When starved of nutrients, some bacterial species develop metabolically dormant spores that can persist in a viable state in the environment for several years. The outermost layers of spores are of particular interest since (i) these represent the primary site for interaction with the environment and (ii) the protein constituents may have biotechnological applications. The outermost layer, or exosporium, in Bacillus megaterium QM B1551 spores is of interest, as it is morphologically distinct from the exosporia of spores of the pathogenic Bacillus cereus family. In this work, we provide evidence that structurally important protein constituents of the Bacillus megaterium exosporium are different from those in the Bacillus cereus family. We also show that one of these proteins, when purified, can assemble to form sheets of exosporium-like material. This is significant, as it indicates that spore-forming bacteria employ different proteins and mechanisms of assembly to construct their external layers.
KEYWORDS: Bacillus, spore coat
ABSTRACT
The exosporium of Bacillus megaterium QM B1551 spores is morphologically distinct from exosporia observed for the spores of many other species. Previous work has demonstrated that unidentified genes carried on one of the large indigenous plasmids are required for the assembly of the Bacillus megaterium exosporium. Here, we provide evidence that pBM600-encoded orthologues of the Bacillus subtilis CotW and CotX proteins, which form the crust layer in spores of that species, are structural components of the Bacillus megaterium QM B1551 spore exosporium. The introduction of plasmid-borne cotW and orthologous cotX genes to the PV361 strain, which lacks all indigenous plasmids and produces spores that are devoid of an exosporium, results in the development of spores with a rudimentary exosporium-type structure. Additionally, purified recombinant CotW protein is shown to assemble at the air-water interface to form thin sheets of material, which is consistent with the idea that this protein may form a basal layer in the Bacillus megaterium QM B1551 exosporium.
IMPORTANCE When starved of nutrients, some bacterial species develop metabolically dormant spores that can persist in a viable state in the environment for several years. The outermost layers of spores are of particular interest since (i) these represent the primary site for interaction with the environment and (ii) the protein constituents may have biotechnological applications. The outermost layer, or exosporium, in Bacillus megaterium QM B1551 spores is of interest, as it is morphologically distinct from the exosporia of spores of the pathogenic Bacillus cereus family. In this work, we provide evidence that structurally important protein constituents of the Bacillus megaterium exosporium are different from those in the Bacillus cereus family. We also show that one of these proteins, when purified, can assemble to form sheets of exosporium-like material. This is significant, as it indicates that spore-forming bacteria employ different proteins and mechanisms of assembly to construct their external layers.
INTRODUCTION
In response to nutrient exhaustion, bacteria of the orders Bacillales and Clostridiales initiate sporulation, a tightly regulated process of cellular differentiation that results in the formation of a metabolically dormant endospore (1). Virtually all spores share the same general morphological features, in which a membrane-bound protoplast forms a central core that is enveloped by two layers of structurally distinct peptidoglycans. In turn, a second membrane and then a multilayered proteinaceous coat surround the outer layer of cortical peptidoglycan. The primary function of the coat is to protect the interior of the spore from chemical and enzymatic attack, whereas the cortical peptidoglycan is essential for maintenance of the reduced hydration state of the spore core and, by inference, for metabolic dormancy (2, 3).
The greatest variation in spore morphology is observed in an outermost structure referred to as the exosporium, which is present in many—but not all—species, and which can adopt a range of architectures and surface features in different species (4–6). The best studied of these exosporial structures belong to spores of the Bacillus cereus family, which includes Bacillus anthracis, the causative agent of anthrax. The exosporium in B. cereus family spores is a thin, sheath-like structure that is typically significantly larger and more flexible than the densely packed spore within. Its biological function(s) has not been unequivocally defined; however, a series of electron crystallographic studies have revealed that it has an ordered structure punctuated with an array of what appear to be openings (7–10). These openings support the often-stated idea that the exosporium, as the spore's interface with the environment, serves in one capacity as a molecular sieve (8).
Spores of several strains of Bacillus megaterium have also been the focus of studies on exosporium morphology and composition (11–16). The species is comprised of many strains, which form a clade within the Bacillus genus that is quite distinct from both the B. subtilis and B. cereus clades. The QM B1551 strain in particular is marked by a distinctive close-fitting exosporium, which when viewed under the electron microscope appears to comprise two identical hemispheres, which meet to form a prominent ridge that envelops the circumference of the spore. Similarly, in contrast to B. cereus spores, which are entirely surrounded by a nap of short hair-like fibers, the exosporium of B. megaterium QM B1551 spores has a nap localized to only a single pole of the spore (although in both species the fibers are formed from orthologous collagen-like BclA proteins) (11). A recent study revealed that key structural components of the B. megaterium QM B1551 exosporium are encoded on one, and possibly on two, of seven plasmids indigenous to this strain (11). Whereas the identities of the plasmids are known—plasmid pBM600 is certainly indispensable to exosporium formation, while plasmid pBM500 may also be important—the identities of key structural genes have yet to be established. As reported in the same study, only a limited number of orthologues of known B. cereus family exosporium genes were identified as being present in the B. megaterium QM B1551 genome. These included the nap proteins BclA and BclB (encoded on pBM500) and the BxpB protein (also known as ExsFA; encoded on pBM600); the latter is required for attachment of the BclA fibers that form the B. cereus family exosporial nap (17, 18), but it appeared to be dispensable for both exosporium assembly and nap attachment in B. megaterium spores (11).
In the current study, we have investigated the role of pBM600-encoded orthologues of the B. subtilis CotW and CotX proteins in B. megaterium spores. These proteins form part of the outermost layer of B. subtilis spores, the so-called crust, which may represent a rudimentary exosporium-type structure in this species (19, 20). Here, we report on the expression and localization of these proteins in B. megaterium QM B1551 spores and provide evidence that they may form the basal layer or key structural components of the exosporium.
RESULTS
Identification of cotW, cotX1, and cotX2 in B. megaterium QM B1551.
Proteins located in the outermost layers of B. subtilis spores, including in the crust, are encoded in two adjacent operons comprising the cotVWXYZ genes (21). BLASTP searches conducted with the CotV through CotZ protein sequences against the B. megaterium genome yielded orthologues of only the CotW and CotX proteins (orthologues of B. subtilis crust and B. cereus family exosporium proteins encoded in the B. megaterium QM B1551 genome are detailed in Table S1 in the supplemental material). The B. megaterium cotW gene is located on the antisense strand of plasmid pBM600 and has the locus identifier BMQ_pBM60030. The sequence encodes a predicted protein of 91 amino acids, with a predicted molecular mass of 10.2 kDa. The predicted protein shares 35% sequence identity at the amino acid level with B. subtilis CotW.
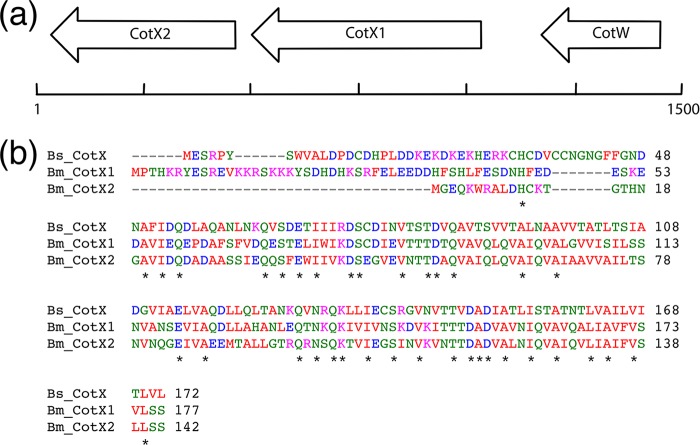
Open reading frames for two orthologues of CotX were identified immediately downstream of cotW (Fig. 1a) on the same antisense strand of plasmid pBM600 (locus identifiers BMQ_pBM60029, provisionally referred to as CotX1, and BMQ_pBM60028, which is referred to as CotX2). CotX1 is predicted to comprise 177 amino acids, has a predicted molecular mass of 19.9 kDa, and shares 34% sequence identity at the amino acid level with B. subtilis CotX (Fig. 1b). The predicted CotX2 protein is truncated at the N terminus with respect to CotX1 and is predicted to comprise 142 amino acids, with a molecular mass of 15.2 kDa. The CotX2 protein shares 38% sequence identity with B. subtilis CotX and 53% with B. megaterium CotX1. Searches for orthologues of B. subtilis CotV, CotY, and CotZ did not result in any hits, indicating that either these proteins are below the cutoff employed for BLAST searches or they are not encoded within the B. megaterium QM B1551 genome.
FIG 1.
(a) Genetic organization of cotX2 (BMQ_pBM60028), cotX1 (BMQ_pBM60029), and cotW (BMQ_pBM60030) on plasmid pBM600 of B. megaterium QM B1551. DNA sequence and reverse transcription-PCR (RT-PCR) analyses indicate that cotW is monocistronic, whereas cotX1 and cotX2 are organized as an operon comprising both genes. (b) Clustal Omega sequence alignment of B. subtilis (Bs) CotX with B. megaterium (Bm) CotX1 and CotX2. Asterisks denote conserved residues.
Transcriptional analysis of cotW, cotX1, and cotX2 in B. megaterium QM B1551.
Reverse transcription-PCR (RT-PCR) was employed to determine whether the cotW, cotX1, and cotX2 genes are transcribed during sporulation. RNA was collected at various time points during sporulation and then converted to cDNA, which was used in RT-PCRs with gene-specific primers. The resultant RT-PCR profiles indicate that all three genes are transcribed during sporulation (see Fig. S1 in the supplemental material). Transcription of cotW is initiated shortly after entry to the stationary phase and appears to peak approximately 5 to 6 h into sporulation. Examination of the gene's upstream region revealed a possible sigma E (σE) consensus sequence (7 out of 8 residues match the consensus −35 sequence, and 3 out of 8 match the −10 sequence [22]), indicating early expression in the mother cell. Additionally, a potential rho-independent terminator sequence (GCGCCTTTTTTAGGCGT) was identified immediately downstream of cotW (free energy of stem-loop region, −8.8 kcal/mol), indicating that this gene is monocistronic.
Both cotX1 and cotX2 were transcribed throughout sporulation, with apparent maxima from 6 to 8 h into sporulation. Faint bands detected prior to the time defined as entry to the stationary phase presumably reflect asynchronous sporulation. A perfect match with the −10 region of the sigma K (σK) consensus sequence (22) was identified at an appropriate location upstream of the putative start codon for cotX1, which is commensurate with late-stage transcription in the mother cell (4). The close proximity of the cotX1 and cotX2 genes, which are separated by 37 nucleotides, is indicative of genetic organization as an operon. RT-PCR experiments using primers that spanned the 5′ end of cotX1 and the 3′ end of cotX2 provided evidence for a single mRNA that encompassed both genes, indicating that the CotX structural genes are transcribed as a single transcriptional unit (Fig. S1).
Expression and localization of CotW and CotX fluorescence fusion proteins in B. megaterium QM B1551.
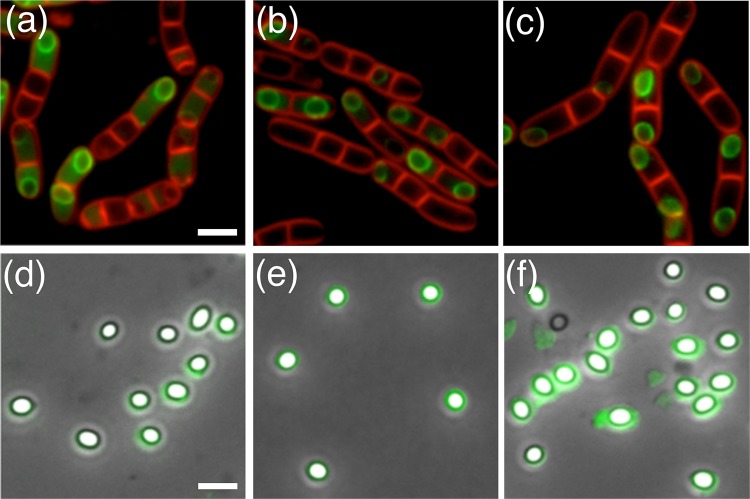
A series of B. megaterium strains were designed to express CotW, CotX1, and CotX2 as green fluorescent protein (GFP) fusion proteins with a view to examining, via fluorescence microscopy, the expression and localization of these proteins during sporulation. In the first set of experiments, cotW-gfp, cotX1-gfp, and cotX2-gfp constructs were introduced, independently, to wild-type B. megaterium cells and colonies that had integrated the respective plasmids at the cloned loci selected for analysis by fluorescence microscopy. Fluorescence associated with expression of the CotW-GFP protein was evident in the mother cell 2 to 3 h into sporulation and prior to the development of refractile forespores (Fig. 2; see also Fig. S2a in the supplemental material). The protein was observed to form a fluorescent shell around the developing spore at or around the same time as the development of refractility, with no obvious preference for initiation of localization at either of the poles (Fig. S2b). Fluorescence intensity was observed to increase in both the mother cell and forespore surface as sporulation proceeded (Fig. S2c). However, although the mature spores retained some fluorescence, the intensity was noticeably reduced compared to levels associated with spores immediately prior to mother cell lysis (Fig. 2d; see also Fig. S2d).
FIG 2.
Fluorescence micrographs of sporulating B. megaterium QM B1551 (a) cotW-gfp cells, (b) gfp-cotX1 cells, and (c) cotW-cotX1-cotX2-gfp cells. Fluorescence micrographs overlaid with phase-contrast images are shown for B. megaterium QM B1551 (d) cotW-gfp spores, (e) gfp-cotX1 spores, and (f) cotW-cotX1-cotX2-gfp spores. Bar, 2.5 μm.
Since the CotX1-GFP C-terminal fusion protein failed to localize to the spore surface during sporulation (data not shown), a strain engineered to express an N-terminal fusion protein (CotX1-GFPN) from a low-copy-number episomal plasmid was constructed (JM121). Fluorescence associated with expression of the CotX1-GFPN protein was first observed at the mid to late stages of sporulation, by which time the forespore had become refractile (Fig. 2; see also Fig. S3 in the supplemental material). In contrast to CotW-GFP, only very faint fluorescence was observed throughout the mother cell cytoplasm, with the protein instead appearing to localize immediately to the developing forespore. The mature spores were observed to retain relatively bright fluorescence, although this was of a somewhat nonuniform and patchy distribution.
The timing of expression of the CotX2-GFP protein was similar to that of the CotX1 protein, as one would expect for genes that are organized in an operon. As with CotX1-GFPN, fluorescence was observed principally around the developing forespore and not in the mother cell cytoplasm (Fig. 2; see also Fig. S4 in the supplemental material), perhaps indicating that both CotX proteins have a strong affinity for the developing spore surface. As sporulation proceeded, CotX2-GFP formed a discrete ring around the developing forespore, resulting in bright green fluorescent shells, which were retained in the mature spores. Expression of CotW-GFP and CotX2-GFP from an episomal plasmid was examined for consistency with the CotX1-GFPN construct. In both cases fluorescence was observed but was noticeably dimmer than that observed when expressed from native loci, perhaps as a result of competition for binding sites with the native proteins (data not shown).
Expression and localization of CotW and CotX fluorescence fusion proteins in B. megaterium PV361.
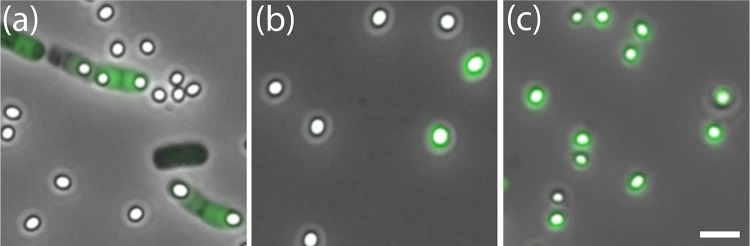
The episomal plasmid-based constructs described above permitted complementation-type experiments to be conducted in B. megaterium PV361, which is isogenic with strain QM B1551 but which lacks all indigenous plasmids and has no exosporium (11). Accordingly, the cotW-gfp, cotX1-gfp, and cotX2-gfp plasmids were introduced to the PV361 strain with a view to ascertaining whether any of these proteins could localize and assemble to the developing forespore in an exosporium-deficient strain (Table 1). The pattern of expression of CotW-GFP in the PV361 background was similar to that observed in the wild-type background, although the fluorescence signal was noticeably enhanced (Fig. 3). The fusion protein was visible as bright green, diffuse fluorescence in the mother cell throughout sporulation, although it was not observed to localize specifically around the forespore. The mature spores lacked any observable fluorescence, indicating that the protein was unable to localize or adhere to the spore surface, perhaps as a result of this construct lacking the cotX1 and cotX2 genes. It is possible also that the GFP moiety may interfere with localization of the CotW-GFP fusion protein to the spore.
TABLE 1.
Strains of B. megaterium and plasmids used in this study
| Strain, transformant, or plasmid | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| QM B1551 | Wild-type strain | P. S. Vary |
| PV361 | Plasmidless isogenic variant of QM B1551 | P. S. Vary |
| QM B1551 transformants | ||
| JM106 | cotW::pVLG6 (gfp) Cmr | This study |
| JM107 | cotX1::pVLG6 (gfp) Cmr | This study |
| JM108 | cotX2::pVLG6 (gfp) Cmr | This study |
| JM118 | pHT315-cotW-gfp MLSr | This study |
| JM119 | pHT315-cotW-cotX1-gfp MLSr | This study |
| JM120 | pHT315-cotW-cotX1-cotX2-gfp MLSr | This study |
| JM121 | pHT315-gfp-cotX1 MLSr (i.e., encoding GFP at the N terminus of CotX1) | This study |
| PV361 transformants | ||
| JM127 | pHT315-cotW-gfp MLSr | This study |
| JM128 | pHT315-cotW-cotX1-gfp MLSr | This study |
| JM129 | pHT315-gfp-cotX1 MLSr (i.e., encoding GFP at the N terminus of CotX1) | This study |
| JM130 | pHT315-cotW-cotX1-cotX2-gfp MLSr | This study |
| AG136 | pHT315-cotX2-gfp MLSr | This study |
| AG137 | pHT315-gfp-cotX2 MLSr (i.e., encoding GFP at the N terminus of CotX2) | This study |
| AG138 | pHT315-cotX1-cotX2-gfp MLSr | This study |
| JM131 | pHT315-cotW MLSr | This study |
| JM132 | pHT315-cotX1 MLSr | This study |
| JM133 | pHT315-cotX2 MLSr | This study |
| JM134 | pHT315-cotX1-cotX2 MLSr | This study |
| JM135 | pHT315-cotW-cotX1-cotX2 MLSr | This study |
| Plasmids | ||
| pVLG6 | E. coli-Bacillus shuttle plasmid designed to create gfp fusions; orits, Ampr, Cmr, gfp | 37 |
| pHT315 | Low-copy-no. episomal plasmid derived from B. thuringiensis; Ampr, MLSr | 38 |
| pBadcLIC-GFP | LIC vector for E. coli, gfp, Ampr | 36 |
Cmr, chloramphenicol resistance; MLSr, macrolide-lincosamide-streptogramin B resistance; Ampr, ampicillin resistance.
FIG 3.
Fluorescence micrographs overlaid with phase-contrast images are shown for B. megaterium PV361 (a) cotW-gfp sporulating cells and mature spores, (b) gfp-cotX1 spores, and (c) cotW-cotX1-cotX2-gfp spores. Bar, 2.5 μm.
Fluorescence associated with the CotX1-GFPN protein was apparent in the PV361 background after completion of engulfment and the development of forespore refractility, similar to that observed previously when introduced to wild-type cells (Fig. 3; see also Fig. S5 in the supplemental material). Fluorescent foci were observed to localize to the developing forespore in addition to the diffuse green cytoplasm. As sporulation progressed, the fluorescence intensity increased, and the protein was observed to completely envelop the developing spore. Intriguingly, the fluorescent shell was retained on the majority of mature spores, although some were observed to have no fluorescence. These observations indicate that the CotX1-GFPN fusion protein was able to localize to the spore surface in the presumed absence of an exosporium, although it does not appear to have been anchored sufficiently on at least a proportion of the population. PV361 cells complemented with cotX2-gfp, in the first instance in tandem with the cotW and cotX1 structural genes, were able to express and assemble the fusion protein around the developing forespore (Fig. 3). Spores of this strain mostly retained intense fluorescence, indicating that the fusion protein was attached securely to the surface of the exosporium-deficient spores. In contrast, no fluorescence was observed in sporulating cells or mature spores of PV361 cells containing plasmid-borne cotX2 placed under the control of the native promoter sequence, either when present individually or when located downstream of cotX1 (data not shown). Similar observations were made whether CotX2 was designed with N- or C-terminal GFP fusions. Hence, it seems that the CotW and CotX1 proteins are required for the correct expression and assembly of CotX2 to the spore surface.
Spore immunolabeling.
In order to ascertain whether the various GFP fusion proteins were accessible to antisera, and therefore potentially surface exposed, immunolabeling of spores of the various strains was conducted with anti-GFP antisera (Fig. S6). Of the wild-type background strains bearing fusion proteins, significant cross-reactivity with anti-GFP antisera was detected in CotX1-GFPN and CotX2-GFP spores. This was the case whether the proteins were expressed from the native locus or from episomal plasmids. CotW-GFP spores, which showed only a weak fluorescent signal, reacted to the antisera at levels comparable to the low-level nonspecific binding observed in wild-type and SleL-GFP spores (SleL is located in the inner coat [23]). Accordingly, these data indicate that both CotX orthologues are surface exposed in B. megaterium QM B1551 spores, raising the possibility that they are components of the exosporium. Similar results were obtained in immunolabeling experiments involving GFP fusions to CotW, CotX1, and CotX2 in the PV361 background (see Fig. S6 in the supplemental material). Again, only the CotX-GFP spores cross-reacted significantly with the antisera, indicating that even in the presumed absence of an exosporium, B. megaterium CotX proteins are localized to the spore surface. Notably, overlays of phase-contrast and fluorescence images show that the fluorescent signal is localized to the light gray region—perhaps the exosporium or interspace—that surrounds phase-bright spores in the wild-type background (Fig. S6b and c). However, in the PV361 background complemented with cotW-cotX1-cotX2-gfp, the fluorescent signal appears to be more closely associated with the surface of the phase-bright spores (Fig. S6f).
Electron microscopy analyses.
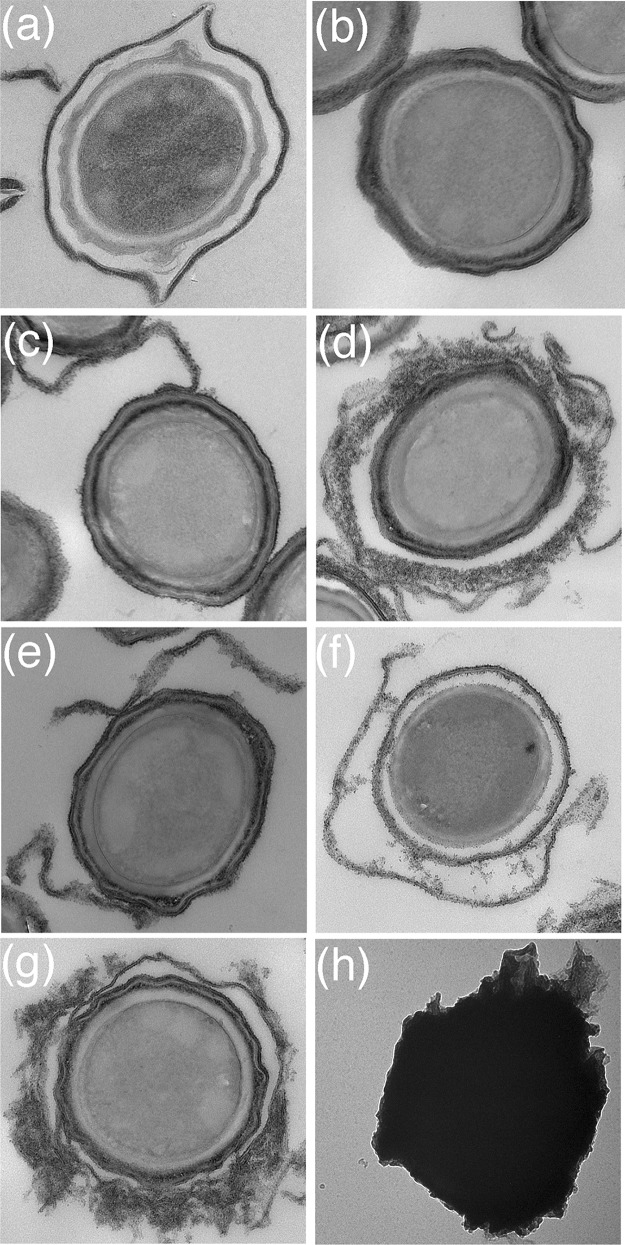
The observation that both CotX1 and CotX2 proteins appeared to be surface localized in wild-type and exosporiumless genetic backgrounds raised the possibility that these proteins are perhaps components of the exosporium and outer coat. Alternatively, they may contribute directly or otherwise to the assembly of an exosporium-type structure in the PV361 background. To investigate these hypotheses, electron microscopy studies were conducted on PV361 spores complemented with plasmid-borne copies of the cotW, cotX1, and cotX2 genes and presumed associated regulatory sequences. Micrographs of sectioned spores (Fig. 4) reveal that spores of strains individually complemented with cotW, cotX1, and cotX2, respectively, share typical PV361 spore morphologies. However, in each case, cables and/or sheets of material with a granular appearance were visible and appeared to be associated with the outer spore surface. This material is particularly evident in spores complemented with cotX1, which in some cases entirely envelops the spore. Additionally, the material appears to form a layer that is distinct from the outer spore coat, giving the appearance of an exosporium-coat interspace. Whereas similar structures are observed in spores of PV361 complemented with both cotX genes, the assembly of what most closely aligns with a rudimentary exosporium-type structure is most evident in spores complemented with both cotX genes and cotW (JM135). In this case, most spores show the additional layer of material, which again is largely separated from the spore coat by a distinct interspace. Thin-section transmission electron micrographs of these cells at different stages of sporulation are shown in Fig. S7 in the supplemental material.
FIG 4.
Thin-section transmission electron micrographs of (a) B. megaterium QM B1551 spores and (b) exosporium-deficient B. megaterium PV361 spores. Panels c through g show spores of strains in the PV361 background complemented with plasmid-borne copies of (c) cotW, (d) cotX1, (e) cotX2, (f) cotX1-cotX2, and (g) cotW-cotX1-cotX2. Panel h shows an intact, negatively stained PV361 cotW-cotX1-cotX2 spore, which is encased in an exosporium-like sheath.
Electron microscopy (EM) analysis of negatively stained intact spores generally supports the observations derived from sectioned spores (Fig. 4). Most of the spores complemented with individual genes of interest have smooth surfaces, reminiscent of those of the parental PV361 spores, although the EM grids show evidence of sheets and fragments of granular material. This has presumably been shed from the spore surface. Spores of the triple-complemented strain, however, are encased in sheaths of what we assume to be similar material, which appears to be akin to a rudimentary exosporium structure. Furthermore, fluorescence microscopy analyses conducted with PV361 spores complemented with all three genes indicate that this apparent exosporium-type material is resistant to shear associated with passage through a cell disrupter and sonication (see Fig. S8 in the supplemental material).
Heterologous expression and self-assembly of B. megaterium CotW.
Results presented above indicate that the B. megaterium CotW and CotX proteins have an important role in the assembly of the spore exosporium and may even be major structural components of this outermost layer of the spore. Literature reports on the crystalline nature of the exosporium of other species of Bacillus, principally in B. cereus/anthracis (7, 8) but also alluded to in early reports on B. megaterium spores (24, 25), imply that major structural proteins of the exosporium should have properties of self-assembly. Indeed, this has been demonstrated recently for the ExsY protein in B. cereus family spores (26), while heterologously expressed B. subtilis coat proteins have also been shown to assemble into various macromolecular assemblies (26). Accordingly, genes encoding B. megaterium CotW, CotX1, and CotX2 were cloned both with and without C-terminal GFP and His10 moieties into arabinose-inducible vectors for expression in Escherichia coli. Expression of the various recombinant proteins was examined under variable arabinose concentrations and at temperatures ranging from 16 to 37°C. Of the proteins and conditions tested, expression of CotW-GFP appeared most successful when examined by fluorescence microscopy, with bright green fluorescence and few inclusion bodies being observed in the E. coli cytoplasm when induced with 0.001% arabinose at 30°C. In contrast, the CotX proteins failed to express to any appreciable level in E. coli, with or without the GFP fusion, and also when examined under conditions designed for coexpression (data not shown).
Initial tests for self-assembly of the CotW protein (i.e., without the GFP moiety) were conducted by applying 5 μl of purified CotW (45 mg/ml) onto a glass microscope slide and incubating it at 37°C until dry. A drop of water was then placed on the dried material, and the slide was examined by phase-contrast microscopy. These analyses revealed the presence of sheet- or film-like material, which was particularly evident at the air/water interface (Fig. 5a). The proteinaceous film was observed to increase in size while viewed under the microscope, indicating that B. megaterium CotW can self-assemble into larger macromolecular structures. Additionally, staining the CotW protein with thioflavin T revealed bright green fluorescence, indicating that the protein has a beta-rich structure (see Fig. S9 in the supplemental material).
FIG 5.
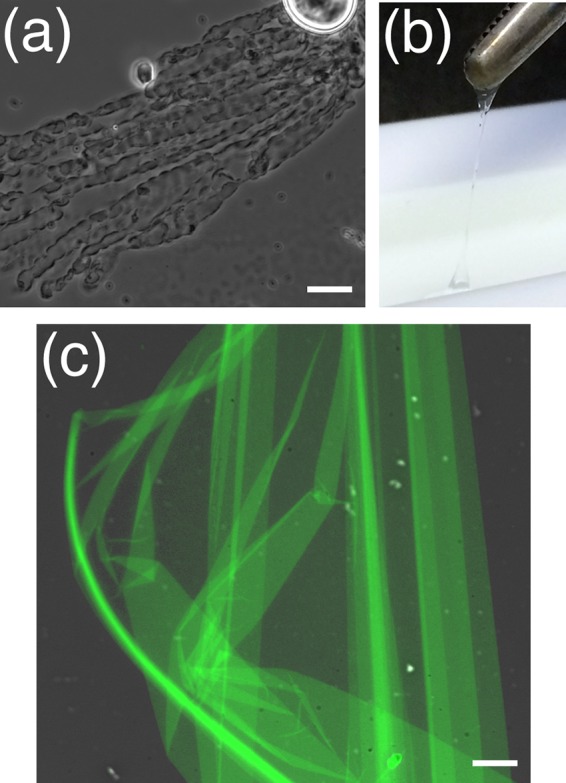
Assembly of recombinant B. megaterium CotW and CotW-GFP into macroscopic structures. (a) Phase-contrast micrograph of CotW sheet formation at the air-water interphase. (b) Removal of a CotW-GFP film from the air-water interphase with tweezers. (c) Overlay of fluorescence and phase-contrast micrographs of assembled CotW-GFP. Bars, 25 μm (a) and 15 μm (c).
The procedure was then scaled up and investigated further, this time using the CotW-GFP fusion protein, where 2.5 mg of purified protein was added to 300 ml of water in a Langmuir trough. The protein was incubated overnight under ambient conditions, and upon examination, it was revealed to have formed a thin film at the air-water interface that could be removed and gathered into a fiber with tweezers (Fig. 5b). When examined under the fluorescence microscope, the substance was observed to consist of a very fine sheet of green fluorescent material (Fig. 5c), providing strong evidence that the films were fabricated from self-assembled CotW-GFP protein.
DISCUSSION
The current paper examines the role of orthologues of the B. subtilis spore crust proteins CotW and CotX in B. megaterium QM B1551 spores. These pBM600-encoded proteins were the focus of attention, since previous work indicated that genes essential for the presence of an exosporium in B. megaterium QM B1551 spores are located on this plasmid (11). Additionally, aside from BxpB and BclA/B, orthologues of known B. cereus exosporium proteins appear to be absent from the B. megaterium genome. Notably, only orthologues of CotW and CotX (of which there are two) appear to be encoded in the B. megaterium genome, as opposed to the cotVWXYZ gene cluster in B. subtilis. The apparent absence of key exosporium and/or outer-spore coat genes identified from other species in the B. megaterium genome presumably accounts for the significant morphological differences evident in B. megaterium spores compared to those of the aforementioned species.
The study has revealed that all three genes—cotW, cotX1 and cotX2—are transcribed in the mother cell during sporulation, with the resultant proteins localizing to the spore during development. Despite strong fluorescence in the mother cell, mature CotW-GFP spores display only weak fluorescence, indicating that the fusion protein may not assemble correctly on the spore or perhaps that it has a dependency on the presence of one or both of the CotX proteins (the cotW-gfp construct probably has a polar effect on expression of the downstream cotX genes). Immunofluorescent labeling experiments conducted with anti-GFP antisera indicate that both CotX proteins—but not CotW—are located on the surface of wild-type QM B1551 spores and on spores of complemented exosporiumless PV361-derived strains. An exterior location for the CotX proteins with respect to CotW is consistent with ellipsoid localization microscopy measurements on the location of these proteins in B. megaterium spores (23).
Perhaps the most significant findings reported here are derived from electron microscopy analyses, namely, that the introduction of the cotW and cotX genes to the plasmidless PV361 strain resulted in the formation of a rudimentary exosporium-type structure around the resultant spores. The development of this structure, which is clearly distinguishable by EM analysis from the spore coat, appears most advanced when cotW, cotX1, and cotX2 are all present, although spores produced by PV361 strains complemented with individual cotW or cotX genes also have cables and sheets of granular material associated with the outermost layers of the spores. The observation that the CotW and CotX proteins appear capable of forming a rudimentary exosporium indicates that these proteins have a structural role in the B. megaterium exosporium, which is consistent with previous suggestions that the B. subtilis spore crust represents a vestigial exosporium (27–29). Orthologues of CotW and CotX are also encoded in the genomes of several B. cereus strains but are absent from B. anthracis (27), indicating that they do not function as structural components of exosporia across the genus. Similarly, the B. subtilis CotY crust protein and its paralogue ExsY, both of which are absent from the B. megaterium genome, appear to have crucial structural roles in the exosporia of B. cereus and B. anthracis (9, 30, 31).
Unfortunately, despite several attempts, it has not yet proved possible to create cotW or cotX null strains in the QM B1551 background in order to further investigate the role of the products of these genes in exosporium assembly. However, immunofluorescence labeling experiments provide some evidence that the cables/sheets of exosporium material are comprised of the CotX proteins. Similarly, the CotW protein was observed to form intact sac-like structures in complemented PV361 spores when examined by negative stain transmission electron microscopy (TEM), which indicates that this protein forms a component of the basal layer of the B. megaterium exosporium. The ability of purified recombinant CotW to self-assemble into extended sheet-like structures provides further evidence to support this hypothesis. Equally, the ability to self-assemble into macromolecular structures does not necessarily mean that CotW is a part of the exosporium, since a number of B. subtilis spore coat proteins have been shown to assemble into a variety of macromolecular structures (26).
Regardless, the relatively disordered appearance of the exosporium-type structure in cotW-cotX-complemented PV361 spores is significantly different from the compact, walnut-like exosporium evident in wild-type B. megaterium QM B1551 spores. It seems certain, therefore, that additional as-yet-unidentified proteins have important roles in the assembly and structure of the B. megaterium exosporium. Candidate proteins include BMQ_2983, which was identified from bioinformatic analyses as being cysteine rich, a property that is generally important in promoting self-assembly to higher-ordered structures via disulfide bond formation (9). Intriguingly, both CotX proteins studied in this work contain only a single cysteine residue, whereas CotW contains none, indicating that assembly to higher-ordered structures is driven by other mechanisms.
Finally, a number of earlier studies speculate that the exosporium of B. megaterium QM B1551 spores contains a galactosamine-6-phosphate polymer skeleton that confers rigidity to the structure (14, 25, 32). A number of genes that may have a role in the synthesis of such a polymer, including BMQ_pBM60014 and BMQ_pBM60015, are carried, typically in clusters, on plasmid pBM600. Hence, it may well be that carbohydrate components have a role in defining the morphology of exosporia in B. megaterium, and perhaps in other species. This possibility and the role of plasmid-borne genes in exosporium development are areas we consider worthy of future investigation.
MATERIALS AND METHODS
Bacterial strains and preparation of spores.
B. megaterium strains employed in this study, which were all isogenic with the QM B1551 strain, were cultured routinely at 30°C on LB medium supplemented where appropriate with antibiotics (Table 1). Transformation was achieved using standard polyethylene glycol-mediated procedures, followed by recovery on hypertonic medium supplemented with antibiotics (33). Strains that had integrated pVLG6 plasmids to create gfp fusions were isolated, and the construct was verified by PCR, essentially as described previously (34). Spores were prepared by nutrient exhaustion in supplemented nutrient broth and subsequently purified by repeated rounds of centrifugation and resuspension of spore pellets in ice-cold water (33).
Molecular biology procedures.
Transcriptional analysis from loci of interest was carried out by RT-PCR using gene-specific primers designed to amplify ∼400-bp DNA fragments (35). Two approaches were used to create strains bearing translational gfp fusions to genes of interest. In the first, open reading frames (ORFs) or genomic regions of interest were amplified by PCR, using oligonucleotides that incorporated appropriate restriction sites at the 5′ and 3′ ends. PCR amplicons were then purified, digested, and ligated with the integration vector pVLG6, which had been restricted with the same enzymes. Ligation mixtures were used to transform E. coli cells to carbenicillin resistance, with the resultant plasmids being used subsequently to transform B. megaterium cells to chloramphenicol resistance. Strains that had undergone homologous recombination to integrate plasmids at cloned loci were identified by PCR after incubation at nonpermissive temperature (42°C) in the presence of antibiotics. The second approach involved the use of the low-copy-number episomal pHT315 vector. Here, PCR was used to amplify ORFs of interest from B. megaterium QM B1551 genomic DNA and the gfp gene, which were then assembled using the Klenow assembly method (https://openwetware.org/wiki/Klenow_Assembly_Method:_Seamless_cloning). In most cases, gfp was positioned to encode in-frame C-terminal fusion proteins, although in strains JM121, JM129, and AG137, gfp preceded the respective cotX ORFs to create N-terminal fusion proteins. The various PCR amplicons were ligated with linearized pHT315, and the resultant plasmids were used to transform B. megaterium to erythromycin and lincomycin resistance. A similar pHT315-based approach was used to introduce cotW, cotX, and variants thereof, minus the gfp moiety, to create strains intended for electron microscopy analyses. Oligonucleotides used in this work are detailed in Table 2. Escherichia coli DH5α, cultured in LB medium supplemented with 50 μg/ml carbenicillin, was used for cloning and plasmid propagation procedures.
TABLE 2.
Oligonucleotide primers used in this work
| Primer | Sequence (5′–3′)a |
|---|---|
| Primers used to construct pVLG6-gfp fusion plasmids | |
| cotW For | GCGAAGCTTATGATAGATACATATAGTATTATGAC |
| cotW Rev | GGCGAATTCCTTTGGTTTTGTTTTTGGAG |
| cotX1 For | GCGAAGCTTATGCCAACTCATAAACGTTATG |
| cotX1 Rev | GGCCTGCAGAGAACTAAGGACAGAAACAAATAC |
| cotX2 For | GCGAAGCTTATGGGTGAACAAAAATGGAGAGC |
| cotX2 Rev | GGCGAATTCTGAGCTAAGTAGAGAAACAA |
| Primers used to create pHT315-based complementation plasmids | |
| cotW For | CCGATCGGATCCTAAAACAAACTAGTGGATAATGTC |
| cotW Rev | CCGATCGGATCCTTACTTTGGTTTTGTTTTTGGAGAAGTGGCC |
| cotX1 For | CCGATCGGATCCCCTCTTAAAAGCGCCTTTTTTAGGCG |
| cotX1 Rev | CCGATCGGATCCTTAAGAACTAAGGACAGAAACAAATACTGC |
| cotX2 Rev | CCGATCGGATCCTTATGAGCTAAGTAGAGAAACAAATATTGCG |
| cotX1 Inverse Rev | GGAAAGGAGTGTTAATCATGGGTGAACAAAAATGGAGAGCACTAGACC |
| cotX2 Inverse For | GATTAACACTCCTTTCCTTATATGAAATAATATATG |
| gfp Rev | CCGATCGGATCCTTATTTGTATAGTTCATCCATGCC |
| Primers used to construct pHT315-gfp-cotX1 | |
| cotX1 Inverse For | GGGGATCAGGTGGTGGAGGCTCACCAACTCATAAACGTTATGAGTCTCGT |
| PcotX1 Inverse Rev | GTGAAAAGTTCTTCTCCTTTACTCATGATTAACACTCCTTTCCTTATATG |
| gfp For | CATATAAGGAAAGGAGTGTTAATCATGAGTAAAGGAGAAGAACTTTTC |
| N-gfp Rev(2) | CTCCACCACCTGATCCCCCACCGCCAGATCCGCCACCTCCTTTGTATAGTTCATCCATGCCATGTGTAATCC |
| Primers used to construct pHT315-cotX2-gfp | |
| pHT315-PcotX2 For | CTATGACCATGATTACGCCAAGCTTTTCTGATTCCTCTTAAAAGCGCC |
| PcotX2 Rev | GTGCTCTCCATTTTTGTTCACCCATGATTAACACTCCTTTCCTTATATG |
| PcotX2 For | CATATAAGGAAAGGAGTGTTAATCATGGGTGAACAAAAATGGAGAGCAC |
| cotX2-gfp Rev(1) | CCAGTGAAAAGTTCTTCTCCTTTACTGCCTCCGCCTTTTGCTGCTGCTTCTCCGCCTCCTGAGCTAAGTAGAGAAACAAATATTG |
| cotX2-gfp For(1) | TTGTTTCTCTACTTAGCTCAGGAGGCGGAGAAGCAGCAGCAAAAGGCGGAGGCAGTAAAGGAGAAGAACTTTTCACTGG |
| pHT315-gfp Rev | GTTGTAAAACGACGGCCAGTGAATTCTTATTTGTATAGTTCATCCATGCC |
| Primers used to construct pHT315-cotX1-cotX2-gfp | |
| pHT315-PcotX1-cotX2 For | GACCATGATTACGCCAAGCTTTTCTGATTCCTCTTAAAAGCGCC |
| cotX2-gfp Rev(2) | GTTCTTCTCCTTTACTGCCTCCGCCTTTTGCTGCTGCTTCTCCGCCTCCTGAGCTAAGTAGAGAAACAAATATTGCG |
| cotX2-gfp For(2) | GTTTCTCTACTTAGCTCAGGAGGCGGAGAAGCAGCAGCAAAAGGCGGAGGCAGTAAAGGAGAAGAACTTTTCACTGGAG |
| pHT315-gfp Rev | GTTGTAAAACGACGGCCAGTGAATTCTTATTTGTATAGTTCATCCATGCC |
| Primers used to create pBADcLIC constructs | |
| cotW LIC For | ATGGGTGGTGGATTTGCTATAGATACATATAGTATTATGACA |
| cotW LIC Rev | TTGGAAGTATAAATTTTCCTTTGGTTTTGTTTTTGGAGAAGTGG |
Restriction enzyme sites are underlined.
Anti-GFP assays.
Spores (optical density at 600 nm [OD600] = 10) were incubated with gentle agitation in phosphate-buffered saline (PBS; pH 7.4) containing 2% (wt/vol) bovine serum albumin (BSA) for 1 h at 4°C. Spores were harvested by centrifugation (4,000 × g for 5 min), resuspended in PBS-BSA, and incubated with 500-fold-diluted anti-GFP antibody (ab290; Abcam, Cambridge, UK) for 40 min, followed by 3 washes with PBS-BSA. Antibody-labeled spores were subsequently centrifuged and resuspended in PBS-BSA before incubation with 500-fold-diluted Dylight-594 conjugated anti-rabbit IgG antibody (ab96885; Abcam) for 40 min. Labeled spores were subjected to three rounds of washing with PBS and then analyzed by fluorescence microscopy.
Cloning, expression, and purification of recombinant CotW and CotW-GFP.
The cotW ORF, minus the stop codon, was amplified by PCR using B. megaterium QM B1551 genomic DNA as a template and primers that facilitate ligation-independent cloning (36). The PCR amplicon was subsequently purified and cloned into the pBADcLIC-GFP expression plasmid, which adds GFP and His10 moieties to the C terminus of the expressed protein. A variant that lacked the GFP moiety but retained the His10 tag was also prepared. Protein expression was conducted using E. coli TOP10 cells in 600 ml of LB medium supplemented with 50 μg/ml carbenicillin. Cultures were incubated at 37°C, 225 rpm, and protein expression was induced with arabinose (final concentration, 0.001% [wt/vol]) when the cell density had attained an OD600 of 0.6. The cells were permitted to grow for 5 h prior to harvesting by centrifugation at 9,000 × g for 15 min at 4°C. Cell pellets were washed once with sterile water (4,000 × g for 10 min at 4°C), and the pellet was stored at −20°C until further use. Recombinant spore proteins were purified by affinity chromatography. Briefly, cell pellets from 1.2 liters of expression culture were resuspended in 8 ml of binding buffer (20 mM sodium phosphate [pH 7.4], 500 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 20 mM imidazole) and passed three times through a One Shot cell disrupter operated at 20 kPsi. Cell lysates were centrifuged at 15,000 rpm for 20 min at 4°C before passing the supernatant through a Millex-HV syringe-driven filter unit (0.45-μm polyvinylidene difluoride membrane) and loading onto a 1-ml HisTrap excel column fitted to an ÄKTA Pure instrument (GE Healthcare, Uppsala, Sweden). The protein was eluted in the same buffer containing 500 mM imidazole and then buffer-exchanged and concentrated into 20 mM sodium phosphate (pH 7.4) and 150 mM NaCl, using a 10-kDa MWCO Amicon centrifugal filter unit (Merck Millipore, Watford, UK). The purity of recombinant proteins was assessed at all stages by SDS-PAGE.
Assembly of recombinant CotW and CotW-GFP.
Purified recombinant B. megaterium CotW protein (45 mg/ml) was deposited (5 μl) onto a microscope slide and incubated at 37°C until dry. Deionized water (3 μl) was added, and the sample was then examined by phase-contrast microscopy. For larger-scale experiments, 2.5 mg of purified CotW-GFP was added to 300 ml of deionized water in a Langmuir trough and incubated overnight at room temperature. The polymerized protein was carefully removed from the surface using tweezers, deposited onto a glass slide, and analyzed by fluorescence microscopy. Thioflavin T assays were conducted by adding 10 μl of thioflavin T solution to CotW samples dried on microscope slides and then incubating for 10 min at 37°C. BSA was used as a negative control. Samples were then washed with deionized water and examined by phase-contrast and fluorescence microscopy.
Microscopy.
Fluorescence and thin-section transmission electron microscopy (TEM) analyses were conducted as described previously (34).
Supplementary Material
ACKNOWLEDGMENTS
We have no competing financial interests. J.M. was the recipient of an EPSRC Doctoral Training grant.
We gratefully acknowledge the assistance of Jeremy Skepper (University of Cambridge), Caitlin Brumby, Svetomir Tzokov, and Per Bullough (University of Sheffield) with electron microscopy work, as well as David Bailey's assistance with cloning procedures.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01734-18.
REFERENCES
- 1.Tan IS, Ramamurthi KS. 2014. Spore formation in Bacillus subtilis. Environ Microbiol Rep 6:212–225. doi: 10.1111/1758-2229.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev 64:548–572. doi: 10.1128/MMBR.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerhardt P, Marquis RE. 1989. Spore thermoresistance mechanisms, p 43–46. In Smith I, Slepecky RA, Setlow P (ed), Regulation of prokaryotic development—structural and functional analysis of bacterial sporulation and germination. American Society for Microbiology, Washington, DC. [Google Scholar]
- 4.McKenney PT, Driks A, Eichenberger P. 2013. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol 11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henriques AO, Moran CP. 2007. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 6.Stewart GC. 2015. The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol Mol Biol Rev 79:437–457. doi: 10.1128/MMBR.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerhardt P, Ribi E. 1964. Ultrastructure of the exosporium enveloping spores of Bacillus cereus. J Bacteriol 88:1774–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball DA, Taylor R, Todd SJ, Redmond C, Couture-Tosi E, Sylvestre P, Moir A, Bullough PA. 2008. Structure of the exosporium and sublayers of spores of the Bacillus cereus family revealed by electron crystallography. Mol Microbiol 68:947–958. doi: 10.1111/j.1365-2958.2008.06206.x. [DOI] [PubMed] [Google Scholar]
- 9.Terry C, Jiang S, Radford DS, Wan Q, Tzokov S, Moir A, Bullough PA. 2017. Molecular tiling on the surface of a bacterial spore—the exosporium of the Bacillus anthracis/cereus/thuringiensis group. Mol Microbiol 104:539–552. doi: 10.1111/mmi.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kailas L, Terry C, Abbott N, Taylor R, Mullin N, Tzokov SB, Todd SJ, Wallace BA, Hobbs JK, Moir A, Bullough PA. 2011. Surface architecture of endospores of the Bacillus cereus/anthracis/thuringiensis family at the subnanometer scale. Proc Natl Acad Sci U S A 108:16014–16019. doi: 10.1073/pnas.1109419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manetsberger J, Hall EA, Christie G. 2015. Plasmid-encoded genes influence exosporium assembly and morphology in Bacillus megaterium QM B1551 spores. FEMS Microbiol Lett 362:fnv147. doi: 10.1093/femsle/fnv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanzilli M, Donadio G, Addevico R, Saggese A, Cangiano G, Baccigalupi L, Christie G, Ricca E, Isticato R. 2016. The exosporium of Bacillus megaterium QM B1551 is permeable to the red fluorescence protein of the coral Discosoma sp. Front Microbiol 7:1752. doi: 10.3389/fmicb.2016.01752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaman TC, Pankratz HS, Gerhardt P. 1972. Ultrastructure of the exosporium and underlying inclusions in spores of Bacillus megaterium strains. J Bacteriol 109:1198–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koshikawa T, Beaman TC, Pankratz HS, Nakashio S, Corner TR, Gerhardt P. 1984. Resistance, germination, and permeability correlates of Bacillus megaterium spores successively divested of integument layers. J Bacteriol 159:624–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishihara T, Takubo Y, Kawamata E, Koshikawa T, Ogaki J, Kondo M. 1989. Role of outer coat in resistance of Bacillus megaterium spore. J Biochem 106:270–273. doi: 10.1093/oxfordjournals.jbchem.a122843. [DOI] [PubMed] [Google Scholar]
- 16.Takubo Y, Okuda M, Takemura I, Haruna F, Sawatari A, Nishihara T, Kondo M. 1989. Characterization and deposition of the proteins in the outermost layer of Bacillus megaterium spore. Microbiol Immunol 33:527–538. doi: 10.1111/j.1348-0421.1989.tb02003.x. [DOI] [PubMed] [Google Scholar]
- 17.Sylvestre P, Couture-Tosi E, Mock M. 2005. Contribution of ExsFA and ExsFB proteins to the localization of BclA on the spore surface and to the stability of the Bacillus anthracis exosporium. J Bacteriol 187:5122–5128. doi: 10.1128/JB.187.15.5122-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steichen CT, Kearney JF, Turnbough CL. 2005. Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J Bacteriol 187:5868–5876. doi: 10.1128/JB.187.17.5868-5876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura D, Kuwana R, Takamatsu H, Watabe K. 2011. Proteins involved in formation of the outermost layer of Bacillus subtilis spores. J Bacteriol 193:4075–4080. doi: 10.1128/JB.05310-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenney PT, Driks A, Eskandarian HA, Grabowski P, Guberman J, Wang KH, Gitai Z, Eichenberger P. 2010. A distance-weighted interaction map reveals a previously uncharacterized layer of the Bacillus subtilis spore coat. Curr Biol 20:934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Ichikawa H, Halberg R, Kroos L, Aronson AI. 1994. Regulation of the transcription of a cluster of Bacillus subtilis spore coat genes. J Mol Biol 240:405–415. doi: 10.1006/jmbi.1994.1456. [DOI] [PubMed] [Google Scholar]
- 22.Haldenwang WG. 1995. The sigma factors of Bacillus subtilis. Microbiol Mol Biol Rev 59:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manetsberger J, Manton JD, Erdelyi MJ, Lin H, Rees D, Christie G, Rees EJ. 2015. Ellipsoid localization microscopy infers the size and order of protein layers in Bacillus spore coats. Biophys J 109:2058–2066. doi: 10.1016/j.bpj.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawasaki C, Nishihara T, Kondo M. 1969. Ultrastructure and its relation to the fractions isolated from spore coat of Bacillus megaterium. J Bacteriol 97:944–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nasu M, Ogaki J, Nishihara T, Nakanishi T, Ichikawa T, Kondo M. 1984. Occurence of galactosamine-6-phosphate as a constituent of Bacillus megaterium spore coat. Microbiol Immunol 28:181–188. doi: 10.1111/j.1348-0421.1984.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 26.Jiang S, Qiang W, Krajcikova D, Tang J, Tzokov SB, Barak I, Bullough PA. 2015. Diverse supramolecular structures formed by self-assembling proteins of the Bacillus subtilis spore coat. Mol Microbiol 97:347–359. doi: 10.1111/mmi.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson I, Sorokin A, Kapatral V, Reznik G, Bhattacharya A, Mikhailova N, Burd H, Joukov V, Kaznadzey D, Walunas T, D'Souza M, Larsen N, Pusch G, Liolios K, Grechkin Y, Lapidus A, Goltsman E, Chu L, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N, Ivanova N. 2005. Comparative genome analysis of Bacillus cereus group genomes with Bacillus subtilis. FEMS Microbiol Lett 250:175–184. doi: 10.1016/j.femsle.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Sousa J, Silva M, Balassa G. 1976. An exosporium-like outer layer in Bacillus subtilis spores. Nature 263:53–54. doi: 10.1038/263053a0. [DOI] [PubMed] [Google Scholar]
- 29.Waller LN, Fox N, Fox KF, Fox A, Price RL. 2004. Ruthenium red staining for ultrastructural visualization of a glycoprotein layer surrounding the spore of Bacillus anthracis and Bacillus subtilis. J Microbiol Methods 58:23–30. doi: 10.1016/j.mimet.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 30.Johnson MJ, Todd SJ, Ball DA, Shepherd AM, Sylvestre P, Moir A. 2006. ExsY and CotY are required for the correct assembly of the exosporium and spore coat of Bacillus cereus. J Bacteriol 188:7905–7913. doi: 10.1128/JB.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boydston JA, Yue L, Kearney JF, Turnbough CL. 2006. The ExsY protein is required for complete formation of the exosporium of Bacillus anthracis. J Bacteriol 188:7440–7448. doi: 10.1128/JB.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishikawa J-I, Tada M, Takubo Y, Nishihara T, Kondo M. 1987. Occurrence in Bacillus megaterium of uridine 5′-diphospho-N-acetylgalactosamine and uridine 5′-diphosphogalactosamine in the biosynthesis of galactosamine-6-phosphate polymer. J Bacteriol 169:1338–1340. doi: 10.1128/jb.169.3.1338-1340.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, Ustok FI, Johnson CL, Bailey DMD, Lowe CR, Christie G. 2013. Investigating the functional hierarchy of Bacillus megaterium PV361 spore germinant receptors. J Bacteriol 195:3045–3053. doi: 10.1128/JB.00325-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manetsberger J, Hall EA, Christie G. 2014. BMQ_0737 encodes a novel protein crucial to the integrity of the outermost layers of Bacillus megaterium QM B1551 spores. FEMS Microbiol Lett 358:162–169. doi: 10.1111/1574-6968.12520. [DOI] [PubMed] [Google Scholar]
- 35.Ramirez-Peralta A, Gupta S, Butzin XY, Setlow B, Korza G, Leyva-Vazquez MA, Christie G, Setlow P. 2013. Identification of new proteins that modulate the germination of spores of Bacillus species. J Bacteriol 195:3009–3021. doi: 10.1128/JB.00257-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geertsma ER, Poolman B. 2010. Production of membrane proteins in Escherichia coli and Lactococcus lactis, p 17–38. In Mus-Veteau I. (ed), Heterologous expression of membrane proteins, vol. 601 Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 37.Lewis PJ, Marston AL. 1999. GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227:101–109. doi: 10.1016/S0378-1119(98)00580-0. [DOI] [PubMed] [Google Scholar]
- 38.Arantes O, Lereclus D. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115–119. doi: 10.1016/0378-1119(91)90495-W. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.