This work describes a 7-month follow-up of the excretion by different routes of Coxiella burnetii genotype SNP1/MST13 in a herd of goats that suffered high rate of abortions (81%), generating high environmental contamination. Some of the workers and visitors who accessed the farm were infected, with fever as the main symptom but a low incidence of pneumonia. The detected strain (SNP1/MST13 genotype) turned out to be very aggressive in goats. The viability of C. burnetii was demonstrated in the environment of the farm at the time of abortions, but 2 months after the last parturition, no viable bacteria were detected. These results highlighted the importance of implementing good biosafety measures at farms and avoiding the entrance of visitors to farms several months after the end of the kidding period.
KEYWORDS: Coxiella burnetii, goats, SNP, MST, viability, dust
ABSTRACT
This study describes a Q fever outbreak in a herd of 77 Alpine goats which suffered a high rate of abortions (81% [58/72]) in January 2017 and presents the results of monitoring the contamination and viability of Coxiella burnetii in the farm environment several months after the outbreak. Over the course of 7 months, we studied bacterial shedding by 35 dams with abortions to monitor C. burnetii infection dynamics and the duration of excretion. The highest bacterial shedding load was observed in vaginal mucus, followed by in feces and in milk. Conversely, the duration of C. burnetii shedding was longer through feces (5 months after abortion) than milk (3 months). C. burnetii DNA was detected throughout the study in aerosol samples periodically collected indoors and outdoors from the animal premises. Mouse inoculation and culture in Vero cells demonstrated the presence of viable isolates in dust collected from different surfaces inside the animal facilities during the period of time with the highest number of abortions but not in dust collected 2, 3, and 4 months after the last parturition. Some workers and visitors were affected by Q fever, with attack rates of 78% (7/9) and 31% (4/13), respectively. Affected people mostly showed fever and seroconversion, along with myalgia and arthralgia in two patients and pneumonia in the index case. The genotype identified in animal and environmental samples (SNP1/MST13) turned out to be very aggressive in goats but caused only moderate symptoms in people. After the diagnosis of abortion by Q fever in goats, several control measures were implemented at the farm to prevent contamination inside and outside the animal facilities.
IMPORTANCE This work describes a 7-month follow-up of the excretion by different routes of Coxiella burnetii genotype SNP1/MST13 in a herd of goats that suffered high rate of abortions (81%), generating high environmental contamination. Some of the workers and visitors who accessed the farm were infected, with fever as the main symptom but a low incidence of pneumonia. The detected strain (SNP1/MST13 genotype) turned out to be very aggressive in goats. The viability of C. burnetii was demonstrated in the environment of the farm at the time of abortions, but 2 months after the last parturition, no viable bacteria were detected. These results highlighted the importance of implementing good biosafety measures at farms and avoiding the entrance of visitors to farms several months after the end of the kidding period.
INTRODUCTION
Coxiella burnetii is an intracellular bacterium of the class Gammaproteobacteria that causes Q fever, a widespread zoonosis. Domestic ruminants are considered the most important source of infection for humans (1–3). Although 60% of human infections are asymptomatic, C. burnetii can cause a simple febrile syndrome or a febrile syndrome accompanied by pneumonia and/or hepatic involvement after an incubation period of 2 to 3 weeks (4). Q fever is endemic in the Basque Country (northern Spain), and the notification of cases to the System of Microbiological Information (Sistema de Información Microbiológica [SIM]) for an epidemiological assessment is compulsory. In areas of endemicity, occasional outbreaks of Q fever occur in people, and the attack rate depends on the exposure of the nonimmune population to the infection source. Most human Q fever outbreaks have been related to goats and sheep (1, 5, 6), and direct or indirect contact with infected herds through the inhalation of contaminated aerosols is the most common source of infection (4). In fact, some authors stated that a single C. burnetii is sufficient for infection and to produce clinical disease (7, 8). When C. burnetii infection appears in a herd, control measures must be quickly implemented, and biosafety is essential to prevent the propagation of infection outside animal facilities. These measures include the collection and destruction of placentas and fetuses, the correct management of manure, the application of general hygiene procedures, including periodic disinfections, and vaccination, among others (1, 6).
Q fever in small ruminants has been mostly associated with late abortions, stillbirths, and the delivery of weak offspring (9). Q fever abortions in caprine herds are more important than in sheep flocks, affecting sometimes up to 90% of females (10). After an abortion or normal birth, the infected animal sheds the bacteria through the placenta and fetal fluids, which contain millions of bacteria (11) and represent a high-risk infectious material. In addition, infected animals shed the bacteria through vaginal fluids, feces, milk, and urine. Some studies stated that the most important route for C. burnetii shedding in goats was the milk (12), whereas in an experimental infection, C. burnetii was excreted through feces and vaginal mucus up to 95 days postpartum, with a shorter period of excretion through milk (13). Bacterial shedding by different routes together with the movement of the animals inside the stable generates the formation of C. burnetii-contaminated aerosols, which pose a risk for people that have access to an infected farm during the kidding period (14–16). In addition, C. burnetii can be displaced several kilometers by the wind, affecting people living far from the infected farms (5) and thus hampering the epidemiological investigation of the source of infection.
Although Coxiella can survive in the environment for long periods of time (17, 18), scarce data are available about the viability of C. burnetii in the farm environment after an outbreak of Q fever in ruminants (15). Knowledge of the duration of C. burnetii viability in animal premises is essential to elaborate the recommendations for farmers concerning the minimum periods of time before handling the manure, receiving visitors at the farm, etc. This paper describes a 7-month follow-up of the excretion of C. burnetii by different routes in a herd of goats that suffered a high rate of abortions, generating high environmental contamination and affecting workers and visitors of the farm. The results of the epidemiological investigation of the outbreak in the herd and the environment are presented, along with the identification of the genotype of C. burnetii involved and its viability in dust collected from the farm. Also, a description of the Q fever cases in workers and visitors of the farm is provided. The control measures taken after the outbreak are detailed and discussed.
RESULTS
Confirmation of Q fever abortion in the goat herd and implementation of control measures.
The goat herd suffered an outbreak of abortions at the end of gestation, which peaked between 18 January and 31 January (39 abortions) and had affected 81% of pregnant females (58 of 72) by the end of the season. Q fever was diagnosed on the basis of the observation of C. burnetii-compatible bacteria in placenta and vaginal fluids (Stamp staining) and the detection of C. burnetii DNA in these samples. Additionally, the histological study of the placentas showed foci of necrosis in the cotyledon villi, accompanied by an inflammatory infiltrate and the presence of large numbers of bacteria in the cytoplasm of the trophoblasts. Also, the presence of an inflammatory infiltrate composed mainly of lymphocytes and neutrophils was found in the walls of arterioles. The presence of other bacteria causing abortions was ruled out. Eleven of the twelve serum samples from goats with recent abortions analyzed by enzyme-linked immunosorbent assays (ELISAs) had antibodies against C. burnetii.
When C. burnetii was diagnosed as the cause of the abortions (26 January), control measures were immediately implemented and access by visitors was forbidden. Personal hygiene measures were imposed, including the use of exclusive clothing and footwear when work was conducted within the animal premises. The control measures included antibiotic treatments (double treatments with oxytetracycline) for pregnant animals and the confinement of the herd within the premises to prevent placentas and fluids from being expelled outside the farm. The placentas were collected with gloves and placed in watertight containers, and lime was added to the surfaces for the inactivation of C. burnetii. The manure was not removed from the animal facilities until 4 months after the last parturition, and afterwards, the premises were cleaned and disinfected with Virkon. Three-month-old replacements were vaccinated and revaccinated (18 kids) with inactivated phase I vaccine (Coxevac; Ceva Santé Animale). Finally, the milk for cheese production was pasteurized.
C. burnetii shedding and serological response in goats.
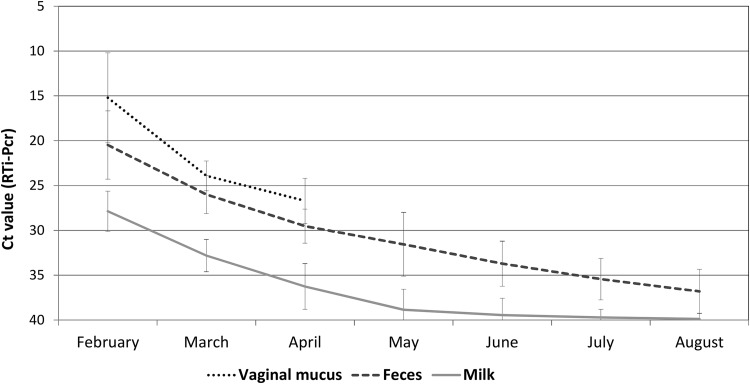
C. burnetii shedding was studied for 7 months (February to August 2017) in 35 of 58 goats with recent abortions. The bacterial load detected was higher in vaginal fluids and feces than in milk (as inferred by cycle threshold [CT] values). In May, 4 months after the peak in the number of abortions, the amount of bacterial excretion was still significant in feces, whereas it was low in milk (Fig. 1). The evolution of C. burnetii DNA shedding in bulk-tank milk (BTM) samples followed the same decreasing trend and became negative in August (CT >40), at the end of the study (data not shown).
FIG 1.
Dynamics and duration of Coxiella burnetii shedding (mean CT ± standard deviation [SD]) through different excretion routes in 35 goats with recent abortions.
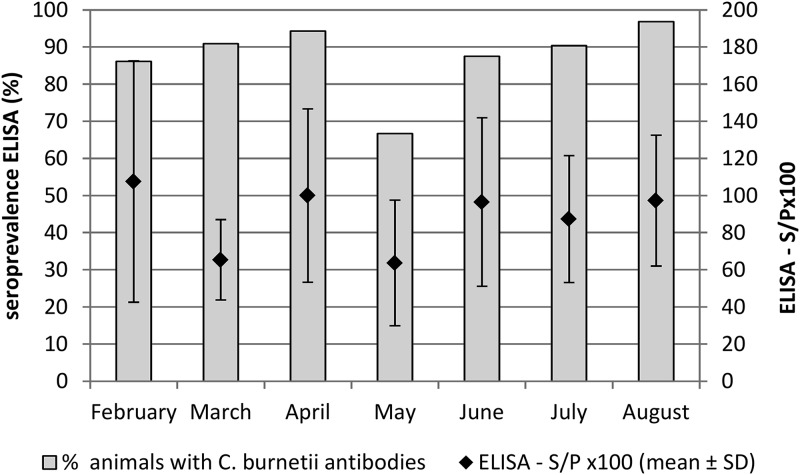
The seroprevalence estimated in individual milk samples by ELISAs was high, and most of the examined goats seroconverted. Antibodies persisted at least for 7 months after abortion (Fig. 2).
FIG 2.
Evolution of seroprevalence against C. burnetii in the 35 goats with recent abortions studied.
C. burnetii in environmental samples.
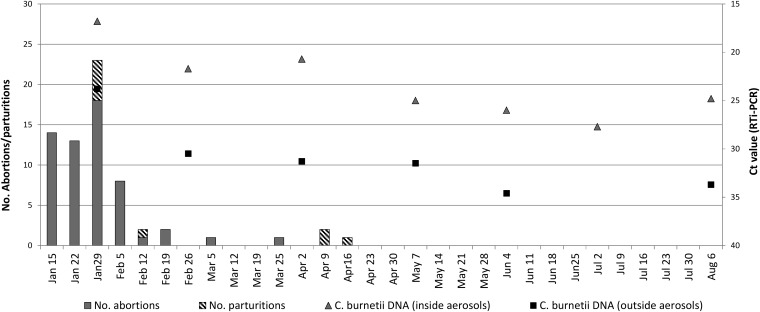
The bacterial load (expressed in CT values) found in aerosols was high during the peak in the number of abortions, as shown in Fig. 3. C. burnetii was detected in aerosols taken outside the animal facilities during all the samplings but always with lower bacterial loads than those detected within the farm. Coxiella DNA was also detected in all dust samples collected in the 4 samplings from different surfaces of the animal facilities. CT values obtained by real-time PCR (RTi-PCR) amplification of the IS1111 gene were always low, indicating a high bacterial burden. The highest load (CT, 18.9) corresponded to the sample collected immediately after the peak in the number of abortions in February, whereas the bacterial burden in dust samples taken in later samplings was lower (CT, 22.1 to 24.2).
FIG 3.
Dynamics of C. burnetii DNA in aerosols taken inside and outside animal premises and relationship with abortions and normal parturitions. Bars indicate weekly accumulated numbers (left y axis) and symbols indicate C. burnetii load (right y axis). No aerosol samples were collected outside the premises in July, due to rain.
SNP and MST genotyping.
Genotyping analyses of 4 samples of vaginal fluids from goats with recent abortions and 2 dust samples identified single nucleotide polymorphism (SNP) and multispacer sequence type (MST) SNP1/MST13.
Viability of C. burnetii in dust.
The spleens from four mice inoculated with the dust homogenate collected in February were positive by RTi-PCR, which showed lower CT values (30.6 and 32.6) in the two mice euthanized 21 days postinfection (p.i.) than in those euthanized 7 days p.i. (CT, 35.3 and 35.9). The number of C. burnetii genome equivalents (GE) recovered in the spleen of one mouse euthanized 21 days p.i. was slightly higher than the GE injected (Table 1), suggesting that C. burnetii was viable and the bacteria multiplied in vivo. In the homogenates of dust collected 2, 3, and 4 months after the last parturition, C. burnetii was not detected in the spleens of mice even when targeting the multiple-copy IS1111 gene. No relevant splenomegaly was detected in mice (Table 1). Vero cells cultured with homogenates of spleens obtained 21 days p.i. from mice inoculated with dust collected during the peak abortion period (C. burnetii RTi-PCR-positive spleens) were positive for C. burnetii, whereas there was no bacterial growth from cells cultured with homogenates obtained from mice on day 7 p.i. The bacterial loads (GE) recovered at each passage are shown in Table 1.
TABLE 1.
Results of the viability study of C. burnetii in dust using BALB/c and cultivation in Vero cell lines
| Collection date (no. of months since last parturition) | BALB/c mouse inoculation |
Spleen wt/body wt | Culture in cell lines (Vero E6) (GE/ml) |
Viable C. burnetii | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of GEa injected | Days p.i.b | No. of GE in spleen | Inoculated | Day 6 p.i. | 1st passage | 2nd passage | 3rd passage | |||
| February (0) | 87.5 × 103 | 7 | 66.7 × 103 | 0.0032 | 8.8 × 102 | 0.0 | 0.0 | 0.0 | 0.0 | No |
| 7 | 93.2 × 103 | 0.0040 | 0.0 | 0.0 | No | |||||
| 21 | 80.9 × 103 | 0.0040 | 4.9 × 103 | 5.0 × 103 | 4.5 × 104 | 2.7 × 105 | 2.9 × 105 | Yes | ||
| 21 | 79.3 × 104 | 0.0054 | 3.6 × 104 | 1.2 ×105 | 2.3 × 105 | 1.7 × 105 | 4.7 × 105 | Yes | ||
| June (2) | 100 × 103 | 7 | 0.0 | 0.0037 | NDc | NAd | NA | NA | NA | No |
| 7 | 0.0 | 0.0039 | ND | NA | NA | NA | NA | No | ||
| 21 | 0.0 | 0.0045 | ND | NA | NA | NA | NA | No | ||
| 21 | 0.0 | 0.0043 | ND | NA | NA | NA | NA | No | ||
| July (3) | 100 × 103 | 7 | 0.0 | 0.0046 | ND | NA | NA | NA | NA | No |
| 7 | 0.0 | 0.0041 | ND | NA | NA | NA | NA | No | ||
| 21 | 0.0 | 0.0049 | ND | NA | NA | NA | NA | No | ||
| 21 | 0.0 | 0.0041 | ND | NA | NA | NA | NA | No | ||
| August (4) | 100 × 103 | 7 | 0.0 | 0.0041 | ND | NA | NA | NA | NA | No |
| 7 | 0.0 | 0.0042 | ND | NA | NA | NA | NA | No | ||
| 21 | 0.0 | 0.0044 | ND | NA | NA | NA | NA | No | ||
| 21 | 0.0 | 0.0046 | ND | NA | NA | NA | NA | No | ||
| Negative control (PBS) | NA | 7 | 0.0 | 0.0038 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | No |
| 7 | 0.0 | 0.0033 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | No | ||
| 21 | 0.0 | 0.0037 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | No | ||
| 21 | 0.0 | 0.0033 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | No | ||
GE, genome equivalents of C. burnetii determined by quantitative real-time PCR targeting IS1111.
p.i., postinfection.
ND, not done.
NA, not applicable.
Q fever in farm workers and visitors.
After the initial case of pneumonia by Q fever in one of the workers, an epidemiological survey was conducted on the group of farm workers, i.e., 5 men and 3 women between 22 and 57 years old. All of them participated in different tasks on the farm, and although two of them were mostly in charge of the goats, the other 6 workers could not rule out sporadic visits to the stable after early December 2016. Five of the eight workers had a self-limited febrile illness with onset of symptoms between mid-December and mid-January, and two of them presented also with myalgia and arthralgia for 2 to 3 days. At the time of the survey (end of January), they were all asymptomatic. Taking into account the first case, the patient with pneumonia, a total of 6 workers had symptoms compatible with Q fever. Serological results showed responses indicative of acute infection in 7 workers; all met the definition of “case,” 3 of them without clinical symptoms (Table 2). Despite having clinically compatible symptoms, one worker was considered a “no-case,” as he tested negative for phase II IgM. The PCRs were negative in all cases. The attack rate in this group was 78% (7/9).
TABLE 2.
Epidemiological and serological data compiled from level 1 risk group
| Sex | Age (yrs) | Access to the stable | Symptoms | Serology of blood extractionb |
Case | Attack rate (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| 1st |
2nd |
||||||||
| IgMc | IgG | IgMc | IgG | ||||||
| Farm workers | 78 | ||||||||
| Malea | 57 | No | Yes | 1/128 | 1/128 | 1/256 | 1/1024 | Yes | |
| Male | 57 | No | Yes | Neg | Neg | Neg | Neg | No | |
| Male | 59 | No | No | 1/1024 | 1/512 | NDd | ND | Yes | |
| Male | 34 | Yes | Yes | Neg | 1/512 | Neg | 1/1024 | No | |
| Female | 35 | No | Yes | 1/64 | 1/64 | 1/128 | 1/512 | Yes | |
| Male | 38 | No | Yes | 1/1024 | 1/2048 | ND | ND | Yes | |
| Female | 29 | No | Yes | 1/128 | 1/2048 | ND | ND | Yes | |
| Male | 22 | Yes | No | 1/512 | 1/128 | ND | ND | Yes | |
| Female | 43 | Yes | No | Neg | Neg | 1/512 | 1/128 | Yes | |
| Visitors | 31 | ||||||||
| Female | 18 | Yes | Yes | Neg | Neg | Neg | Neg | No | |
| Male | 21 | Yes | No | Neg | Neg | Neg | Neg | No | |
| Male | 18 | Yes | No | Neg | Neg | ND | ND | No | |
| Male | 25 | Yes | Yes | Neg | Neg | Pos | 1/256 | Yes | |
| Male | 18 | Yes | Yes | Neg | Neg | Neg | Neg | No | |
| Male | 18 | Yes | No | Neg | Neg | Neg | Neg | No | |
| Male | 18 | Yes | No | Pos | Neg | Pos | 1/128 | Yes | |
| Male | 20 | Yes | Yes | Neg | Neg | Pos | 1/128 | Yes | |
| Male | 18 | Yes | No | Neg | Neg | Neg | Neg | No | |
| Male | 19 | Yes | No | Pos | Neg | Pos | Neg | No | |
| Male | 19 | Yes | No | Neg | Neg | Pos | 1/256 | Yes | |
| Male | 30 | Yes | Yes | Neg | Neg | Neg | Neg | No | |
| Male | 35 | Yes | No | Neg | Neg | Neg | Neg | No | |
Index case.
Neg, negative; Pos, positive.
IgM determined by IFAT (farm workers) or indirect chemiluminescent immunoassay (visitors).
ND, not done.
An epidemiological survey was also conducted in mid-February on 12 students and one teacher, aged 18 to 35 years, from an agricultural school that visited the farm on 16 January. Five students had fever, with the onsets of symptoms between the end of January and the beginning of February. All were asymptomatic at the time of the survey (14 February). The serological study showed responses indicative of acute infection in 4 of them, but 2 of them did not have any symptoms. In this group, the attack rate was 31% (4/13) (Table 2). The PCRs were negative in all cases.
The group of people included in level 2 consisted of 34 members from 2 families (24 male and 10 female individuals between 5 and 73 years old), with residences outside the municipality where the farm was located but who spent weekends and/or vacations in houses near the farm. A telephone survey conducted in mid-February confirmed that they had not had direct contact with the goats. No risk factors (immunosuppression, pregnancy, valvulopathy, or vascular prosthesis) were identified in this group, but they were advised to visit the doctor if suffering from fever, flu, or any symptoms compatible with Q fever. They were also asked to report any news concerning any worsening conditions of their health status. At the time of writing the manuscript, no news was reported.
DISCUSSION
In Europe, most Q fever outbreaks in humans originate from small ruminants after the inhalation of aerosols contaminated with C. burnetii (1, 4). Herein, we present an outbreak of Q fever associated with a goat herd, of which 78% suffered abortions in a short period of time, generating an aerosol-borne spread of C. burnetii that contributed to the infection of the farm workers and visitors who entered the animal premises during the kidding period. The follow-up of 35 goats with recent abortions provided useful epidemiological information on the dynamics of C. burnetii excretion through different routes and the duration of bacterial shedding. Until recently, milk was considered, in terms of duration, the main excretion route of Coxiella in goats (12). This study has demonstrated that the bacterial load shed through milk is lower than the quantity shed through vaginal mucus or feces, and the shedding period is shorter in milk. Thus, 3 months after abortion, C. burnetii was not present in milk, whereas C. burnetii shedding in feces, albeit at low levels (CT, <35), was still detected 5 months after an abortion. Similarly, an experimental infection in goats with the strain causing a large outbreak in The Netherlands (13) also identified feces and vaginal mucus as the major shedding routes. Taking into account similar studies carried out in sheep flocks (19, 20), it seems that C. burnetii excretion patterns and the duration of shedding are very similar in goats and sheep.
Bacteria released to the environment, together with the movement of the animals inside the barn, generate high levels of environmental contamination for long periods of time, which, as shown in this study, are sometimes longer than the duration of the kidding period. The presence of C. burnetii DNA in aerosols highlighted the risk of inhalation of the bacterium by people who entered the affected farm. Therefore, among other biosafety measures, access for visitors should be prohibited during the parturition period (16, 21), as C. burnetii in the environment, even at low numbers, poses a public health risk (22). As expected, the highest bacterial load in air was detected in the first sampling after the peak in the number of abortions; afterwards, there was a progressive but slow decrease, with CT levels inside the barn remaining below 30 until the end of the experiment. The biological significance of PCR-positive aerosols is not clear, as the detection of C. burnetii DNA in environmental samples does not necessarily imply that these bacteria are viable. There are many studies that investigated the presence of C. burnetii DNA in the environment (23–26), but only a few studied its viability (15, 24, 27). The laboratory methods to investigate C. burnetii viability include cell culture, the use of animal models, and in vitro methods (ethidium monoazide-PCR [EMA-PCR]) (28). The assessment of bacterial viability in environmental samples is not easy, as they are contaminated with a variety of microbes. Mice have the ability to clear the contamination in this type of sample, and C. burnetii can be recovered from the spleen 21 days after inoculation (24). In this study, mice were experimentally inoculated with dust samples that had low CTs by RTi-PCR, indicating that they contained high Coxiella loads (29). The combination of the inoculation of BALB/c mice and the cultivation of spleen homogenates in Vero cells showed that C. burnetii was viable in the dust collected at the time of the abortions (February). C. burnetii was not detected from mice inoculated with dust samples collected 2, 3, and 4 months after the last parturition, suggesting that the bacteria were no longer viable. The time C. burnetii remains viable in dust has not been fully investigated, but the present results agree with some reports stating that it can be viable for 2 months (1). This is valuable information for farms with sheep and goat herds that receive visitors (schools, tourists, etc.) as an in-farm activity for extra income and will help to establish the minimum period during which affected farms should not receive visitors. However, it is necessary to investigate if all Coxiella genotypes remain viable for the same period of time, a key aspect to establish correct control measures.
Additional biosafety measures are essential to prevent the spread of infection. The management of manure is a crucial aspect in infection spread control, because aerosols containing C. burnetii can be generated while removing the bedding. Thus, in the outbreak in The Netherlands, removing the manure from the stable within 30 days after the ending of kidding was prohibited (21). In the studied farm, manure was kept inside the stable for 4 months after the last parturition to avoid environmental contamination. Fetuses and placentas were immediately removed and treated with lime in a watertight container. The trade or transport of animals was prohibited and the pasteurization of milk was imposed by public health authorities. These farming practices contributed to reducing the risks for the local population, as no new cases were detected in the municipality.
The Q fever outbreak had attack rates of 78% among farm workers and 31% in the group of students who spent 2 to 3 h inside and around the farm. All blood samples collected from human patients were PCR negative, probably because C. burnetii DNA becomes undetectable in serum when a serological response develops (30); therefore, it was not possible to isolate C. burnetii for genotyping. Fortunately, the samples collected from the animals and the farm environment were positive and some of them could be genotyped. The genotype identified, SNP1/MST13, was previously found in humans from Spain, Portugal, and France (4) and also in sheep and cattle from the region (31). The clinical presentation of C. burnetii infection depends on both the virulence of the infecting strain and the specific risk factors of the patient (4). Here, despite the high air contamination with C. burnetii inside and outside the animal premises at the time of the abortions, none of the workers or visitors needed hospitalization. Similarly, the group of people in level 2 that frequented the area during weekends or vacations did not report any compatible symptoms, despite the aerosol contamination detected outdoors. Although this would suggest that the SNP1/MST13 genotype causes milder clinical symptoms in humans, risk factors specific to the patient or the region (endemic versus nonendemic) cannot be disregarded. On the other hand, the infection of goats with genotype SNP1/MST13 resulted in a severe outbreak that resulted in a rate of abortions that almost reached 80%, suggesting that this is a very virulent genotype for goats. The source of infection in the affected herd could not be established, as the herd produced its own replacements and did not share grazing pastures with other domestic ruminants. Likely, the infection came from purchased food/hay or wildlife. Interestingly, most of the human cases were concentrated between mid-December and mid-January, before the abortions began in the goat herd. It is known that pregnant infected goats can shed C. burnetii through feces weeks before abortion or parturition occurs (11), representing the possible moment and source of infection for the affected people. In this study, most of the goats with recent abortions seroconverted, indicating that the SNP1/MST13 genotype induced a high humoral response with long-lasting antibodies, as demonstrated by the observation that 7 months after the abortions, 97% of the goats still had antibodies. This high seroprevalence, together with the vaccination plan applied to replacement kids, albeit not completely, should reduce significantly the percentage of abortions in subsequent farrowing periods, as suggested by other authors (32, 33).
In conclusion, once Q fever appears in a herd, control measures should be maintained for a long period of time and access for visitors prohibited at least for 2 months after the end of parturition. This study is an example of a holistic One Health approach in which medical and veterinary institutions collaborate in the investigation of zoonotic outbreaks involving people and animals, with the aim of protecting the health of the citizens and the consumers.
MATERIALS AND METHODS
Case presentation.
On 27 January 2017, the general practitioner from a municipality in Gipuzkoa (Basque Country, northern Spain) notified the epidemiology surveillance unit of a case of Q fever. A 57-year-old male presented with an acute respiratory infection with the onset of symptoms in the middle of December 2016. The radiological study confirmed pneumonia, and the patient was treated with amoxicillin-clavulanic acid. C. burnetii infection was confirmed by phase II IgM and IgG detection. The patient worked and lived at a dairy goat farm. He mentioned that in the previous weeks, there had been 20 abortions in the herd and that the veterinarian attending the farm had collected and sent to the laboratory different samples for diagnosis. The patient reported that, besides him, another 10 people were living at the farm, including 8 adults and two children aged 1 and 5 years. He also mentioned that on 16 January, a group of 13 visitors (12 students and one teacher) from an agricultural school visited the farm, spending 2 to 3 h inside and outside the animal facilities. In addition, in the surroundings of the farm, there were cottages occupied during the weekends and holiday periods by two different families. The occupational health authority (OSALAN) was also informed on the same day. A multidisciplinary group that included microbiologists, veterinarians, occupational health technicians, and epidemiologists was gathered to investigate the case and design an epidemiological investigation.
Epidemiological study at the dairy goat farm.
The worker affected by Q fever (index case) worked at a dairy goat herd composed of 77 Alpine breed goats. The herd had no contact with other flocks and the owner did not buy animals from other flocks (produced its own replacements). The females were usually mated in autumn, in September and November, and the kidding season extended from February to April. After kidding, the dams had a long lactation period before being retained for subsequent parturition. The animals were fed by grazing supplemented with hay and concentrate. Different types of dairy products, including yogurts and fresh and cured cheese made with raw milk, were produced at the farm.
Sampling and laboratory analyses for abortifacient agents.
On 24 January (3 days before the case notification), the farm veterinarian had sent to the Department of Animal Health of NEIKER 1 placenta, 6 vaginal swabs, and 12 serum samples after observing an outbreak of abortions at the end of the gestational period (21 abortions since 15 January). Bacteriological analyses, consisting of Stamp staining (placenta) and bacteriological cultures (vaginal fluids and placenta) in general media (blood agar, MacConkey agar, and Farrell selective agar), were performed to rule out causes of bacterial abortion. A histopathological study of the placenta was also performed to evaluate lesions associated with infectious abortifacients. If the histopathological study revealed lesions compatible with Q fever, the placenta samples were also analyzed by real-time PCR (RTi-PCR) for the detection of Coxiella DNA (34). Serum samples were analyzed by ELISAs for the detection of antibodies against C. burnetii (LSIVET ruminant milk/serum Q fever kit; Thermo Fisher Scientific), Pestivirus (Border disease) (IDEXX Laboratories, ME, USA), and Chlamydophila abortus (complement fixation test).
Follow-up of C. burnetii infection in goats and the farm environment.
After the diagnosis of Q fever abortion by bacteriological, molecular, and histopathological methods, control measures were recommended to the farmer by the farm veterinarian and the animal health authorities. To estimate the infection prevalence within the herd and the duration of C. burnetii shedding in infected goats, the farm was visited monthly from February to August. The samples collected included vaginal fluids (first 3 visits) and milk and feces taken from 35 goats with recent abortions, along with a bulk-tank milk (BTM) sample per visit. Environmental samples consisted of aerosol samples taken monthly inside and outside the animal premises with an air sampler (MD8 Airscan air sampler; Sartorius) and dust samples taken in duplicates from 10 different surfaces inside the animal premises at the first visit (2 February) and 2, 3, and 4 months after the last parturition (21 April). Animal and environmental samples were processed for DNA extraction as previously described (23) and analyzed by RTi-PCR targeting the IS1111 gene (34). A commercial internal amplification control (IAC; TaqMan exogenous internal positive control; Thermo Fisher Scientific) was included in the assay to monitor for PCR inhibitors.
To evaluate seroprevalence against C. burnetii, individual milk samples were centrifuged and milk sera were tested for Q fever antibodies with an ELISA (LSIVET ruminant milk/serum Q fever kit; Thermo Fisher Scientific) according to the protocol recommended by the manufacturer for milk samples. A cocktail of antigen phases I and II was used in this assay to detect total anti-C. burnetii immunoglobulin G antibodies (IgG). An index ratio of the optical density of the tested milk serum to the optical density of the positive control (S/P) was calculated according to the manufacturer's instructions. Milk samples with S/P indices of ≤0.3 were considered negative, while samples with an S/P of >0.3 were considered positive.
Genotyping.
A selection of animal and environmental samples with positive RTi-PCR results and low CT values was genotyped by multispacer sequence typing (MST) (35) with some modifications described elsewhere (36) and by SNP discrimination using RTi-PCR (37).
Studies of C. burnetii viability.
These studies were carried out in biosafety level 3 (BSL3) building facilities and consisted of experimental inoculations of mice and culture in Vero cells. Once permissions were obtained from the ethical and animal welfare committee (Bizkaiko Foru Aldundia, document 3/2017, registration 15.328, 22 February 2017), 20 6-week-old BALB/c male mice were included in the experiment to test 4 samples along with a negative control, with 4 replicates. The samples consisted of four homogenates of pooled dust, collected during the above-mentioned periods and prepared according to procedures already published (24). The quantification of C. burnetii genome equivalents (GE) in each homogenate was carried out by quantitative real-time PCR (qPCR) using 5 μl of DNA (in triplicates) and specific primers and a probe targeting the IS1111 gene (34). In each qPCR run, a standard curve was generated using 10-fold serial dilutions of a known concentration of Nine Mile (RSA439) phase II strain of C. burnetii DNA. Four aliquots of 400 μl were prepared from each homogenate of pooled dust, containing 87.5 × 103 to 100.0 × 103 C. burnetii GE. These 4 aliquots were inoculated intraperitoneally in 4 mice each; negative-control mice (n = 4) were inoculated intraperitoneally with 400 μl phosphate-buffered saline (PBS). Mice were euthanized on days 7 postinoculation (p.i.) (2 mice) and 21 days p.i. (2 mice), and the spleens were removed. The level of splenomegaly was determined from the ratio of the spleen weight to the body weight. The spleens were stored at −80°C for further analyses.
Half of the spleen from each mouse was processed for DNA extraction and real-time PCR amplification. Briefly, the spleen was mixed with Tris-EDTA (TE) buffer (10 mM Tris base, 1 mM EDTA, pH 8) and homogenized with a TissueLyser (Qiagen Hilden, Germany). Two hundred microliters of the mixture was treated with 180 μl of buffer ATL and digested with 20 μl of proteinase K (8 mg/ml) for 3 h at 55°C before DNA extraction with a QIAmp DNA blood minikit (Qiagen, Hilden, Germany). The presence of C. burnetii was assessed by RTi-PCR targeting the IS1111 gene (34), and positive samples were subjected to qPCR (34), as mentioned above, to quantify the number of C. burnetii GE detected in spleen and compare it with the number of GE inoculated.
African green monkey epithelial cells, VERO C1008 (Vero 76, clone E6, Vero E6 ATCC CRL-1586), were grown in T75 culture flasks containing Dulbecco's modified Eagle medium (DMEM; Lonza Biologics Porriño S.L., Pontevedra, Spain) supplemented with 1% l-glutamine (Lonza Biologics Porriño S.L., Pontevedra, Spain), 0.4% nonessential amino acids (Sigma-Aldrich Quimica, S.L., Madrid, Spain), and 10% fetal bovine serum (FBS; Sigma-Aldrich Quimica, S.L., Madrid, Spain) at 37°C in 5% CO2. The other half of the qPCR-positive spleen was homogenized with 700 μl DMEM and 2% FBS in a TissueLyser. One hundred microliters of each homogenate was placed on shell vials (SV) containing Vero cells and centrifuged at 600 × g for 1 h at 20°C. Then, 700 μl of fresh DMEM with 2% FBS was added to the SV and incubated at 37°C in 5% CO2 for 6 days. After harvesting C. burnetii from SV on day 6 p.i., three passages of 1,000 μl of harvested cells were transferred at weekly intervals to T25 culture flasks containing a Vero layer. At day 6 p.i. and before each passage, 200 μl was collected for DNA extraction and qPCR, according to the procedures described above. Cultures with unchanged or increased GE during the procedure were considered to be positive. Uninfected control cells were kept close to infected cells to rule out possible cross-contaminations.
Epidemiological investigation of the human outbreak.
An epidemiological investigation, including the people working at the farm, the visitors, and other populations at risk, was carried out. The primary health care center of the municipality was contacted and requested to collect information regarding additional patients with clinical symptoms compatible with C. burnetii infection.
Two levels of risk were considered with different degrees of exposure to infection. Level 1 included people with a high risk of exposure at the farm from early December 2016: workers living at the farm (9 adults) and visitors (the group of students and teacher). An epidemiological questionnaire and a serological study were conducted for this group. Level 2 included people who stayed on weekends or vacations at the two cottages in the surrounding area. The group of vacationers was contacted by telephone to be informed of the situation and to identify individuals who belong to level 1 (people who had had access to the stable) and/or suffer other risk factors (immunosuppression, pregnancy, valvulopathy, etc.). The 1- and 5-year-old children whose parents worked at the farm but showed no symptoms during the study period were also included in level 2.
Laboratory investigation.
Blood samples were obtained from the level 1 group for serological determination of C. burnetii phase II IgM and IgG antibodies. To evaluate seroconversion, a second blood sample was obtained 3 to 4 weeks later. The blood samples from farm workers were analyzed by an indirect immunofluorescence antibody test (IFAT) in the microbiology laboratory of the Hospital Universitario de Donostia-San Sebastian, considering as positive a phase II IgM of ≥1/256 and a phase II IgG of ≥1/128 (C. burnetii IFA IgG and IFA IgM; Vircell S.L., Spain). PCR for the detection of C. burnetii DNA was also performed with the first blood extraction sample (30, 38). Blood samples from the visiting student group were analyzed in the microbiology laboratory of the Hospital Universitario de Basurto (Bilbao), and the serological methods used included an indirect chemiluminescent immunoassay (C. burnetii Virclia IgM monotest; Vircell S.L., Spain) for the detection of phase II IgM antibodies (expressed as positive, negative, or doubtful) and an IFAT for phase II IgG considering titers of ≥1/128 as positive (C. burnetii IFA IgG; Vircell S.L., Spain).
A confirmed human case was defined as a person who belonged to the group of risk level 1, had access to the farm after early December 2016, and had a positive result in laboratory analyses (PCR positive, positive results in the first determination of phase II IgG or IgM, or seroconversion, always with phase II IgM positive), with or without compatible clinical symptoms.
ACKNOWLEDGMENTS
This study was supported by the Spanish National Institute for Agricultural and Food Research and Technology (INIA; project RTA 2013-00051-C2-00) and the European Regional Development Fund (ERDF). R.Á.-A. is the recipient of a predoctoral fellowship from INIA (FPI-2015).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank the staff of the farm for their collaboration during the sampling visits. We also thank Eva Alonso (Epidemiology Service from Public Health Department, Basque Government) for her helpful comments.
The authors declare no conflict of interest.
REFERENCES
- 1.EFSA Panel on Animal Health and Welfare. 2010. Scientific opinion on Q fever. EFSA J 8:1595. doi: 10.2903/j.efsa.2010.1595. [DOI] [Google Scholar]
- 2.Georgiev M, Afonso A, Neubauer H, Needham H, Thiery R, Rodolakis A, Roest H, Stark K, Stegeman J, Vellema P, van der Hoek W, More S. 2013. Q fever in humans and farm animals in four European countries, 1982 to 2010. Euro Surveill 18:20407. doi: 10.2807/ese.18.08.20407-en. [DOI] [PubMed] [Google Scholar]
- 3.Porter SR, Czaplicki G, Mainil J, Guatteo R, Saegerman C. 2011. Q fever: current state of knowledge and perspectives of research of a neglected zoonosis. Int J Microbiol 2011:248418. doi: 10.1155/2011/248418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eldin C, Melenotte C, Mediannikov O, Ghigo E, Million M, Edouard S, Mege JL, Maurin M, Raoult D. 2017. From Q fever to Coxiella burnetii infection: a paradigm change. Clin Microbiol Rev 30:115–190. doi: 10.1128/CMR.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tissot-Dupont H, Amadei MA, Nezri M, Raoult D. 2004. Wind in November, Q fever in December. Emerg Infect Dis 10:1264–1269. doi: 10.3201/eid1007.030724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Brom R, van Engelen E, Roest HI, van der Hoek W, Vellema P. 2015. Coxiella burnetii infections in sheep or goats: an opinionated review. Vet Microbiol 181:119–129. doi: 10.1016/j.vetmic.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Madariaga MG, Rezai K, Trenholme GM, Weinstein RA. 2003. Q fever: a biological weapon in your backyard. Lancet Infect Dis 3:709–721. doi: 10.1016/S1473-3099(03)00804-1. [DOI] [PubMed] [Google Scholar]
- 8.Tigertt WD, Benenson AS, Gochenour WS. 1961. Airborne Q fever. Bacteriol Rev 25:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agerholm JS. 2013. Coxiella burnetii associated reproductive disorders in domestic animals–a critical review. Acta Vet Scand 55:13. doi: 10.1186/1751-0147-55-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer NC, Kierstead M, Key DW, Williams JC, Peacock MG, Vellend H. 1983. Placentitis and abortion in goats and sheep in Ontario caused by Coxiella burnetii. Can Vet J 24:60–61. [PMC free article] [PubMed] [Google Scholar]
- 11.Arricau-Bouvery N, Souriau A, Lechopier P, Rodolakis A. 2003. Experimental Coxiella burnetii infection in pregnant goats: excretion routes. Vet Res 34:423–433. doi: 10.1051/vetres:2003017. [DOI] [PubMed] [Google Scholar]
- 12.Rodolakis A, Berri M, Hechard C, Caudron C, Souriau A, Bodier CC, Blanchard B, Camuset P, Devillechaise P, Natorp JC, Vadet JP, Arricau-Bouvery N. 2007. Comparison of Coxiella burnetii shedding in milk of dairy bovine, caprine, and ovine herds. J Dairy Sci 90:5352–5360. doi: 10.3168/jds.2006-815. [DOI] [PubMed] [Google Scholar]
- 13.Roest HJ, van Gelderen B, Dinkla A, Frangoulidis D, van Zijderveld FG, Rebel J, van Keulen L. 2012. Q fever in pregnant goats: pathogenesis and excretion of Coxiella burnetii. PLoS One 7:e48949. doi: 10.1371/journal.pone.0048949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bruin A, van der Plaats RQ, de Heer L, Paauwe R, Schimmer B, Vellema P, van Rotterdam BJ, van Duynhoven YT. 2012. Detection of Coxiella burnetii DNA on small-ruminant farms during a Q fever outbreak in the Netherlands. Appl Environ Microbiol 78:1652–1657. doi: 10.1128/AEM.07323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kersh GJ, Fitzpatrick KA, Self JS, Priestley RA, Kelly AJ, Lash RR, Marsden-Haug N, Nett RJ, Bjork A, Massung RF, Anderson AD. 2013. Presence and persistence of Coxiella burnetii in the environments of goat farms associated with a Q fever outbreak. Appl Environ Microbiol 79:1697–1703. doi: 10.1128/AEM.03472-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whelan J, Schimmer B, de Bruin A, van Beest Holle MR, van der Hoek W, ter Schegget R. 2012. Visits on “lamb-viewing days” at a sheep farm open to the public was a risk factor for Q fever in 2009. Epidemiol Infect 140:858–864. doi: 10.1017/S0950268811001427. [DOI] [PubMed] [Google Scholar]
- 17.Gürtler L, Bauerfeind U, Blumel J, Burger R, Drosten C, Groner A, Heiden M, Hildebrandt M, Jansen B, Offergeld R, Pauli G, Seitz R, Schlenkrich U, Schottstedt V, Strobel J, Willkommen H. 2014. Coxiella burnetii—pathogenic agent of Q (Query) fever. Transfus Med Hemother 41:60–72. doi: 10.1159/000357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reimer LG. 1993. Q fever. Clin Microbiol Rev 6:193–198. doi: 10.1128/CMR.6.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Astobiza I, Barandika JF, Hurtado A, Juste RA, García-Pérez AL. 2010. Kinetics of Coxiella burnetii excretion in a commercial dairy sheep flock after treatment with oxytetracycline. Vet J 184:172–175. doi: 10.1016/j.tvjl.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 20.Joulié A, Laroucau K, Bailly X, Prigent M, Gasqui P, Lepetitcolin E, Blanchard B, Rousset E, Sidi-Boumedine K, Jourdain E. 2015. Circulation of Coxiella burnetii in a naturally infected flock of dairy sheep: shedding dynamics, environmental contamination, and genotype diversity. Appl Environ Microbiol 81:7253–7260. doi: 10.1128/AEM.02180-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneeberger PM, Wintenberger C, van der Hoek W, Stahl JP. 2014. Q fever in the Netherlands - 2007–2010: what we learned from the largest outbreak ever. Med Mal Infect 44:339–353. doi: 10.1016/j.medmal.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Brooke RJ, Kretzschmar ME, Mutters NT, Teunis PF. 2013. Human dose response relation for airborne exposure to Coxiella burnetii. BMC Infect Dis 13:488. doi: 10.1186/1471-2334-13-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astobiza I, Barandika JF, Ruiz-Fons F, Hurtado A, Povedano I, Juste RA, Garcia-Perez AL. 2011. Coxiella burnetii shedding and environmental contamination at lambing in two highly naturally infected dairy sheep flocks after vaccination. Res Vet Sci 91:e58–e63. doi: 10.1016/j.rvsc.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Kersh GJ, Wolfe TM, Fitzpatrick KA, Candee AJ, Oliver LD, Patterson NE, Self JS, Priestley RA, Loftis AD, Massung RF. 2010. Presence of Coxiella burnetii DNA in the environment of the United States, 2006 to 2008. Appl Environ Microbiol 76:4469–4475. doi: 10.1128/AEM.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bruin A, Janse I, Koning M, de Heer L, van der Plaats RQ, van Leuken JP, van Rotterdam BJ. 2013. Detection of Coxiella burnetii DNA in the environment during and after a large Q fever epidemic in the Netherlands. J Appl Microbiol 114:1395–1404. doi: 10.1111/jam.12163. [DOI] [PubMed] [Google Scholar]
- 26.de Rooij MM, Borlee F, Smit LA, de Bruin A, Janse I, Heederik DJ, Wouters IM. 2016. Detection of Coxiella burnetii in ambient air after a large Q fever outbreak. PLoS One 11:e0151281. doi: 10.1371/journal.pone.0151281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kersh GJ, Priestley RA, Hornstra HM, Self JS, Fitzpatrick KA, Biggerstaff BJ, Keim P, Pearson T, Massung RF. 2016. Genotyping and axenic growth of Coxiella burnetii isolates found in the United States environment. Vector Borne Zoonotic Dis 16:588–594. doi: 10.1089/vbz.2016.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori M, Boarbi S, Michel P, Bakinahe R, Rits K, Wattiau P, Fretin D. 2013. In vitro and in vivo infectious potential of Coxiella burnetii: a study on Belgian livestock isolates. PLoS One 8:e67622. doi: 10.1371/journal.pone.0067622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eldin C, Angelakis E, Renvoise A, Raoult D. 2013. Coxiella burnetii DNA, but not viable bacteria, in dairy products in France. Am J Trop Med Hyg 88:765–769. doi: 10.4269/ajtmh.12-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneeberger PM, Hermans MH, van Hannen EJ, Schellekens JJ, Leenders AC, Wever PC. 2010. Real-time PCR with serum samples is indispensable for early diagnosis of acute Q fever. Clin Vaccine Immunol 17:286–290. doi: 10.1128/CVI.00454-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Astobiza I, Tilburg JJ, Pinero A, Hurtado A, Garcia-Perez AL, Nabuurs-Franssen MH, Klaassen CH. 2012. Genotyping of Coxiella burnetii from domestic ruminants in northern Spain. BMC Vet Res 8:241. doi: 10.1186/1746-6148-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berri M, Rousset E, Champion JL, Russo P, Rodolakis A. 2007. Goats may experience reproductive failures and shed Coxiella burnetii at two successive parturitions after a Q fever infection. Res Vet Sci 83:47–52. doi: 10.1016/j.rvsc.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 33.de Cremoux R, Rousset E, Touratier A, Audusseau G, Nicollet P, Ribaud D, David V, Le Pape M. 2012. Coxiella burnetii vaginal shedding and antibody responses in dairy goat herds in a context of clinical Q fever outbreaks. FEMS Immunol Med Microbiol 64:120–122. doi: 10.1111/j.1574-695X.2011.00893.x. [DOI] [PubMed] [Google Scholar]
- 34.Schets FM, de Heer L, de Roda Husman AM. 2013. Coxiella burnetii in sewage water at sewage water treatment plants in a Q fever epidemic area. Int J Hyg Environ Health 216:698–702. doi: 10.1016/j.ijheh.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Glazunova O, Roux V, Freylikman O, Sekeyova Z, Fournous G, Tyczka J, Tokarevich N, Kovacava E, Marrie TJ, Raoult D. 2005. Coxiella burnetii genotyping. Emerg Infect Dis 11:1211–1217. doi: 10.3201/eid1108.041354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurtado A, Alonso E, Aspiritxaga I, Lopez E, Ocabo IB, Barandika JF, Fernandez-Ortiz DE, Murua JI, Urbaneja F, Alvarez-Alonso R, Jado I, Garcia-Perez AL. 2017. Environmental sampling coupled with real-time PCR and genotyping to investigate the source of a Q fever outbreak in a work setting. Epidemiol Infect 145:1834–1842. doi: 10.1017/S0950268817000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huijsmans CJ, Schellekens JJ, Wever PC, Toman R, Savelkoul PH, Janse I, Hermans MH. 2011. Single-nucleotide-polymorphism genotyping of a Coxiella burnetii during a Q fever outbreak in The Netherlands. Appl Environ Microbiol 77:2051–2057. doi: 10.1128/AEM.02293-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klee SR, Tyczka J, Ellerbrok H, Franz T, Linke S, Baljer G, Appel B. 2006. Highly sensitive real-time PCR for specific detection and quantification of Coxiella burnetii. BMC Microbiol 6:2. doi: 10.1186/1471-2180-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]