S. oneidensis is among the first- and the best-studied metal-reducing bacteria, with great potential in bioremediation and biotechnology. However, many questions regarding mechanisms closely associated with those applications, such as iron homeostasis, including iron uptake, export, and regulation, remain to be addressed. Here we show that Feo is a primary player in iron uptake in addition to the siderophore-dependent route. The investigation also resolved a few puzzles regarding the unexpected phenotypes of the putA mutant and lactate-dependent iron uptake. By elucidating the physiological roles of these two important iron uptake systems, this work revealed the breadth of the impacts of iron uptake systems on the biological processes.
KEYWORDS: iron uptake, Shewanella, siderophore, lactate, Feo system
ABSTRACT
Shewanella oneidensis is an extensively studied bacterium capable of respiring minerals, including a variety of iron ores, as terminal electron acceptors (EAs). Although iron plays an essential and special role in iron respiration of S. oneidensis, little has been done to date to investigate the characteristics of iron transport in this bacterium. In this study, we found that all proteins encoded by the pub-putA-putB cluster for putrebactin (S. oneidensis native siderophore) synthesis (PubABC), recognition-transport of Fe3+-putrebactin across the outer membrane (PutA), and reduction of ferric putrebactin (PutB) are essential to putrebactin-mediated iron uptake. Although homologs of PutA are many, none can function as its replacement, but some are able to work with other bacterial siderophores. We then showed that Fe2+-specific Feo is the other primary iron uptake system, based on the synthetical lethal phenotype resulting from the loss of both iron uptake routes. The role of the Feo system in iron uptake appears to be more critical, as growth is significantly impaired by the absence of the system but not of putrebactin. Furthermore, we demonstrate that hydroxyl acids, especially α-types such as lactate, promote iron uptake in a Feo-dependent manner. Overall, our findings underscore the importance of the ferrous iron uptake system in metal-reducing bacteria, providing an insight into iron homeostasis by linking these two biological processes.
IMPORTANCE S. oneidensis is among the first- and the best-studied metal-reducing bacteria, with great potential in bioremediation and biotechnology. However, many questions regarding mechanisms closely associated with those applications, such as iron homeostasis, including iron uptake, export, and regulation, remain to be addressed. Here we show that Feo is a primary player in iron uptake in addition to the siderophore-dependent route. The investigation also resolved a few puzzles regarding the unexpected phenotypes of the putA mutant and lactate-dependent iron uptake. By elucidating the physiological roles of these two important iron uptake systems, this work revealed the breadth of the impacts of iron uptake systems on the biological processes.
INTRODUCTION
Iron (Fe) is an essential element for virtually all living organisms because it participates in an array of biological processes as an extremely versatile prosthetic component for proteins (1). However, iron acquisition is a challenge for life in general, because in oxic environments iron exists in the extremely insoluble ferric (Fe3+) form (2). To survive and compete, microorganisms have evolved multiple strategies to obtain iron, in various forms, from the surroundings. The most diverse and broadly distributed iron uptake mechanisms used by bacteria are those involved in siderophore-dependent iron acquisition. Siderophores are small-molecule organic chelators with a very high affinity for Fe3+ which are biosynthesized under low-iron conditions (3). Once produced, these molecules are exported into the environment, where they efficiently interact with iron to form an iron-siderophore complex, which is shuttled back into the cell via specific transport pathways (4). In Gram-negative bacteria, iron-siderophore complexes are recognized and imported into the periplasm by TonB-dependent siderophore receptors (TBSRs) in the outer membrane (OM), a process depending on the energy transduced by the TonB-ExbB-ExbD system located in the inner membrane (IM) (5). Subsequently, iron-siderophore complexes are generally translocated across the IM by the activity of ABC transporters or permeases into the cytoplasm, where Fe3+ is reduced to ferrous iron (Fe2+) and released from the complex (6). In some cases, dissociation of iron-siderophore complex occurs in the periplasm either by modification of the siderophore scaffold or by reduction of Fe3+ into Fe2+ (6).
Bacteria can also import Fe2+. In contrast to Fe3+, Fe2+ usually exists in its free form and can be taken up directly by transport systems (7). To date, a variety of Fe2+ acquisition systems in bacteria have been characterized. While most of these systems are species or strain specific and promiscuous with respect to divalent metal ions, the Feo system is widely distributed and solely dedicated to the transport of Fe2+ (8). The majority (89%) of bacterial Feo systems are composed of two subunits, FeoA and FeoB, but variations exist, including three subunits (FeoA, FeoB, and FeoC) that are present only in some gammaproteobacterial species such as Escherichia coli and a single fused FeoA/FeoB protein (8). FeoA is a small-molecule hydrophilic protein which has been shown to be required for Feo function in several bacteria by promoting the formation of the Feo complex (9–13). In contrast, FeoB is a large protein with an N-terminal, cytoplasmic domain regulating transport and a C-terminal polytopic transmembrane domain that functions as a permease or a GTP-gated channel for Fe2+ transport (14, 15).
Shewanella oneidensis, a representative of dissimilatory metal-reducing bacterial species, is a Gram-negative facultative gammaproteobacterium capable of respiring a variety of chemicals, including iron ores, as electron acceptors (EAs) (16). S. oneidensis is rich in iron-containing proteins, which are largely accountable for the respiratory versatility, and consequently requires iron at levels higher than those required by the model bacterium E. coli (17). For iron uptake, S. oneidensis synthesizes and secretes putrebactin, the only siderophore produced naturally (18). Putrebactin is an unsaturated macrocyclic dihydroxamic acid synthesized from putrescine by three proteins, PubA, PubB, and PubC (19). The putrebactin-specific TBSR and ferric putrebactin reductase are predicted to be PutA and PutB, respectively, encoded by genes clustered with the pubABC operon (20, 21). With respect to iron physiology, PutA has been found to be crucial to iron acquisition under low-iron conditions and its loss affects expression of the pubABC operon (20). However, the physiological effects of the putrebactin loss on iron uptake are rather minor; as a result, an as-yet-unidentified siderophore has been proposed to function in the absence of putrebactin (22, 23). Moreover, the importance of PutB for iron physiology has been found to be negligible, implying the presence of alternative reductases for releasing iron from iron-siderophore complexes (22).
The pub-putA-putB clusters are conserved in sequenced Shewanella species and strains (see Fig. S1A in the supplemental material), and PutB is homologous to characterized ferric hydroxamate reductases of other bacteria (22). These data suggest that the Pub proteins, PutA, and PutB constitute an iron uptake pathway specific for putrebactin. We therefore propose that the inconsistent influences of Pub proteins, PutA, and PutB on the iron physiology of S. oneidensis are probably a result of interference of other iron transport systems, whose contributions to iron uptake are altered in the strains lacking Pub and Put proteins. In this study, we found that the Feo system, the other primary iron uptake system in S. oneidensis, is largely accountable for the differences in the phenotypes of pub and put mutants. In its absence, all proteins encoded by the genes in the pub-putA-putB cluster are essential to putrebactin-mediated iron uptake, validating the idea that these proteins are components of the putrebactin-mediated iron uptake pathway. We further showed that the Feo system is also the route through which hydroxyl acids, such as lactate, promote iron uptake in S. oneidensis.
RESULTS
PutA appears to be the only TBSR that is critical for iron uptake in S. oneidensis.
Given that the impaired iron uptake of the ΔputA strain can be reversed by addition of Fe3+ (20), it is natural to propose that alternative Fe3+ import routes exist in S. oneidensis, albeit they are less effective. For identification, we first looked into other TBSRs; there are 8 additional such proteins encoded in the S. oneidensis genome (S. oneidensis 0798 [SO_0798], SO_1156, SO_1482, SO_3914, SO_4422, SO_4516, IrgA [SO_4523], and SO_4743) (see Table S1 in the supplemental material). BLASTp analysis results for PutA revealed modest sequence similarities (E values ranging from e−24 to e−9) between these proteins except SO_4516 (E value, 0.14) and PutA. In addition, these annotated TBSRs, including PutA, are generally similar in size, ranging from 663 to 815 amino acid (aa) residues in length, with the majority being 720 ± 15 aa.
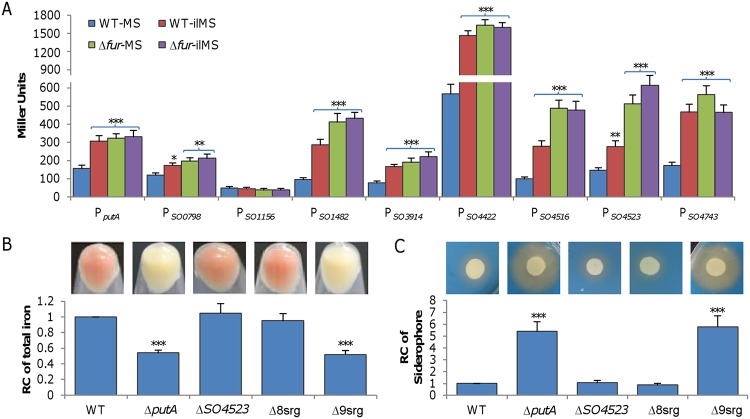
Expression levels of iron transport systems are generally responsive to changes in intracellular iron concentrations via the activity of Fur (ferric uptake regulator), which plays a key role in maintaining iron homeostasis (24). In line with this, for all of these TBSRs except SO_1156 and PutA, Fur boxes have been identified in the region upstream of their coding genes (21). To provide direct evidence for their implication in iron uptake, we examined the impact of iron and of the fur deletion on expression of all TBSR genes. DNA fragments of ∼400 bp upstream of the operons for these receptor genes, all of which are predicted to be transcribed independently (25), were amplified and placed in front of the E. coli lacZ gene within integrative vector pHGR03 (26). The resulting lacZ reporter vectors were introduced into the wild-type and Δfur strains, and activities of the promoters were assayed in cells grown to the mid-log phase (optical density at 600 nm [OD600], ∼0.3) in MS (KCl, 1.34 mM; NaH2PO4, 5 mM; Na2SO4, 0.7 mM; NaCl, 52 mM; piperazine-N,N′-bis[2-ethanesulfonic acid] [PIPES], 3 mM; NH4Cl, 28 mM; sodium lactate, 30 mM; MgSO4, 1 mM; CaCl2, 0.27 mM; FeCl4, 3.6 μM, pH 7.0) and iron-limited MS (ilMS) media (Fig. 1A). Upon growth in MS medium, all promoters under test except PSO1156 were significantly activated by the fur deletion compared to the wild type. While the presence and absence of the Fur box in the promoter region perfectly explained the phenomena seen with the Fur box-containing promoters and PSO1156, respectively, the response of PputA was exceptional because a Fur box was lacking. Similarly, only PSO1156 was not responsive to iron limitation (under conditions of growth in ilMS medium), a condition allowing derepression of Fur-controlled genes. Expectedly, the effect of the fur deletion on all of these promoters was independent of the iron level. These data indicate that TBSRs are subject to regulation by iron in general.
FIG 1.
PutA appears to be the only TBSR that is critical for iron uptake in S. oneidensis. (A) Expression of TBSR genes in wild-type and Δfur strains grown in MS and ilMS (iron-limited MS) analyzed by an integrative lacZ reporter. Cells of the mid-log phase were prepared as described in Materials and Methods, and the activity of the indicated promoters was assayed. Asterisks indicate statistically significant differences for each strain compared to the sample grown in MS (*, P < 0.05; **, P < 0.01; ***, P < 0.001 [throughout the study]). (B) Total iron in the wild-type (WT) and ΔputA strains and representative TBSR mutants. Cultures (OD600 of ∼0.6) of indicated strains grown in LB were pelleted for photography and then subjected to determination of the iron content. Δ8srg, a strain in which all TBSR genes but putA are deleted; Δ9srg, a strain in which all TBSR genes are deleted. (C) Siderophore production in the wild-type, ΔputA, and representative TBSR mutant strains. Cultures (OD600 of ∼0.6) of indicated strains were adjusted to the same OD and divided, and one part was applied onto LB-CAS plates and incubated for 16 h before photography was performed (upper panel). The other part was used for quantification of siderophore contents by the CAS assay of the supernatants (lower panel). For the data in panels B and C, the reaction mixtures were first adjusted according to the protein levels of samples, and then the averaged levels of the mutants were normalized to that in the wild-type strain, which was set to 1, giving relative content (RC). Statistics values were deduced on the basis of comparisons to the wild type. All experiments were performed at least three times, and the data are presented either as means ± SEM or as values representative of similar results.
To determine the involvement of these TBSRs in iron uptake, in-frame deletion mutants for each of them were constructed. In contrast to the ΔputA strain, which has the whitish culture (WC) phenotype resulting from low iron content (20), deletion strains for all other TBSR genes were indistinguishable from the wild-type strain, displaying the signature reddish-brown culture color of Shewanella (Fig. 1B; see also Fig. S2). Consistently, iron levels in these mutants were comparable to that in the wild type. This result indicates that the impacts of TBSRs other than PutA on iron uptake are insignificant. To test whether the presence of multiple TBSRs together would make a difference, we removed the TBSR coding genes in a stepwise manner. Deletion of all TBSR genes but putA (Δ8srg) did not significantly affect the culture color or iron levels (Fig. 1B). The additional removal of putA from the Δ8srg strain, however, turned the culture color from reddish to whitish and substantially reduced the iron content, the same results as were seen with the putA single-knockout strain. Moreover, we measured siderophore levels in these mutants using the Chromeazurol S (CAS) assay. While the Δ8srg strain resembled the wild-type strain in siderophore levels, the additional removal of the putA gene drastically increased siderophore production (Fig. 1C). Notably, the difference in siderophore levels between the strains lacking PutA and all 9 TBSRs (Δ9srg) was insignificant. It is worth mentioning that the reddish-brown culture strains grew similarly to the wild type whereas the WC strains displayed modestly impaired growth because of the deficiency in c-type cytochromes (see Fig. S2A in the supplemental material) as revealed before (20). These data, taken together, indicate that PutA is the only TBSR for putrebactin and that the importance of the other TBSRs in putrebactin-mediated iron uptake is negligible, although many of them are responsive to iron in S. oneidensis.
Deletion of pubABC, putA, or putB results in different phenotypes.
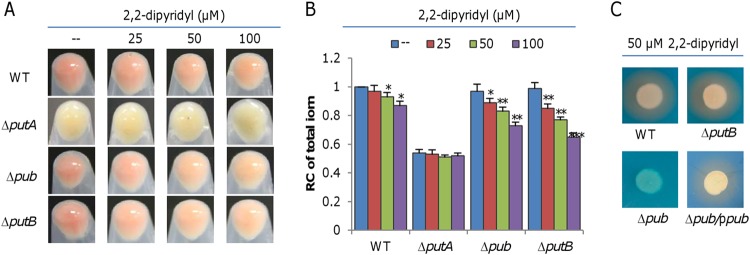
It is well known that iron-siderophore uptake by TBSRs is highly selective and is sometimes even a stereoseletive process, with each iron-siderophore complex having a specific TBSR (27, 28). The unreplaceability of PutA by other TBSRs supports the idea of a functional linkage of the Pub proteins, PutA, and PutB in putrebactin-meidated iron uptake. Encouraged by this, we set out to validate the inconsistent effects of Pub proteins, PutA, and PutB on iron uptake. To this end, we attempted to knock out putB and the entire pub operon (pubABC) to compare the resulting mutants with the ΔputA strain. Unlike the WC ΔputA strain, both the Δpub (ΔpubABC) and ΔputB strains grown in LB exhibited an reddish-brown culture phenotype, and consistently, these two mutants had normal iron levels and growth compared to the wild-type strain (Fig. 2A; see also Fig. 2B and S3B). The CAS assay revealed that the Δpub strain lost the ability to produce putrebactin irrespective of the culturing media used and that this defect was corrected by expression of the operon in trans (Fig. 2C). Interestingly, unlike the Δpub and ΔputA strains, the ΔputB strain was not different from the wild-type strain with respect to siderophore production (Fig. 2C). An expression analysis confirmed that the activity of the pub promoter was not significantly affected by the PutB loss (Fig. S3).
FIG 2.
Characteristics of the pub-putA-putB cluster mutants. (A) Culture colors of indicated strains. Cultures (OD600 of ∼0.6) of indicated strains grown in LB were pelleted for photography without or with membrane-permeable iron chelator 2,2-dipyridyl at various concentrations. (B) Total iron levels in indicated strains. Data corresponding to relative contents of iron were obtained, and data were processed as described for Fig. 1. Statistics values were deduced for each strain using comparisons to the sample grown in LB without 2,2-dipyridyl. (C) Siderophore production of indicated strains grown in LB with 50 μM 2,2-dipyridyl. Δpub/ppub, the Δpub strain carrying a copy of the pub operon in pHGE-Ptac for complementation. Data represent results obtained with 0.1 mM IPTG in expression assays. All experiments were performed at least three times, and data are presented either as means ± SEM or as values representative of similar results.
Siderophore-mediated iron uptake is particularly effective when iron availability is limited. Thus, we compared the effects of the pub, putA, and putB deletions on culture colors and iron content when cells were grown in LB with iron chelator 2,2-dipyridyl at various concentrations (Fig. 2A). With respect to both characteristics, the putA mutant was expectedly not significantly affected by the addition of 2,2-dipyridyl (Fig. 2A and B). However, the pub and putB mutants, along with the wild-type strain, became paler with increases in the 2,2-dipyridyl concentrations; clearly, both mutants were more sensitive to 2,2-dipyridyl addition than the wild-type strain, indicating that the loss of PutB does have an impact on iron uptake. Thus, it is clear that components of the siderophore-mediated iron uptake system, including biosynthesis, transport, and reduction, differ from one another in their levels of physiological impact on iron uptake.
FeoAB is the other primary iron uptake system.
Given that putrebactin is the only siderophore that S. oneidensis produces (18), it is certain that the loss of PubABC annuls the siderophore-dependent iron uptake. Moreover, as the Δpub strain is not impaired in iron uptake, there must be alternative systems accountable for iron uptake in the absence of siderophore. According to the genome annotation (29), S. oneidensis possesses a Feo system, which is the major Fe2+ transport system and is composed of FeoA and FeoB encoded by the feo operon (SO_1783-SO_1784) (Fig. S1B).
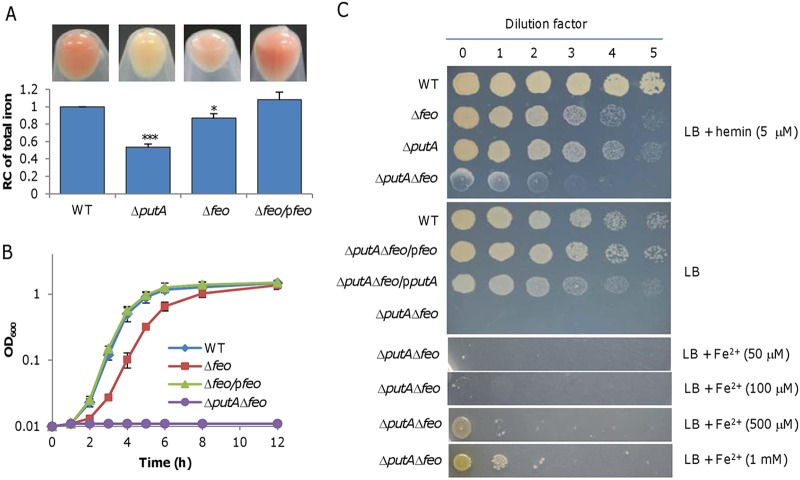
To assess the importance of the Feo system in iron uptake, we constructed a feoAB deletion strain (Δfeo). Rhe Δfeo cells grown in LB had a lighter culture color and lower total iron content than the wild-type strain, albeit the level was significantly higher than that seen with the ΔputA strain (Fig. 3A). In line with the low iron content, this mutant displayed impaired growth in LB which was able to be fully rescued by genetic complementation (Fig. 3B). The growth defect resulting from the Feo loss appeared to be more severe than that caused by the putA deletion (Fig. S2C). Subsequently, we intended to remove the feo genes from the ΔputA strain with the established method (30). Despite multiple attempts, no ΔputA Δfeo colonies were obtained after the resolution (theoretically generating a population of a 50:50 mixture of the mutant and wild-type cells), the last step of the mutagenesis procedure performed on LB plates. On the basis of our experience (31), the failure implies that the siderophore- and Feo-dependent iron uptake systems are synthetically lethal under the experimental conditions used. By using LB plates containing hemin as an iron source, we obtained the intended mutant, which grew extremely poorly. Expectedly, no growth for the ΔputA Δfeo strain was observed either on LB plates or in liquid LB for 48 h (Fig. 3B and C). Complementation with either putA or feo enabled growth, confirming that the inability to grow in LB was due to the intended mutations.
FIG 3.
The Feo system is the other primary iron uptake system in S. oneidensis. (A) Culture color phenotype and iron levels. Cultures (OD600 of ∼0.6) of indicated strains grown in LB were pelleted for photography. Relative contents of iron were obtained, and data were processed as described for Fig. 1. Statistics values were deduced for each strain using comparisons to the WT. (B) Growth of indicated strains in LB. (C) Growth on LB plates with indicated additives. Complementation of the feo and putA deletion mutant was performed with the vector carrying IPTG-inducible promoter Ptac. IPTG concentration, 0.1 mM. All other strain experiments were performed with empty vector. Experiments were performed at least three times, and data are presented either as means ± SEM or as values representative of similar results.
Recently, FicI (SO_3966), a protein of the Mg2+ transporter E (MgtE) family, has been demonstrated to be a secondary Fe2+ importer in S. oneidensis and to function only under conditions with high Fe2+ concentrations (32). We therefore hypothesized that the ΔputA Δfeo strain may be able to grow with elevated iron levels. Indeed, although growth was not observed with 100 μM Fe2+ in 48 h, it was evident in the presence of 0.5 mM Fe2+ and was further improved with a 1 mM concentration (Fig. 3C). Despite this, growth of the ΔputA Δfeo strain remained severely defective, with a lag phase of ∼10 h (Fig. S2D). Additionally, we found that the removal of Feo did not affect siderophore production (Fig. S4A). Furthermore, we demonstrated that the ΔputA ΔfeoA and ΔputA ΔfeoB strains were also unable to grow on LB plates, indicating that both subunits of Feo are essential to function (Fig. S4B). These data, taken together, enabled us to conclude that Feo is the other primary iron uptake system in S. oneidensis.
All genes in the pubABC-putA-putB cluster are essential for putrebactin-mediated iron uptake.
The depletion of Pub proteins or PutB did not elicit any noticeable phenotype, indicating the possibility that functional replacements for these proteins may be encoded in the genome. Given the synthetic lethality of PutA and Feo, we predicted that Feo would also form synthetic lethal pairs with Pub and PutB unless their functional substituents existed. To investigate this, we attempted to remove pub and putB genes from the Δfeo strain on LB plates supplemented with hemin. The resulting deletion strains, mutants Δpub Δfeo and ΔputB Δfeo, failed to show detectable growth on LB plates in 48 h (Fig. 4A). The CAS assay confirmed that the Δpub Δfeo strain lost the ability to produce putrebactin and that the ΔputB Δfeo strain was normal in that respect (Fig. 4B). The synthetically lethal phenotypes of these double mutants are attributable to the additional removal of pub and putB genes, as genetic complementation experiments performed with the corresponding genes were successful (Fig. 4A). In addition, we deleted pubA, pubB, and pubC individually from the Δfeo strain to test whether the intermediates of the putrebactin biosynthesis pathway could function as siderophores. All of the resulting strains were found to be indistinguishable from the Δpub Δfeo strain in this respect (Fig. S5). On the basis of these data, we conclude that all genes of the pubABC-putA-putB cluster are essential for putrebactin-dependent iron uptake in S. oneidensis.
FIG 4.
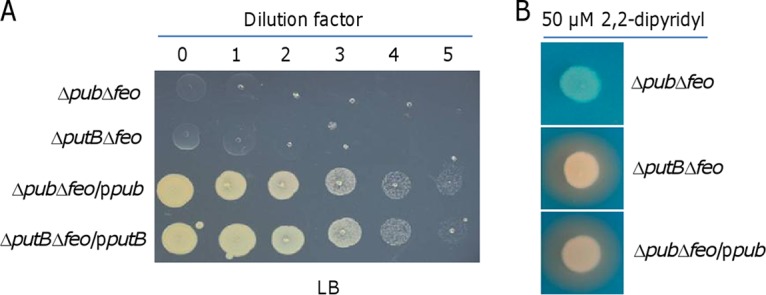
pubABC and putB are essential for putrebactin-mediated iron uptake. (A) Growth of indicated strains on LB plates. (B) Siderophore production of indicated strains grown in LB with 50 μM 2,2-dipyridyl. Complementation of the pub and putB deletions was performed with the vector carrying IPTG-inducible promoter Ptac. IPTG concentration, 0.1 mM. All experiments were performed at least three times, and data are representative of experiments with similar results.
S. oneidensis is able to take up iron with siderophores produced by some other bacteria.
Although TBSRs other than PutA are unable to recognize putrebactin, it is possible that some of them may work with siderophores released from other microbes in the environment for iron uptake, a scenario reported for many bacterial genera, such as Vibrio and Pseudomonas (3). To test this, we employed a cross-feeding assay to examine whether spent culture supernatant of other bacteria could rescue the synthetically lethal phenotype of the Δpub Δfeo strain.
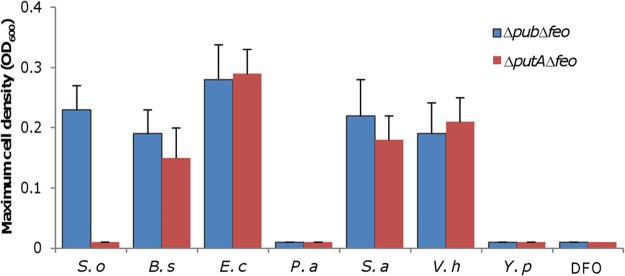
The bacterial species tested included E. coli, Pseudomonas aeruginosa, Vibrio harveyi, Yersinia pseudotuberculosis, Bacillus subtilis, and Staphylococcus aureus, and the siderophores that they produce are shown in Fig. S6 (3). All of these bacteria were cultivated under iron starvation conditions (LB with 50 μM 2,2-dipyridyl) to ensure siderophore production (Fig. S6). The wild-type and Δpub Δfeo strains were inoculated into LB mixed with 0.5 ml of the cell-free supernatants of the spent cultures for each bacterium, and growth was monitored. Growth of the wild-type strain was observed in the supernatants of all cultures under test (data not shown), indicating that none of the spent medium critically inhibited growth of S. oneidensis. Similarly, the Δpub Δfeo strain was able to grow in the supernatants of the S. oneidensis ΔputA culture, which was used as the positive control (Fig. 5). In contrast, in the same supernatants there was no growth with the ΔputA Δfeo strain, serving as the negative control. With the bacteria under test, the growth phenotypes of the Δpub Δfeo and ΔputA Δfeo strains were identical; both were able to grow in the supernatants of E. coli, V. harveyi, B. subtilis, and S. aureus but not in the supernatants of other species (Fig. 5). Moreover, we tested the effect of a commercially available siderophore, desferrioxamine (DFO), on growth of these two strains. The result showed that DFO at concentrations of 0.5 to 100 μM could not support growth under the experimental conditions (Fig. 5). On the basis of these data, it appears that S. oneidensis has receptors for siderophores from E. coli, V. harveyi, B. subtilis, and S. aureus.
FIG 5.
S. oneidensis is able to take up iron with siderophores produced by some other bacteria. Cultures of the indicated bacteria grown in LB to the stationary phase with 50 μM 2,2-dipyridyl were centrifuged. A 500-μl volume of the supernatant from each bacterium was added to 1.5 ml fresh LB for cultivation of indicated strains. For DFO, LB concentrations of 5 to 100 μM was tested. Growth was monitored, and data corresponding to the maximum cell density are presented. S. o, S. oneidensis; B. s, B. subtilis; E. c, E. coli; P. a, P. aeruginosa; S. a, S. aureus; V. h, V. harveyi; Y. p, Y. pseudotuberculosis. The experiments were performed at least three times, and data are presented as means ± SEM.
Overproduced putrebactin dictates the WC phenotype of the ΔputA strain.
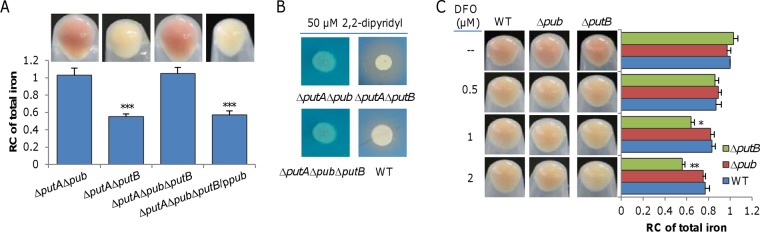
The WC phenotype of the ΔputA strain is puzzling given that the Δpub and ΔputB strains were normal with respect to culture color. As the first step to solve this, we constructed Δpub ΔputA and ΔputA ΔputB strains. Surprisingly, the culture colors of these two mutants grown in LB were not the same; the ΔputA Δpub strain was reddish-brown whereas the ΔputA ΔputB strain was whitish (Fig. 6A). We then deleted all genes in this cluster together and found that the resulting mutant (Δpub ΔputA ΔputB) was reddish-brown in LB. As the expression of the pub operon in trans converted the Δpub ΔputA ΔputB strain to whitish, it is conclusive that the pub genes are required for the WC phenotype.
FIG 6.
Overproduced putrebactin dictates the WC phenotype of the ΔputA strain. (A) The culture color phenotype and iron levels. Cultures (OD600 of ∼0.6) of indicated strains grown in LB were pelleted for photography. Relative contents of iron were obtained, and data were processed as described for Fig. 1. Statistics values were deduced for each strain using comparisons to the ΔputA Δpub strain. (B) Siderophore production of indicated strains grown in LB with 50 μM 2,2-dipyridyl. (C) Effect of DFO on culture color and iron levels of indicated strains. Cultures (OD600 of ∼0.6) of indicated strains grown in LB with DFO at various concentrations were pelleted and photographed. Relative contents of iron were obtained, and data were processed as described for Fig. 1. Statistics values were deduced for each strain using comparisons to the WT grown under the same conditions. All experiments were performed at least three times, and data are presented either as means ± SEM or as values representative of similar results.
In the ΔputA strain, putrebactin is constitutively produced at elevated levels. Since the Feo system is dominantly responsible for iron uptake of the ΔputA strain, we reasoned that the free iron levels in the environment decreased because of chelation of overproduced putrebacterin, leading to the WC phenotype. This notion was supported by the finding that the ΔputA ΔputB strain overproduced siderophore, resembling the ΔputA strain in that respect (Fig. 6B). For further confirmation, we examined effects of DFO, a siderophore that (as shown above) cannot be imported into the cell, on the culture color of relevant strains (Fig. 5). It was immediately seen that the impact of DFO on culture colors of the wild-type and Δpub and ΔputB strains was more evident (Fig. 6C) than that seen with membrane-permeable 2,2-dipyridyl (33) as shown in Fig. 2A. At 0.5 μM, DFO whitened the cultures of all three strains and decreased their iron content significantly. Apparently, the ΔputB strain was more sensitive to the siderophore than the wild-type strain, as, in the presence of 2 μM DFO, it turned out to be indistinguishable from the ΔputA strain in that respect. The difference in the responses of the wild-type and ΔputB strains to DFO can be reasonably explained by the accumulation of putrebactin in the ΔputB culture because the two strains produce putrebacterin at comparable levels but the latter strain loses putrebactin-dependent iron uptake activity. On the basis of this finding, we propose that excessive putrebactin is largely accountable for the WC phenotype of the ΔputA strain.
Hydroxy acids facilitate iron import through the Feo system in S. oneidensis.
We have previously shown that lactate facilitates iron uptake of the ΔputA strain through a mechanism that is independent of the presence of lactate permeases (20). In the presence of l-lactate at a concentration of 8 mM or above (d-lactate is much less effective), the ΔputA culture grown in LB is red. Consistently, grown in defined medium MS with lactate, the ΔputA strain and the wild-type strain are similar in culture color (20). This intriguing finding is critically relevant to Shewanella physiology as lactate had been used as the carbon source and electron donor for defined media in most of the published studies.
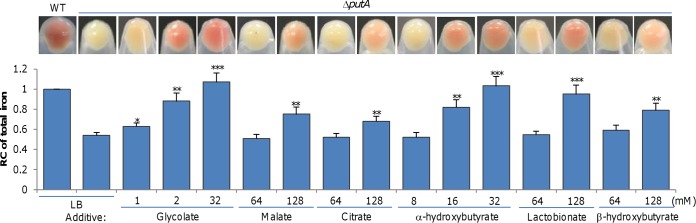
To decipher the mystery, we grew the ΔputA strain in LB supplemented with a variety of carboxylic acids at 32 mM, including formate, acetate, pyruvate, glycolate, propionate, malate, and citrate, some of which have been used before as carbon sources to support growth of S. oneidensis (34–37). Lactate-like effects on the ΔputA culture color were observed only with glycolate (Fig. 7). With concentrations of up to 128 mM, both malate and citrate exhibited a noticeable influence on the culture color whereas the others remained ineffective (Fig. 7). As citrate has the highest stability constant for both Fe2+ and Fe3+ among these acids (38), the iron-chelating capacity does not seem critical. Glycolate, malate, and citrate, as well as lactate, differ from the remaining acids in that they are classified as α-hydroxy acids (Fig. S7), implying that that characteristic may be critical for the effect. This notion was supported by the finding that β-hydroxybutyrate was much less effective than α-hydroxybutyrate in facilitating iron uptake. Furthermore, we compared the effects of three α-hydroxy acids which differ from one another in size. While glycolate at 2 mM was sufficient to turn the color of the ΔputA culture to reddish, to achieve the similar effects, α-hydroxybutyrate and lactobionate required 32 and 128 mM, respectively (Fig. 7), indicating that small-molecule α-hydroxy acids are more effective.
FIG 7.
Alpha-hydroxy acids facilitate iron import through the Feo system in S. oneidensis. Data corresponding to the culture color phenotype and iron levels are presented. Cultures (OD600 of ∼0.6) of indicated strains grown in LB were pelleted for photography. Relative contents of iron were obtained, and data were processed as described for Fig. 1. Statistics values were deduced for each strain using comparisons to the ΔputA strain grown in LB. All experiments were performed at least three times, and data are presented either as means ± SEM or as values representative of similar results.
To test whether the iron uptake promoted by α-hydroxy acids is dependent on Feo, the Δpub ΔfeoAB strain was inoculated into LB containing 32 mM lactate. For incubations lasting up to 48 h, no growth was observed (data not shown). The same results were obtained from all other chemicals tested above, even with concentrations of up to 128 mM. Thus, we conclude that hydroxy acids, especially α-type hydroxy acids, are able to facilitate iron uptake, a process relying on the Feo system.
DISCUSSION
The primary objective of this study was to address the questions associated with the siderophore-dependent iron uptake in S. oneidensis that were raised but left unanswered in our previous reports (20, 21). We have performed a genetic analysis of predicted components of iron uptake pathways, generating the following three contributions to the current understanding of the subject. First, the recognition of putrebactin is specifically mediated by PutA although the bacterium possesses an array of siderophore receptors which may interact with siderophores produced by other bacteria. Second, the Feo system is another major route for iron uptake. Third, hydroxyl acids, especially α-type hydroxyl acids, facilitate iron uptake via the Feo system.
On the basis of the findings reported previously and determined in this study, a model of iron uptake in S. oneidensis was proposed (Fig. 8). Although we previously proposed on the basis of phenotypic analysis of the putA mutant that PutA is the only receptor for putrebactin in S. oneidensis (20), direct evidence is lacking. As the mutant devoid of all other putative siderophore receptors was normal in its iron uptake and as putrebactin is the only siderophore that S. oneidensis produces naturally (18), it is clear that PutA is the sole protein responsible for recognition and uptake of ferric putrebactin. Despite this, the alternative siderophore receptors may still be involved in iron uptake, as most of them are responsive to iron with respect to expression. The responsiveness is mediated by the regulator Fur, given the presence of the Fur box in their promoter regions (21). More importantly, the notion gains support from the observation that S. oneidensis is able to utilize siderophores produced by some other bacteria to acquire iron for growth (Fig. 8). However, there is a caveat with respect to this observation: all major iron uptake systems must be removed because they obscure the physiological influences of siderophore receptors other than PutA.
FIG 8.
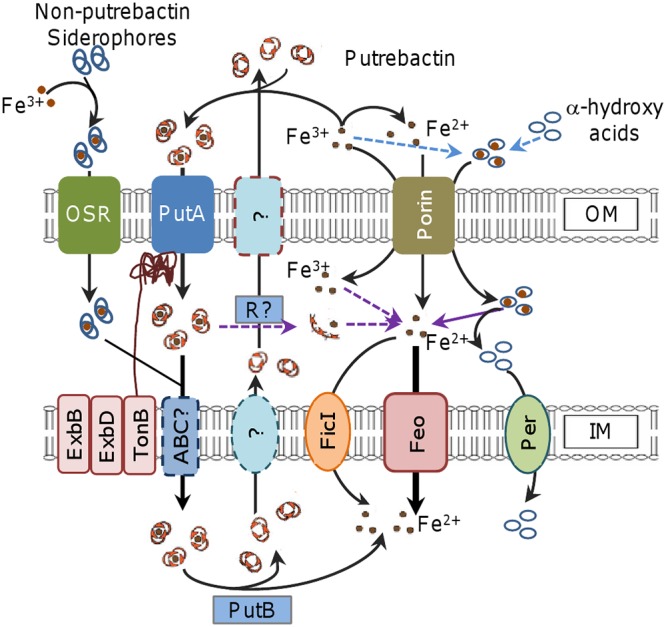
Model for iron uptake in S. oneidensis. Fe3+ in the environment can be reduced to Fe2+ and scavenged by putrebactin produced and secreted by S. oneidensis to form Fe3+-putrebactin complexes. The complex enters the periplasm through PutA. Fe3+ can also be captured by some other bacterial siderophores present in the environment and enters the cell through other siderophore receptors (OSR). Through the activity of either unknown ABC transporters (ABC?) or reductases (R?), the complex is imported into the cytoplasm or releases Fe2+, respectively. In the cell, Fe2+ is released from the complex after the reduction catalyzed by PutB. Transportation of Fe2+ across the IM is primarily mediated by Feo, with the assistance of FicI when its concentrations are high. Alpha-hydroxy acids probably compete with siderophores for Fe3+ for formation of iron-carrying complexes, which likely enter the periplasm via porin, where Fe3+ is reduced and released. The resulting Fe3+ crosses the IM via Feo. Alpha-hydroxy acids can be imported in large amounts through specific permeases (Per); in the case of lactate, such permeases include LctP1 and LctP2, which are not involved in lactate-dependent iron uptake.
It is frequently seen that multiple iron uptake systems are encoded in a bacterial genome, but the simultaneous loss of the major routes generally results in synthetic lethality under normal growth conditions (39–41). On the basis of this feature, we identified Feo as the other primary iron uptake system that is present in addition to that depending on putrebactin (Fig. 8). Feo is dedicated to transport of ferrous ions and functions under both anoxic and oxic conditions, although it is induced in anoxic and low-pH environments, which allow iron to exist in the soluble ferrous form (8, 42). S. oneidensis is among the best-studied dissimilarity metal-reducing bacteria, capable of reducing Fe3+ to Fe2+, even in the presence of oxygen (16, 43). Naturally, the contribution of the Feo system to iron uptake is likely unusually significant. This indeed is the case given that the loss of Feo results in an evident defect in growth whereas the lack of the siderophore-dependent route (the pub mutant) does not affect growth or affects it at most marginally. A reasonable explanation for the differences in the growth defects of the feo and pub mutants is that the feo mutant imports iron at a rate significantly lower than that seen with the pub mutant.
Using the strain in which both major iron uptake systems were absent, we have been able to identify xenosiderophores that S. oneidensis can utilize. The cross-feeding assays revealed that the supernatants of the E. coli, B. subtilis, and S. aureus cultures were able to support growth of the strain missing all of the feo and pub genes. E. coli produces two siderophores, enterobactin and aerobactin, which belong to catecholate-type and mixed-type siderophores, respectively (44). In S. oneidensis, SO_4523 (IrgA) is annotated as the TBSR for enterobactin and is highly homologous to E. coli FepA and P. aeruginosa PfeA (BLASTp E values, 9e−54 and 7e−60, respectively), which recognize and bind to ferric enterobactin (45, 46). Similarly to S. oneidensis, P. aeruginosa is equipped with a large number of TBSRs for siderophores and does not produce enterobactin by itself (47). Hence, it is very likely that IrgA is the TBSR for enterobactin in S. oneidensis. We do not yet know the TBSRs of S. oneidensis for siderophores produced by B. subtilis and S. aureus. IrgA may also work with bacillibactin (produced by B. subtilis), which is structurally similar to enterobactin (Fig. 8), but it does not recognize any of the siderophores produced by S. aureus (48–50). We are working to identify S. oneidensis TBSRs for these xenosiderophores.
The depletion of Feo also enabled us to validate the essentiality of PubABC and PutB to putrebactin-dependent iron uptake (Fig. 8). This resolves the conflicting phenotypes resulting from the deletion of the pub, putA, and putB genes, supporting the notion of a functional linkage of the proteins encoded by the pubABC-putA-putB cluster. The WC phenotype of the putA mutant is attributable to enhanced putrebactin production, a scenario that also occurs with the addition of membrane-impermeative DFO. In both cases, the consequence is that iron levels further decrease by chelation, preventing the Feo system from taking up iron at rates that meet the physiological needs. The loss of PutB, however, does not have such an effect because putrebactin is not overproduced. This seems reasonable because the putB mutant retains the ability to retrieve siderophores released into the surroundings.
One of the most striking findings in our previous report was that the putA mutant displays the reddish-brown culture phenotype when grown in defined MS medium (20). The factor that accounts for the phenotype is lactate, which has been used as the default carbon source in defined media for cultivation of Shewanella species (51). In this study, we showed that lactate, as well as many hydroxyl acids, facilitates iron uptake in S. oneidensis (Fig. 8). Apparently, the chelating capacity of these hydroxyl acids does not dictate the process because citrate (whose stability constants for Fe2+ and Fe3+ are 3.2 and 11.85, respectively), the strongest iron chelator among these hydroxyl acids, is rather weak in facilitating iron uptake (38). In parallel, the nature of the acids is also not the determining factor as formate, acetate, pyruvate, and propionate do not work. Rather, it is clear that two characteristics determine the efficiency of these acids in their promotion of iron uptake: acids with an α-hydroxyl group are more efficient than those with a β-hydroxyl group, and for acids of the same configuration, the smaller the molecule, the more effective. As the WC phenotype of the putA mutant is due to increased putrebactin production, it is conceivable that these small-molecule acids achieve this by chelating and delivering iron into the periplasm. Once in the periplasm, ferric iron appears to be reduced to the ferrous form, which is then transported into the cytoplasm by the Feo system. It is worth mentioning that the route by which these hydroxyl acids promote iron uptake is unlikely to be the pathway for their transport across the IM in a large quantity. For example, to support growth, lactate is imported via the activity of lactate permeases LctP1 and LctP2 (20).
In our previous reports, the Fur regulon was predicted with newly determined Fur box sequences (21). Unlike the pub operon, putA does not appear to be under the direct control of Fur. However, our data reveal that expression of the pub operon and expression of the putA operon take place in concert in response to iron levels and Fur depletion. How this occurs is unknown, but it may be linked to siderophore levels. We speculate that when cells are subjected to conditions of iron scarcity, Fur is inactivated and the pub operon is derepressed, leading to siderophore production, which in turn upregulates expression of the putA gene. However, this merits further investigation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Sequences of the primers used in this study are given in Table 2. All chemicals are from Sigma-Aldrich Co. unless otherwise noted. E. coli and S. oneidensis were grown aerobically in LB (Difco, Detroit, MI) at 37°C and 30°C for genetic manipulation. When appropriate, the growth medium was supplemented with the following: 2,6-diaminopimelic acid (DAP), 0.3 mM; ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; gentamicin, 15 μg/ml.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Description | Source/reference |
|---|---|---|
| Strains | ||
| S. oneidensis | ||
| MR-1 | Wild type | Laboratory stock |
| HG0798 | ΔSO_0798 mutant derived from MR-1 | This study |
| HG1156 | ΔSO_1156 mutant derived from MR-1 | This study |
| HG1482 | ΔSO_1482 mutant derived from MR-1 | This study |
| HG1783 | ΔfeoA mutant derived from MR-1 | This study |
| HG1784 | ΔfeoB mutant derived from MR-1 | This study |
| HG1783-4 | Δfeo mutant derived from MR-1 | This study |
| HG3030 | ΔpubA mutant derived from MR-1 | 20 |
| HG3031 | ΔpubB mutant derived from MR-1 | 20 |
| HG3032 | ΔpubC mutant derived from MR-1 | 20 |
| HG3030-2 | Δpub mutant derived from MR-1 | 20 |
| HG3033 | ΔputA mutant derived from MR-1 | 20 |
| HG3034 | ΔputB mutant derived from MR-1 | 20 |
| HG3914 | ΔSO_3914 mutant derived from MR-1 | This study |
| HG4422 | ΔSO_4422 mutant derived from MR-1 | This study |
| HG4516 | ΔSO_4516 mutant derived from MR-1 | This study |
| HG4523 | ΔSO_4523 mutant derived from MR-1 | This study |
| HG4743 | ΔSO_4743 mutant derived from MR-1 | This study |
| HG3033-Feo | ΔputA Δfeo mutant derived from MR-1 | This study |
| HG3034-Feo | ΔputB Δfeo mutant derived from MR-1 | This study |
| HGPub-Feo | Δpub Δfeo mutant derived from MR-1 | This study |
| HGPut-Feo | ΔputA Δpub ΔputB mutant derived from MR-1 | This study |
| HG-8TBSR | Δ8srg mutant derived from MR-1 | This study |
| HG-9TBSR | Δ9srg mutant derived from MR-1 | This study |
| Other bacteria | ||
| E. coli MG1655 | Wild type | Laboratory stock |
| E. coli DH5α | Host strain for routine cloning | Laboratory stock |
| E. coli WM3064 | Donor strain for conjugation; ΔdapA | W. Metcalf, UIUC |
| P. aeruginosa PAO1 | Wild type | 52 |
| V. harveyi BB120 | Wild type | 53 |
| Y. pseudotuberculosis YpIII | Wild type | 54 |
| B. subtilis 642 | Wild type | 55 |
| S. aureus ATCC 25923 | Wild type | 56 |
| Plasmids | ||
| pHGM01 | Apr Gmr Cmr suicide vector | 30 |
| pHGE-Ptac | IPTG-inducible Ptac expression vector | 57 |
| pHGEI01 | Integrative lacZ reporter vector | 26 |
| pBBR-Cre | Helper vector for antibiotic marker removal | 58 |
| pHGEI01-PSO0789 | For measuring PSO0789 activity | This study |
| pHGEI01-PSO1156 | For measuring PSO1156 activity | This study |
| pHGEI01-PSO1482 | For measuring PSO1482 activity | This study |
| pHGEI01-PSO3914 | For measuring PSO3914 activity | This study |
| pHGEI01-PSO4422 | For measuring PSO4422 activity | This study |
| pHGEI01-PSO4516 | For measuring PSO4516 activity | This study |
| pHGEI01-PSO4523 | For measuring PSO4523 activity | This study |
| pHGEI01-PSO4743 | For measuring PSO4743 activity | This study |
| pHGEI01-PputA | For measuring PputA activity | 20 |
| pHGEI01-Ppub | For measuring Ppub activity | This study |
| pHGE-Ptac-pub | Vector for inducible expression of pub | This study |
| pHGE-Ptac-feo | Vector for inducible expression of feo | This study |
| pHGE-Ptac-putA | Vector for inducible expression of putA | This study |
| pHGE-Ptac-putB | Vector for inducible expression of putB | This study |
Ap, ampicillin; Cm, chloramphenicol; Gm, gentamicin; UIUC, University of Illinois Urbana—Champaign.
TABLE 2.
Primers used in this study
| Primer | Primer sequences |
|---|---|
| In-frame deletion | |
| HG0789-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTAGGGAACTGTCACATTGGCACC |
| HG0789-M5I | GGTCCGGGTTCGCTATCTATTTGCACGGATCACTTTGTCGCC |
| HG0789-M3I | ATAGATAGCGAACCCGGACCGGCGATATCCACTCTGGCCGAT |
| HG0789-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGGTGGCGTGGGCGGACTGTTCTT |
| HG1156-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTCAAGCCATATTATGGCGCGGCA |
| HG1156-M5I | GGTCCGGGTTCGCTATCTATATCACAACACACAGGTATTGCC |
| HG1156-M3I | ATAGATAGCGAACCCGGACCAACCGCCAATCTGCTCGCGCAA |
| HG1156-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGGTCTGTCATGGCATTAATCA |
| HG1482-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG1482-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG1482-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG1482-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG1783-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG1783-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG1783-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG1783-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG1784-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG1784-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG1784-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG1784-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG3030-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG3030-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG3030-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG3030-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG3031-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG3031-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG3031-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG3031-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG3032-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG3032-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG3032-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG3032-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG3034-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG3034-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG3034-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG3034-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG3914-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG3914-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG3914-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG3914-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG4422-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG4422-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG4422-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG4422-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG4516-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG4516-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG4516-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG4516-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG4523-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG4523-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG4523-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG4523-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| HG4743-M5O | GGGGACAAGTTTGTACAAAAAAGCAGGCTGCATGCGATTGGCGATTATCTG |
| HG4743-M5I | GGTCCGGGTTCGCTATCTATACGCGGCCAATTTGCCACACATA |
| HG4743-M3I | ATAGATAGCGAACCCGGACCAGTGTAATGATGCCTATTCTGC |
| HG4743-M3O | GGGGACCACTTTGTACAAGAAAGCTGGGTGATCAATCAAGTAATTAGACAC |
| Controlled expression | |
| Pub-CEF | GGGAATTCGTTGGAAGAAAAAGAAATACTCTGG |
| Pub-CER | CGGGATCCCCCGACATCTCTGTGACTGAATCC |
| Feo-CEF | GGGAATTCGTGCTAGAAGCATATCGTAAAC |
| Feo-CER | CGGGATCCCGCTGGCTTTTGCATTTTATATC |
| PutB-CEF | GGGAATTCATGCCAAAGTTACGATCAGC |
| PutB-CER | CGGGATCCGCGACGATGCGAGAGATAACA |
| lacZ reporters | |
| Ppub-F | GGGAATTCCAGGGAACTGTCACATTGGCACC |
| Ppub-R | CGGGATCCAGGTCTAACTATATTGCCAGTA |
| P0789-F | GGGAATTCGGCATGCGATTGGCGATTATCTG |
| P0789-R | GGGAATTCCAAACTTCCTTTGCACCTTTTT |
| P1156-F | GGGAATTCCCTTAGTATTAGGCGTCGGGTTT |
| P1156-R | CGGGATCCGGATAGACTCCTAATTTAAATT |
| P1482-F | CGGGATCCCAAGCCATATTATGGCGCGGCA |
| P1482-R | CGGGATCCAATCAAGGCATTGTAGTGAATG |
| P3914-F | GGGAATTCGGCATGCGATTGGCGATTATCTG |
| P3914-R | GGGAATTCCAAACTTCCTTTGCACCTTTTT |
| P4422-F | GGGAATTCCCTTAGTATTAGGCGTCGGGTTT |
| P4422-R | CGGGATCCGGATAGACTCCTAATTTAAATT |
| P4516-F | CGGGATCCCAAGCCATATTATGGCGCGGCA |
| P4516-R | CGGGATCCAATCAAGGCATTGTAGTGAATG |
| P4523-F | GGGAATTCGGCATGCGATTGGCGATTATCTG |
| P4523-R | GGGAATTCCAAACTTCCTTTGCACCTTTTT |
| P4743-F | GGGAATTCGGCATGCGATTGGCGATTATCTG |
| P4743-R | GGGAATTCCAAACTTCCTTTGCACCTTTTT |
For physiological characterization, both LB and MS defined medium containing 0.02% (wt/vol) vitamin-free Casamino Acids and 30 mM sodium lactate as the electron donor were used in this study, and consistent results were obtained (59). Fresh medium was inoculated with overnight cultures grown from a single colony at a 1:100 dilution, and growth was determined by recording the optical density at 600 nm (OD600) of cultures.
In-frame mutant construction and complementation.
In-frame deletion strains were constructed using the att-based fusion PCR method as described previously (30). In brief, two fragments flanking the genes of interest were amplified by PCR with outside primers (O; Table 2) containing attB and gene-specific sequences and with inside primers (I; Table 2) containing complementary sequences and gene-specific sequences and were then linked by a second round of PCR with outside primers. The fused fragments were introduced into plasmid pHGM01 using Gateway BP clonase II enzyme mix (Invitrogen) according to the manufacturer's instructions. The resulting vectors were maintained in E. coli DAP auxotroph WM3064 and subsequently transferred into relevant S. oneidensis strains via conjugation. Integration of the deletion constructs into the chromosome was selected by resistance to gentamicin and confirmed by PCR. Verified transconjugants were grown in LB in the absence of NaCl and plated on LB supplemented with 10% sucrose. Gentamicin-sensitive and sucrose-resistant colonies were screened by PCR for intended deletions. Mutants were verified by sequencing the mutated region.
Plasmid pHGE-Ptac was used for genetic complementation of the mutants (57). Genes of interest generated by PCR were placed under the control of isopropyl-β-d-1-thiogalactoside (IPTG)-inducible promoter Ptac within pHGE-Ptac. After verification by sequencing, the resultant vectors were transferred into the relevant strains via conjugation.
Analysis of gene expression.
The activity of promoters was assessed using a single-copy integrative lacZ reporter system as described previously (26). A DNA fragment of ∼400 bp containing the sequence upstream of the coding region for each gene under test was amplified and cloned into pHGEI01 reporter vector and verified by sequencing. The resultant vector was then transferred by conjugation into relevant S. oneidensis strains, in which it integrated into the chromosome, and the antibiotic marker was then removed by an established approach (58). Cells grown to the mid-log phase under conditions specified in the text and/or figure legends were collected, β-galactosidase activity was determined by monitoring color development at 420 nm using a Synergy 2 Pro200 multi-detection microplate reader (Tecan), and the results are presented as Miller units (26).
Culture color assay and quantification of intracellular total iron.
Cells grown in LB to the late log phase (OD600 of ∼0.8) were collected by centrifugation. After being photographed for culture color, the pellet was washed with phosphate-buffered saline (PBS, pH 7.4) and adjusted to an OD600 of ∼0.6, and quantification of total iron was carried out with the established method (60). In brief, aliquots of 50 ml were mixed with 5 ml of 50 mM NaOH, sonicated on ice, and centrifuged at 5,000 × g for 10 min. The cell lysates (100 μl) were then mixed with 100 μl 10 mM HCl plus 100 μl iron-releasing reagent (a freshly mixed solution of equal volumes of 1.4 M HCl and 4.5% [wt/vol] KMnO4) and treated at 60°C for 2 h. After cooling, the iron detection reagents (6.5 mM ferrozine–6.5 mM neocuproine–2.5 M ammonium acetate–1 M ascorbic acid–water) were added. The absorbance of samples at 550 nm was measured 30 min later. The standard curve was depicted using FeCI3 at concentrations of up to 300 μM.
Siderophore assays.
In order to assess siderophore production and secretion, S. oneidensis strains were grown in LB without or with 50 μM 2,2-dipyridyl to the stationary phase and cell-free culture supernatants were obtained by centrifugation. Siderophore levels within the supernatants were quantified using the Chromeazurol S (CAS) assay (61). For visualization, S. oneidensis strains grown on agar plates under the same conditions were subjected to direct detection of siderophores (61).
Viability assay.
S. oneidensis strains grown to the mid-log phase were adjusted to approximately 108 CFU/ml followed by 10-fold serial dilutions. A 5-μl volume of each dilution was spotted onto LB plates without or with relevant chemicals. The plates were incubated at 30°C for at least 24 h before being read.
Cross-feeding assays.
For the detection of other bacterial siderophores that S. oneidensis can use, cross-feeding assays were performed. E. coli, P. aeruginosa, V. harveyi, Y. pseudotuberculosis, B. subtilis, and S. aureus were inoculated into LB containing the iron chelator 2,2′-dipyridyl at 50 μM. When cultures entered the stationary phase, cells were removed by centrifugation and the supernatant was collected from each bacterial culture. The test strains, S. oneidensis mutants Δpub Δfeo and ΔputA Δfeo, were inoculated into the mixture of 1.5 ml LB and a 0.5-ml volume of one of the supernatants, and growth was monitored by recording OD600 values.
Other analyses.
Homologues of proteins of interest were identified via a BLASTp search of the NCBI's nonredundant protein database, using the amino acid sequence as the query. Student's t test was performed for experimental data by using Prism software (GraphPad Software, San Diego, CA) unless otherwise noted. Experiments were performed multiple times (indicated in the figure legends) independently. Values were presented as means ± standard errors of the means (SEM).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by National Natural Science Foundation of China (41476105) and Natural Science Foundation of Zhejiang Province (LZ17C010001).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01752-18.
REFERENCES
- 1.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Melton ED, Swanner ED, Behrens S, Schmidt C, Kappler A. 2014. The interplay of microbially mediated and abiotic reactions in the biogeochemical Fe cycle. Nat Rev Microbiol 12:797–808. doi: 10.1038/nrmicro3347. [DOI] [PubMed] [Google Scholar]
- 3.Hider RC, Kong X. 2010. Chemistry and biology of siderophores. Nat Prod Rep 27:637–657. doi: 10.1039/b906679a. [DOI] [PubMed] [Google Scholar]
- 4.Crosa JH, Walsh CT. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. 2010. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol 64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schalk IJ, Guillon L. 2013. Fate of ferrisiderophores after import across bacterial outer membranes: different iron release strategies are observed in the cytoplasm or periplasm depending on the siderophore pathways. Amino Acids 44:1267–1277. doi: 10.1007/s00726-013-1468-2. [DOI] [PubMed] [Google Scholar]
- 7.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 8.Lau CKY, Krewulak KD, Vogel HJ. 2016. Bacterial ferrous iron transport: the Feo system. FEMS Microbiol Rev 40:273–298. doi: 10.1093/femsre/fuv049. [DOI] [PubMed] [Google Scholar]
- 9.Lau CKY, Ishida H, Liu Z, Vogel HJ. 2013. Solution structure of Escherichia coli FeoA and its potential role in bacterial ferrous iron transport. J Bacteriol 195:46–55. doi: 10.1128/JB.01121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H, Lee H, Shin D. 2012. The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem Biophys Res Commun 423:733–738. doi: 10.1016/j.bbrc.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 11.Perry RD, Mier I, Fetherston JD. 2007. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. BioMetals 20:699. doi: 10.1007/s10534-006-9051-x. [DOI] [PubMed] [Google Scholar]
- 12.Stevenson B, Wyckoff EE, Payne SM. 2016. Vibrio cholerae FeoA, FeoB, and FeoC interact to form a complex. J Bacteriol 198:1160–1170. doi: 10.1128/JB.00930-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weaver EA, Wyckoff EE, Mey AR, Morrison R, Payne SM. 2013. FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB. J Bacteriol 195:4826–4835. doi: 10.1128/JB.00738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seyedmohammad S, Fuentealba Natalia A, Marriott Robert A, Goetze Tom A, Edwardson JM, Barrera Nelson P, Venter H. 2016. Structural model of FeoB, the iron transporter from Pseudomonas aeruginosa, predicts a cysteine lined, GTP-gated pore. Biosci Rep 36:e00322. doi: 10.1042/BSR20160046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petermann N, Hansen G, Schmidt CL, Hilgenfeld R. 2010. Structure of the GTPase and GDI domains of FeoB, the ferrous iron transporter of Legionella pneumophila. FEBS Lett 584:733–738. doi: 10.1016/j.febslet.2009.12.045. [DOI] [PubMed] [Google Scholar]
- 16.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JLM, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 17.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A, Hess M, Omelchenko MV, Kostandarithes HM, Makarova KS, Wackett LP, Fredrickson JK, Ghosal D. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- 18.Soe CZ, Codd R. 2014. Unsaturated macrocyclic dihydroxamic acid siderophores produced by Shewanella putrefaciens using precursor-directed biosynthesis. ACS Chem Biol 9:945–956. doi: 10.1021/cb400901j. [DOI] [PubMed] [Google Scholar]
- 19.Kadi N, Arbache S, Song L, Oves-Costales D, Challis GL. 2008. Identification of a gene cluster that directs putrebactin biosynthesis in Shewanella species: PubC catalyzes cyclodimerization of N-hydroxy-N-succinylputrescine. J Am Chem Soc 130:10458–10459. doi: 10.1021/ja8027263. [DOI] [PubMed] [Google Scholar]
- 20.Dong Z, Guo S, Fu H, Gao H. 2017. Investigation of a spontaneous mutant reveals novel features of iron uptake in Shewanella oneidensis. Sci Rep 7:11788. doi: 10.1038/s41598-017-11987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu H, Liu L, Dong Z, Guo S, Gao H. 2018. Dissociation between iron and heme biosynthesis is largely accountable for respiration defects of Shewanella oneidensis fur mutants. Appl Environ Microbiol 84:e00039-. doi: 10.1128/AEM.00039-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fennessey CM, Jones ME, Taillefert M, DiChristina TJ. 2010. Siderophores are not involved in Fe(III) solubilization during anaerobic Fe(III) respiration by Shewanella oneidensis MR-1. Appl Environ Microbiol 76:2425–2432. doi: 10.1128/AEM.03066-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouzuma A, Hashimoto K, Watanabe K. 2012. Roles of siderophore in manganese-oxide reduction by Shewanella oneidensis MR-1. FEMS Microbiol Lett 326:91–98. doi: 10.1111/j.1574-6968.2011.02444.x. [DOI] [PubMed] [Google Scholar]
- 24.Fillat MF. 2014. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 546:41–52. doi: 10.1016/j.abb.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 25.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu H, Jin M, Ju L, Mao Y, Gao H. 2014. Evidence for function overlapping of CymA and the cytochrome bc1 complex in the Shewanella oneidensis nitrate and nitrite respiration. Environ Microbiol 16:3181–3195. doi: 10.1111/1462-2920.12457. [DOI] [PubMed] [Google Scholar]
- 27.Greenwald J, Nader M, Celia H, Gruffaz C, Geoffroy V, Meyer JM, Schalk IJ, Pattus F. 2009. FpvA bound to non-cognate pyoverdines: molecular basis of siderophore recognition by an iron transporter. Mol Microbiol 72:1246–1259. doi: 10.1111/j.1365-2958.2009.06721.x. [DOI] [PubMed] [Google Scholar]
- 28.Hoegy F, Lee X, Noel S, Rognan D, Mislin GLA, Reimmann C, Schalk IJ. 2009. Stereospecificity of the siderophore pyochelin outer membrane transporters in fluorescent pseudomonads. J Biol Chem 284:14949–14957. doi: 10.1074/jbc.M900606200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol 20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 30.Jin M, Jiang Y, Sun L, Yin J, Fu H, Wu G, Gao H. 2013. Unique organizational and functional features of the cytochrome c maturation system in Shewanella oneidensis. PLoS One 8:e75610. doi: 10.1371/journal.pone.0075610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou G, Yin J, Chen H, Hua Y, Sun L, Gao H. 2013. Combined effect of loss of the caa3 oxidase and Crp regulation drives Shewanella to thrive in redox-stratified environments. ISME J 7:1752–1763. doi: 10.1038/ismej.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett BD, Redford KE, Gralnick JA. 2018. MgtE homolog FicI acts as a secondary ferrous iron importer in Shewanella oneidensis strain MR-1. Appl Environ Microbiol 84:e01245-17. doi: 10.1128/AEM.01245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S, Imlay JA. 2003. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J Bacteriol 185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt KA, Flynn JM, Naranjo B, Shikhare ID, Gralnick JA. 2010. Substrate-level phosphorylation is the primary source of energy conservation during anaerobic respiration of Shewanella oneidensis strain MR-1. J Bacteriol 192:3345–3351. doi: 10.1128/JB.00090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flynn CM, Hunt KA, Gralnick JA, Srienc F. 2012. Construction and elementary mode analysis of a metabolic model for Shewanella oneidensis MR-1. Biosystems 107:120–128. doi: 10.1016/j.biosystems.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Pinchuk GE, Geydebrekht OV, Hill EA, Reed JL, Konopka AE, Beliaev AS, Fredrickson JK. 2011. Pyruvate and lactate metabolism by Shewanella oneidensis MR-1 under fermentation, oxygen limitation, and fumarate respiration conditions. Appl Environ Microbiol 77:8234–8240. doi: 10.1128/AEM.05382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo S, Guo W, Nealson KH, Feng X, He Z. 2016. 13C Pathway analysis for the role of formate in electricity generation by Shewanella oneidensis MR-1 using lactate in microbial fuel cells. Sci Rep 6:20941. doi: 10.1038/srep20941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith RM, Martell AE. 1989. Carboxylic acids, p 299–359. Critical stability constants: second supplement. doi: 10.1007/978-1-4615-6764-6_12 Springer Science, Boston, MA. [DOI] [Google Scholar]
- 39.Perry RD, Mier I, Fetherston JD. 2007. Roles of the Yfe and Feo transporters of Yersinia pestis in iron uptake and intracellular growth. BioMetals 20:699–703. doi: 10.1007/s10534-006-9051-x. [DOI] [PubMed] [Google Scholar]
- 40.Peng ED, Wyckoff EE, Mey AR, Fisher CR, Payne SM. 2016. Nonredundant roles of iron acquisition systems in Vibrio cholerae. Infect Immun 84:511–523. doi: 10.1128/IAI.01301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sankari S, O'Brian MR. 2016. The Bradyrhizobium japonicum ferrous iron transporter FeoAB is required for ferric iron utilization in free living aerobic cells and for symbiosis. J Biol Chem 291:15653–15662. doi: 10.1074/jbc.M116.734129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kammler M, Schön C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol 175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan J, Chen Y, Zhou G, Chen H, Gao H. 2013. Investigation of roles of divalent cations in Shewanella oneidensis pellicle formation reveals unique impacts of insoluble iron. Biochim Biophys Acta 1830:5248–5257. doi: 10.1016/j.bbagen.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 44.Wilson BR, Bogdan AR, Miyazawa M, Hashimoto K, Tsuji Y. 2016. Siderophores in iron metabolism: from mechanism to therapy potential. Trends Mol Med 22:1077–1090. doi: 10.1016/j.molmed.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat Struct Biol 6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 46.Gasser V, Baco E, Cunrath O, August PS, Perraud Q, Zill N, Schleberger C, Schmidt A, Paulen A, Bumann D, Mislin GLA, Schalk IJ. 2016. Catechol siderophores repress the pyochelin pathway and activate the enterobactin pathway in Pseudomonas aeruginosa: an opportunity for siderophore–antibiotic conjugates development. Environ Microbiol 18:819–832. doi: 10.1111/1462-2920.13199. [DOI] [PubMed] [Google Scholar]
- 47.Cornelis P, Dingemans J. 2013. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol 3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ghssein G, Brutesco C, Ouerdane L, Fojcik C, Izaute A, Wang S, Hajjar C, Lobinski R, Lemaire D, Richaud P, Voulhoux R, Espaillat A, Cava F, Pignol D, Borezée-Durant E, Arnoux P. 2016. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 352:1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 49.Drechsel H, Freund S, Nicholson G, Haag H, Jung O, Zähner H, Jung G. 1993. Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. Biometals 6:185–192. doi: 10.1007/BF00205858. [DOI] [PubMed] [Google Scholar]
- 50.Meiwes J, Fiedler HP, Haag H, Zähner H, Konetschny-Rapp S, Jung G. 1990. Isolation and characterization of staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicus DSM 20459. FEMS Microbiol Lett 55:201–205. doi: 10.1111/j.1574-6968.1990.tb13863.x. [DOI] [PubMed] [Google Scholar]
- 51.Pinchuk GE, Rodionov DA, Yang C, Li X, Osterman AL, Dervyn E, Geydebrekht OV, Reed SB, Romine MF, Collart FR, Scott JH, Fredrickson JK, Beliaev AS. 2009. Genomic reconstruction of Shewanella oneidensis MR-1 metabolism reveals a previously uncharacterized machinery for lactate utilization. Proc Natl Acad Sci U S A 106:2874–2879. doi: 10.1073/pnas.0806798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin J, Mao Y, Ju L, Jin M, Sun Y, Jin S, Gao H. 2014. Distinct roles of major peptidoglycan recycling enzymes in β-lactamase production in Shewanella oneidensis. Antimicrob Agents Chemother 58:6536–6543. doi: 10.1128/AAC.03238-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bassler BL, Greenberg EP, Stevens AM. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol 179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu S, Peng Z, Cui B, Wang T, Song Y, Zhang L, Wei G, Wang Y, Shen X. 2014. FliS modulates FlgM activity by acting as a non-canonical chaperone to control late flagellar gene expression, motility and biofilm formation in Yersinia pseudotuberculosis. Environ Microbiol 16:1090–1104. doi: 10.1111/1462-2920.12222. [DOI] [PubMed] [Google Scholar]
- 55.Gao HC, Jiang X, Pogliano K, Aronson AI. 2002. The E1 beta and E2 subunits of the Bacillus subtilis pyruvate dehydrogenase complex are involved in regulation of sporulation. J Bacteriol 184:2780–2788. doi: 10.1128/JB.184.10.2780-2788.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X, Qian C, Fang H, Wen Y, Zhou J, Zhan Z, Ding R, Li O, Gao H. 2011. Paenimacrolidin, a novel macrolide antibiotic from Paenibacillus sp F6-B70 active against methicillin-resistant Staphylococcus aureus. Microb Biotechnol 4:491–502. doi: 10.1111/j.1751-7915.2010.00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo Q, Dong Y, Chen H, Gao H. 2013. Mislocalization of Rieske protein PetA predominantly accounts for the aerobic growth defect of tat mutants in Shewanella oneidensis. PLoS One 8:e62064. doi: 10.1371/journal.pone.0062064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fu H, Chen H, Wang J, Zhou G, Zhang H, Zhang L, Gao H. 2013. Crp-dependent cytochrome bd oxidase confers nitrite resistance to Shewanella oneidensis. Environ Microbiol 15:2198–2212. doi: 10.1111/1462-2920.12091. [DOI] [PubMed] [Google Scholar]
- 59.Shi M, Wan F, Mao Y, Gao H. 2015. Unraveling the mechanism for the viability deficiency of Shewanella oneidensis oxyR null mutant. J Bacteriol 197:2179–2189. doi: 10.1128/JB.00154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. 2004. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem 331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 61.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.