The data presented here suggest a newly discovered function of outer membrane vesicles (OMV) that are produced from the outer membrane of the bacterial species Lysobacter enzymogenes strain C3. We show that these OMV can be released from the surface of the cells to deliver antibiotics to target fungal organisms as a mechanism of killing or growth inhibition. Understanding the role of OMV in antibiotic delivery can generally lead to improved strategies for dealing with antibiotic-resistant organisms. These results also add to the evidence that some bacterially produced antibiotics can be discovered and purified using methods designed for isolation of nanoscale vesicles. Information on these systems can lead to better identification of active molecules or design of delivery vehicles for these molecules.
KEYWORDS: antifungal agents, host-pathogen interactions, outer membrane vesicles
ABSTRACT
Lysobacter enzymogenes C3 is a predatory strain of Gram-negative gliding bacteria that produces antifungal antibiotics by the polyketide synthetic pathway. Outer membrane vesicles (OMV) are formed as a stress response and can deliver virulence factors to host cells. The production of OMV by C3 and their role in antifungal activity are reported here. Vesicles in the range of 130 to 150 nm in diameter were discovered in the cell-free supernatants of C3 cultures. These OMV contain molecules characteristic of bacterial outer membranes, such as lipopolysaccharide and phospholipids. In addition, they contain chitinase activity and essentially all of the heat-stable antifungal activity in cell supernatants. We show here that C3 OMV can directly inhibit growth of the yeast Saccharomyces cerevisiae as well as that of the filamentous fungus Fusarium subglutinans. The activity is dependent on physical contact between OMV and the cells. Furthermore, fluorescent lipid labeling of C3 OMV demonstrated transfer of the membrane-associated probe to yeast cells, suggesting the existence of a mechanism of delivery for membrane-associated molecules. Mass spectrometric analysis of C3 OMV extracts indicates the presence of molecules with molecular weights identical to some of the previously identified antifungal products of C3. These data together suggest that OMV act as an important remote mobile component of predation by Lysobacter.
IMPORTANCE The data presented here suggest a newly discovered function of outer membrane vesicles (OMV) that are produced from the outer membrane of the bacterial species Lysobacter enzymogenes strain C3. We show that these OMV can be released from the surface of the cells to deliver antibiotics to target fungal organisms as a mechanism of killing or growth inhibition. Understanding the role of OMV in antibiotic delivery can generally lead to improved strategies for dealing with antibiotic-resistant organisms. These results also add to the evidence that some bacterially produced antibiotics can be discovered and purified using methods designed for isolation of nanoscale vesicles. Information on these systems can lead to better identification of active molecules or design of delivery vehicles for these molecules.
INTRODUCTION
Lysobacter enzymogenes strain C3 (referred to here as C3) was originally identified as Stenotrophomonas maltophilia (1), and broad-range antifungal activity has been associated with its predatory lifestyle. Because this activity acts against pathogenic fungi that attack various crop species, C3 has been under development as a biocontrol organism to combat several plant diseases incited by fungi (2, 3). This antifungal activity may also involve molecules that can act as therapeutic agents for human fungal diseases (4). C3 is known to produce bioactive molecules through the polyketide synthase/nonribosomal peptide synthetase (PKS/NRPS) pathway (3). Some of these molecules, particularly polycyclic tetramate macrolactams, are stable to heat, can be obtained by extraction with organic solvents, and have been demonstrated to kill or inhibit the growth of various specific fungi. This activity was originally identified as a complex that could be purified by ammonium sulfate precipitation, and it was designated heat-stable antifungal factor (HSAF) (3). Dihydromaltophilin (3, 5) was initially identified as one of the major C3 antifungal antibiotics in HSAF, and it is now essentially synonymous with HSAF. In addition, other polycyclic tetramate macrolactam products of C3, such as alteramide A and B, have been identified with apparently different antifungal specificities and mechanisms of activity (6). The process by which HSAF is delivered to fungal cells has not been elucidated, although it has been suggested that one or more of the well-defined type I to type IV secretory complexes may mediate secretion or injection into host cells (7).
Gram-negative bacteria are also known to produce outer membrane vesicles (OMV) under certain circumstances. These vesicles bud from the outer membrane, often as a stress response (8–13), and can contain a number of different components, including toxins (14), signaling molecules (15), genetic material (16–18), and self-defense compounds, such as enzymes that can degrade antibiotics (19). Accumulating evidence indicates that OMV play a major role in host-pathogen interactions (20, 21). In some species, OMV harbor small-molecule anti-host antibiotics (22, 23), while in others the antihost activities are primarily enzymatic (9, 24). In particular, vesicle production has been previously documented in Lysobacter sp. strain XL1, showing the association of host-lytic enzymes with the vesicles (25, 26). However, the production of OMV by the C3 biocontrol strain has not been previously investigated.
We report here that C3 grown in liquid culture forms what appear to be outer membrane vesicles (OMV) that contain most if not all of the heat-stable antifungal activity associated with supernatants from C3 cells. C3 OMV were found to be approximately 130 to 150 nm in diameter, and they contain phospholipids and lipopolysaccharide, DNA, and a collection of associated proteins, including a chitinase activity. Heat-stable antifungal activity against a filamentous fungus and yeast was observed, and experiments exploring the mechanism of transfer to these host cells are presented. Purified OMV extracts yielded molecules with mass spectra expected for known C3 PKS/NRPS pathway products with antifungal activity. These data demonstrate vesicular packaging of intrinsically synthesized small-molecule antibiotics by this predatory species.
RESULTS
The C3 strain produces outer membrane vesicles.
The presence of OMV in the cell-free supernatants of C3 was tested under several conditions. Putative OMV could be obtained either by directly sedimenting cell-free supernatant to obtain an OMV pellet or by ultrafiltration of the supernatants through a 300-kDa-cutoff filter (equivalent to a spherical protein particle cutoff of approximately 7 to 8 nm). In the latter case, the material collected from the filter was then sedimented and washed for further analysis. Additionally, minimal medium (MM) or 10% tryptic soy broth (TSB) was found to support C3 OMV production. Vesicle material could only be obtained in late log phase or stationary-phase culture and not in mid-log-phase culture.
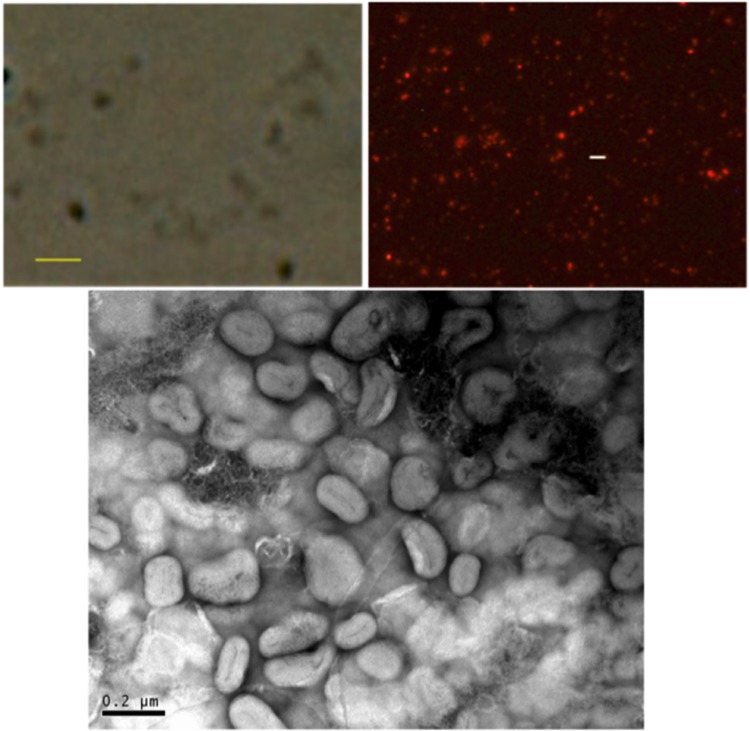
Observation of the washed, resuspended material by light microscopy revealed particles that were apparently less than 1 μm (Fig. 1). It was possible to label these particles with a well-characterized probe, diIC18 (see Materials and Methods), which resulted in labeled submicron particles that could be observed by fluorescence microscopy (Fig. 1). Importantly, an enhancement in fluorescence of the probe was observed (approximately 5- to 10-fold; data not shown), consistent with the behavior expected for insertion into a hydrophobic membrane environment. These data suggested that the observed particles were indeed vesicles with a delimiting membrane, consistent with OMV. Samples were also analyzed by negative-stain transmission electron microscopy (EM), showing apparently aggregated particles/vesicles with diameters in the 100- to 150-nm range (Fig. 1). Aggregation, fusion, and/or other distortions of the negatively charged vesicles may be an artifact of the negative-stain EM preparation, perhaps as a result of the presence of polyvalent positive uranyl ions in the procedure.
FIG 1.
Microscopic images of vesicular material from C3 cell culture supernatants. (Top left) Phase contrast image (800×) of unlabeled vesicles. Bar, 1 μm. (Top right) Fluorescence photomicrograph (800×) of vesicles labeled with diIC18. Bar represents 2 μm. (Bottom) Negative-stain electron microscopic images of C3 OMV preparation.
Additionally, OMV from washed pellets were sedimented on a discontinuous iodixanol gradient (equal volumes of 15, 25, 40, and 54%) and migrated into a visible band at the interface between 15 and 25% iodixanol (approximately 1.18 and 1.3 g/cm3, respectively; data not shown). This density is consistent with the buoyant densities of previously identified OMV of other bacteria (27, 28).
Size and charge of the particulate samples were also analyzed by dynamic light scattering and electrophoretic mobility, respectively. The data for three separate preparations revealed a homogeneous and narrow size distribution with a z average size of approximately 135 nm, a low polydispersity index (PDI) of approximately 0.09, and a notable lack of particles of other sizes (Fig. 2). Zeta potential measurements indicated a negative surface potential of approximately −10 mV.
FIG 2.
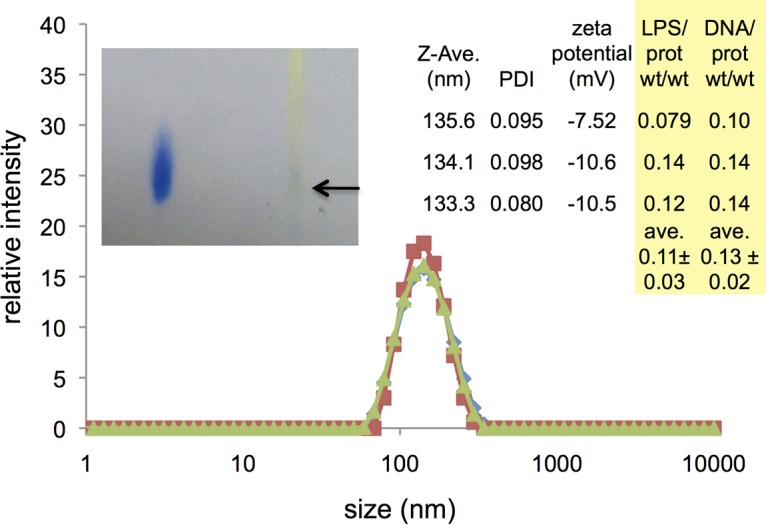
Size, charge and composition of three OMV preparations. Graph represents the distribution of vesicle sizes detected by dynamic light scattering, with a peak in z average intensity at approximately 135 nm. (Right inset) The z average size for each sample and polydispersity index (PDI), as well as zeta potential. Data for estimated LPS/protein and DNA/protein ratios are also shown for each preparation. (Left inset) Evidence for phospholipids from a Bligh-Dyer extract of C3 OMV as detected with thin-layer chromatography with Zinzadze reagent staining. Left sample: standard POPG [1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)]; Rf, ∼0.41. Right sample: Bligh-Dyer extract of C3 OMV where arrow denotes blue phosphate positive spot; Rf, ∼0.42.
Outer membrane vesicles of bacteria would also be expected to contain lipids characteristic of the cell envelope. C3 cells, like those of most Gram-negative bacteria, display lipopolysaccharide (LPS) on their surfaces. Therefore, the presence of LPS in these vesicles and its relationship to the protein content were tested in the same three preparations (see Fig. 2). For spherical vesicles of approximately 135 nm in diameter, the expected LPS/protein ratio based strictly on geometrical considerations would fall in the range of 0.08 to 0.66 (see Materials and Methods), depending on whether any protein is encapsulated in the interior of the vesicle, as well as on the membrane protein content. These preparations of C3 OMV contained an estimated average ratio of 0.11 ± 0.03 g LPS/g protein, based on an Escherichia coli LPS standard and the Bradford protein assay. These data are consistent with a considerable amount of protein, both in the membrane and encapsulated inside the vesicles (see calculations in Materials and Methods). Additionally, Bligh-Dyer extracts (29) of OMV preparations that were analyzed by thin-layer chromatography appeared to show components consistent with phospholipids commonly found in bacterial outer membranes, such as phosphatidylglycerol or phosphatidylethanolamine (Fig. 2, inset). The presence of outer membrane lipids further corroborates the identity of the isolated material as OMV.
Protein content of the vesicles was analyzed by polyacrylamide gel electrophoresis, Western blotting immunoassay, and an assay for chitinase activity. Gel electrophoresis showed major protein bands, documented in Table 1 (data not shown). Chitinase activity was consistently detected in washed OMV preparations from chitin-induced cultures as monitored by a fluorogenic substrate assay (see Materials and Methods). Treatment of OMV with proteinase K did not entirely eliminate chitinase activity, indicating that at least some portion of the chitinase may be encapsulated inside the OMV (Table 1). Freeze-thaw (10 times) and washing of these OMV by repeated sedimentation also resulted in retrieval of approximated 20% of the total OMV-associated chitinase activity. These data suggest the possibility of a membrane-associated form of chitinase.
TABLE 1.
Preliminary characterization of components of C3 OMVa
| OMV component | Content identification | Entrapped material |
|---|---|---|
| Protein | Major bands: 28, 31, 37, 53, and 70 kDa; chitinase activity observed | ∼80–89% chitinase activity protected from proteinase K digestion |
| Nucleic acids | 0.13 g DNA/g protein | ∼100% inside (not accessible to DNase) |
| Lipids | 0.11 g LPS/g protein; phospholipids present |
Experimental details are given in Materials and Methods.
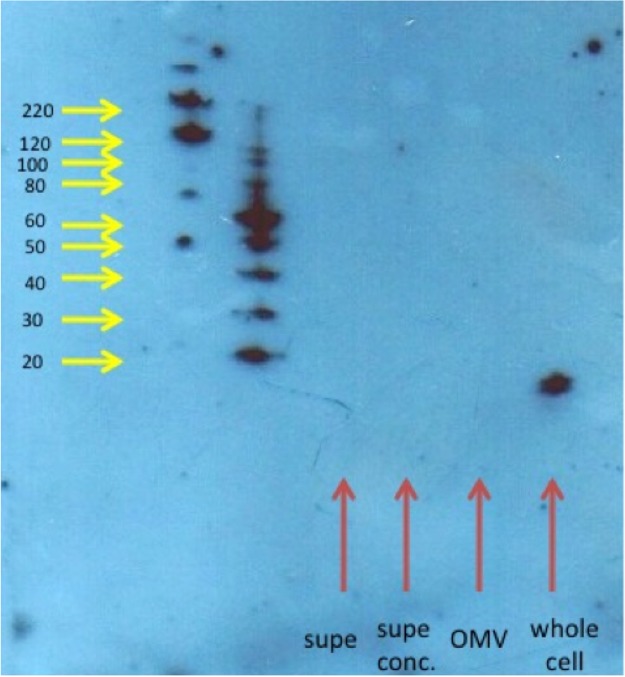
OMV preparations were also tested by Western blotting for the presence of PilA, the major component of Lysobacter pili. No evidence for the existence of PilA was found in OMV preparations, while a strong band could be seen in an equal protein concentration of whole-cell preparations (Fig. 3). These data confirm that C3 OMV are truly a result of the budding of specific portions of the outer membrane and not just outer membrane fragments that may retain pili.
FIG 3.
Western blot analysis of C3 OMV, whole C3 cells, and supernatants from C3 OMV. An 8 to 25% gradient acrylamide gel was used to separate samples, and the blot was probed with an antibody against a peptide within the PilA protein. The expected molar mass of PilA is 14 kDa. Molecular weights of standards are shown with yellow arrows. Samples are shown with red arrows. Equal amounts of protein from OMV and whole cells were loaded on the gel.
Nucleic acids are also commonly found to be associated with OMV, both externally and internally (16–18, 27). DNA was detected in C3 OMV preparations by standard ethanol precipitation procedures, and the presence of DNA was also observed via a fluorogenic DNA-binding probe (see Materials and Methods and Fig. 2, right side). When intact OMV were treated with DNase I, most of the DNA in the OMV preparations was inaccessible to the enzyme, indicating encapsulation of most DNA in the interior of the vesicles. Although the presence of a small amount of RNA cannot be ruled out, treatment of nucleic acids isolated from OMV with RNase-free DNase did not yield detectable material.
In summary, a particulate fraction with a well-defined z average diameter of approximately 135 nm is obtained from C3 culture supernatants under limiting nutrient conditions. The existence of membrane lipids, enhancement of membrane probe fluorescence, and observation of encapsulated material and other components common to OMV of Gram-negative bacteria strongly suggest that OMV are a common product of L. enzymogenes C3.
Antifungal activity is associated with OMV.
A yeast growth assay (Saccharomyces cerevisiae strain S30) was used as the primary test for antifungal activity of fractions prepared from C3 culture. Activity against Fusarium subglutinans was also tested in some cases (see below). Initially, extraction methods similar to those previously reported to retrieve HSAF from cell supernatants were used (30). Specifically, ethyl acetate extracts of the cell-free supernatants were obtained, dried, and redissolved in a smaller volume of ethanol and tested in yeast growth assays, as described in Materials and Methods. Antifungal activity was observed from late log or stationary-phase C3 supernatants, while no activity (or OMV) was found in mid-log-phase supernatants (data not shown).
Cell-free supernatants were also fractionated by ultrafiltration through a 300-kDa-cutoff filter. This resulted in an observable film of material on the filter surface. When the filter material was resuspended in an equal volume of medium (compared to the filtrate) and extracted with ethyl acetate as above, the antifungal activity assay showed that >90% of the activity was recoverable from the surface of the filter (Fig. 4). The activity present in resuspended OMV material could be further sedimented by high-speed centrifugation (140,000 × g). The washed pellets containing OMV were then also analyzed for protein content and activity. These data also showed that all detectable activity was associated with the OMV (see below).
FIG 4.
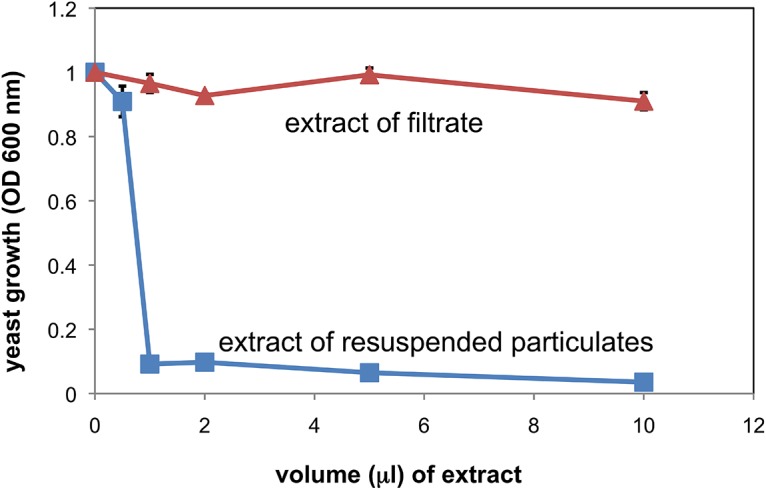
Yeast growth inhibitory activity of extract samples from filtrates and resupended OMV. Resuspended OMV and filtrate fractions (300-kDa cutoff) were extracted under identical conditions, dried, and redissolved in equal volumes of ethanol. Ethanol aliquots were added to dilute yeast cultures (S30 strain) in the volumes indicated, and overnight growth was monitored by turbidity at 600 nm (n = 3 ± standard deviation [SD]; some error bars smaller than symbols; P < 0.05 for all samples of >0.5 μl). More details are given in Materials and Methods.
The nature of the antifungal activity in these OMV pellets was further investigated. Figure 5 shows the dose response in terms of OMV protein concentration. We found that active OMV could be isolated from cultures in MM as well as from those in 10% TSB, but in MM, addition of colloidal chitin to the culture medium was necessary to obtain OMV with maximal specific activity. OMV from chitin-induced cultures also showed chitinase activity. When the OMV samples were heat treated at 70°C for 30 min, all of the yeast growth-inhibitory activity remained intact (Fig. 5), whereas all chitinase activity was destroyed (not shown). Therefore, it can be concluded that the observed antifungal activity is solely due to heat-stable factors that are likely to be small molecules, at least under the conditions of these experiments. Additionally, OMV were isolated from a mutant strain of C3 in which the PKS/NRPS pathway is disrupted (designated HSAF−). These OMV showed no antifungal activity. These data, taken together, strongly suggested that C3 OMV package heat-stable antifungal molecules, such as dihydromaltophilin, and that the observed antifungal activity is solely due to these molecules.
FIG 5.
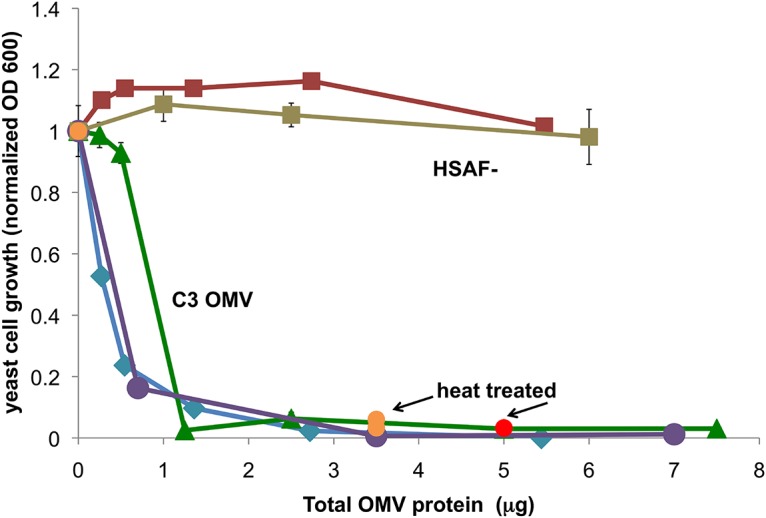
Specific activity of OMV preparations in yeast growth assays. Two separate preparations of C3 OMV were isolated from culture supernatants grown in MM (blue and violet, “C3 OMV”). Orange data points represent OMV that were heat treated at 70°C before testing for activity. OMV isolated from C3 HSAF− mutant cells grown in MM are shown by squares (HSAF−, red-brown). For preparations from 10% TSB, activity curves are shown by green triangles for C3 wild type and brown squares for HSAF− mutant (n = 3 ± SD; some error bars smaller than symbols). The activity of heat-treated C3 OMV from the same 10% TSB preparation is shown at 5 μg total protein as a red circle (n = 3 ± SD; error bars smaller than symbol). No activity was observed in samples of similar volume from the OMV-free supernatants of these preparations. Data are representative of more than 20 preparations of C3 OMV, which all showed full activity at 5 μg of total protein.
Further characterization and confirmation of OMV association of antifungal activity were performed using isopycnic sedimentation on iodixanol density medium. Based on the observation of the density behavior of OMV samples mentioned above, samples were loaded onto pure 25% iodixanol to test whether more dense antifungal protein particles might have been mixed with the relatively light vesicles. In this case, a band of OMV material could be visually observed near the top of the 25% iodixanol solution after high-speed centrifugation. When samples were prelabeled with diIC18, the fluorescence of labeled OMV also migrated to a position near the top of the 25% iodixanol (not shown). Correspondingly, yeast antifungal activity also remained near the top of the 25% iodixanol, as did a large portion of the protein (Fig. 6). Therefore, it would appear that the C3 antifungal activity is exclusively associated with light fractions, again consistent with OMV localization.
FIG 6.
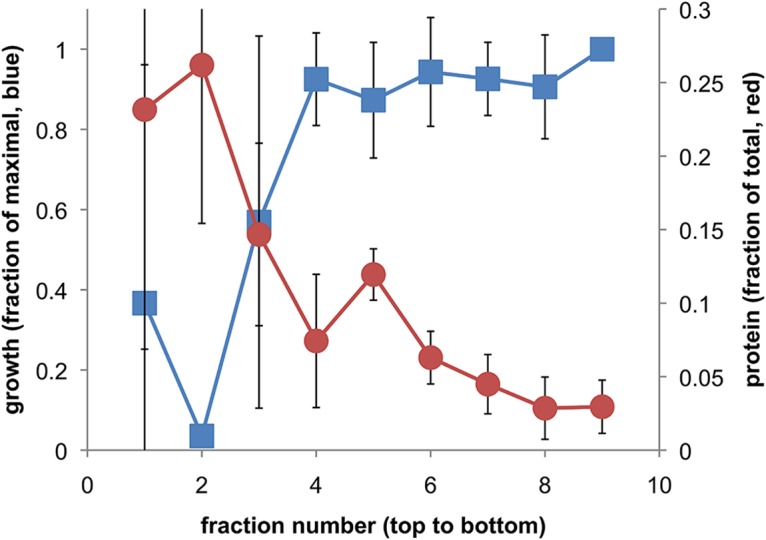
Density-based fractionation of antifungal activity and protein concentration of OMV samples. OMV samples in TBS loaded on 25% iodixanol were recovered after centrifugation in the fractions shown, where 1 to 10 run from the top to the bottom of the centrifuge tube, corresponding to least to most dense material. The protein concentration and effect of each fraction on yeast cell growth were determined and plotted on a fractional scale (n = 3 separate tubes ± SD; some error bars are smaller than symbols; for fraction 2 yeast cell growth, P < 0.05 versus fractions 4 to 9; P = 0.09 for fraction 3; no significance established for fraction 1).
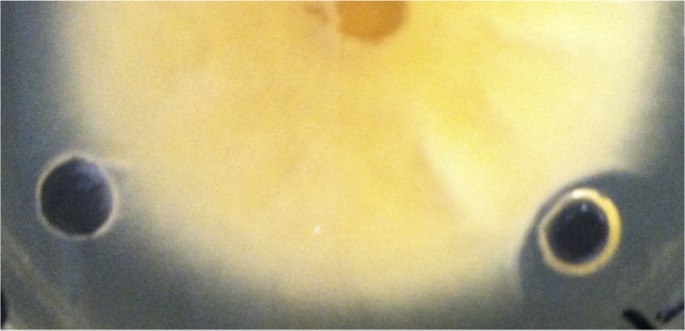
C3 is known to exert antibiotic activity against filamentous fungi. Therefore, OMV preparations were also tested against a filamentous fungus, F. subglutinans. This species was grown on agar plates, and the activities of OMV preparations were tested by placing the samples in wells in the agar. As can be seen by the lack of mycelia around the OMV well, Fusarium growth was also inhibited by the C3 OMV (Fig. 7). Although the format of the assay was quite different, the OMV protein concentration required to inhibit Fusarium growth was generally in a range similar to that of the concentrations needed to inhibit yeast growth.
FIG 7.
Growth inhibition of mycelia of Fusarium subglutinans by OMV. Fusarium culture was grown from the center of an agar plate, and wells in the agar containing control buffer (left) or C3 OMV (right) were evaluated. Clear zone around the right-hand well indicates growth inhibition.
OMV-mediated delivery of antifungal compounds to yeast cells.
A corollary of OMV localization of antifungal activity is the hypothesis that there is a mechanism by which OMV can deliver the relevant compounds to yeast and other fungal cells. The requirements for delivery would depend critically on how antifungal antibiotics are associated with the OMV. The effect of freeze-thawing on the OMV activity was used as an initial crude test of localization. If activity is localized in a water-soluble form in the interior of the vesicles, freeze-thawing should destabilize the membranes and release the encapsulated molecules or complexes. Freeze-thawing (10 times) and washing of the OMV did not affect the ability of OMV to inhibit yeast growth (data not shown), suggesting that the active molecules may be membrane bound.
If activity is membrane bound and not easily diffusible through aqueous media, delivery may require close contact between OMV and host cells. A filter-based activity assay was used to test this concept. Yeast cells and C3 OMV were placed on opposite sides of a 100-kDa filter (“trans filter”) or on the same side (“cis filter”), and the ability of the yeast cells to grow was measured. Short (5.5-h) and long (overnight) incubation times were used. At either incubation time, the 100-kDa filter did not allow diffusion of active molecules from OMV to reach the yeast cells so as to inhibit their growth, resulting in a relatively high turbidity of the trans-filter yeast culture (optical densities at 600 nm [OD600] = 0.176 ± 0.037 at 5.5 h and 0.228 ± 0.0007 overnight). Under the same conditions, incubation of OMV and host yeast cells on the same side of the membrane strongly inhibited the growth of the cells (OD600 = 0.010 ± 0.012 for 5.5 h of incubation). These data suggest that OMV activity against yeast cells depends on direct contact between the OMV and cells.
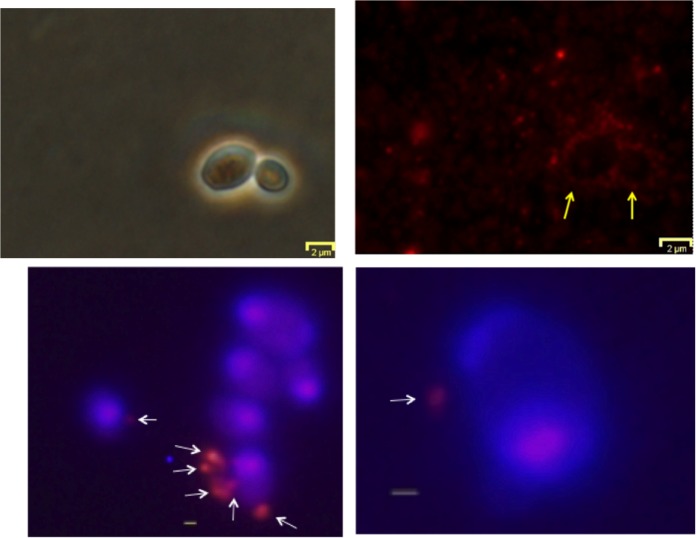
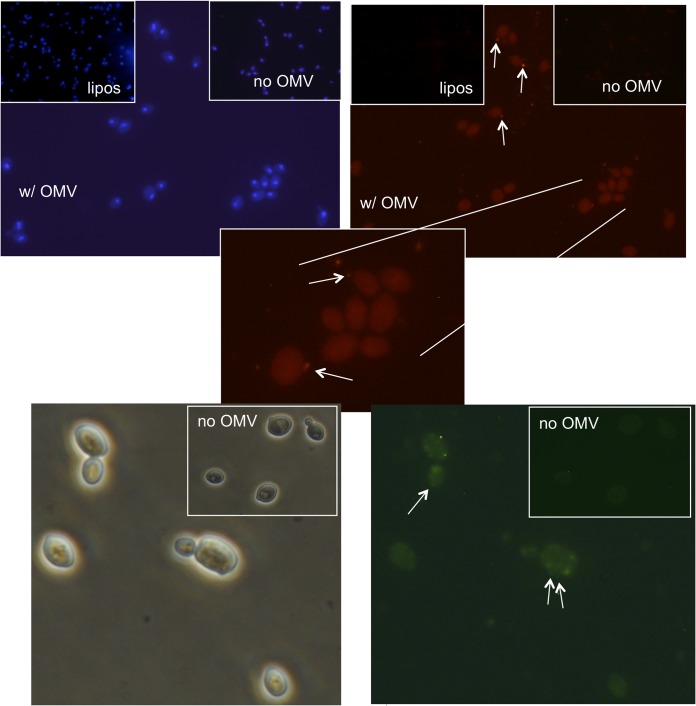
To assess the potential for contact-mediated transfer of antifungal activity, OMV binding to yeast cells was also studied using diIC18-labeled OMV. After incubation, OMV fluorescence was found to be associated with pellets of washed yeast cells in significant amounts (56% ± 0.7% of total OMV fluorescence versus 7% ± 4% in the absence of the yeast cells). Binding was also assessed by observation of S30 cells by fluorescence microscopy. Yeast cells, which were incubated with diIC18-labeled OMV and observed before washing, often showed a concentration of vesicles around the surface of the cells (Fig. 8). Similarly, OMV labeled with an amine-reactive red fluorescent probe also appeared to bind to some yeast cells that had been washed by sedimentation, although many fewer OMV remained attached to cells. (Fig. 8).
FIG 8.
Binding of OMV to yeast cells. Top panels depict diIC18-labeled OMV (red fluorescence) interacting with yeast cells without washing. Top left and right panels are phase-contrast and fluorescence versions of the same view. Yellow arrows indicate the position of two yeast cells. Bottom panels: Hoechst 33342-labeled yeast cells (blue) were incubated with Alexa Fluor 555-NHS-labeled OMV (red) and then washed by centrifugation. Overlays of fluorescent images are shown. Gray bar, 500 nm; white arrows indicate putative OMV or clusters of OMV bound to the cells.
Further studies with OMV labeled with membrane-localizing probes showed data consistent with transfer of these hydrophobic molecules to yeast cell membranes (Fig. 9). When OMV labeled with the probes diIC18 or diOC18 were allowed to interact with the yeast cells for a period of time and washed extensively, a dim, diffuse fluorescence across the whole cell was observed. In contrast, little or no transfer of the diIC18 probe was observed in experiments with liposomes of similar size to OMV. As noted above, a small fraction of these cells appeared to have OMV that are bound and apparently intact. However, essentially all of the cells displayed diffuse fluorescence, which was significantly above background (as shown). It can therefore be proposed that this fluorescence may represent transfer of the OMV membrane-localized probes to membranes or other structures within the yeast cells. One interpretation of this result is that the OMV have some mechanism by which to transfer molecules from their membranes through the cell wall of the yeast cells and into the plasma membrane. Further investigation will be necessary to establish this paradigm.
FIG 9.
Apparent transfer of hydrophobic labels from C3 OMV to yeast cells. Yeast cells were labeled with Hoechst 33342 (upper photos, blue) or unlabeled (lower photos). OMV were labeled with diIC18 (red fluorescence, upper photos) or diOC18 (green fluorescence, lower photos) and allowed to interact with yeast cells. After washing to remove unbound OMV, yeast cells were observed by fluorescence or phase-contrast microscopy. Insets in the upper right of each photo are shown to compare background fluorescence of samples with no added fluorescently labeled OMV. Insets in the upper left of the top photos compare yeast cells incubated with liposomes labeled with diIC18, composed of a 1:1 ratio of phosphatidylglycerol and phoshpatidylcholine and of similar size to OMV. An expanded portion of one of the upper photos is shown in the middle panel. Arrows point to putative OMV bound to yeast cells. All fluorescent photos with and without OMV are compared under identical photomicrographic parameters and identical image enhancement parameters (brightness and contrast).
Identification of candidate molecules for antifungal activity.
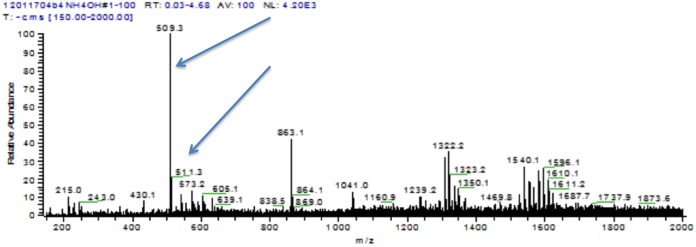
In an attempt to search for previously identified antifungal species, extracts were partially purified by thin-layer chromatography and subjected to analysis by mass spectrometry. When C3 OMV were extracted with ethyl acetate, the concentrated extracts in methanol revealed a complex mixture of molecules with no apparently dominant species. Further purification was possible by a micropreparative thin-layer chromatography procedure. A UV-positive band was extracted from the plates and demonstrated to have yeast growth-inhibitory activity. Analysis of an extracted band by mass spectrometry gave the data shown in Fig. 10.
FIG 10.
Negative-ion m/z spectrum of TLC-purified antifungal activity from C3 OMV. A UV-positive band was obtained from a TLC separation of an OMV extract. The extracted band was dissolved in methanol for analysis by electrospray ionization-mass spectroscopy, as shown above. Arrows highlight peaks at m/z 509.3 and 511.3 (discussed in text). The peak at m/z 863.1 is an artifact that also appears in pure solvent spectra.
Because the thin-layer chromatograph (TLC) purification was performed in a strongly basic solvent, it was expected that negative ion products might be prominent in the analysis. In the negative ion mode, a prominent peak was observed at m/z 509, possibly consistent with alteramide B, with a smaller peak at m/z 511, possibly consistent with dihydromaltophilin (arrows in Fig. 10; see Discussion). Two series of peaks are also seen in the ranges of m/z 1,332 and 1,596. These groups of peaks appear to have multiplets separated by mass units of 14, the mass of CH2 units.
In the positive ion scans, some of the same peaks can be observed but apparently as disodium forms of the negative ions. For instance, peaks at m/z 555 and 558 may correspond to peaks at 509 and 512 in the negative ion scans (data not shown). The potential identities of these compounds are discussed further below.
DISCUSSION
The existence of extracellular bacterial vesicles has been known for some time (8). However, the identification of OMV as a potential part of the antifungal biocontrol activity of C3 fits a body of recent research elucidating the importance of OMV production as an extracellular mechanism of delivery of active molecules to host cells (14–26). Therefore, a role for OMV in the action of Lysobacter species fits an emerging paradigm in the field. The OMV identified here have some expected characteristics. For instance, the ratio of LPS to protein in these preparations is particularly telling. The value observed for C3 (0.11 g/g) falls within a calculated range based on geometry and size (see Materials and Methods) and is also consistent with other measurements of this ratio in other OMV. For instance, OMV from Neisseria and Vibrio species have been reported to have ratios of 0.06 to 0.08 and 0.08 to 0.09 (31, 32), respectively. These relatively low ratios suggest that large amounts of protein are encapsulated in the interior or located on the surface of OMV. The apparent encapsulation of a substantial amount of chitinase activity in the interior of C3 OMV is consistent with this speculation.
OMV can act as long-distance mediators of antifungal activity that would allow the bacterial cells to avoid fungal defenses while exerting a toxic effect on the target. However, the ability of C3 OMV to reach and deliver associated molecules to fungal cells brings up other questions. Specifically, the cell wall of fungi may be expected to inhibit the contact and transfer of molecules from vesicles. It cannot be fully ruled out that there is a mechanism by which the OMV can disrupt the cell wall. Our experiments with proteinase K digestion of OMV suggest that at least some portion of chitinase activity is located on the outside of the OMV, perhaps on the surface, where it could mediate cell wall digestion. However, heat treatment, which destroyed chitinase activity and may do the same for other enzymes, did not affect the ability of OMV to deliver antibiotic activity under the conditions of these experiments. Although enzymatic activity may still prove to play a role, other mechanisms may need to be explored. Vesicular traffic across the cell wall of some species of fungi has been reported (33, 34). Therefore, actual movement of OMV through the cell wall could be possible, although no evidence of this exists in the particular case of the experiments presented here.
Furthermore, it is not clear if the observation of antifungal activity in OMV under these conditions actually reflects the situation in a natural environment or when in contact with the host cells (35). On the one hand, the observation of OMV under nutrient stress would seem to be consistent with conditions expected in natural environments, where nutrient broths do not exist. However, it is unclear when OMV would be released and how they might be directed to host cells, unless there is a mechanism of preferential binding. Previous data from other species have suggested that OMV can prime host cells for bacterial binding, which could apply in this case (36). Furthermore, it cannot be ruled out at this point that the observed production of OMV is really the consequence of a different mechanism of attack of the host cell that simply manifests as vesiculation of Lysobacter cells in broth culture. For instance, outer membrane bulging and protrusion without going all the way to vesicle production may be a normal event when C3 cells make contact with host fungi.
Although the mass spectrometry analysis alone does not positively identify any particular active molecule, the results strongly suggest two particular molecules. Alteramide B is the only C3 antifungal product specifically shown to possess significant activity against S. cerevisiae in previous work (6). The observed negative ion m/z of 509 is consistent with a single proton extraction from alteramide B. Therefore, it is a likely candidate as an active molecule in these OMV. A small peak was also observed in the negative ion scan at m/z 511. This species could be dihydromaltophilin, which has a molar mass of 512 g/mol in its un-ionized form. Finally, a comment can be made about a series of peaks seen in the range of m/z 1,332 and 1,596. These groups of peaks appear to have multiplets separated by mass units of 14, the expected mass of a CH2 unit. These m/z ratios could represent the existence of dimer ion clusters of phospholipids with a single negative charge, as has been observed previously (37). Because lipopolysaccharides are known to contain acyl groups of various lengths, these multiplets may also represent lipid A species. Interestingly, the overall mass of these species could be consistent with that of partially deacylated lipid A, based on comparison to the lipid A mass in other Gram-negative bacteria (38). Deacylation of lipid A and LPS has been proposed to constitute part of a mechanism of OMV biogenesis in other bacterial species (38).
The fact that C3 OMV carry these small-molecule antibiotics is a particularly important result. Although C3 antifungal antibiotics were previously thought to exist in the form of a protein-containing complex, identification of the relevant complexing entity as vesicular in nature is novel. The result has consequences in terms of the ongoing efforts to develop these antibiotics to treat plant or animal fungal diseases. At one level, the ability to concentrate and sequester the antibiotic activity of interest by a simple filtration process may help to speed the efforts to scale up and purify the relevant molecules. Additionally, the very fact that the antifungal molecules are associated with OMV suggests that strategies to increase OMV production may also enhance antibiotic yields. On the other hand, the fact that the HSAF− mutant still produces OMV suggests that OMV and antibiotic production may not be tightly coupled. Further investigation would be needed to understand this point. In addition, the actual means of association of the antibiotic molecules with OMV membranes requires more study. One key parameter to understand is whether these antibiotics associate directly with the lipid bilayer of OMV, as suggested for quorum signaling molecules (15), or with a membrane-associated protein. This localization may point the way to the best formulation strategies to ultimately accompany therapeutic development.
MATERIALS AND METHODS
Growth of bacteria and vesicle isolation.
OMV were prepared from cell-free culture supernatants of the C3 strain of Lysobacter enzymogenes or, in some cases, from the K19 mutant strain designated HSAF− (3), in which the polyketide synthase of the HSAF polyketide pathway has been deleted by mutation. OMV were obtained from one of two media, 10% tryptic soy broth or minimal medium (MM). The latter is based on the medium used previously to obtain OMV from Lysobacter sp. XL1 (25). In some cases, MM was supplemented with 1% colloidal chitin. After overnight growth in one of these media (30°C), bacterial cells were removed by centrifugation at 20,000 × g for 20 min (25). The remaining supernatants were either used directly for solvent extractions (see below) or processed as follows to obtain OMV.
OMV preparation from cell-free supernatants was performed either by direct ultracentrifugation at ∼140,000 × g for 2 h (Beckman SW40Ti swinging bucket rotor; t/k factor ≈ 0.88 min · rpm2 [where k factor is the clearing factor]) or the supernatants were first ultrafiltered through a 300-kDa-cutoff polysulfone membrane (EMD Millipore, Billerica, MA) in a 400-ml stirred cell (Amicon model 8400). Both methods generated a small volume of solid material that was typically resuspended in Tris-buffered saline (TBS) and filtered through a 0.45-μm filter to remove any residual whole cells. Resuspension was followed by a smaller volume ultracentrifugation (Beckman AirFuge, A95 rotor, ∼140,000 × g, 15 min; t/k factor ≈ 1.25 min · rpm2) to obtain a concentrated pellet of OMV. Preparations were then washed by multiple centrifugation and resuspension steps (usually 2 or 3 times). OMV yield was measured by protein content, typically by Bradford assay using bovine serum albumin as a standard. A typical yield from an overnight 300-ml culture in 10% TSB was approximately 0.5 to 0.7 mg of total OMV protein.
OMV were labeled when necessary using one of three different probes, namely, Alexa Fluor 555–N-hydroxysuccinimide (Life Technologies, Grand Island, NY) to label amino groups and two hydrophobic probes, the green fluorescent dioctadecyloxacarbocyanine perchlorate (diOC18) and red fluorescent 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (diIC18). diOC18 in dimethylformamide was added to a vortexed solution of OMV at a ratio of 21 ng per μg of OMV protein (as measured by Bradford assay). Labeled OMV were pelleted and washed after labeling. OMV were labeled with diIC18 in essentially the same way, except that an ethanol solution was used. Alexa Fluor 555-NHS (∼2 μg) was incubated with OMV (∼50 μg protein) in sodium bicarbonate solution for 1 h, followed by washing and resuspension in TBS. Fluorescence measurements were performed in 96-well plates using an Applied Biosystems Cytofuor series 4000 multiwell plate reader.
Liposomes for microscopy experiments [1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine in a 1/1 ratio with 1 mol% diIC18] were prepared (39) by resuspension in of a dry film of lipid (Avanti Polar Lipids, Birmingham, AL) containing the probe, followed by extrusion through a 0.1-μm Nuclepore, polycarbonate track-etch membrane (GE Whatman, Pittsburgh, PA) using a hand extruder (Avanti Polar Lipids, Birmingham, AL).
Samples for negative-stain electron microscopy were prepared by resuspension of a high-speed OMV pellet into a buffer of sodium cacodylate containing 2.5% glutaraldehyde for fixation and washed, followed by staining with uranyl formate. Stained samples were applied to glow-discharged carbon/Formvar-coated 300 mesh copper grids (EM Sciences, Hatfield, PA). Samples were observed using a JEM-100CXII electron microscope (JEOL) with the assistance of the Electron Microscopy Facility, Nelson Biology Laboratory, Rutgers University, New Brunswick, NJ.
OMV particle size was measured using a Zetasizer Nano-ZS particle analyzer (Malvern Instruments, Westborough, MA), with the assistance of Zoltan Szekely and Dan Myers. OMV samples were diluted approximately 10- to 100-fold for analyses, e.g., to approximately 10 to 20 mg/liter of protein in TBS.
LPS assay.
Assay of LPS content was performed using a chromogenic version (ToxinSensor) of the Limulus amebocyte lysate assay (Genscript, Piscataway, NJ). Standard curves based on E. coli LPS were generated to estimate the amount of LPS in C3 OMV samples.
Chitinase activity.
Activity of OMV from cells grown in MM with 1% chitin was measured using a fluorogenic substrate, of 4-methylumbelliferyl β-d-N,N′,N″-triacetylchitotrioside (TC; Sigma-Aldrich, St. Louis, MO). Fluorescence is generated by enzymatic cleavage of the substrate and was measured in a 96-well plate format on an Applied Biosystems Cytofluor 4000 plate reader with excitation wavelength of 350 nm and emission wavelength of 460 nm. The relative fluorescence at defined time points within the linear range of fluorescence development was used to compare the activity of samples. For heat treatment experiments, OMV samples were incubated at 70°C for 30 min before assaying for chitinase activity. Proteinase K (Sigma-Aldrich, St. Louis, MO) was used to assess accessibility of chitinase activity associated with OMV by comparison of incubation for 48 h with proteinase K (∼5 g/liter) at 30°C versus that without. For freeze-thaw experiments, OMV samples were frozen in liquid nitrogen for at least 20 s and then thawed on a warm block in a process repeated 10 times. OMV were washed free of soluble material after this treatment by sedimentation two times.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Samples were prepared by dilution 1:1 with a 2× Laemmli sample buffer containing β-mercaptoethanol. Electrophoresis was performed on a Phast gel system (GE Healthcare, Life Sciences, Pittsburgh, PA) using an 8 to 25% polyacrylamide precast gel and SDS-containing buffer strips. Protein bands were visualized by silver staining. Western blotting was performed using the Phast Gel apparatus and a probe antibody against a peptide sequence (CTYTGGDKESQIPSS) in the PilA protein of C3 (Biomatik, Wilmington, DE), donated by Donald Kobayashi, Rutgers University. Detection was via chemiluminescence resulting from horseradish peroxidase-coupled second antibody and a luminescent substrate (LumiSensor; GenScript, Piscataway, NJ).
DNA assay.
DNA content in OMV preparations was assessed either by standard extraction and precipitation methods involving ethanol or isopropanol or by use of a fluorogenic detection method based on dye binding to DNA. In the former method, DNA was precipitated and resuspended in Tris-EDTA buffer, and content was detected by optical density at 280 nm using a Nanodrop spectrophotometer. The identity of DNA was confirmed by comparing yields of OMV nucleic acids after incubation with DNase I or RNase A. In the fluorogenic assays, samples were incubated with Diamond nucleic acid dye (Promega, Madison, WI), which can cross cell membranes. OMV were incubated with or without DNase I, and then DNA content was measured by fluorescence (excitation/emission, 485/580 nm) to assess protected or encapsulated DNA.
Yeast growth assay.
To measure yeast growth-inhibitory activity of OMV samples, the turbidities of Saccharomyces cerevisiae S30 cell cultures were measured. In initial experiments, approximately 10 μl each of OMVs and overnight S30 culture were incubated for 30 min at 30°C. The cultures were then expanded with 200 μl yeast extract-peptone-dextrose (YPD) medium and incubated for 5.5 h or overnight at 30°C. The optical density of 100 μl of each culture diluted with medium to 1 ml was measured at 600 nm (OD600). In some cases, the assay was modified by direct mixing of OMV and yeast into 200 μl of YPD medium, followed by an overnight incubation. Both protocols gave similar results in terms of OMV activity in most cases. OMV from cells grown in 10% TSB showed similar specific activity to those from cells grown in MM with 1% chitin. For experiments to test contact dependence of OMV effects, small filtration devices (<1 cm diffusion length) with 100-kDa-cutoff filters (Centrex; Schleicher and Schuell) were used and sterilely filled on opposite sides of the filter with solutions as indicated. The chamber with OMV alone or buffer alone contained 100 μl of YPD or Tris-buffered saline with 10 mM glucose. The chamber with yeast cells contained 200 μl of YPD medium. Devices were incubated horizontally with shaking to facilitate liquid contact on both sides of the membrane. After 5.5 h or overnight, 100 μl of the yeast-containing solution was diluted into 1 ml of YPD, and OD600 was measured. For experiments in which organic solvent extracts were being tested, ethanol-dissolved samples were used. The yeast cells were able to tolerate up to 15% (vol/vol) ethanol in YPD medium.
Fusarium growth assay.
Fusarium subglutinans (kindly provided by Marshall Bergen and James White, Rutgers University) was initially cultured in YPD liquid medium and then spread onto YPD agar plates. A 4-mm plug from the plate was excised and laid upside down onto the surface of a fresh YPD plate so that hyphae grew from the center of the plate. After sufficient growth, wells were created approximately 5 mm from the edge of the growing mycelia, and samples (50 μl) were loaded into the wells for testing. Growth in the region of the wells was assessed by visual observation.
OMV binding to yeast cells.
OMV were labeled for yeast binding assays with an ethanolic solution of DiIC18, as described above, at a ratio of 20 ng per μg of OMV protein, with a final ethanol concentration of 0.1% (vol/vol). This mixture was incubated at 37°C for 1 h. Labeled OMV were pelleted (140,000 × g) and resuspended in 400 μl TBS. The S30 strain of the yeast Saccharomyces cerevisiae was grown overnight in YPD medium, collected and washed by sedimentation, and resuspended in TBS with 10 mM glucose. Typically, 10 μl of OMV was mixed with 20 μl of yeast cells diluted into 1 ml of TBS with glucose. Association with yeast was measured by incubation at 30°C, followed by collection of yeast cells by centrifugation at 4,500 × g. Fluorescence of the DiIC18 associated with the yeast pellet was used as a measure of association.
For OMV binding experiments by microscopy, the S30 strain of the yeast Saccharomyces cerevisiae was grown overnight in YPD medium and collected and washed by sedimentation. Ten microliters of the washed overnight culture was incubated with 10 μl of labeled OMV preparations (approximately 50 ng total protein) or liposomes (0.125 mM total lipid) for 2 h and diluted into 1 ml of Tris buffered saline (TBS) with 10 mM glucose, and the yeast cells were collected by centrifugation at 1,000 × g and washed twice. After resuspension in 10 μl of TBS-glucose, the samples were used for fluorescence microscopy. In some cases, samples were observed without dilution and removal of unbound OMV. Samples were observed using an Olympus FSX100 inverted epifluorescence microscope.
Solvent extractions of antifungal activity.
Bligh-Dyer extractions to analyze lipids were performed as described previously (29), with appropriate volume adjustments to accommodate sample size.
Ethyl acetate was used to extract preparations at various stages of purification using variations on previously published methods (30). For the large-volume cell-free supernatants used in analysis of activity in Fig. 4, 125 ml of ultrafiltrate (see above) or a resuspension of the material on the filter surface into 125 ml of 10% TSB was used for extraction. Medium was extracted 3 times with 50 ml of ethyl acetate using a separatory funnel. The collected upper phase of solvent was evaporated and the entire extraction was redissolved into 250 μl of ethanol for activity testing.
Some washed OMV preparations were extracted with ethyl acetate in a smaller-volume format. Preparations were diluted to approximately 100 mg/liter of protein before extraction into an equal volume of ethyl acetate. After removal of solvent, the solids were redissolved into ethanol for activity assays or methanol for mass spectrometry or further purification by thin-layer chromatography.
Thin-layer chromatography.
Samples obtained from Bligh-Dyer extractions to test for OMV phospholipid content (see data in Fig. 2) were concentrated and spotted onto silica gel plates (Analtech Uniplate; 250 μm HF; Sigma-Aldrich, St. Louis, MO) and run in the solvent system chloroform:methanol:ammonium hydroxide (65:25:2.5). Phospholipid standards were 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG) from Avanti Polar Lipids (Birmingham, AL).
For purification of active antifungal molecules by ethyl acetate extraction and analysis by mass spectrometry, the solvent system chloroform:methanol:ammonium hydroxide (65:25:5) was used initially. On preparative scale plates (Analtech Uniplate, 20 by 20 cm, 2,000-μm silica gel GF with UV 254 nm fluorescence), this system yielded a UV-positive band at approximately Rf = 0.11 that could be extracted into methanol, redissolved in ethanol, and demonstrated to possess activity against yeast cells. Subsequently, for mass spectrometry (MS) analysis, samples were purified using a solvent system of chloroform-methanol-20% ammonium hydroxide (65:30:5), which resulted in an Rf of 0.55 for the active compound (MS-grade high-performance thin-layer chromatography silica gel 60 F254; EMD Merck, Burlington, MA). This band was extracted into methanol for analysis by mass spectrometry.
Mass spectrometry was performed on a Finnigan LCQ Duo electrospray ionization instrument (Department of Chemistry, Rutgers University; thanks to Alexei Ermakov for assistance).
OMV LPS/protein calculation.
An estimation of range of possible ratios for LPS/protein was calculated based on assumption of spherical vesicles of about 135 nm in diameter with an outer monolayer consisting of LPS and 50% by weight protein in the membrane.
The maximum LPS/protein value would be in empty vesicles. If the membrane is assumed to be 50% by weight protein, then, of the remaining 50%, more than half is LPS. We assume 33% of total weight is LPS (because of the large LPS molecular weight). So maximum LPS/protein (g/g) is 0.66.
The lowest possible LPS/protein ratio would come from OMV packed with a maximum amount of protein in the interior of the vesicle. An internal radius of 64 nm is assumed with a 3-nm membrane thickness. Then the internal volume is equal to 4/3 × 3.14 × (64 nm)3, or 1.1 × 106 nm3. An estimation of the protein in the membrane can be made from the total membrane volume, which is equal to 4/3 × 3.14 (67 nm)3 − 4/3 × 3.14 (64 nm)3, or 162,000 nm3. So the ratio of protein in the filled vesicle to that in the empty vesicle would be (1.1 + 0.16)/0.16, or 7.9. By this estimate the minimum LPS/protein ratio would be 0.66/7.9, or 0.08. These are clearly very rough estimates, but finding an LPS/protein ratio in this range would give a modicum of confidence in the measurement.
ACKNOWLEDGMENTS
This work was funded by the Byrne Seminar program, Department of Plant Biology, Rutgers Master in Business and Science program, and by personal contributions from Paul Meers.
The C3 strain of L. enzymogenes was donated by the laboratory of Donald Kobayashi, Rutgers University. The K19 mutant of L. enzymogenes C3 was kindly provided by the laboratory of Gary Yuen, University of Nebraska. Equipment and supplies were donated by Transave, Inc., Anthony Scotto, and Barbara A. Zilinskas. Shanee Grant and David Itenberg also contributed to this work.
REFERENCES
- 1.Sullivan RF, Holtman MA, Zylstra GJ, White JF Jr, Kobayashi DY. 2003. Taxonomic positioning of two biological control agents for plant diseases as Lysobacter enzymogenes based on phylogenetic analysis of 16S rDNA, fatty acid composition and phenotypic characteristics. J Appl Microbiol 94:1079–1086. doi: 10.1046/j.1365-2672.2003.01932.x. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi DY, Yuen GY. 2007. The potential of Lysobacter spp. as bacterial biological control agents for plant diseases. CAB Reviews Persp Agr Vet Sci Nutr Nat Res 2:007. [Google Scholar]
- 3.Li S, Jochum CC, Yu F, Zaleta-Rivera K, Du L, Harris SD, Yuen GY. 2008. An antibiotic complex from Lysobacter enzymogenes strain C3: antimicrobial activity and role in plant disease control. Phytopathology 98:695–701. doi: 10.1094/PHYTO-98-6-0695. [DOI] [PubMed] [Google Scholar]
- 4.Panthee S, Hamamoto H, Paudel A, Sekimizu K. 2016. Lysobacter species: a potential source of novel antibiotics. Arch Microbiol 198:839–845. doi: 10.1007/s00203-016-1278-5. [DOI] [PubMed] [Google Scholar]
- 5.Graupner PR, Thornburgh S, Mathieson JT, Chapin EL, Kemmitt GM, Brown JM, Snipes CE. 1997. Dihydromaltophilin; a novel fungicidal tetramic acid containing metabolite from Streptomyces sp. J Antibiot (Tokyo) 50:1014–1019. doi: 10.7164/antibiotics.50.1014. [DOI] [PubMed] [Google Scholar]
- 6.Ding Y, Li Y, Li Z, Zhang J, Lu C, Wang H, Shen Y, Du L. 2016. Alteramide B is a microtubule antagonist of inhibiting Candida albicans. Biochim Biophys Acta 10:2097–2106. doi: 10.1016/j.bbagen.2016.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi DY, Crouch JA. 2009. Bacterial/fungal interactions: from pathogens to mutualistic endosymbionts. Annu Rev Phytopathol 47:63–82. doi: 10.1146/annurev-phyto-080508-081729. [DOI] [PubMed] [Google Scholar]
- 8.Hoekstra D, Van Der Laan JW, De Leij L, Witholt B. 1976. Release of outer membrane fragments from normally growing Escherichia coli. Biochim Biophys Acta 455:889–899. doi: 10.1016/0005-2736(76)90058-4. [DOI] [PubMed] [Google Scholar]
- 9.Kadurugamuwa JL, Beveridge TJ. 1996. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol 178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by gram-negative bacteria is a novel envelope stress response. Mol Microbiol 63:545–558. doi: 10.1111/j.1365-2958.2006.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macdonald IA, Kuehn MJ. 2013. Stress-induced outer membrane vesicle production by Pseudomonas aeruginosa. J Bacteriol 195:2971–2981. doi: 10.1128/JB.02267-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumgarten T, Sperling S, Seifert J, von Bergen M, Steiniger F, Wick LY, Heipieper HJ. 2012. Membrane vesicle formation as multiple stress response mechanism enhances cell surface hydrophobicity and biofilm formation of Pseudomonas putida DOT-T1E. Appl Microbiol Biotechnol 93:837–845. doi: 10.1007/s00253-011-3442-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwechheimer C, Kuehn MJ. 2015. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bielaszewska M, Rüter C, Bauwens A, Greune L, Jarosch K-A, Steil D, Zhang W, He X, Lloubes R, Fruth A, Kim KS, Schmidt MA, Dobrindt U, Mellmann A, Karch H. 2017. Host cell interactions of outer membrane vesicle-associated virulence factors of enterohemorrhagic Escherichia coli O157: intracellular delivery, trafficking and mechanisms of cell injury. PLoS Pathog 13:e1006159. doi: 10.1371/journal.ppat.1006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mashburn-Warren L, Howe J, Garidel P, Richter W, Steiniger F, Roessle M, Brandenburg K, Whiteley M. 2008. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Mol Microbiol 69:491–502. doi: 10.1111/j.1365-2958.2008.06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yaron S, Kolling GL, Simon L, Matthews KR. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl Environ Microbiol 66:4414–4420. doi: 10.1128/AEM.66.10.4414-4420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Renelli M, Matias V, Lo RY, Beveridge TJ. 2004, DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150:2161–2169. doi: 10.1099/mic.0.26841-0. [DOI] [PubMed] [Google Scholar]
- 18.Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. 2011. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother, 55:3084–3090. doi: 10.1128/AAC.00929-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciofu O, Beveridge TJ, Kadurugamuwa J, Walther-Rasmussen J, Hoiby N. 2000. Chromosomal β-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J Antimicrob Chemother 45:9–13. doi: 10.1093/jac/45.1.9. [DOI] [PubMed] [Google Scholar]
- 20.Ellis TN, Kuehn MJ. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuehn MJ, Kesty NC. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev 19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- 22.Berleman JE, Allen S, Danielewicz MA, Remis JP, Gorur A, Cunha J, Hadi MZ, Zusman DR, Northen TR, Witkowska HE, Auer M. 2014. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol 5:474. doi: 10.3389/fmicb.2014.00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schrempf H, Merling P. 2015. Extracellular Streptomyces lividans vesicles: composition, biogenesis and antimicrobial activity. Microb Biotech 8:644–658. doi: 10.1111/1751-7915.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khandelwal P, Banerjee-Bhatnagar N. 2003. Insecticidal activity associated with the outer membrane vesicles of Xenorhabdus nematophilus. Appl Environ Microbiol 69:2032–2037. doi: 10.1128/AEM.69.4.2032-2037.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasilyeva NV, Tsfasman IM, Suzina NE, Stepnaya OA, Kulaev IS. 2008. Secretion of bacteriolytic endopeptidase L5 of Lysobacter sp. XL1 into the medium by means of outer membrane vesicles. FEBS J 275:3827–3835. doi: 10.1111/j.1742-4658.2008.06530.x. [DOI] [PubMed] [Google Scholar]
- 26.Kudryakova I, Shishkova N, Vasilyeva N. 2016. Outer membrane vesicles of Lysobacter sp. XL1: biogenesis, functions, and applied prospects. Appl Microbiol Biotechnol 100:4791–4801. doi: 10.1007/s00253-016-7524-6. [DOI] [PubMed] [Google Scholar]
- 27.Dorward DW, Garon CF, Judd RC. 1989. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol 171:2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bauman SJ, Kuehn MJ. 2006. Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microbes Infect 8:2400–2408. doi: 10.1016/j.micinf.2006.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 30.Qian G, Wang Y, Liu Y, Xu F, He Y-W, Du L, Venturi V, Fan J, Hu B, Liu F. 2013. Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl Environ Microbiol 79:6604–6616. doi: 10.1128/AEM.01841-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyngby J, Olsen LH, Eidem T, Lundanes E, Jantzen E. 2002. Quantification of lipopolysaccharides in outer membrane vesicle vaccines against meningococcal disease. High-performance liquid chromatographic determination of the constituent 3-hydroxy-lauric acid. Biologicals 30:7–13. doi: 10.1006/biol.2001.0285. [DOI] [PubMed] [Google Scholar]
- 32.Leitner DR, Feichter S, Schild-Prüfert K, Rechberger GN, Reidl J, Schild S. 2013. Lipopolysaccharide modifications of a cholera vaccine candidate based on outer membrane vesicles reduce endotoxicity and reveal the major protective antigen. Infect Immun 81:2379–2393. doi: 10.1128/IAI.01382-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casadevall A, Nosanchuk JD, Williamson P, Rodrigues ML. 2009. Vesicular transport across the fungal cell wall. Trends Microbiol 17:158–162. doi: 10.1016/j.tim.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker L, Sood P, Lenardon MD, Milne G, Olson J, Jensen G, Wolf J, Casadevall A, Adler-Moore J, Gowa NAR. 2018. The viscoelastic properties of the fungal cell wall allow traffic of AmBisome as intact liposome vesicles. mBio 9:e02383-17. doi: 10.1128/mBio.02383-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomada S, Sonego P, Moretto M, Engelen K, Petot I, Perazzoli M, Puopolo G. 2017. Dual RNA-Seq of Lysobacter capsici AZ78 – Phytophthora infestans interaction shows the implementation of attack strategies by the bacterium and unsuccessful oomycete defense responses. Environ Microbiol 19:4113–4125. doi: 10.1111/1462-2920.13861. [DOI] [PubMed] [Google Scholar]
- 36.Metruccio MM, Evans DJ, Gabriel MM, Kadurugamuwa JL, Fleiszig SM. 2016. Pseudomonas aeruginosa outer membrane vesicles triggered by human mucosal fluid and lysozyme can prime host tissue surfaces for bacterial adhesion. Front Microbiol 7:871–879. doi: 10.3389/fmicb.2016.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.James PF, Perugini MA, O'Hair RAJ. 2006. Sources of artefacts in the electrospray ionization mass spectra of saturated diacylglycerophosphocholines: from condensed phase hydrolysis reactions through to gas phase intercluster reactions. J Am Soc Mass Spec 17:384–394. doi: 10.1016/j.jasms.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Bohuszewicz O, Liu J, Low HH. 2016. Membrane remodelling in bacteria. J Struct Biol 196:3–14. doi: 10.1016/j.jsb.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hope MJ, Bally MB, Webb G, Cullis PR. 1985. Production of large unilamellar vesicles by a rapid extrusion procedure: characterization of size distribution, trapped and ability to maintain a membrane potential. Biochim Biophys Acta 812:55–65. doi: 10.1016/0005-2736(85)90521-8. [DOI] [PubMed] [Google Scholar]