations
- PCR
Polymerase chain reaction
- PFGE
Pulsed-field gel electrophoresis
- TNIS
Taiwan Nosocomial Infections Surveillance System
Authors’ contributions
THW: performed data analysis and interpretation, and drafted the manuscript. YSL: analysis of data, critical review of article. NYW: Performed drug susceptibility tests and performed PCR and sequencing. CPL and TRY: Designed protocols and supervised this study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by Institutional Review Board, MacKay Memorial Hospital, Taipei, Taiwan. Protocol no. 14MMHIS125.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Teng-Ho Wang, Phone: +886-2-2786-1288, Email: dah49@tpech.gov.tw.
Yi-Shing Leu, Phone: +886-2-2543-3535, Email: lys@mmh.org.tw.
Nai-Yu Wang, Phone: +886-2-2543-3535, Email: wang@mmh.org.tw.
Chang-Pan Liu, Phone: +886-2-2543-3535, Email: joeliu5929@hotmail.com.tw.
Tsong-Rong Yan, Phone: 886-2-2182-2928, Email: tryan@ttu.edu.tw.
References
- 1.Visca P, Seifert H, Towner KJ. Acinetobacter infection--an emerging threat to human health. IUBMB Life. 2011;63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control (Taiwan). Annual Report of Nosocomial Infections Surveillance System in Taiwan 2007 and 2014. Available at: https://www.cdc.gov.tw/english/info.aspx?treeid=00ED75D6C887BB27&nowtreeid=F0131176AA46D5DB&tid=1A8C498AF5F8AF5D. Accessed 1 Jan 2018.
- 3.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 4.Kim UJ, Kim HK, An JH, Cho SK, Park KH, Jang HC. Update on the epidemiology, treatment, and outcomes of carbapenem-resistant Acinetobacter infections. Chonnam Med J. 2014;50:37–44. doi: 10.4068/cmj.2014.50.2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 6.La Scola B, Gundi VA, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol. 2006;44:827–832. doi: 10.1128/JCM.44.3.827-832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006;27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Higgins PG, Lehmann M, Seifert H. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2010;35:305–314. doi: 10.1016/j.ijantimicag.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Higgins PG, Pérez-Llarena FJ, Zander E, Fernández A, Bou G, Seifert H. OXA-235, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob Agents Chemother. 2013;57:2121–2126. doi: 10.1128/AAC.02413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo-beta-lactamases. J Antimicrob Chemother. 2007;59:321–322. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 11.Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Queenan AM, Bush K. Carbapenemases: the versatile beta–lactamases. Clin Microbiol Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YC, Huang YT, Tan CK, Kuo YW, Liao CH, Lee PI, et al. Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: comparison of clinical features, prognostic factors and outcomes. J Antimicrob Chemother. 2011;66:1839–1846. doi: 10.1093/jac/dkr200. [DOI] [PubMed] [Google Scholar]
- 17.Lee HY, Chen CL, Wu SR, Huang CW, Chiu CH. Risk factors and outcome analysis of Acinetobacter baumannii complex bacteremia in critical patients. Crit Care Med. 2014;42:1081–1088. doi: 10.1097/CCM.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 18.Chuang YC, Sheng WH, Li SY, Lin YC, Wang JT, Chen YC, et al. Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with Acinetobacter bacteremia. Clin Infect Dis. 2011;52:352–360. doi: 10.1093/cid/ciq154. [DOI] [PubMed] [Google Scholar]
- 19.Fitzpatrick MA, Ozer E, Bolon MK, Hauser AR. Influence of ACB complex genospecies on clinical outcomes in a U.S. hospital with high rates of multidrug resistance. J Inf Secur. 2015;70:144–152. doi: 10.1016/j.jinf.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wisplinghoff H, Paulus T, Lugenheim M, Stefanik D, Higgins PG, Edmond MB, et al. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J Inf Secur. 2012;64:282–290. doi: 10.1016/j.jinf.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Kuo HY, Yang CM, Lin MF, Cheng WL, Tien N, Liou ML. Distribution of blaOXA-carrying imipenem-resistant Acinetobacter spp. in 3 hospitals in Taiwan. Diagn Microbiol Infect Dis. 2010;66:195–199. doi: 10.1016/j.diagmicrobio.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Lin YC, Hsia KC, Chen YC, Sheng WH, Chang SC, Liao MH, et al. Genetic basis of multidrug resistance in Acinetobacter clinical isolates in Taiwan. Antimicrob Agents Chemother. 2010;54:2078–2084. doi: 10.1128/AAC.01398-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chuang YC, Sheng WH, Lauderdale TL, Li SY, Wang JT, Chen YC, et al. Molecular epidemiology, antimicrobial susceptibility and carbapenemase resistance determinants among Acinetobacter baumannii clinical isolates in Taiwan. J Microbiol Immunol Infect. 2014;47:324–332. doi: 10.1016/j.jmii.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Principe L, Piazza A, Giani T, Bracco S, Caltagirone MS, Arena F, et al. Epidemic diffusion of OXA-23 producing Acinetobacter baumannii isolates in Italy: results of the first cross-sectional countrywide survey. J Clin Microbiol. 2014;52:3004–3010. doi: 10.1128/JCM.00291-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chusri S, Chongsuvivatwong V, Rivera JI, Silpapojakul K, Singkhamanan K, McNeil E, et al. Molecular epidemiology and spatiotemporal analysis of hospital-acquired Acinetobacter baumannii infection in a tertiary care hospital in southern Thailand. J Hosp Infect. 2017;95:53–58. doi: 10.1016/j.jhin.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Hu YF, Hou CJ, Kuo CF, Wang NY, Wu AY, Leung CH, et al. Emergence of carbapenem-resistant Acinetobacter baumannii ST787 in clinical isolates from blood in a tertiary teaching hospital in northern Taiwan. J Microbiol Immunol Infect. 2017;50:640–645. doi: 10.1016/j.jmii.2016.08.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Please contact author for data requests.