We clarified the elusive biological function of cyanophycin in the nondiazotrophic cyanobacterium Synechocystis sp. PCC 6803. Cyanophycin is a dynamic carbon/nitrogen storage polymer (multi-arginyl-l-polyaspartate) that is conditionally present in most cyanobacteria and a few heterotrophic bacteria as cellular inclusion granules. Here, we show that the cyanophycin-synthesizing enzyme CphA in the nonactive state localizes diffusely in the cytoplasm. When cyanophycin synthesis is triggered, active CphA first aggregates into foci and then covers the surface of mature cyanophycin granules, which in vitro requires Mg2+ as a cofactor. Cyanophycin accumulation enables Synechocystis sp. to optimize nitrogen assimilation under nitrogen-poor conditions, in particular when the nitrogen supply fluctuates and during day/night cycles, by allowing continuous nitrogen assimilation and storage. Therefore, cyanophycin provides the wild-type cyanobacterium with a clear fitness advantage over non-cyanophycin-producing cells in natural environments with fluctuating nitrogen supply.
KEYWORDS: CphA, cyanobacteria, cyanophycin, nitrogen storage, nitrogen utilization, synechocystis
ABSTRACT
Cyanophycin is a carbon/nitrogen storage polymer widely distributed in most cyanobacterial strains and in a few heterotrophic bacteria. It is a nonribosomal polypeptide consisting of equimolar amounts of aspartate and arginine. Here, we focused on the physiological function and cell biology of cyanophycin in the unicellular nondiazotrophic cyanobacterium Synechocystis sp. strain PCC 6803. To study the cellular localization of the cyanophycin-synthesizing enzyme CphA during cyanophycin synthesis and degradation, we fused it to green fluorescent protein. When CphA was inactive, it localized diffusely in the cytoplasm. When cyanophycin synthesis was triggered, CphA first aggregated into foci and later localized on the surface of cyanophycin granules. In the corresponding cell extracts, localization of CphA on the cyanophycin granule surface required Mg2+. During cyanophycin degradation, CphA dissociated from the granule surface and returned to its inactive form in the cytoplasm. To investigate the physiological role of cyanophycin, we compared wild-type cells with a CphA-deficient mutant. Under standard laboratory conditions, the ability to synthesize cyanophycin did not confer a growth advantage. To mimic the situation in natural habitats, cells were cultured with a fluctuating and limiting nitrogen supplementation and/or day/night cycles. Under all of these conditions, cyanophycin provided a fitness advantage to the wild type over the mutant lacking cyanophycin. During resuscitation from nitrogen starvation, wild-type cells accumulated cyanophycin during the night and used it as an internal nitrogen source during the day. This demonstrates that cyanophycin can be used as a temporary nitrogen storage to uncouple nitrogen assimilation from photosynthesis.
IMPORTANCE We clarified the elusive biological function of cyanophycin in the nondiazotrophic cyanobacterium Synechocystis sp. PCC 6803. Cyanophycin is a dynamic carbon/nitrogen storage polymer (multi-arginyl-l-polyaspartate) that is conditionally present in most cyanobacteria and a few heterotrophic bacteria as cellular inclusion granules. Here, we show that the cyanophycin-synthesizing enzyme CphA in the nonactive state localizes diffusely in the cytoplasm. When cyanophycin synthesis is triggered, active CphA first aggregates into foci and then covers the surface of mature cyanophycin granules, which in vitro requires Mg2+ as a cofactor. Cyanophycin accumulation enables Synechocystis sp. to optimize nitrogen assimilation under nitrogen-poor conditions, in particular when the nitrogen supply fluctuates and during day/night cycles, by allowing continuous nitrogen assimilation and storage. Therefore, cyanophycin provides the wild-type cyanobacterium with a clear fitness advantage over non-cyanophycin-producing cells in natural environments with fluctuating nitrogen supply.
INTRODUCTION
One of the most important steps in evolution on Earth was the appearance of oxygen in the atmosphere. This dramatic change in the redox state of our planet, called the “great oxidation event,” was triggered by the emergence of cyanobacteria capable of oxygenic photosynthesis (1).
Some of the cyanobacteria acquired the ability to fix atmospheric nitrogen, which allowed them to spread throughout the illuminated biosphere and fertilize it (2). Nitrogen is a necessary macronutrient for all life and therefore constitutes a growth-limiting factor in many terrestrial and aquatic ecosystems (3). Nondiazotrophic cyanobacteria can use a variety of combined nitrogen sources, such as nitrate or ammonia. In the absence of a usable nitrogen source, these cyanobacteria face nitrogen starvation, a situation that arrests anabolic metabolism, followed by a process known as chlorosis, which finally results in a state of dormancy (4). Chlorosis is characterized by the degradation of photosynthetic pigments, which leads to a color change from blue-green to yellow (5). Together with the degradation of light-harvesting phycobilisomes, the cells accumulate carbon-reserve polymers, e.g., glycogen or polyhydroxybutyrate, and arrest the cell cycle (6–8). When nitrogen deprivation is prolonged, cells further reduce their bulk of cellular proteins until they reach a final chlorotic stage, where they maintain a low residual level of photosynthetic activity. In this state, cells are able to survive long periods of starvation. After the addition of a nitrogen source, they are able to regreen and resume growth (9).
The process of resuscitation from nitrogen-starvation-induced chlorosis has been investigated in detail in the unicellular and nondiazotrophic cyanobacterium Synechocystis sp. strain PCC 6803 (here, Synechocystis sp.) (7, 10, 11). The addition of a usable nitrogen source triggers a tightly coordinated resuscitation program, which results in the restoration of the vegetative cell cycle within 48 h. This process can be divided into two major phases. First, the cells turn on glycogen catabolism, which provides energy and carbon skeletons for nitrogen assimilation (10); this is used to first reinstall the basic anabolic machinery, in particular, the translational apparatus (7, 12). After 12 to 16 h, transition to the second phase occurs, where the cells reassemble their photosynthetic apparatus. The cells regreen and engage oxygenic photosynthesis (7). After reaching full photosynthetic activity, the cells enter the vegetative cell cycle. During resuscitation from nitrogen starvation, the carbon/nitrogen storage polymer cyanophycin transiently accumulates (7, 13). Cyanophycin (also known as cyanophycin granule polypeptide) is present in most cyanobacterial species and in a few heterotrophic bacteria (14). It is a nonribosomal polypeptide consisting of equimolar amounts of arginine and aspartate. Every aspartyl moiety of the polyaspartate backbone is linked with an arginine residue via an isopeptide bond (15). In nondiazotrophic cyanobacteria, cyanophycin accumulates when excess nitrogen is supplied and during unbalanced growth that lowers the growth rate (e.g., during sulfate, phosphate, or potassium starvation) (16, 17).
With a C/N ratio of 2:1, cyanophycin is extremely rich in nitrogen and is therefore used as a nitrogen storage compound. This plays an important role in nitrogen-fixing cyanobacteria, in particular in those that differentiate heterocysts to fix nitrogen during the day. The heterocysts accumulate large cyanophycin structures at the contact sites to the vegetative cells, termed the polar nodes; these nodes play a role in nitrogen trafficking between the nitrogen-fixing heterocyst and the vegetative cells of the filament (18). Nonheterocystous strains, such as Cyanothece sp. strain ATCC 51142, synthesize cyanophycin at night during nitrogen fixation, where in the absence of photosynthetic activity, nitrogenase is protected from harmful oxygen. During the day, nitrogen fixation is arrested, and cyanophycin is degraded to mobilize the fixed nitrogen (19, 20).
Cyanophycin synthetase (CphA) builds cyanophycin from arginine and aspartate in an ATP-consuming elongation reaction that requires KCl, MgCl2, and a sulfhydryl reagent (dithiothreitol [DTT] or β-mercaptoethanol) (21). The elongation reaction requires an unknown cyanophycin primer that must consist of at least three Asp-Arg building blocks (22) as a starting point. The primary structure of CphA consists of two regions; both regions contain an active site and an ATP binding site (22, 23). The putative cyanophycin elongation cycle starts at the C terminus of the cyanophycin primer. First, the carboxylic acid group of the polyaspartate backbone is activated by phosphorylation with the γ-phosphoryl group of ATP. Subsequently, one aspartate is bound at the C terminus by its amino group, forming a peptide bond. The intermediate (β-Asp-Arg)n-Asp is then transferred to the second active site of CphA and phosphorylated at the β-carboxyl group of aspartate. Finally, the α-amino group of arginine is linked via an isopeptide bond to the β-carboxyl group of aspartate (22). Cyanophycin accumulates in the form of opaque and light-scattering granules in the cell (6). CphB, an intracellular cyanophycinase, catalyzes the degradation of cyanophycin to β-Asp-Arg dipeptides (24). The last step in cyanophycin catabolism is the cleavage of the β-Asp-Arg dipeptides to monomeric arginine and aspartate, catalyzed by isoaspartyl dipeptidases (25).
Several previous studies have indicated that arginine availability is the main bottleneck of cyanophycin biosynthesis (17, 26–29). The committed step in arginine biosynthesis, the N-acetylglutamate kinase (NAGK) reaction, is regulated by the signal transduction protein PII (30). PII senses the energy status and the C/N ratio by binding 2-oxoglutarate and ATP (31, 32). Binding of PII enhances the catalytic efficiency of NAGK and decreases its feedback inhibition by arginine (33), resulting in increased arginine production, followed by the accumulation of cyanophycin (17, 29).
In the last decades, research on cyanophycin has mainly focused on its potential use in different biotechnological and industrial applications, whereas the biology of cyanophycin, in particular in nondiazotrophic cyanobacteria, remains largely uninvestigated. Previous studies revealed an unexplained transient accumulation of cyanophycin during the outgrowth of dormant Synechocystis sp. cells from nitrogen starvation (7, 13). Here, we aimed at identifying the role of cyanophycin synthesis in the recovery of Synechocystis sp. cells from nitrogen starvation and, more generally, at clarifying its function during fluctuating ambient nitrogen supply.
RESULTS
CphA localization changes during resuscitation from nitrogen starvation.
To investigate the intracellular localization of CphA, we fused cphA to the gene encoding enhanced green fluorescent protein (eGFP) (yielding eGFP fused to the C terminus of CphA) and inserted cphA-eGFP under the control of the native cphA promoter into the Synechocystis sp. shuttle vector pVZ322 (34). The resulting pVZ322-cphA-eGFP plasmid was introduced into Synechocystis sp. by triparental mating (34). The resulting strain, termed Synechocystis sp. strain CphA-eGFP, was used to investigate the cellular localization of CphA using comprehensive statistical analysis of microscopy images.
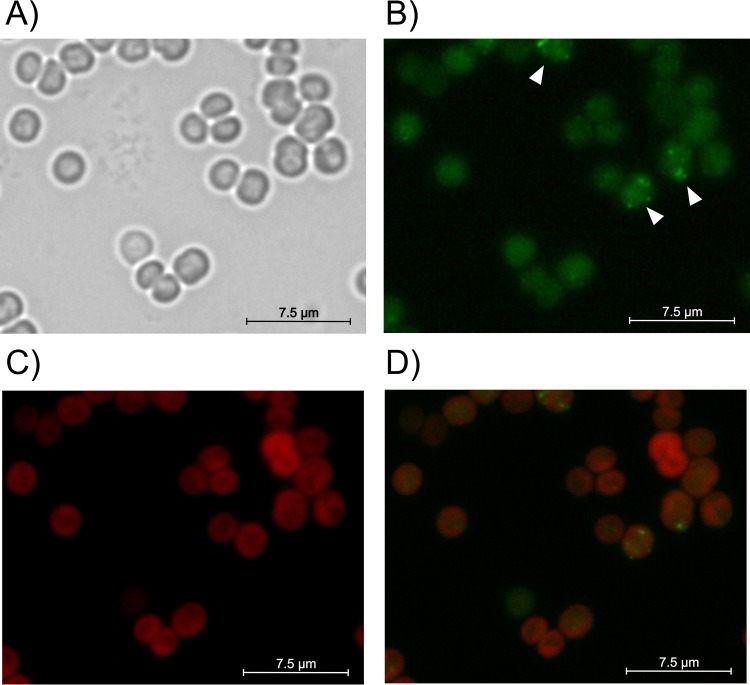
In the majority of nutrient-replete cells (87% ± 3%; n = 486) in the mid-exponential phase of growth (optical density at 750 nm [OD750], approximately 0.6), the GFP signal was uniformly distributed in the cytoplasm and did not colocalize with the fluorescence signal of the thylakoid membranes near the cell periphery (Fig. 1A to D). In the remaining cells (13% ± 3%), distinct foci formed. Accordingly, the cyanophycin content during exponential growth was usually less than 1% of the cell dry mass, and visible cyanophycin granules were absent (6, 17, 35).
FIG 1.
Localization of CphA-eGFP during exponential growth (OD750 of 0.6) of Synechocystis sp. (A) Bright-field image of cells. (B) GFP fluorescence; white arrows point to cells with CphA-eGFP foci. (C) Autofluorescence of thylakoid membranes. (D) Overlay of GFP and thylakoid membrane fluorescence images.
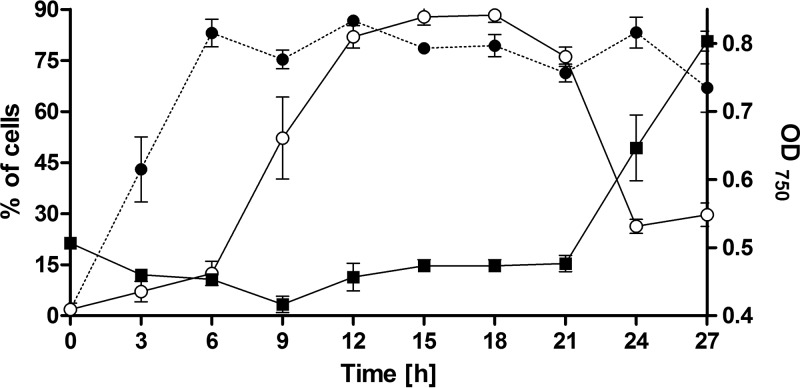
To reveal the localization of CphA during periods of cyanophycin accumulation or degradation, we monitored CphA-eGFP localization after the addition of nitrate to Synechocystis sp. cells that had been starved for nitrogen for 4 days. These conditions are known to induce transient cyanophycin accumulation (7, 13, 29). The cphA gene is constitutively expressed in different nitrogen regimes (7). In agreement, a recent quantitative proteomics analysis of resuscitating nitrogen-starved Synechocystis sp. cells revealed minor changes in the abundance of CphA. It increased by about 10% from cells starved of nitrogen for 21 days to fully recovered exponentially growing cells (11). In nitrogen-starved cells, the GFP signal was almost exclusively found as a diffuse signal in the cytoplasmic space (98% ± 2% of the cells) (see Fig. S1 in the supplemental material). Three hours after the addition of nitrate, the CphA-eGFP strain formed distinct foci in almost half of the cells (Fig. 2). After 6 h and up to 27 h after nitrate addition, the majority of cells contained CphA-eGFP foci (83% ± 7%). As the foci appeared, their number per cell simultaneously increased, with a maximum between 6 and 12 h after nitrate addition (Fig. 2 and S2A); thereafter, the number of foci gradually decreased. The apparent diameter of the CphA-eGFP foci reached a maximum between 12 and 21 h after nitrate addition (Fig. S2B). To correlate the appearance of the CphA-eGFP foci with the occurrence of cyanophycin granules, we stained the granules with the arginine-specific Sakaguchi stain. In bright-field images, the granules are opaque; the Sakaguchi stain enables a sensitive and clear identification of cyanophycin granules (17). Compared to the appearance of CphA-eGFP foci, the appearance of the cyanophycin granules was delayed. Six hours after nitrate addition, the number of cells containing cyanophycin granules increased dramatically, reaching a plateau of 87.8% ± 4.2% of the cells at 15 h after nitrate addition. Eighteen hours after the addition of nitrate, both the number of cells with visible cyanophycin granules and the diameter of the foci decreased (Fig. 2 and S2B). As the cyanophycin granules degraded, the cells simultaneously started to grow again (Fig. 2).
FIG 2.
Number of CphA-eGFP foci and cyanophycin granules during 27 h of resuscitation of Synechocystis sp. cultures that were nitrogen starved for 4 days. Cells were resuscitated by adding 17.3 mM nitrate to starved cultures. Measurements are means of three biological replicates. Filled circles, percentage of cells with visible CphA-eGFP foci (n ≥ 200 cells per time point); open circles, percentage of cells with cyanophycin granules visualized with the arginine-specific Sakaguchi stain (n ≥ 150 cells per time point); filled squares, growth curve.
During recovery from nitrogen starvation, cyanophycin is produced transiently, and the maximal level of accumulated cyanophycin was relatively low compared to other conditions that trigger cyanophycin accumulation (13, 17). Potassium starvation subjects cyanobacterial cells to an intense and immediate stress, which results in massive cyanophycin accumulation (17). This motivated us to analyze CphA localization under such conditions. Upon induction of potassium starvation, CphA relocalized from an initial diffuse cytoplasmic localization into distinct foci and later clearly localized on the surface of the cyanophycin granules, forming a halo-like structure (Fig. S3). Cyanophycin granules of larger diameter were amorphous and not spherical (Fig. S3). The proportion of cells with cyanophycin granules and CphA-eGFP foci increased during potassium starvation (Fig. S4A). Shifting the potassium-starved cells back to standard BG-11 medium restored the growth within approximately 24 h, concomitantly with the degradation of the cyanophycin granules. During cyanophycin degradation, it appeared in some cases that CphA-eGFP no longer colocalized with the granules (Fig. S3). The proportion of cells with cyanophycin granules and CphA-eGFP foci decreased accordingly. After 24 h, only about 20% of the cells contained cyanophycin granules. Simultaneously, 70% of the cells contained CphA-eGFP foci, while the remaining cells show the CphA-eGFP signal exclusively distributed in the cytoplasmic space (Fig. S4B). Furthermore, the average size of the granules decreased, while the number of granules per cell increased (Fig. S5), indicating that the granules disaggregate into smaller particles.
CphA localization on the cyanophycin granule surface requires Mg2+.
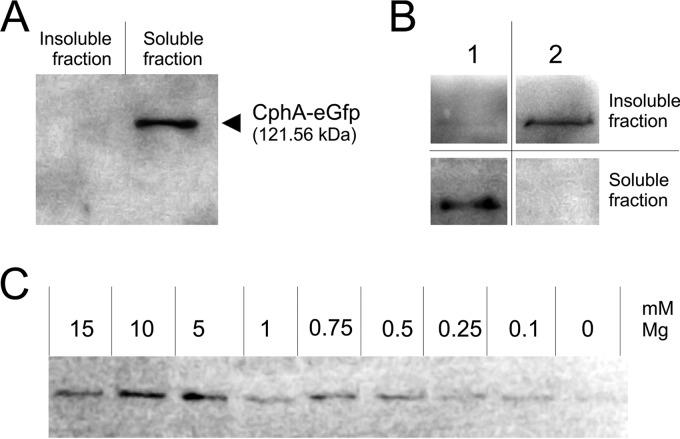
The above-mentioned results showed that CphA is homogenously distributed in the cytoplasm as long as no cyanophycin synthesis occurs, and when cyanophycin accumulation is triggered, CphA relocalizes into foci and subsequently attaches to the surface of growing cyanophycin granules. In vitro, the elongation reaction of CphA requires cyanophycin primers, arginine, aspartate, MgCl2, ATP, and a sulfhydryl reagent (β-mercaptoethanol or DTT) (21). To determine how these components affect the localization of CphA-eGFP in cell extracts, we analyzed cell lysates using anti-eGFP antibodies and immunoblots. Cell extracts were prepared from potassium-starved Synechocystis sp. cells that produced CphA-eGFP and separated into soluble and insoluble fractions, with the insoluble fractions containing the cyanophycin granules. We determined whether CphA remained associated with granules in the presence of different buffer components.
In potassium-starved Synechocystis sp. cells, CphA-eGFP appeared to be localized on the surface of the cyanophycin granule in vivo (Fig. S3). When these cells were lysed in a buffer containing 50 mM Tris-HCl (pH 7.4) and 4 mM EDTA, CphA was detected only in the soluble fraction (Fig. 3A). In contrast, when cells were lysed in a buffer containing 50 mM Tris-HCl (pH 8.2), 20 mM MgCl2, and 20 mM KCl, CphA appeared in the insoluble fraction, which indicated that it remained granule associated, because cyanophycin granules remain in the insoluble fraction, owing to their insolubility at neutral pH (Fig. 3B). To test whether MgCl2 is responsible for the persistent association of CphA with granules in cell extracts, cells were lysed in the same buffer containing different MgCl2 concentrations (Fig. 3C). In the absence of Mg2+, only residual amounts of CphA were detected in the insoluble fraction. Increasing amounts of Mg2+ led to increased amounts of CphA in the insoluble fraction, with an apparent optimal concentration of 10 mM Mg2+ for in vitro association with cyanophycin granules (Fig. 3C).
FIG 3.
Immunoblot detection of CphA-eGFP using anti-GFP primary antibodies. Cell extracts of potassium-starved Synechocystis sp. cells were fractionated into soluble and insoluble fractions. (A) Cells lysed in a buffer containing 50 mM Tris-HCl (pH 7.4) and 4 mM EDTA. (B) Comparison of potassium-starved Synechocystis cells lysed in the presence of 4 mM EDTA (same buffer mentioned above) (lane 1) and a buffer without EDTA containing 50 mM Tris-HCl (pH 8.2), 20 mM MgCl2, and 20 mM KCl (21) (lane 2). (C) Insoluble fraction of cells lysed in Tris-KCl buffer containing different MgCl2 concentrations.
Lack of CphA has no influence on the response to nitrogen starvation.
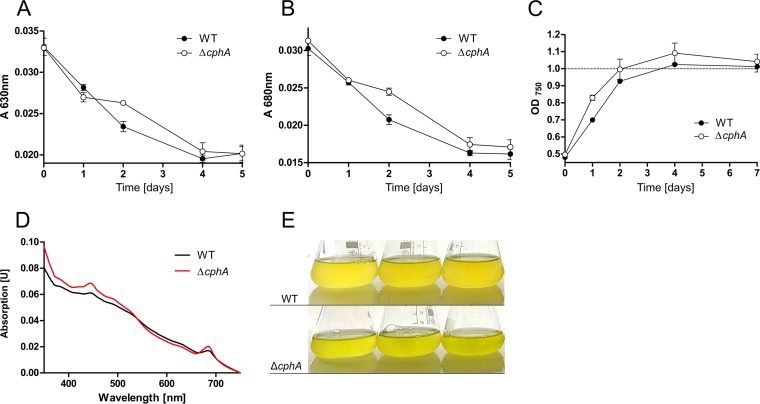
To gain further insights into the physiological role of cyanophycin, we generated a cphA deletion mutant in which the slr2002 (cphA) open reading frame was replaced with a kanamycin resistance cassette. Complete segregation of the mutant was confirmed by PCR (Fig. S6). The inability of the mutant to accumulate cyanophycin granule peptide (CGP) was confirmed by microscopy and cyanophycin extraction (data not shown). To investigate a possible influence of cyanophycin deficiency on the nitrogen starvation response, Synechocystis sp. wild-type and ΔcphA mutant cells were grown to an OD750 of 0.4 to 0.6, washed, and resuspended in BG-11 lacking a combined nitrogen source. This treatment induces nitrogen chlorosis, which is visible as the degradation of the photosynthetic apparatus (9). We monitored the degradation of pigments during chlorosis by recording the UV-Vis spectra of the cells. In the course of chlorosis, the absorbance at 630 nm (phycobiliproteins; Fig. 4A) and at 680 nm (chlorophyll a/b; Fig. 4B) decreased in wild-type and ΔcphA mutant cultures similarly. After induction of nitrogen starvation, wild-type and ΔcphA mutant cells divided one more time, as indicated by the doubling of the OD750 of the culture (Fig. 4C). After prolonged nitrogen starvation for 13 days, cells of both the wild type and the ΔcphA mutant completed chlorosis, and the cultures appeared yellowish (Fig. 4E). However, the pigment composition of long-term starved cultures of the ΔcphA mutant and wild type slightly differed; the peak heights at 440 nm and 680 nm in UV-Vis spectra in the ΔcphA mutant were higher, which indicated a higher residual amount of chlorophyll a/b (Fig. 4D).
FIG 4.
Changes in pigmentation and growth during nitrogen starvation of Synechocystis sp. wild type and a ΔcphA mutant under continuous light. Nitrogen starvation was induced by resuspending washed exponentially growing cells in BG-11 medium lacking a nitrogen source to an OD750 of 0.5. The degradation of pigments during chlorosis was monitored by recording UV-Vis spectra, which were normalized to cultures of the same optical density at 750 nm. Shown are the means of three biological replicates. (A) Course of absorbance of phycobiliproteins at 630 nm. (B) Course of absorbance of chlorophyll a/b at 680 nm. (C) Growth curve (OD750) of Synechocystis sp. during nitrogen starvation. (D) UV-Vis spectrum of Synechocystis sp. wild type and ΔcphA mutant cells that were nitrogen starved for 13 days. Spectra were normalized to cultures of the same optical density at 750 nm. (E) Three biological replicates of cultures of Synechocystis sp. wild type and ΔcphA mutant starved of nitrogen for 13 days.
Cyanophycin synthesis delays resuscitation from nitrogen starvation under standard laboratory conditions.
As cyanophycin transiently accumulates during resuscitation from nitrogen starvation (7), we focused on this process to investigate the physiological role of cyanophycin. To determine whether cyanophycin synthesis influences the resuscitation process, Synechocystis sp. wild-type and ΔcphA mutant cells were first nitrogen starved for 2 weeks, and then resuscitation was initiated by the addition of 5 mM nitrate, adjusting the cultures to an OD750 of 0.5. The cultures were incubated under continuous light with shaking. The time course of regreening was monitored by recording the UV-Vis spectra of the cultures.
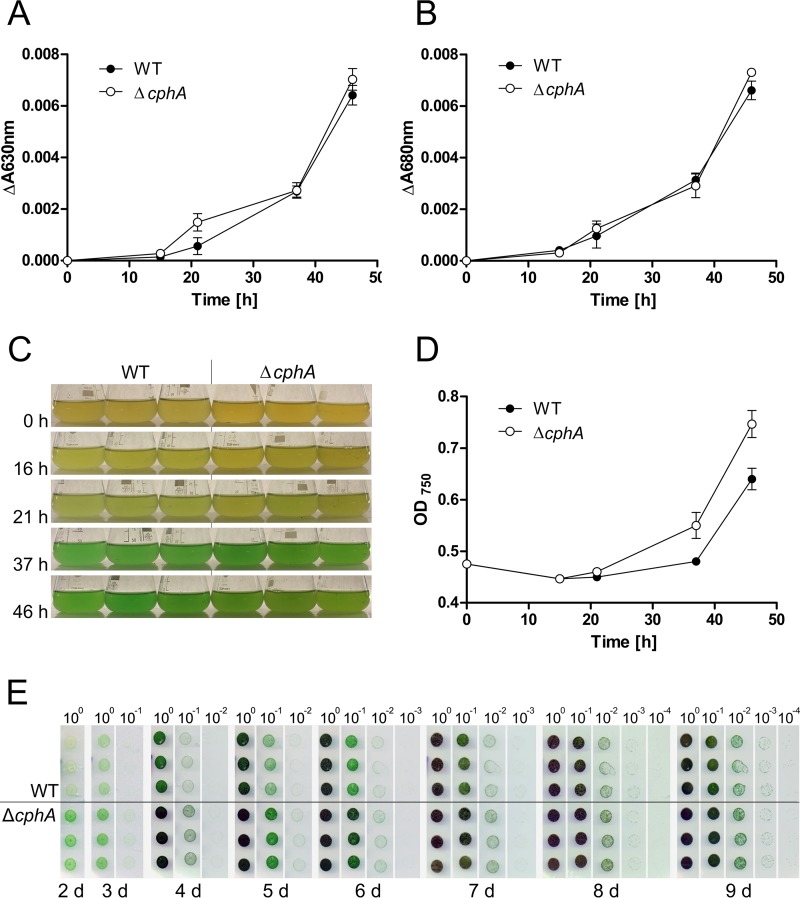
Twelve hours after the addition of nitrate to the chlorotic cultures, the absorbance at 630 nm (phycobiliproteins; Fig. 5A) and at 680 nm (chlorophyll a/b; Fig. 5B) began to increase. The wild-type and ΔcphA mutant cultures regreened similarly (Fig. 5A to C). However, the ΔcphA mutant had a growth advantage over the wild type in liquid medium (as revealed by the optical density of the culture; see Fig. 5D). Thirty-six hours after the addition of nitrogen, the difference in the optical density amounts to 0.07 OD750 units. After 46 h, the ΔcphA mutant reached an OD750 of 0.747 ± 0.045 compared to the wild type, with 0.64 ± 0.036, which is a significant difference (P = 0.0329). This growth disadvantage of the wild type turned out even more clearly on solid medium (as revealed by the drop plate method; see Fig. 5E). In the drop plate method, the area of the ΔcphA mutant was green already after 2 days, which indicated a return to the vegetative cell cycle and growth; the wild type lagged behind in the regreening process. This growth advantage of the ΔcphA mutant over the wild type continued until day 8 of recovery, at which time the two strains showed a similar degree of recovery.
FIG 5.
Resuscitation from 2-weeks-nitrogen-starved Synechocystis sp. wild-type and ΔcphA cultures in BG-11 liquid medium and on agar under continuous light. (A and B) Absorbance of phycobiliproteins at 630 nm (A) and absorbance of chlorophyll a/b at 680 nm (B) during regreening. Values shown are the means of three biological replicates. (C) Cultures of resuscitating wild type (WT) and the ΔcphA mutant; 0 h, induction of resuscitation. (D) Growth curve (OD750) of wild-type and the ΔcphA mutant cultures during resuscitation. Values shown are the means of three biological replicates. (E) Drop plate method of resuscitating wild-type and ΔcphA mutant cells on BG-11 agar plates containing 17.3 mM nitrate (standard concentration in BG-11 medium). Cultures were adjusted to an OD750 of 1.0, diluted in a 10-fold dilution series, and cultivated under continuous light. Photos were taken starting after 2 days of cultivation. Assays were performed using three biological replicates per strain. d, days.
Cyanophycin accumulation helps overcome a fluctuating or limiting nitrogen supply.
The growth disadvantage of the cyanophycin-forming wild-type cells during resuscitation from nitrogen chlorosis was puzzling. We questioned whether this disadvantage was due to unnatural laboratory conditions (nitrogen-rich medium and continuous light). Therefore, we nitrogen starved Synechocystis wild-type and ΔcphA mutant cells for 9 days and compared their resuscitation with a fluctuating and/or limiting nitrogen supplementation by a modification of the drop plate method (Fig. 6), which allows nitrogen supplementation to be changed without imposing additional stress to the cells that occurs with harvesting, washing, and resuspending cells in another medium.
FIG 6.
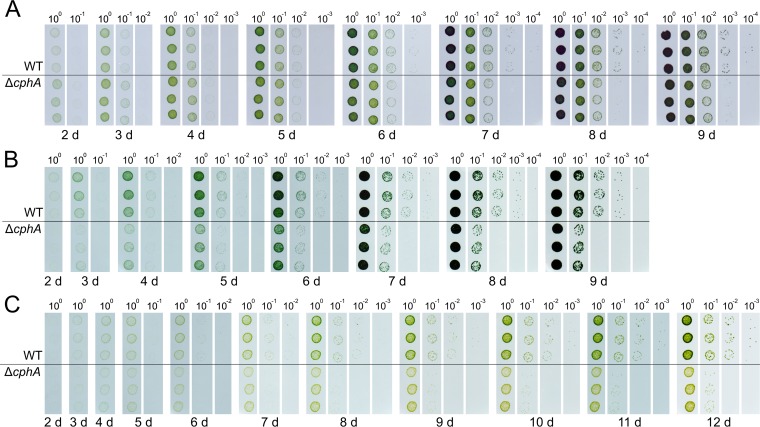
Synechocystis sp. wild-type and ΔcphA mutant cells starved of nitrogen for 9 days and resuscitated under continuous light on BG-11 agar plates supplying fluctuating or limiting nitrogen levels. Chlorotic cultures were adjusted to an OD750 of 1.0 and diluted in series. To enable fluctuating nitrogen supplementation, each cell suspension was dropped onto a transfer membrane, which was periodically moved to another plate containing a different nitrogen concentration. Photos were taken starting after 2 days of cultivation. Assays were performed using three biological replicates per strain. (A) Cells were exposed to 17.3 mM nitrate for 6 h per day and to no nitrogen for 18 h. (B) Cells were continuously exposed to a low concentration of nitrate (1.73 mM); to avoid nitrogen starvation, the transfer membrane was moved daily to a fresh plate containing 1.73 mM nitrate. (C) Both nitrogen limitation and fluctuating nitrogen availability were combined by exposing the cells to 1.73 mM nitrate for 4 h per day.
When chlorotic cells were exposed under continuous light to 6-h periods of nitrogen excess (17.3 mM nitrate) per day (6 h on BG-11 with nitrate and 18 h on nitrogen-free BG-11), the ΔcphA mutant grew slightly faster than the wild type during the first 3 days (Fig. 6A). After the fourth day, the wild type and ΔcphA mutant reached the same culture density. After 7 days, the wild type displayed a slight growth advantage. After 9 days, the wild type clearly had a growth advantage over the ΔcphA mutant.
To test resuscitation with continuous but low nitrogen supplementation, chlorotic cells were exposed under continuous light to only 1.73 mM nitrate (10% of the standard concentration) and transferred to fresh plates containing 1.73 mM nitrate every day. Colonies of green cells of both strains appeared after 3 days (Fig. 6B), but the wild type showed a clear growth advantage over the ΔcphA mutant throughout the experiment (Fig. 6B).
In the next experiment, we combined nitrogen limitation and fluctuating nitrogen availability. A limiting amount of nitrogen (1.73 mM nitrate) was available for only 4 h per day under continuous light. Under these conditions, the wild type showed the clearest growth advantage over the ΔcphA mutant (Fig. 6C). Furthermore, a difference in the colors of the cultures of the two strains became apparent. Cultures of the ΔcphA mutant were more yellow than those of the wild type, which indicated that the ΔcphA cells could not fully recover their photosynthetic pigments and remained in a semichlorotic state.
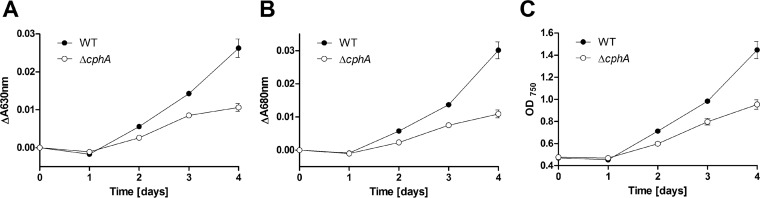
To determine whether the growth advantage under fluctuating nitrogen concentrations of wild-type cells over the ΔcphA mutant could also be observed in liquid culture, we provided nitrogen-starved liquid cultures of Synechocystis sp. wild-type and the ΔcphA mutant with 10 mM nitrate for 4 h, followed by 20 h with no nitrogen source; this cycle was repeated for 4 days (Fig. 7). After 2 days, both strains began to regreen, but the wild type regreened faster than the ΔcphA mutant (Fig. 7A and B). After 4 days, the growth advantage of the wild type was clearly visible, since the optical density of the wild-type culture increased 3-fold, whereas that of the ΔcphA mutant increased only 2-fold (Fig. 7C).
FIG 7.
Synechocystis sp. wild-type and ΔcphA mutant cells starved of nitrogen for 9 days and resuscitated under continuous light in liquid medium supplying fluctuating nitrogen concentrations. Values are the means of three biological replicates. Chlorotic cultures were adjusted to an OD750 of 0.5. Resuscitation was induced by adding 10 mM nitrate. After 4 h of nitrogen availability, the cells were washed and resuspended in nitrogen-free BG-11 medium. This routine was repeated over 4 days. The progress of regreening was documented by measuring the absorbance of phycobiliproteins at 630 nm (A) and chlorophyll a/b at 680 nm (B). (C) Growth curve of the two strains.
Cyanophycin accumulated in the night can trigger resuscitation during the day.
Photosynthetic organisms in nature have to deal with day-night changes and a fluctuating and limiting nutrient supplementation. We hypothesized that cyanophycin that accumulates in nondiazotrophic cyanobacteria in the night could be used as an intracellular nitrogen source during the day. To test this hypothesis, we combined nitrogen limitation and fluctuating nitrogen supplementation with day/night periods.
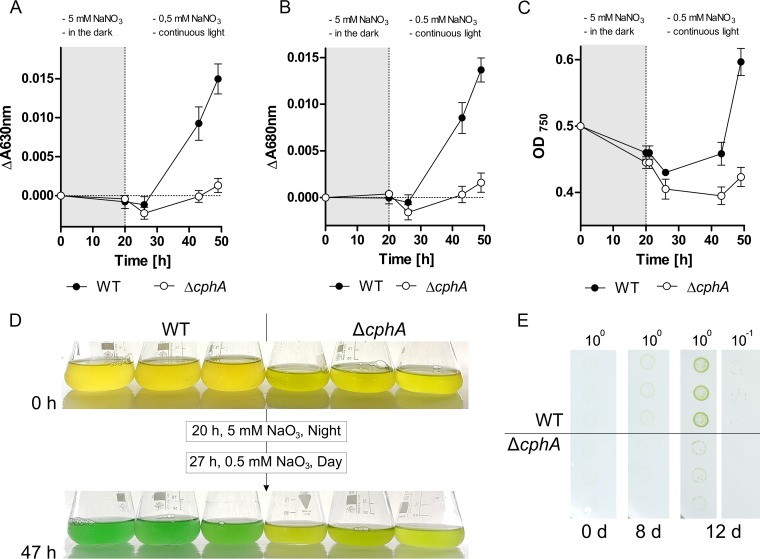
Nitrogen-starved liquid cultures of Synechocystis sp. wild-type and the ΔcphA mutant were exposed to 5 mM nitrate for 20 h without light. After this dark period, the cells were washed and resuspended into fresh medium containing a small amount of nitrate (0.5 mM) and exposed to light. After the 20-h dark period, the wild type accumulated cyanophycin to 3.7% ± 0.6% of the cell dry mass, but the pigmentation of the wild type and ΔcphA mutant was still at its initial low level (Fig. 8A and B). Shortly after light exposure in low-nitrate medium, the wild type started to synthesize pigments, but the ΔcphA mutant was not able to regreen (Fig. 8A, B, and D). Forty-seven hours after the first addition of nitrate, wild-type cultures appeared green and cultures began to grow (Fig. 8C and D), whereas the ΔcphA mutant was still chlorotic. We further analyzed the growth behavior with a modified version of the drop plate method, in which Synechocystis sp. wild-type and ΔcphA mutant cells starved of nitrogen for 1 week were grown on plates with fluctuating nitrogen supply and with day/night cycles (16-h day/8-h night). A small amount of nitrate (1.7 mM) was provided only during the night. Under these conditions, the overall growth of both strains was very low. Nevertheless, the wild type showed a clear growth advantage over the ΔcphA mutant; growth of the wild type and mutant was detected after 8 and 12 days of cultivation, respectively (Fig. 8E).
FIG 8.
Synechocystis sp. wild-type and ΔcphA mutant strains starved of nitrogen for 1 week and resuscitated in liquid medium and on agar plates with fluctuating nitrogen supply and day/night cycles. Starved cultures of wild-type and the ΔcphA mutant were adjusted to an OD750 of 0.5, and resuscitation was initiated by adding 5 mM nitrate in the absence of light. After 20 h of nitrogen availability in the dark, the cells were washed and resuspended in BG-11 medium containing residual amounts of nitrate (0.5 mM) and placed in the light. The progress of regreening was documented by measuring the absorbance of phycobiliproteins at 630 nm (A) and chlorophyll a/b at 680 nm (B). (C) Growth curve of the two strains. Gray areas in panels A to C indicate the night phase with nitrogen availability; white areas indicate the day phase with nitrogen limitation. Values are the means of three biological replicates. (D) Three liquid cultures of resuscitating Synechocystis sp. wild-type and the ΔcphA mutant. (E) Modified drop plate method of Synechocystis sp. strains starved of nitrogen for 1 week, with three biological replicates per strain. Starved cultures were adjusted to an OD750 of 1.0 and diluted 10-fold in series, and the dilutions were dropped onto a transfer membrane. The cells on the membrane were exposed to a small amount of nitrogen (1.73 mM) once per day for 8 h in the absence of light.
DISCUSSION
CphA localization switch indicates active and inactive forms.
Our results revealed different localizations of the inactive and active forms of CphA in cells of Synechocystis species. Under conditions in which no cyanophycin synthesis occurs, such as during exponential growth or nitrogen starvation, CphA is inactive and mainly located in the cytoplasm. Immediately after the induction of cyanophycin synthesis, CphA aggregated in foci that were randomly distributed in the cell. Formation of the CphA foci may be a consequence the priming of the CphA enzyme. The primers could induce self-aggregation.
CphA in its primed state is ready to elongate the cyanophycin primer. During this elongation process, the CphA foci increase in size and become visible as cyanophycin granules. As cyanophycin accumulates, the numbers of foci and granules per cell continuously decrease, possibly as initial granules fuse to form larger aggregates. This behavior would also explain the amorphous structure of the large granules that we observed when the cellular cyanophycin content was high and which we also observed previously in electron micrographs of a cyanobacterial cyanophycin-overproducer strain (17). The observed ring-like appearance of the fluorescence signal around large cyanophycin granules in our study suggests that during cyanophycin accumulation, CphA-eGFP covers the surface of the granule.
Previous studies have shown that CphA activity in vitro requires cyanophycin primers, arginine, aspartate, MgCl2, ATP, and a sulfhydryl reagent (21). Our results indicate that Mg2+ is strictly required for the association of CphA to the cyanophycin granules. The primary structure of CphA is composed of two regions (23), both of which show sequence similarities to ATP-dependent ligases of two superfamilies. Sequence alignments of the d-alanine–d-alanine ligase DdIB with cyanobacterial CphA show that key residues involved in binding of the ATP-Mg2+ complex are conserved in CphA (22). Accordingly, Mg2+ may stabilize the active conformation of CphA which is able to bind the C terminus of the cyanophycin substrate. In the absence of Mg2+, CphA cannot maintain its enzymatic activity, which could lead to dissociation of the substrate and consequently to release from the granule surface.
Cyanophycin accumulation is triggered in the presence of combined nitrogen sources by several growth-limiting conditions that lead to growth arrest, and cyanophycin degradation can be triggered by restoring the growth of the arrested cells. During cyanophycin degradation, the localization of CphA-eGFP changed from the surface of the cyanophycin granule to the cytoplasm. This suggests that as cyanophycin degrades, CphA dissociates from the granule surface and distributes in an inactive form throughout the cytoplasm, where it remains silent until cyanophycin synthesis is triggered again by growth-arresting conditions. A model of the cyanophycin accumulation and degradation cycle during nitrogen-induced chlorosis and resuscitation is depicted in Fig. 9.
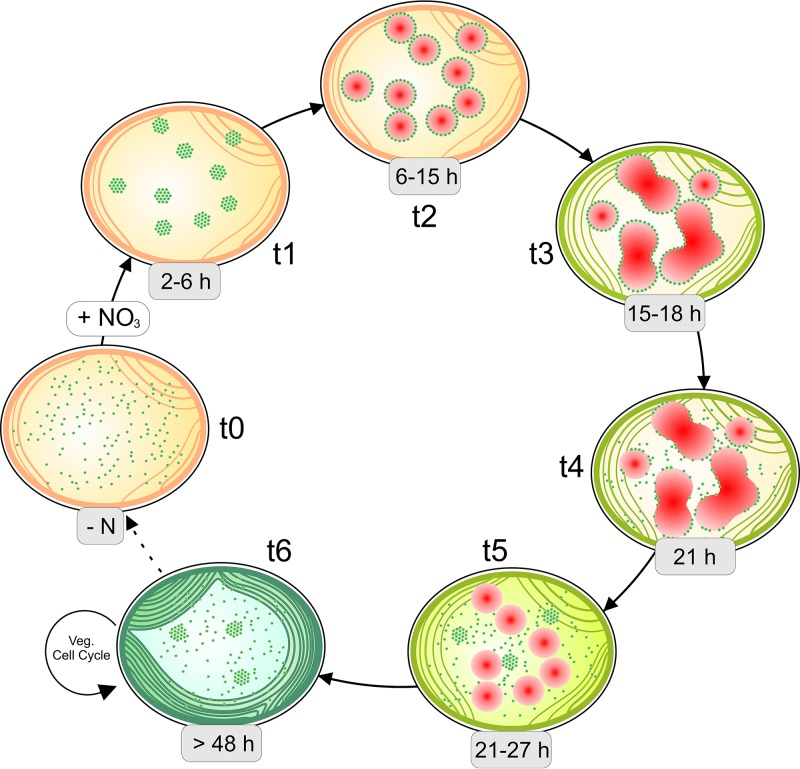
FIG 9.
Hypothetical cycle of cyanophycin accumulation and degradation during resuscitation from nitrogen starvation. t0, when cyanophycin does not accumulate, such as during nitrogen starvation, CphA (green dots) is distributed in the cytoplasm; t1, 2 to 6 h after the addition of nitrogen, CphA aggregates in foci; t2, 6 to 15 h after the addition of nitrogen, the first cyanophycin granules (red) appear, with CphA localized on the surface of the growing granules; t3, 15 to 18 h after nitrogen addition, adjacent granules merge when they collide, building amorphous granules with CphA on their surface; t4, 21 h after nitrogen addition, when cyanophycin begins to degrade, CphA begins to dissociate from the granule surface; t5, 21 to 27 h after nitrogen addition, the granules become smaller, and CphA is distributed in the cytoplasm; t6, after the cell completes resuscitation after more than 48 h, it starts to divide, with CphA mainly distributed in the cytoplasm. Veg, vegetative.
Our previous transcriptome study of nitrogen-depleted Synechocystis sp. cells has shown that the transcript levels of cphA in cells starved for nitrogen and of exponentially growing cells are very similar (1.2-fold upregulated during 14 days nitrogen starvation compared to nitrogen-supplemented cells). However, during resuscitation from nitrogen starvation, the cphA transcript level 24 h after the addition of nitrogen is approximately 1.7-fold higher than in exponentially growing cells (7). In agreement, quantitative proteomics revealed that the level of CphA after 21 days of nitrogen starvation is slightly reduced compared to nitrate-supplemented cells and goes back to initial values during recovery (11).
The synthesis of cyanophycin is probably tuned by the cellular level of arginine, which increases under growth-limiting conditions because of lowered protein biosynthesis. High concentrations of arginine surpass the Km of CphA for arginine and could therefore trigger the biosynthesis of cyanophycin (29). Whether CphA is subjected to additional activity control remains to be elucidated.
Cyanophycin-deficient mutant has a growth advantage under laboratory conditions.
The ability to synthesize cyanophycin is widespread among cyanobacteria and other eubacteria. With a C/N ratio of 2:1, cyanophycin is perfectly suited as a nitrogen storage compound. However, the physiological significance of cyanophycin for nondiazotrophic cyanobacteria remained largely unknown. Here, we showed that in the nondiazotrophic unicellular cyanobacterium Synechocystis sp., the deletion of cphA, which causes the inability to produce cyanophycin, has no impact on the nitrogen starvation response. The wild type and the ΔcphA mutant responded to nitrogen starvation similarly, namely, with chlorosis. However, the ΔcphA mutant reproducibly retained slightly higher residual pigment content in the chlorotic state. Merritt et al. (36) reported in nitrogen-limited cells of Synechocystis sp. the presence of a cyanophycin-like polymer that contains glutamic acid instead of arginine (36). Such a compound could be synthesized by a side-reaction of CphA and could affect the degradation of pigments in a so-far-unknown manner.
Several earlier studies have reported the transient accumulation of cyanophycin in cyanobacteria during resuscitation from nitrogen starvation (7, 13). Allen et al. (1980) reported an immediate synthesis of cyanophycin with a peak of 5 to 6% of the cell dry weight 8 to 12 h after the addition of NaNO3 to nitrogen-starved Synechocystis sp. PCC 6308 cultures (13). Here, we show that under standard laboratory conditions with continuous light and nitrogen excess, the ΔcphA mutant showed a clear growth advantage over the wild type both in liquid medium and on agar plates. The synthesis of cyanophycin costs 1.3 ± 0.1 mol ATP per mol incorporated amino acid in vitro (37). Without using the benefit of cyanophycin as a storage compound, the synthesis of cyanophycin is only a burden to the cells, and consequently, the cyanophycin-deficient ΔcphA mutant has an advantage over the wild type. This is the case under artificial laboratory cultivation conditions, which provide an excess of nitrogen and light.
Cyanophycin is beneficial under natural conditions.
In nature, microorganisms have to deal with a fluctuating and limiting nitrogen availability (3). We hypothesized that cyanophycin accumulation is a strategy to overcome temporal limitations in nitrogen availability. Growth and resuscitation experiments of Synechocystis sp. wild-type and the ΔcphA mutant confirmed this hypothesis. The ability to store nitrogen in the form of cyanophycin was beneficial during resuscitation under a fluctuating or limiting nitrogen supplementation, and the cells have a growth advantage over the cyanophycin-deficient mutant. When nitrogen fluctuation and limitation are combined, the cyanophycin-deficient mutant was not able to fully recover from chlorosis.
To further mimic natural conditions, we tested day/night cycles in addition to a fluctuating and/or limiting nitrogen supply. In the unicellular diazotrophic cyanobacterium Cyanothece sp. ATCC 51142 and the filamentous cyanobacterium Trichodesmium sp., cyanophycin acts as temporary nitrogen storage to enable the coexistence of nitrogen fixation and photosynthesis in the same cell (19, 20). These cyanobacteria fix nitrogen in the night and store the fixed nitrogen in the form of cyanophycin. During the day, when cells carry out photosynthesis, cyanophycin is degraded to mobilize the fixed nitrogen. Based on this behavior in diazotrophic cyanobacteria, we hypothesized that cyanophycin is involved in nitrogen storage also in nondiazotrophic cyanobacteria at night. Indeed, when we combined fluctuating nitrogen supplementation and day/night cycles, the resuscitation of Synechocystis sp. wild-type cells proceeded as fast as in continuous light, with growth restoration after 48 h. In contrast, the cyanophycin-deficient mutant was not able to resuscitate in this time period.
The first phase of resuscitation from nitrogen chlorosis occurs in a light-independent mode, where the cells catabolize glycogen using the parallel operating Entner-Doudoroff and oxidative pentose phosphate pathways (10). Only when the photosynthetic apparatus is reinstalled after approximately 12 to 16 h does metabolism switch back from the heterotrophic mode to a mixotrophic mode and finally to the fully autotrophic mode. Our data suggest that most of the nitrogen that is required for regreening can be assimilated in the initial heterotrophic phase and stored as cyanophycin. The stored cyanophycin makes internal nitrogen available to the cells during the second phase of resuscitation, which can then proceed, even if the ambient nitrogen supply is again low.
Nitrate assimilation by cyanobacteria is tightly regulated at both the transcriptional and posttranslational levels (38, 39). Nitrate uptake is mediated by the bispecific nitrate/nitrite transporter encoded by the nrtABCD genes. This operon is only expressed in the absence of ammonium, and the nitrate/nitrite transporter is rapidly and reversibly inhibited by the addition of ammonia (38–40). The PII signal transduction protein appears to be involved in this process, since a PII-deficient mutant shows no ammonium-responsive inhibition of nitrate and nitrite uptake (41). In vegetative growing cells, in addition to the requirement for ammonium absence, the operation of the nitrate/nitrite transporter also requires CO2 fixation, because the inhibition of photosynthetic CO2 fixation results in the simultaneous inhibition of nitrate uptake (42). Newly assimilated nitrogen is incorporated in the central metabolism via the glutamine synthetase (GS) and glutamate synthase (GOGAT) cycle. GS activity is tightly regulated by light/dark transitions; induction of the inhibitory factor 7 (IF7) and IF17 in the dark causes GS to switch off (43–45). This leads to the inhibition of nitrogen assimilation in the dark, both at the level of nitrate transport and by inhibition of the ammonium-assimilating GS. Our results showed that cyanophycin accumulates upon nitrate addition to chlorotic cells in the dark, which requires active nitrate assimilation. This contradicts the dark switch-off nitrate assimilation in vegetative cells. It is possible that the PII signaling protein is directly involved in this behavior. Nonphosphorylated PII mediates a light-dependent inhibition of nitrate utilization (41, 46). However, during nitrogen starvation, PII is highly phosphorylated and remains in the highly phosphorylated state during the first period of resuscitation (11, 47). In this state, PII is probably unable to inhibit nitrate uptake. In agreement, Reyes et al. (48) have shown that the inactivation of GS in the dark is attenuated under nitrogen depletion, which allows the cells to utilize nitrogen in the dark after a nitrogen starvation period (48). The GS inhibitory factors IF7 and IF17 are regulated by the global nitrogen control transcriptional factor NtcA, which is in turn controlled by 2-oxoglutarate levels and the PII-PipX network (49–51). Apparently, during nitrogen starvation, where NtcA is activated by elevated levels of 2-oxoglutarate and by binding PipX, the nocturnal derepression of the gifA and gifB genes appears to be abrogated (52, 53).
The occurrence of cyanophycin in cyanobacteria has been known for more than 100 years. However, fundamental questions on its biological function or the benefit of cyanophycin accumulation remained largely uninvestigated. Altogether, our study sheds new light on the process of cyanophycin accumulation and reveals new insights into the biological function of cyanophycin in nondiazotrophic cyanobacteria. Interestingly, artificial laboratory conditions do not provide any fitness advantage for cyanophycin-producing cells, and in a fluctuating environment, CphA becomes beneficial. As the cphA gene is constitutively expressed in Synechocystis sp., the cells are programmed to overcome fluctuating nitrogen supply and transient periods of starvation in a constantly changing environment.
MATERIALS AND METHODS
Cultivation conditions.
Standard procedures for cloning in Escherichia coli NEB 10-beta (NEB) and E. coli XL1-Blue (Stratagene) were followed. Strains were grown in LB medium at 37°C with constant shaking at 300 rpm.
Synechocystis sp. strains were cultivated photoautotrophically in BG-11 medium supplemented with 5 mM NaHCO3 (54) at 27°C with constant shaking at 120 rpm and illumination at 40 to 50 microeinsteins. Growth was monitored by measuring the optical density at 750 nm. BG-11 agar plates contained 1.5% (wt/vol) Bacto agar (Difco), 0.3% (wt/vol) sodium thiosulfate pentahydrate, and 10 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)-NaOH (pH 8) (Roth). Antibiotics were added to the medium when required.
For the induction of starvation conditions, cells of exponentially growing cultures (OD750, 0.4 to 0.6) were harvested, washed, and resuspended in BG-11 medium lacking a specific nutrient. For nitrogen starvation, the cells were resuspended in BG-11 medium without a nitrogen source. For resuscitation from nitrogen starvation, chlorotic cultures were adjusted to an OD750 of 0.5, harvested, and resuspended in BG-11 containing a nitrogen source. To introduce potassium starvation, cells were harvested, washed, and resuspended in BG-11 medium containing Na2HPO4 in an amount equimolar to that of the K2HPO4 it replaced. Regeneration from potassium starvation was induced by harvesting, washing, and resuspending cells in BG-11 medium containing a potassium source.
For the drop plate method, Synechocystis sp. cultures were adjusted to an OD750 of 1. The cultures were diluted 10-fold in series in BG-11 medium lacking nitrogen. Drops (5 μl) of five dilutions (100 to 10−4) were placed on BG-11 agar plates containing specific nitrate concentrations. When cells were to be shifted to different solid media, the drops were placed on mixed-cellulose ester transfer membranes (pore size, 0.45 μm; HATF; Merck Millipore). The transfer membranes were placed on different BG-11 agar plates at the intervals specified in the figure legends. To minimize carryover of the residual nitrate from the initial plate, the transfer membranes were placed on nitrogen-free BG-11 plates for 10 min. The plates were incubated at 27°C with illumination at 40 to 50 microeinsteins.
Construction of a ΔcphA mutant and a cphA-egfp fusion.
PCR fragments were generated using high-fidelity Q5 polymerase (NEB) and oligonucleotides with overlapping regions. Genomic Synechocystis sp. DNA or plasmids served as the templates. The primers, plasmids, and strains used in this study are listed in Tables 1, 2, and 3, respectively.
TABLE 1.
Oligonucleotides used in this study
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pJET1.2 | Cloning vector | Thermo Scientific |
| pVZ322 | Broad-host-range vector | 36 |
| pJET ΔcphA | cphA (ORF slr2002) knockout plasmid | This study |
| pVZ322 cphA-egfp | eGFP fused to the C terminus of cphA | This study |
TABLE 3.
Strains used in this study
| Strain | Description | Reference or source |
|---|---|---|
| E. coli RP4 | Conjugation strain | 36 |
| E. coli NEB 10-beta | Cloning strain | NEB |
| E. coli XL1-Blue | Cloning strain | Stratagene |
| Synechocystis sp. strain PCC 6803 | Wild type | Pasteur Culture Collection |
| CphA-eGFP | Wild type transformed with pVZ322 cphA-egfp | This study |
| ΔcphA mutant | Chromosomal deletion of cphA (ORF slr2002) | This study |
To generate a ΔcphA mutant, 500-bp upstream and downstream genomic regions of slr2002 (cphA) were amplified using the oligonucleotides up_for/up_rev and down_for/down_rev. The kanamycin resistance gene (816 bp) was amplified from pVZ322 (34) using primers kan_for and kan_rev. Upstream and downstream fragments and the kanamycin resistance cassette were fused and incorporated into the linear pJET1.2/blunt cloning vector (Thermo Scientific) by isothermal single-reaction DNA assembly, according to Gibson et al. (55). The resulting construct (pJET ΔcphA) was introduced into competent E. coli NEB 10-beta by transformation. The correct length (1,816 bp) of the insert was confirmed via colony PCR using the primers provided in the CloneJET PCR cloning kit (pJet_seq_for and pJet_seq_rev) (Thermo Scientific). Synechocystis sp. was transformed with pJET ΔcphA via natural competence (56). Transformants were selected on BG-11 agar plates supplemented with 50 μg/ml kanamycin. Complete segregation was confirmed via PCR using primers seg_for, kan_for, and down_rev (Fig. S6).
For the cphA-egfp fusion, cphA including its promoter region was amplified using primers cphA_for and cphA_rev. Subsequently, the enhanced green fluorescent protein gene egfp derived from plasmid pCESL19 (57) was amplified using oligonucleotides gfp_for and gfp_rev. The cphA and egfp amplicons were fused and incorporated into XbaI-digested pVZ322 via DNA assembly according to Gibson et al. (55). The resulting plasmid, pVZ322 cphA-egfp, was introduced into competent E. coli XL1-Blue (Stratagene) cells by transformation. The sequence integrity of the plasmid was verified by sequencing using primers pVZ322_seq_for and pVZ322_seq_rev. Synechocystis sp. was transformed with pVZ322 cphA-egfp by triparental mating (34), and transformants were selected on BG-11 agar plates supplemented with 50 μg/ml kanamycin and 5 μg/ml gentamicin.
Microscopy and staining.
Cells were observed by fluorescence microscopy using a Leica DM5500B microscope with a ×100/1.3 oil objective. The GFP signal was detected with a BP470 40-nm excitation filter and a BP525 50-nm emission filter. Cyanobacterial autofluorescence was detected with a filter cube with excitation filter BP535/50 and suppression filter BP610/75. Images were acquired with a Leica DFC360FX black-and-white camera. Captured black-and-white pictures were colored using the Leica Application Suite Software (LAS AF) provided by Leica Microsystems. Bright-field images were exposed for 5 ms, and fluorescence images were exposed for 100 ms. Images, including those for counting fluorescent foci and measuring the diameter, were evaluated with the Leica Application Suite Software.
Cyanophycin granules were visualized using a staining method based on the arginine-selective Sakaguchi reaction, according to Watzer et al. (17). Photographs were taken with a Leica DM2500 microscope using a ×100/1.3 oil objective. Images were acquired with a Leica DFC420C color camera and Leica Application Suite Software.
During all microscopy studies, microscope slides covered with a dried 2% (wt/vol) agarose solution were used to immobilize the cells.
Protein extract preparation.
Potassium-starved Synechocystis sp. cells were harvested by centrifugation and resuspended in a buffer containing 50 mM Tris-HCl (pH 7.4), 4 mM EDTA, 1 mM DTT, and 0.5 mM benzamidine, or optionally in a buffer containing 50 mM Tris-HCl (pH 8.2), 20 mM MgCl2, 20 mM KCl, and 1 mM DTT (21). Cells were lysed using FastPrep-24 (MP Biomedicals) with 0.1-mm glass beads at a speed of 6.0 m/s for 20 s with five repeats and 5 min of resting after every repeat. Soluble and insoluble fractions were separated by centrifugation at 25,000 × g for 25 min at 4°C. The protein concentration was determined using the Bradford assay (58).
SDS-PAGE and immunoblotting.
Proteins were separated by SDS-PAGE on a 12% polyacrylamide gel according to Sambrook and Russell (59). Total protein (10 μg) was loaded on each lane. For immunoblot detection of CphA-eGFP, proteins were blotted onto a methanol-activated polyvinylidene difluoride (PVDF) membranes, as described previously (60). Membranes were blocked with 10% (wt/vol) milk powder in TBS buffer (50 mM Tris-HCl [pH 7.4], 75 mM NaCl) overnight. Afterwards, the membranes were transferred in 1% (wt/vol) milk powder in TBS buffer containing 1:2,500 diluted rabbit anti-GFP antibody (chromatin immunoprecipitation [ChIP] grade ab290; Abcam) and incubated for 2 h at ambient temperature. Unbound primary antibodies were removed by washing the membranes three times with TBS buffer. Anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase (anti-rabbit polyclonal goat antibody; Sigma-Aldrich) diluted 1:1,000 in 1% (wt/vol) milk powder in TBS buffer was applied to the membranes and incubated for 30 min at ambient temperature. Afterwards, the membranes were washed three times with TBS buffer to remove unbound secondary antibodies. Bands were visualized using the Lumi-Light detection system (Roche Diagnostics). Luminograms were taken with the Gel Logic 1500 imaging system (Kodak) with the associated software.
Cyanophycin extraction and quantification.
Cyanophycin was extracted according to Watzer et al. (17) and quantified by determining the amount of arginine in the extracted sample using the modified Sakaguchi reaction, according to Messineo (61). The determined amount of cyanophycin was normalized to the cell dry mass. The cell dry mass was determined by centrifuging 10 ml of culture and washing and drying the pellet for 4 h at 60°C in a rotational vacuum concentrator. The dried pellets were weighed on an analytical balance.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the DFG (Fo195/9-2) and the research training group GRK 1708.
We thank Alicia Engelbrecht and Waldemar Hauf for help in constructing the CphA-eGFP strain, Daniel Hörömpöli for technical assistance in immunoblot analysis, and Karen A. Brune, Niels Neumann, Sofia Doello, and Moritz Koch for carefully reading the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01298-18.
REFERENCES
- 1.Blankenship RE. 2017. How cyanobacteria went green. Science 355:1372–1373. doi: 10.1126/science.aam9365. [DOI] [PubMed] [Google Scholar]
- 2.Herrero A, Flores E. 2008. The cyanobacteria: molecular biology, genomics, and evolution. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 3.Vitousek PM, Howarth RW. 1991. Nitrogen limitation on land and in the sea: how can it occur? Biogeochemistry 13:87–115. doi: 10.1007/BF00002772. [DOI] [Google Scholar]
- 4.Schwarz R, Forchhammer K. 2005. Acclimation of unicellular cyanobacteria to macronutrient deficiency: emergence of a complex network of cellular responses. Microbiology 151:2503–2514. doi: 10.1099/mic.0.27883-0. [DOI] [PubMed] [Google Scholar]
- 5.Allen MM, Smith AJ. 1969. Nitrogen chlorosis in blue-green algae. Arch Mikrobiol 69:114–120. doi: 10.1007/BF00409755. [DOI] [PubMed] [Google Scholar]
- 6.Allen MM. 1984. Cyanobacterial cell inclusions. Annu Rev Microbiol 38:1–25. doi: 10.1146/annurev.mi.38.100184.000245. [DOI] [PubMed] [Google Scholar]
- 7.Klotz A, Georg J, Budinska L, Watanabe S, Reimann V, Januszewski W, Sobotka R, Jendrossek D, Hess WR, Forchhammer K. 2016. Awakening of a dormant cyanobacterium from nitrogen chlorosis reveals a genetically determined program. Curr Biol 26:2862–2872. doi: 10.1016/j.cub.2016.08.054. [DOI] [PubMed] [Google Scholar]
- 8.Görl M, Sauer J, Baier T, Forchhammer K. 1998. Nitrogen-starvation-induced chlorosis in Synechococcus PCC 7942: adaptation to long-term survival. Microbiology 144:2449–2458. doi: 10.1099/00221287-144-9-2449. [DOI] [PubMed] [Google Scholar]
- 9.Sauer J, Schreiber U, Schmid R, Volker U, Forchhammer K. 2001. Nitrogen starvation-induced chlorosis in Synechococcus PCC 7942. Low-level photosynthesis as a mechanism of long-term survival. Plant Physiol 126:233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doello S, Klotz A, Makowka A, Gutekunst K, Forchhammer K. 2018. A specific glycogen mobilization strategy enables awakening of dormant cyanobacteria from chlorosis. Plant Physiol 177:594–603. doi: 10.1104/pp.18.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spät P, Klotz A, Rexroth S, Maček B, Forchhammer K. 2018. Chlorosis as a developmental program in cyanobacteria: the proteomic fundament for survival and awakening. Mol Cell Proteomics doi: 10.1074/mcp.RA118.000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klotz A, Forchhammer K. 2017. Glycogen, a major player for bacterial survival and awakening from dormancy. Future Microbiol 12:101–104. doi: 10.2217/fmb-2016-0218. [DOI] [PubMed] [Google Scholar]
- 13.Allen MM, Hutchison F. 1980. Nitrogen limitation and recovery in the cyanobacterium Aphanocapsa 6308. Arch Microbiol 128:1–7. doi: 10.1007/BF00422297. [DOI] [Google Scholar]
- 14.Füser G, Steinbüchel A. 2007. Analysis of genome sequences for genes of cyanophycin metabolism: identifying putative cyanophycin metabolizing prokaryotes. Macromol Biosci 7:278–296. doi: 10.1002/mabi.200600207. [DOI] [PubMed] [Google Scholar]
- 15.Simon RD, Weathers P. 1976. Determination of the structure of the novel polypeptide containing aspartic acid and arginine which is found in cyanobacteria. Biochim Biophys Acta 420:165–176. doi: 10.1016/0005-2795(76)90355-X. [DOI] [PubMed] [Google Scholar]
- 16.Allen MM, Hutchison F, Weathers PJ. 1980. Cyanophycin granule polypeptide formation and degradation in the cyanobacterium Aphanocapsa 6308. J Bacteriol 141:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watzer B, Engelbrecht A, Hauf W, Stahl M, Maldener I, Forchhammer K. 2015. Metabolic pathway engineering using the central signal processor PII. Microb Cell Fact 14:192. doi: 10.1186/s12934-015-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burnat M, Herrero A, Flores E. 2014. Compartmentalized cyanophycin metabolism in the diazotrophic filaments of a heterocyst-forming cyanobacterium. Proc Natl Acad Sci U S A 111:3823–3828. doi: 10.1073/pnas.1318564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman LA, Meunier P, Colon-Lopez MS. 1998. Diurnal rhythms in metabolism: a day in the life of a unicellular, diazotrophic cyanobacterium. Photosynth Res 58:25–42. doi: 10.1023/A:1006137605802. [DOI] [Google Scholar]
- 20.Finzi-Hart JA, Pett-Ridge J, Weber PK, Popa R, Fallon SJ, Gunderson T, Hutcheon ID, Nealson KH, Capone DG. 2009. Fixation and fate of C and N in the cyanobacterium Trichodesmium using nanometer-scale secondary ion mass spectrometry. Proc Natl Acad Sci U S A 106:6345–6350. doi: 10.1073/pnas.0810647106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon RD. 1976. The biosynthesis of multi-l-arginyl-poly(l-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim Biophys Acta 422:407–418. doi: 10.1016/0005-2744(76)90151-0. [DOI] [PubMed] [Google Scholar]
- 22.Berg H, Ziegler K, Piotukh K, Baier K, Lockau W, Volkmer-Engert R. 2000. Biosynthesis of the cyanobacterial reserve polymer multi-l-arginyl-poly-l-aspartic acid (cyanophycin): mechanism of the cyanophycin synthetase reaction studied with synthetic primers. Eur J Biochem 267:5561–5570. doi: 10.1046/j.1432-1327.2000.01622.x. [DOI] [PubMed] [Google Scholar]
- 23.Ziegler K, Diener A, Herpin C, Richter R, Deutzmann R, Lockau W. 1998. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartate (cyanophycin). Eur J Biochem 254:154–159. doi: 10.1046/j.1432-1327.1998.2540154.x. [DOI] [PubMed] [Google Scholar]
- 24.Richter R, Hejazi M, Kraft R, Ziegler K, Lockau W. 1999. Cyanophycinase, a peptidase degrading the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartic acid (cyanophycin): molecular cloning of the gene of Synechocystis sp. PCC 6803, expression in Escherichia coli, and biochemical characterization of the purified enzyme. Eur J Biochem 263:163–169. [DOI] [PubMed] [Google Scholar]
- 25.Hejazi M, Piotukh K, Mattow J, Deutzmann R, Volkmer-Engert R, Lockau W. 2002. Isoaspartyl dipeptidase activity of plant-type asparaginases. Biochem J 364:129–136. doi: 10.1042/bj3640129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elbahloul Y, Krehenbrink M, Reichelt R, Steinbüchel A. 2005. Physiological conditions conducive to high cyanophycin content in biomass of Acinetobacter calcoaceticus strain ADP1. Appl Environ Microbiol 71:858–866. doi: 10.1128/AEM.71.2.858-866.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leganés F, Fernandez-Pinas F, Wolk CP. 1998. A transposition-induced mutant of Nostoc ellipsosporum implicates an arginine-biosynthetic gene in the formation of cyanophycin granules and of functional heterocysts and akinetes. Microbiology 144:1799–1805. doi: 10.1099/00221287-144-7-1799. [DOI] [PubMed] [Google Scholar]
- 28.Elbahloul Y, Steinbüchel A. 2006. Engineering the genotype of Acinetobacter sp. strain ADP1 to enhance biosynthesis of cyanophycin. Appl Environ Microbiol 72:1410–1419. doi: 10.1128/AEM.72.2.1410-1419.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maheswaran M, Ziegler K, Lockau W, Hagemann M, Forchhammer K. 2006. PII-regulated arginine synthesis controls accumulation of cyanophycin in Synechocystis sp. strain PCC 6803. J Bacteriol 188:2730–2734. doi: 10.1128/JB.188.7.2730-2734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinrich A, Maheswaran M, Ruppert U, Forchhammer K. 2004. The Synechococcus elongatus PII signal transduction protein controls arginine synthesis by complex formation with N-acetyl-l-glutamate kinase. Mol Microbiol 52:1303–1314. doi: 10.1111/j.1365-2958.2004.04058.x. [DOI] [PubMed] [Google Scholar]
- 31.Fokina O, Chellamuthu VR, Forchhammer K, Zeth K. 2010. Mechanism of 2-oxoglutarate signaling by the Synechococcus elongatus PII signal transduction protein. Proc Natl Acad Sci U S A 107:19760–19765. doi: 10.1073/pnas.1007653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forchhammer K. 2008. PII signal transducers: novel functional and structural insights. Trends Microbiol 16:65–72. doi: 10.1016/j.tim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Maheswaran M, Urbanke C, Forchhammer K. 2004. Complex formation and catalytic activation by the PII signaling protein of N-acetyl-l-glutamate kinase from Synechococcus elongatus strain PCC 7942. J Biol Chem 279:55202–55210. doi: 10.1074/jbc.M410971200. [DOI] [PubMed] [Google Scholar]
- 34.Wolk CP, Vonshak A, Kehoe P, Elhai J. 1984. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc Natl Acad Sci U S A 81:1561–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon RD. 1973. Measurement of the cyanophycin granule polypeptide contained in the blue-green alga Anabaena cylindrica. J Bacteriol 114:1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merritt MV, Sid SS, Mesh L, Allen MM. 1994. Variations in the amino acid composition of cyanophycin in the cyanobacterium Synechocystis sp. PCC 6308 as a function of growth conditions. Arch Microbiol 162:158–166. [DOI] [PubMed] [Google Scholar]
- 37.Aboulmagd E, Oppermann-Sanio FB, Steinbüchel A. 2000. Molecular characterization of the cyanophycin synthetase from Synechocystis sp. strain PCC6308. Arch Microbiol 174:297–306. doi: 10.1007/s002030000206. [DOI] [PubMed] [Google Scholar]
- 38.Flores E, Herrero A. 1994. Assimilatory nitrogen metabolism and its regulation, p 487–517. In Bryant DA. (ed), The molecular biology of cyanobacteria. Springer Netherlands, Dordrecht, The Netherlands. [Google Scholar]
- 39.Flores E, Herrero A. 2005. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem Soc Trans 33:164–167. doi: 10.1042/BST0330164. [DOI] [PubMed] [Google Scholar]
- 40.Kikuchi H, Aichi M, Suzuki I, Omata T. 1996. Positive regulation by nitrite of the nitrate assimilation operon in the cyanobacteria Synechococcus sp. strain PCC 7942 and Plectonema boryanum. J Bacteriol 178:5822–5825. doi: 10.1128/jb.178.19.5822-5825.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee HM, Flores E, Herrero A, Houmard J, Tandeau de Marsac N. 1998. A role for the signal transduction protein PII in the control of nitrate/nitrite uptake in a cyanobacterium. FEBS Lett 427:291–295. doi: 10.1016/S0014-5793(98)00451-7. [DOI] [PubMed] [Google Scholar]
- 42.Romero JM, Lara C, Guerrero MG. 1985. Dependence of nitrate utilization upon active CO2 fixation in Anacystis nidulans: a regulatory aspect of the interaction between photosynthetic carbon and nitrogen-metabolism. Arch Biochem Biophys 237:396–401. doi: 10.1016/0003-9861(85)90291-7. [DOI] [PubMed] [Google Scholar]
- 43.Marqués S, Merida A, Candau P, Florencio FJ. 1992. Light-mediated regulation of glutamine-synthetase activity in the unicellular cyanobacterium Synechococcus sp. PCC 6301. Planta 187:247–253. doi: 10.1007/BF00201947. [DOI] [PubMed] [Google Scholar]
- 44.García-Domínguez M, Reyes JC, Florencio FJ. 1999. Glutamine synthetase inactivation by protein-protein interaction. Proc Natl Acad Sci U S A 96:7161–7166. doi: 10.1073/pnas.96.13.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galmozzi CV, Fernandez-Avila MJ, Reyes JC, Florencio FJ, Muro-Pastor MI. 2007. The ammonium-inactivated cyanobacterial glutamine synthetase I is reactivated in vivo by a mechanism involving proteolytic removal of its inactivating factors. Mol Microbiol 65:166–179. doi: 10.1111/j.1365-2958.2007.05773.x. [DOI] [PubMed] [Google Scholar]
- 46.Kloft N, Forchhammer K. 2005. Signal transduction protein PII phosphatase PphA is required for light-dependent control of nitrate utilization in Synechocystis sp. strain PCC 6803. J Bacteriol 187:6683–6690. doi: 10.1128/JB.187.19.6683-6690.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forchhammer K, Tandeau de Marsac N. 1995. Phosphorylation of the PII protein (glnB gene product) in the cyanobacterium Synechococcus sp. strain PCC 7942: analysis of in vitro kinase activity. J Bacteriol 177:5812–5817. doi: 10.1128/jb.177.20.5812-5817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reyes JC, Crespo JL, Garciadominguez M, Florencio FJ. 1995. Electron-transport controls glutamine-synthetase activity in the facultative heterotrophic cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 109:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Espinosa J, Rodriguez-Mateos F, Salinas P, Lanza VF, Dixon R, de la Cruz F, Contreras A. 2014. PipX, the coactivator of NtcA, is a global regulator in cyanobacteria. Proc Natl Acad Sci U S A 111:E2423–E2430. doi: 10.1073/pnas.1404097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vázquez-Bermúdez MF, Herrero A, Flores E. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett 512:71–74. doi: 10.1016/S0014-5793(02)02219-6. [DOI] [PubMed] [Google Scholar]
- 51.Paz-Yepes J, Flores E, Herrero A. 2003. Transcriptional effects of the signal transduction protein PII (glnB gene product) on NtcA-dependent genes in Synechococcus sp. PCC 7942. FEBS Lett 543:42–46. [DOI] [PubMed] [Google Scholar]
- 52.Tanigawa R, Shirokane M, Maeda S, Omata T, Tanaka K, Takahashi H, Takahashi H. 2002. Transcriptional activation of NtcA-dependent promoters of Synechococcus sp. PCC 7942 by 2-oxoglutarate in vitro. Proc Natl Acad Sci U S A 99:4251–4255. doi: 10.1073/pnas.072587199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-Domínguez M, Reyes JC, Florencio FJ. 2000. NtcA represses transcription of gifA and gifB, genes that encode inhibitors of glutamine synthetase type I from Synechocystis sp. PCC 6803. Mol Microbiol 35:1192–1201. [DOI] [PubMed] [Google Scholar]
- 54.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1–61. [Google Scholar]
- 55.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 56.Grigorieva G, Shestakov S. 1982. Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol Lett 13:367–370. doi: 10.1111/j.1574-6968.1982.tb08289.x. [DOI] [Google Scholar]
- 57.Muro-Pastor AM, Olmedo-Verd E, Flores E. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC 7120. FEMS Microbiol Lett 256:171–177. doi: 10.1111/j.1574-6968.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 58.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 59.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed, vol 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 60.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76:4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Messineo L. 1966. Modification of Sakaguchi reaction: spectrophotometric determination of arginine in proteins without previous hydrolysis. Arch Biochem Biophys 117:534–540. doi: 10.1016/0003-9861(66)90094-4. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.