The ability of sporeformers to remain dormant in a desiccated state is of concern from a safety and spoilage perspective in dairy powder. Traditional culturing techniques are slow and provide little information without further investigation. We describe the identification of mesophilic sporeformers present in powders produced over 1 year, using novel shotgun metagenomic sequencing. This method allows detection and identification of possible pathogens and spoilage bacteria in parallel. Strain-level analysis and functional gene analysis, such as identification of toxin genes, were also performed. This approach has the potential to be of great value with respect to the detection of spore-forming bacteria and could allow a processor to make an informed decision surrounding process changes to reduce the risk of spore contamination.
KEYWORDS: DNA sequencing, dairy, mesophilic, metagenomics, powder, sporeformers, whey
ABSTRACT
Spoilage and pathogenic spore-forming bacteria are a major cause of concern for producers of dairy products. Traditional agar-based detection methods employed by the dairy industry have limitations with respect to their sensitivity and specificity. The aim of this study was to identify low-abundance sporeformers in samples of a powdered dairy product, whey powder, produced monthly over 1 year, using novel culture-independent shotgun metagenomics-based approaches. Although mesophilic sporeformers were the main target of this study, in one instance thermophilic sporeformers were also targeted using this culture-independent approach. For comparative purposes, mesophilic and thermophilic sporeformers were also tested for within the same sample using culture-based approaches. Ultimately, the approaches taken highlighted differences in the taxa identified due to treatment and isolation methods. Despite this, low levels of transient, mesophilic, and in some cases potentially pathogenic sporeformers were consistently detected in powder samples. Although the specific sporeformers changed from one month to the next, it was apparent that 3 groups of mesophilic sporeformers, namely, Bacillus cereus, Bacillus licheniformis/Bacillus paralicheniformis, and a third, more heterogeneous group containing Brevibacillus brevis, dominated across the 12 samples. Total thermophilic sporeformer taxonomy was considerably different from mesophilic taxonomy, as well as from the culturable thermophilic taxonomy, in the one sample analyzed by all four approaches. Ultimately, through the application of shotgun metagenomic sequencing to dairy powders, the potential for this technology to facilitate the detection of undesirable bacteria present in these food ingredients is highlighted.
IMPORTANCE The ability of sporeformers to remain dormant in a desiccated state is of concern from a safety and spoilage perspective in dairy powder. Traditional culturing techniques are slow and provide little information without further investigation. We describe the identification of mesophilic sporeformers present in powders produced over 1 year, using novel shotgun metagenomic sequencing. This method allows detection and identification of possible pathogens and spoilage bacteria in parallel. Strain-level analysis and functional gene analysis, such as identification of toxin genes, were also performed. This approach has the potential to be of great value with respect to the detection of spore-forming bacteria and could allow a processor to make an informed decision surrounding process changes to reduce the risk of spore contamination.
INTRODUCTION
Milk and resultant dairy products can become contaminated by bacteria from a number of sources, including production and processing facility contaminants. Soil, bedding, feed, feces, and teat surfaces all harbor bacteria that can transfer to raw milk (1, 2), and milking and housing practices are a potential contributor to raw milk contamination (3). Although many of the bacteria present in milk are killed by traditional processing techniques, bacterial spores can survive heat treatment and desiccation (1, 4). Furthermore, within processing facilities, microbial biofilms formed on equipment surfaces can be persistent. These are frequently resistant to cleaning, and cells, including spore-forming bacteria, from these biofilms can slough off to contaminate products (5, 6). Furthermore, spores, regardless of their origin, can withstand further processing and remain in a dormant form in powdered dairy products thereafter (7, 8).
On the basis of culture-based analyses, sporeformers identified frequently in dairy powders include representatives of the genera Bacillus, Geobacillus, Anoxybacillus, Lysinibacillus, Brevibacillus, and Paenibacillus (9, 10), as well as Clostridium (11, 12). Some of these sporeformers, including Bacillus licheniformis, Bacillus subtilis, Bacillus amyloliquefaciens, members of Bacillus cereus sensu lato (13), and Geobacillus species (14), are associated with spoilage of dairy products. Also, some members of B. cereus sensu lato have the potential to cause food poisoning. More specifically, B. cereus can cause diarrheal or emetic food poisoning. Diarrheal food poisoning is caused by enterotoxins, i.e., nonhemolytic enterotoxin (Nhe), cytotoxin K (CytK) or hemolysin BL (HBL), with symptom onset occurring 8 to 16 h after ingestion (15, 16). Emetic food poisoning is caused by the emetic toxin cereulide (Ces), which is produced by a nonribosomal peptide synthetase and results in vomiting within a few hours of ingestion (16). Toxin-producing Clostridium species, such as Clostridium botulinum and Clostridium perfringens (7), are also a concern but are less common in dairy powders than bacteria of the class Bacilli (17).
Despite safety and economic concerns, there is little legislation governing the total numbers of permissible sporeformers in dairy powders. However, processors and customers generally set strict guidelines to ensure high standards. U.S. powders destined for international customers have limits of <500 CFU · g−1 thermophilic sporeformers and <1,000 CFU · g−1 mesophilic sporeformers (18). These limits reflect the fact that traditional culture-based detection and enumeration methods for spore-forming bacteria rely on a variety of temperature treatments to differentiate between heat-resistant and highly heat-resistant sporeformers, incubation temperatures (to differentiate among psychrotolerant, mesophilic, and thermophilic sporeformers), and incubation conditions (to differentiate between aerobic and anaerobic sporeformers) (17, 18). A number of selective and chromogenic agars may also be employed to select for and/or identify pathogenic groups, such as Clostridium species and B. cereus group (19, 20). Further biochemical or molecular methods can be subsequently utilized to identify the species isolated (1).
The heavy reliance on culture-based assays has been highlighted as a concern in recent years. These methods are labor intensive, requiring many medium types, sample treatments, incubation temperatures, and conditions per sample to get an overview of the total numbers of different groups of bacteria present. These need to be followed by additional testing to identify or confirm identity of the species present and, where necessary, further analysis to determine if species present contain toxin genes (21).
Here, shotgun metagenomic DNA sequencing (22) is employed to identify bacteria present in dairy powders, as an extension of the recent application of the technology to identify pathogenic strains in a fermented dairy beverage (23) or spoilage microorganisms in cheese (24). Shotgun metagenomics can generate a considerable volume of data, enabling the detection of culturable and nonculturable microorganisms, with accurate identification to species level and sometimes to strain level. It also allows an investigation of specific gene sequences of interest, such as, for example, antimicrobial resistance or toxin-encoding genes (25, 26). However, this analysis remains expensive and poses data analysis-related difficulties due to the frequently large quantity of data generated (22). With a view to beginning to bridge the gap toward the application of this powerful technology to food quality and safety testing, this study employed shotgun metagenomics to test 12 powdered dairy samples produced monthly at the same location over one calendar year to determine the mesophilic spore content of these powders. For the purpose of comparison, one sample (from August [A]) was examined in greater depth to assess the mesophilic spore content (AM) and thermophilic spore content (AT), as well as the culturable mesophilic spore content (AMP) and culturable thermophilic spore content (ATP).
RESULTS
Shotgun metagenomics can be used to identify and determine the functional potential of low-abundance sporeformers present in dairy powders.
A total of 12 whey powder samples, produced monthly at the same location over one calendar year, were sampled upon exit from the spray drier prior to packing and collected for spore analysis. Following spore pasteurization of reconstituted powders, in which samples were incubated at 80°C for 12 min to select for spore-forming bacteria, a total of <400 CFU · g−1 mesophilic and thermophilic sporeformers was enumerated by plating whey solutions on brain heart infusion (BHI) agar and incubating at 30°C and 55°C, respectively, for 24 h. These are within previously described consumer specified limits of <500 CFU · g−1 thermophilic sporeformers and <1,000 CFU · g−1 mesophilic sporeformers (18).
To facilitate more in-depth characterization, sporeformer enrichment was performed and DNA extraction and shotgun metagenomic sequencing were completed. The enrichment approach taken addressed the risk of reads arising from sequencing of DNA from dead cells, but it had the drawback of underrepresenting spores that did not germinate under the conditions employed. The average number of reads after quality filtering and trimming per sample was 1,106,747. Kaiju, Kraken, and MetaPhlAn2 were all used to assess each package's relative ability to taxonomically assign reads, and results from all three tools are presented. Kraken assigned the greatest percentage of reads at the species level. This, coupled with the previously reported possibility of high levels of false positives resulting from Kaiju assignment (27) and the fact that MetaPhlAn2 works off only a subset of marker genes per species (28), is why Kraken was preferentially employed, with a filter threshold of 0.2 to increase precision without detrimentally impacting sensitivity. Furthermore, to reduce the possibility of false positives (27), taxa were included only if present at a minimum of 1% relative abundance in at least one sample; otherwise, reads were assigned as “others.”
B. cereus was found to be the dominant species in 7 of the 12 monthly mesophilic sporeformer-enriched samples, i.e., those from January, February, March, May, July, October, and November. Bacillus thuringiensis, a member of B. cereus sensu lato, was the next most abundant species in these samples (Fig. 1). Among the other mesophilic sporeformer-enriched samples, B. licheniformis was present as the dominant species in April and August. It was also present in the September and December samples but was not dominant. B. subtilis was dominant in the September sample but was also detected at lower relative abundances in August and December. B. paralicheniformis was the dominant species in the December sample and was also present, but not dominant, in April, August, and September (Fig. 1). The June sample was dominated by Brevibacillus brevis and Streptococcus thermophilus. S. thermophilus was also detected at lower levels in other samples, despite not being a sporeformer. All species identified as dominant were confirmed by Kaiju- and MetaPhlAn2-derived results (see Fig. S1 and S2 in the supplemental material).
FIG 1.
Relative abundance of the most abundant species (shotgun metagenomic reads assigned by Kraken with a 0.2 filter threshold and minimum of 1% relative abundance in at least one sample) in samples enriched for mesophilic sporeformers.
Spearman correlations were utilized to evaluate the population relationships in samples with taxonomic analysis and functional gene analysis. Spearman correlations are used for nonnormally distributed data that are either skewed or ordinal. Statistical analysis was performed on correlations, and asymptotic P values were generated using rcorr in the R package Hmisc and corrected with Benjamini-Hochberg corrections for multiple comparisons. Significant negative correlations (P < 0.05) between B. cereus and B. licheniformis and between B. cereus and B. paralicheniformis were noted. Significant positive correlations (P < 0.05) were seen between B. licheniformis and B. paralicheniformis, Bacteroides vulgatus and Bacteroides fragilis, B. vulgatus and others, and B. fragilis and others. As expected, many sporulation-associated functional gene groups were significantly positively correlated (P < 0.05), including those for dormancy and sporulation with those for virulence disease/defense, regulation/cell signaling, iron acquisition/metabolism, and cell wall/capsule formation (see Fig. S3 in the supplemental material).
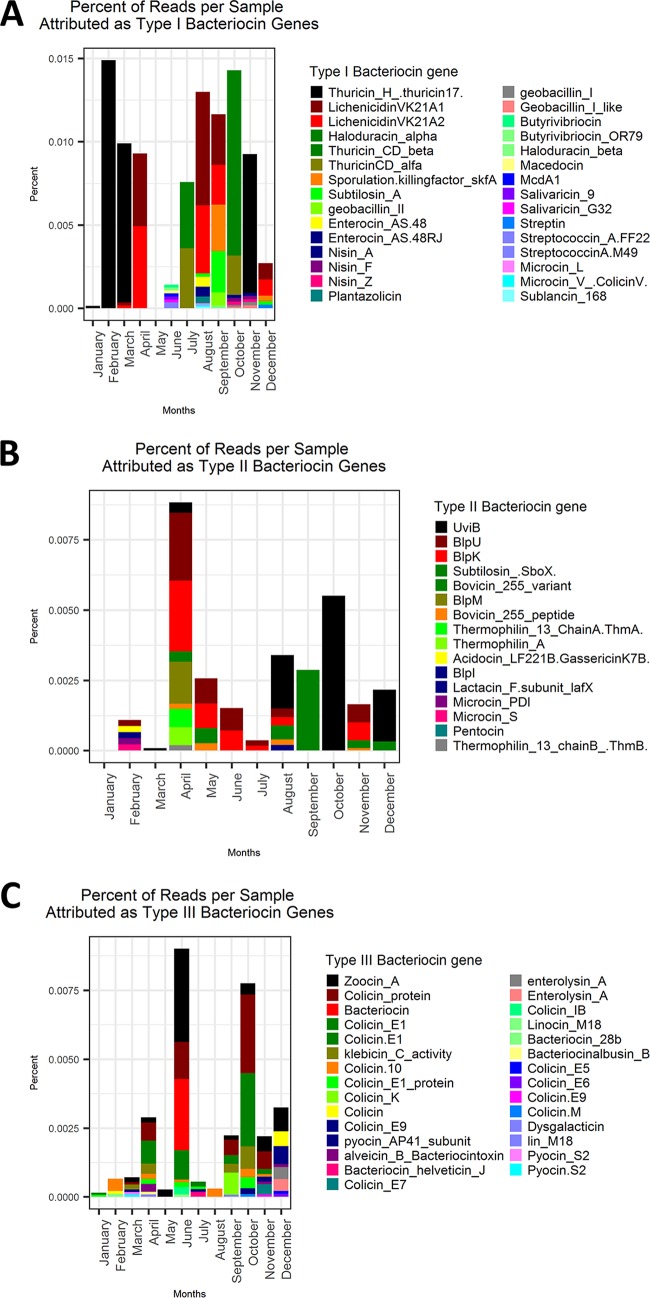
FASTA reads were aligned to the Bagel 3 bacteriocin database using DIAMOND to determine if bacteriocin production potential could be influencing current and future relationship dynamics. This showed a number of potential type I, type II, and type III bacteriocin genes in each sample, with the highest relative abundance per sample going to type I bacteriocin genes (Fig. 2). Bacteriocin genes of note included two lichenicidin peptide genes present in all samples containing B. licheniformis, genes for sporulation killing factor (skfA) and subtilosin seen in samples containing B. subtilis, and thuricin genes in samples containing B. thuringiensis (Fig. 2A).
FIG 2.
Percentage of total reads per sample attributed to bacteriocin genes found in the Bagel 3 database. (A) Percentage of reads attributed to type I bacteriocin genes. (B) Percentage of reads attributed to type II bacteriocin genes. (C) Percentage of reads attributed to type III bacteriocin genes.
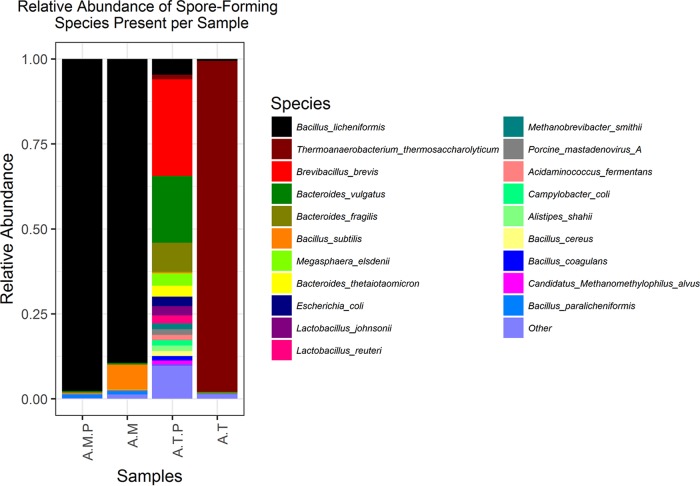
The August sample was selected for a parallel culture-based analysis and was enriched for culturable mesophilic sporeformers by culturing on BHI agar and incubating at 30°C for 48 h, following 48 h enrichment of a spore-pasteurized sample at 30°C. This was labeled August plate-cultured mesophilic sporeformer-enriched sample (AMP). An additional aliquot of the same spore-pasteurized sample was enriched for thermophilic sporeformers by incubating at 55°C for 48 h; this was labeled August thermophilic sporeformer-enriched sample (AT). Subsequently, this sample was plated on BHI agar and incubated at 55°C for 48 h. This was labeled August plate-cultured thermophilic sporeformer-enriched sample (ATP). The original August mesophilic sporeformer-enriched sample was relabeled AM for the purpose of comparison. In the case of the plate-cultured samples (AMP and ATP), colonies were scraped off, and DNA was extracted in all instances and sequenced as previously described. Comparative analysis revealed that the AMP and AM samples had very similar profiles, highlighting that plating did not bias results (Fig. 3). However, the taxonomic profiles of the AT and ATP samples were very different; i.e., the dominant species present in the AT sample was Thermoanaerobacterium thermosaccharolyticum, whereas the equivalent postplating sample (ATP) was comprised of B. brevis and Bacteroides species, as well as a variety of other species at low abundance (Fig. 3).
FIG 3.
Relative abundance of the most abundant species (assigned by Kraken with a 0.2 filter threshold and minimum 1% relative abundance in at least one sample) in the August sample subjected to different enrichment temperatures and conditions.
Beta diversity analysis highlights that samples clustered according to the dominant species present.
Bray-Curtis beta diversity analysis of mesophilic sporeformer reads showed that the 12 samples clustered into 3 distinct groups (individual data points within each group being colocated; Fig. 4A). One cluster consisted of samples that contained B. cereus (i.e., January, February, March, May, July, October, and November); a second cluster consisted of samples from the months April, August, September, and December, which all contained a high relative abundance of B. licheniformis; and a third cluster corresponded to the June sample, which had a high relative abundance of B. brevis (Fig. 4A).
FIG 4.
Alpha and beta diversity metrics for mesophilic sporeformer-enriched samples and August samples subject to different incubation temperatures and conditions. (A) Bray-Curtis weighted classical multidimensional scaling (MDS) of mesophilic sporeformer-enriched samples. (B) Simpson alpha diversity of mesophilic sporeformer-enriched samples. (C) Bray-Curtis weighted classical multidimensional scaling (MDS) of August samples. (D) Simpson alpha diversity of August samples under different conditions.
Alpha diversity analysis did not reveal any notable pattern other than the observation that the June, September, and December samples had the highest diversities (Fig. 4B). As might be expected from taxonomic results, beta diversity analysis of the August sample, which was enriched in a variety of ways, showed that the AM and AMP samples were least dissimilar, whereas the AT and ATP samples differed from each other and from the AM and AMP samples (Fig. 4C). Among these samples, alpha diversity in the ATP sample was higher than those of all others (Fig. 4D).
Toxin gene analysis revealed the presence of potentially toxigenic B. cereus.
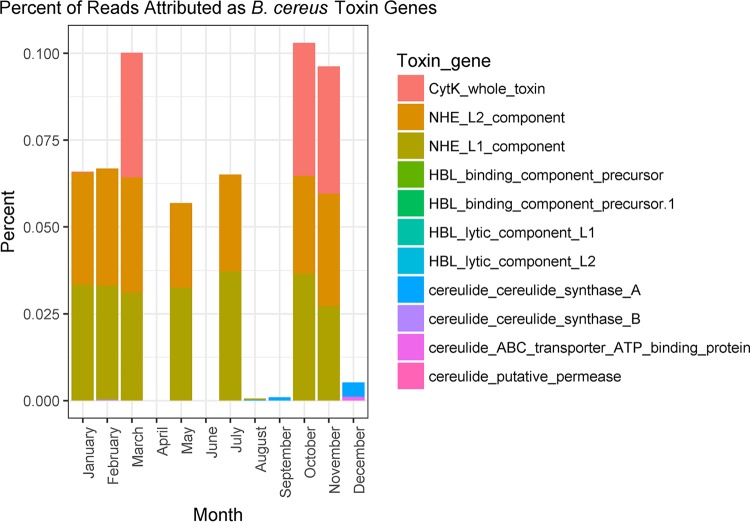
Further analysis was performed to determine if the B. cereus strains detected in some samples might be capable of causing emetic and enterotoxic food poisoning. For this purpose, toxin genes were aligned to sequence reads and verified by aligning them to assembled contigs. The genes used and the genome sequences from which they originated are shown in Table 1. Of the 11 toxin genes screened for among all 12 mesophilic enriched samples, the two nonhemolytic enterotoxin (Nhe) genes nhe L1 and nhe L2 were detected in all 7 samples previously shown to contain high relative abundances of B. cereus. The cytotoxin K-encoding gene was detected in 3 samples (from March, October, and November). The presence of toxin genes in the sequence reads was verified by alignment and visualization using Mauve. Toxin gene sequences were aligned to sample contigs, which were assembled using the IDBA-UD algorithm (see Fig. S4 in the supplemental material). Among samples in which all 3 toxin genes were present, toxin genes accounted for close to 0.1% of reads, and when just the two nhe genes were present, they accounted for close to 0.06% of reads (Fig. 5).
TABLE 1.
Toxin genes aligned to sequence reads
| Toxin | Toxin gene | Gene identifier | Genome | Description |
|---|---|---|---|---|
| Cytotoxin K | cytK | gi|30018278:c1092459-1091449 | Bacillus cereus ATCC 14579 chromosome | BC1110 (cytotoxin K) |
| Nonhemolytic enterotoxin | nhe | gi|30018278:1765248-1766408 | Bacillus cereus ATCC 14579 chromosome | BC1809 (nonhemolytic enterotoxin lytic component L2) |
| Nonhemolytic enterotoxin | nhe | gi|30018278:1766446-1767654 | Bacillus cereus ATCC 14579 chromosome | BC1810 (nonhemolytic enterotoxin lytic component L1) |
| Hemolysin BL | hbl | gi|30018278:c3062258-3060858 | Bacillus cereus ATCC 14579 chromosome | BC3101 (hemolysin BL binding component precursor) |
| Hemolysin BL | hbl | gi|30018278:c3063761-3062634 | Bacillus cereus ATCC 14579 chromosome | BC3102 (hemolysin BL binding component precursor) |
| Hemolysin BL | hbl | gi|30018278:c3065018-3063798 | Bacillus cereus ATCC 14579 chromosome | BC3103 (hemolysin BL lytic component L1) |
| Hemolysin BL | hbl | gi|30018278:c3066399-3065080 | Bacillus cereus ATCC 14579 chromosome | BC3104 (hemolysin BL lytic component L2) |
| Cereulide | cesA | gi|190015498:c35141-25017 | Bacillus cereus strain AH187 plasmid pCER270 | CesA (cereulide synthetase A) |
| Cereulide | cesB | gi|190015498:c25003-16958 | Bacillus cereus strain AH187 plasmid pCER270 | CesB (cereulide synthethase B) |
| Cereulide | cesC | gi|190015498:c16813-15917 | Bacillus cereus strain AH187 plasmid pCER270 | CesC (ABC transporter ATP binding protein) |
| Cereulide | cesD | gi|190015498:c15900-15094 | Bacillus cereus strain AH187 plasmid pCER270 | CesD (putative permease) |
FIG 5.
Percentage of total reads per sample attributed to B. cereus toxin genes.
Strain-level analysis revealed the absence of evidence for persistent contamination.
As the samples that contained high relative abundances of B. cereus appeared to have different toxin profiles, it was decided to carry out PanPhlAn-based strain analysis to more accurately determine if one strain was dominant across these samples, potentially indicating strain persistence in the processing facility or transient colonization by different strains. Sequence reads for all samples were aligned to a pangenome created from 32 complete B. cereus genomes downloaded from the NCBI database. This analysis established that the presence/absence of B. cereus was consistent with that in previous Kraken analysis. PanPhlAn showed that the 7 samples containing B. cereus contained reads that clustered with 5 strains of B. cereus by Euclidean distance, as visualized using GraPhlAn (Fig. 6). The percentage match from sequence reads to the 32 genomes used in the pangenome was also examined (Table 2). Overall, as shown in Table 2, none of the strains identified in the 7 samples are a complete match to any of the 32 strains with which they were compared. Similarly, none of the strains identified in the 7 samples appear to be exactly the same, with the January and February samples being most alike (Fig. 6).
FIG 6.
GraPhlAn visualization of PanPhlAn clustering of 7 samples containing B. cereus with representative strains of the pangenome. The 7 samples contain reads that cluster with at least 5 distinct strains.
TABLE 2.
Percentage match of B. cereus strains present in samples of 32 reference strains
| ENA assembly no. | No. of genes | Percentage match to the 32 genomes used in the pangenome |
||||||
|---|---|---|---|---|---|---|---|---|
| January | February | March | May | July | October | November | ||
| GCA_000007825 | 5,554 | 14.3 | 13.9 | 13.3 | 15.9 | 14.4 | 13.4 | 13 |
| GCA_000008005 | 5,338 | 52 | 53.3 | 89.4 | 53 | 52.4 | 69.2 | 85.8 |
| GCA_000011625 | 5,868 | 46.9 | 47.5 | 35.5 | 42.4 | 44.5 | 38.5 | 34.9 |
| GCA_000013065 | 5,521 | 67.6 | 68.6 | 49.9 | 59.2 | 76.7 | 51.7 | 48.8 |
| GCA_000021205 | 5,366 | 14.8 | 14.7 | 14.4 | 16.6 | 15.4 | 14.5 | 14.1 |
| GCA_000021225 | 5,637 | 65.8 | 66.1 | 50.1 | 59.6 | 79.3 | 51.7 | 49.1 |
| GCA_000021305 | 5,749 | 12.8 | 12.5 | 12.4 | 14.1 | 13.2 | 12.5 | 12.2 |
| GCA_000021785 | 5,595 | 47.2 | 47.4 | 36.2 | 43.7 | 43.5 | 38.3 | 35.7 |
| GCA_000022505 | 5,515 | 46 | 46.5 | 37.2 | 42 | 43.2 | 39.5 | 36.6 |
| GCA_000143605 | 5,586 | 47.5 | 48.3 | 36.4 | 43.4 | 44.5 | 38.7 | 35.8 |
| GCA_000239195 | 5,374 | 46.7 | 46.9 | 36.8 | 42.1 | 43.9 | 39.6 | 36.2 |
| GCA_000283675 | 5,575 | 65.9 | 66.7 | 50.2 | 60.1 | 79.9 | 51.2 | 49.1 |
| GCA_000292415 | 5,427 | 53.8 | 55.5 | 69 | 53.4 | 54.5 | 78.5 | 67.2 |
| GCA_000635895 | 5,625 | 13.8 | 13.9 | 13.4 | 15.3 | 14.2 | 13.6 | 13.1 |
| GCA_000724585 | 5,583 | 37.2 | 37 | 40.7 | 43.5 | 37 | 36.5 | 39.4 |
| GCA_000789315 | 5,781 | 45.5 | 45.9 | 41.5 | 54.3 | 47.2 | 42.7 | 40.6 |
| GCA_000832385 | 5,721 | 45 | 47.4 | 37 | 41.7 | 42.1 | 38.7 | 36.4 |
| GCA_000832405 | 5,522 | 45.8 | 46.3 | 36.9 | 41.7 | 43 | 39.1 | 36.2 |
| GCA_000832525 | 5,653 | 47 | 47.2 | 42.7 | 55.9 | 48.5 | 44.2 | 41.7 |
| GCA_000832765 | 5,708 | 46.6 | 46.9 | 36.1 | 43 | 46.8 | 41.4 | 35.5 |
| GCA_000832805 | 5,708 | 46.4 | 47 | 42.1 | 55.4 | 48.1 | 43.6 | 41.1 |
| GCA_000832845 | 5,332 | 51.7 | 52.1 | 44.3 | 61.4 | 53 | 46.4 | 43.3 |
| GCA_000832865 | 6,126 | 41.5 | 42.4 | 33.8 | 38.3 | 39.1 | 35.6 | 33.2 |
| GCA_000833045 | 5,885 | 46.6 | 47.2 | 35.3 | 42.1 | 44.3 | 38.2 | 34.7 |
| GCA_000835185 | 5,707 | 46.5 | 46.8 | 36.1 | 43.1 | 46.7 | 41.4 | 35.6 |
| GCA_000978375 | 5,312 | 14.9 | 14.9 | 14.5 | 16.7 | 15.2 | 14.5 | 14.2 |
| GCA_001277915 | 5,369 | 14.1 | 13.9 | 13.5 | 15.9 | 14.2 | 13.7 | 13.2 |
| GCA_001518875 | 5,669 | 14.3 | 14.4 | 13.4 | 16.1 | 15 | 13.6 | 13.2 |
| GCA_001635915 | 5,948 | 13.1 | 13.4 | 12.8 | 14.6 | 13.1 | 12.5 | 12.5 |
| GCA_001635955 | 5,945 | 13.2 | 13.5 | 12.8 | 14.6 | 13.1 | 12.5 | 12.6 |
| GCA_001635995 | 5,981 | 14 | 14.1 | 13.2 | 15.3 | 14.2 | 13.2 | 13.1 |
| GCA_001721145 | 5,362 | 15.1 | 15.1 | 14.5 | 17.1 | 15.5 | 14.6 | 14.2 |
DISCUSSION
Traditionally, the detection and identification of bacterial sporeformers have involved culturing under different temperatures and conditions (17, 18), together with the use of selective agars to identify pathogenic species (19, 20), followed by further biochemical or molecular approaches for confirmation (1). This study highlights the ability of next-generation shotgun sequencing of enriched samples to identify low-level persistent or transient spore contamination in a dairy powder. Although many of the species identified are similar to those identified in previous studies using traditional detection methods (17), the approach taken here has the potential to rapidly identify these species while allowing strain-level analysis and, as a result, the tracking of persistent microbes in products. It highlights that extremely low levels of potentially pathogenic bacteria can be present, although in this instance, these bacteria are unlikely to be from persistent contamination in the processing facility but rather are thought to represent a transient low-level contamination. Due to the sensitivity of this approach, new guidelines and standards may need to be introduced to ensure that the risks associated with detection of low-level contamination of B. cereus in powders are adequately reflected.
The combined approach of high-temperature treatment coupled with shotgun sequencing employed in this study was selected for a number of reasons. Prior to high-temperature activation, the spore-forming bacteria present in dairy powders are likely to be in a spore form, from which DNA extraction can be difficult (29). Furthermore, in the absence of enrichment, the possibly high level of DNA remaining from deceased bacteria and the presence of host bovine cells in the powders could also be an issue, as DNA from these sources will also be sequenced when untargeted shotgun metagenomic sequencing is employed (22), thereby necessitating the use of additional steps and expensive kits and reagents to deplete DNA from these other sources (30–34). DNA amplification would likely be necessary thereafter due to the low yield of DNA from the low level of sporeformers present in samples. Amplification methods add extra bias, costs, and potential for contamination (34–36). A more targeted, economical approach was used to focus on sporeformers that could potentially germinate in rehydrated powder if given favorable temperatures, without the extra bias and cost of kits. It should be noted that low-speed centrifugation utilized in the approach from which solids were discarded prior to DNA extraction could result in loss of some spore-forming bacteria that have a high affinity for denatured milk protein solids.
This targeted, culture-independent approach was compared with corresponding culture-dependent approaches, using the August sample as a representative test case. The thermophilic sporeformer-enriched August sample (AT) from which DNA was extracted was also subject to culturing on BHI agar at thermophilic temperatures (ATP) before pooling colonies and extracting DNA. DNA from both AT and ATP samples was sequenced (see Fig. S5 in the supplemental material) and compared.
The results were different in that the dominant species identified following enrichment (i.e., without culturing on BHI agar) was T. thermosaccharolyticum, while this species was not detected in the cultured sample, potentially showing bias due to the agar and conditions used. T. thermosaccharolyticum has previously been detected in dried vegetables (37), and more recently, Thermoanaerobacterium spp. have been detected in the core microbiome of raw milk (38). T. thermosaccharolyticum is a thermophilic, anaerobic sporeformer and was previously classified as a member of the Clostridium genus, although it was subsequently reclassified (39). It is a known canned-food spoilage organism (40) that produces hydrogen and causes swelling in canned foods. This ability to efficiently produce hydrogen makes it a potentially important organism for sustainable biohydrogen production (41, 42). The previous nondetection of the species in dairy powders may relate to an inability to grow on the agar substrates conventionally employed by the dairy industry.
A decision was made to further investigate the potential B. cereus sequences identified to eliminate the potential that the taxonomic classifier was misassigning other members of B. cereus sensu lato and B. cereus sensu stricto, as members of this group are notoriously difficult to differentiate (43). Toxin profiling and PanPhlAn analysis confirmed that the strains had the potential to be pathogenic, while also highlighting differences between them. All except one of the samples that contained B. cereus showed complete alignment with both Nhe toxin-encoding genes. The exception was the February sample, from which one-half of the nhe L1 gene was absent. This may be due to misassembly or poor coverage of the genome, or it may reflect a natural mutation in this strain (Fig. S4). It should be noted that alignment to toxin genes infers potential ability to produce toxins but does not conclusively indicate functional toxin presence. Also, a total of 105 to 109 CFU of toxin-producing B. cereus is needed to cause food poisoning (15). The highly sensitive, qualitative nature of shotgun metagenomic analysis suggests that further steps will need to be taken to determine the risk associated with the detection of these and other toxin genes in food samples. PanPhlAn analysis showed that the 7 samples containing B. cereus contained reads that clustered with at least 5 strains of B. cereus by Euclidean distance, suggesting that the strains identified in the samples are different and not as a result of persistent contamination in the powder production facility or elsewhere.
The negative correlation between B. cereus and B. licheniformis/B. paralicheniformis raises the possibility of competition to determine the dominant species and the possible inhibition of the other species. The potential bacteriocin genes detected in all samples may impact the relationship dynamics observed and may have an antagonistic effect on some species currently or in future food products. Lichenicidin is a two-peptide lantibiotic previously shown to be active against pathogenic Gram-positive bacteria, including B. cereus (44), and we noted alignment to two peptides associated with lichenicidin in all four samples containing B. licheniformis. However, potential identification of two peptide genes does not suggest the presence of the whole bacteriocin gene cluster or infer correct posttranslational modifications to produce a functional bacteriocin. Positive correlations among virulence, disease, defense, dormancy, and sporulation highlight the need to be cautious of sporeformers in food products. A greater understanding of their relationships will aid the prevention of spoilage and pathogenic species that cause concern in food processing.
Disadvantages that need to be overcome in order to allow for the routine use of the sequencing technologies employed in this study primarily relate to the cost of analysis, which is currently too expensive for large-scale routine use. Additionally, there are challenges relating to assembly of genomes from shotgun metagenomic sequencing (22) and difficulties arising from insufficient accuracies associated, to different extents, with taxonomic classifiers (27). There are some solutions emerging, whereby new lower-cost, rapid sequencers are arriving on the market, with MinION (45) leading the way toward quick portable detection systems for microorganisms. Through the generation of more reference genome sequences and good-quality shotgun metagenomic sequencing, reference databases and the accuracy of results will improve.
This study highlights monthly diverging contamination patterns in whey powder production, which converged into 3 distinct mesophilic sporeformer population groups from 12 powder samples produced in the same production facility, and shows that the way in which the powders are treated postproduction (namely, incubation temperature postreconstitution) influences which bacteria germinate and become dominant. Shotgun metagenomics is a useful tool to delve deeper into the understanding of sporeformers and their relationships in food processing, although it brings with it its own set of caveats and need for guidelines for use and interpretation of results.
MATERIALS AND METHODS
Sample preparation and enrichment.
Twelve (i.e., one per month for a calendar year) 5-g whey powder samples from a single supplier were suspended aseptically at a concentration of 10% (wt/vol) in sterile 1/4-strength Ringer's solution (Sigma-Aldrich). Each reconstituted sample was incubated at 80°C for 12 min to select for spore-forming bacteria, as previously described (3, 17). An aliquot was then plated onto brain heart infusion (BHI) agar before incubation at mesophilic (30°C) and thermophilic (55°C) sporeformer-suitable temperatures, and CFU · g−1 values were calculated.
Following spore selection, in which 10% (wt/vol) reconstituted samples were incubated at 80°C for 12 min, reconstituted samples were enriched in a manner consistent with that previously employed to select for low-abundance mesophilic sporeformers, but with incubation at 30°C for 48 h as opposed to at 32°C, as previously documented (3, 17). This is shown in Fig. S5 in the supplemental material.
To facilitate an investigation into the extent to which plating altered the identity of the populations of sporeformers detected, powder sourced in August (A) was treated as described above but with incubation at 55°C to select for thermophilic sporeformers (AT). Both of the enriched August samples were plated onto BHI agar before incubation at the corresponding mesophilic (30°C; AMP) and thermophilic (55°C; ATP) temperatures for 48 h (Fig. S5). DNA was extracted from the surface of these agar plates as described below.
DNA extraction and library preparation.
A total of 50 ml of samples that were reconstituted, heat-treated, and enriched by incubation for 48 h at 30°C were centrifuged at 900 × g for 20 min. The resultant pellet was discarded, and the supernatant was centrifuged at 4,500 × g for 20 min. Pellets were washed in 1 ml of sterile 1/4-strength Ringer's solution and centrifuged at 13,000 × g for 2 min. Washing was repeated twice more, centrifuging for 1 min each time at 13,000 × g. DNA was extracted from the resultant pellet as described below. For DNA extraction from a combination of all colonies on the surface of agar plates, 5 ml of 1/4-strength Ringer's solution was added and colonies were washed off using a sterile Lazy-L spreader (Sigma-Aldrich). An aliquot of 4 ml of the resultant fluid was removed and centrifuged at 4,500 × g for 20 min. The resultant pellet was suspended in 1 ml of 1/4-strength Ringer's solution and centrifuged at 13,000 × g for 2 min. The pellet was washed twice more in 1 ml of Ringer's solution and centrifuged at 13,000 × g. Before the final centrifugation, 200 μl was removed from the mix to a sterile tube. This was centrifuged at 13,000 × g for 1 min. This pellet corresponded to 0.8 ml of culture from the original 4 ml.
All pellets were resuspended in 150 μl of 45 mg · ml−1 lysozyme and incubated at 37°C for 30 min. Samples were centrifuged at 13,000 × g for 1 min, and the supernatant was removed. From this point, the MoBio PowerFood DNA isolation kit protocol was followed, including the use of the alternative lysis step for difficult-to-lyse cells. DNA was eluted in 60 μl of 10 mM Tris-HCl and stored at −20°C. DNA was quantified and quality checked using the Qubit double-stranded DNA (dsDNA) high sensitivity assay kit (Bio-Sciences, Dublin, Ireland) and by visualizing on a 1% (wt/vol) agarose gel.
Samples were prepared for metagenomic sequencing according to Illumina Nextera XT library preparation kit guidelines and sequenced on an Illumina MiSeq sequencing platform at the Teagasc DNA Sequencing Facility with a 2 × 250 V2 kit, using standard Illumina sequencing protocols.
Bioinformatics pipeline.
Raw metagenomic shotgun reads were checked for the presence of human and bovine reads filtered on the presence of quality and quantity and then trimmed to 170 bp with a combination of Picard tools (http://broadinstitute.github.io/picard/) and SAMtools (46). Kraken with a filter threshold of 0.2 (47) and SUPER-FOCUS (48) were used to determine microbial composition to species level and biological functions, respectively. MetaPhlAn2 (28) and Kaiju (49) were also utilized to determine microbial composition for comparison to Kraken results. Eleven B. cereus toxin-associated genes (Table 1) were downloaded from the NCBI database and aligned to sequence reads using Bowtie2 (50). Metagenomes were assembled into contigs using IDBA-UD (51), and toxin genes were aligned to these using Bowtie2 (50). Toxin genes were also aligned to contigs using Mauve version 20150226 (52)—more specifically, progressiveMauve (53)—using default parameters (default seed weight, determine locally collinear blocks [LCBs], full alignment, iterative refinement, min LCB weight = default, sum-of-pairs LCB scoring). PanPhlAn (54) was utilized to determine B. cereus strain profiles in each sample. A total of 32 complete B. cereus genome sequences were downloaded from the NCBI database and a pangenome was generated and compared to all samples. PanPhlAn outputs were visualized using GraPhlAn (55). Spearman correlations with Benjamini-Hochberg corrections for multiple comparisons were made in R using the Hmisc package and visualized using corrplot. Diversity was analyzed using the R vegan package, and data visualization was performed using ggplot2. Sequence reads were aligned to the Bagel 3 bacteriocin database (56) using DIAMOND (57) to determine the bacteriocin potential of the bacteria identified.
Accession number(s).
Sequence data have been deposited in the European Nucleotide Archive (ENA) under the study accession number PRJEB24853.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the Department of Agriculture, Food and the Marine (DAFM) under the FIRM project SACCP, reference number 14/F/883. Research in the Cotter lab is also funded by the SFI award “Obesiobiotics” (11/P1/1137).
Thanks to Orla O'Sullivan, Fiona Crispie, and Laura Finnegan of the Food Bioscience Department in Teagasc Moorepark.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01305-18.
REFERENCES
- 1.Gleeson D, O'Connell A, Jordan K. 2013. Review of potential sources and control of thermoduric bacteria in bulk-tank milk. Irish J Agric Food Res 52:217–227. [Google Scholar]
- 2.Doyle CJ, Gleeson D, O'Toole PW, Cotter PD. 2017. Impacts of seasonal housing and teat preparation on raw milk microbiota: a high-throughput sequencing study. Appl Environ Microbiol 83:e02694-. doi: 10.1128/AEM.02694-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller RA, Kent DJ, Boor KJ, Martin NH, Wiedmann M. 2015. Different management practices are associated with mesophilic and thermophilic spore levels in bulk tank raw milk. J Dairy Sci 98:4338–4351. doi: 10.3168/jds.2015-9406. [DOI] [PubMed] [Google Scholar]
- 4.Setlow P. 2003. Spore germination. Curr Opin Microbiol 6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Rodríguez F, Valero A, Carrasco E, García RM, Zurera G. 2008. Understanding and modelling bacterial transfer to foods: a review. Trends Food Sci Technol 19:131–144. doi: 10.1016/j.tifs.2007.08.003. [DOI] [Google Scholar]
- 6.Faille C, Benezech T, Midelet-Bourdin G, Lequette Y, Clarisse M, Ronse G, Ronse A, Slomianny C. 2014. Sporulation of Bacillus spp. within biofilms: a potential source of contamination in food processing environments. Food Microbiol 40:64–74. doi: 10.1016/j.fm.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Doyle CJ, Gleeson D, Jordan K, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2015. Anaerobic sporeformers and their significance with respect to milk and dairy products. Int J Food Microbiol 197:77–87. doi: 10.1016/j.ijfoodmicro.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Gopal N, Hill C, Ross PR, Beresford TP, Fenelon MA, Cotter PD. 2015. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front Microbiol 6:1418. doi: 10.3389/fmicb.2015.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ronimus RS, Parker LE, Turner N, Poudel S, Ruckert A, Morgan HW. 2003. A RAPD-based comparison of thermophilic bacilli from milk powders. Int J Food Microbiol 85:45–61. doi: 10.1016/S0168-1605(02)00480-4. [DOI] [PubMed] [Google Scholar]
- 10.Sadiq FA, Li Y, Liu T, Flint S, Zhang G, He G. 2016. A RAPD based study revealing a previously unreported wide range of mesophilic and thermophilic spore formers associated with milk powders in China. Int J Food Microbiol 217:200–208. doi: 10.1016/j.ijfoodmicro.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 11.Barash JR, Hsia JK, Arnon SS. 2010. Presence of soil-dwelling clostridia in commercial powdered infant formulas. J Pediatr 156:402–408. doi: 10.1016/j.jpeds.2009.09.072. [DOI] [PubMed] [Google Scholar]
- 12.Buehner KP, Anand S, Djira GD. 2015. Prevalence of thermoduric bacteria and spores in nonfat dry milk powders of Midwest origin. J Dairy Sci 98:2861–2866. doi: 10.3168/jds.2014-8822. [DOI] [PubMed] [Google Scholar]
- 13.De Jonghe V, Coorevits A, De Block J, Van Coillie E, Grijspeerdt K, Herman L, De Vos P, Heyndrickx M. 2010. Toxinogenic and spoilage potential of aerobic spore-formers isolated from raw milk. Int J Food Microbiol 136:318–325. doi: 10.1016/j.ijfoodmicro.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Seale RB, Dhakal R, Chauhan K, Craven HM, Deeth HC, Pillidge CJ, Powell IB, Turner MS. 2012. Genotyping of present-day and historical Geobacillus species isolates from milk powders by high-resolution melt analysis of multiple variable-number tandem-repeat loci. Appl Environ Microbiol 78:7090–7097. doi: 10.1128/AEM.01817-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granum PE, Lund T. 1997. Bacillus cereus and its food poisoning toxins. FEMS Microbiol Lett 157:223–228. doi: 10.1111/j.1574-6968.1997.tb12776.x. [DOI] [PubMed] [Google Scholar]
- 16.Ehling-Schulz M, Guinebretiere MH, Monthan A, Berge O, Fricker M, Svensson B. 2006. Toxin gene profiling of enterotoxic and emetic Bacillus cereus. FEMS Microbiol Lett 260:232–240. doi: 10.1111/j.1574-6968.2006.00320.x. [DOI] [PubMed] [Google Scholar]
- 17.Miller RA, Kent DJ, Watterson MJ, Boor KJ, Martin NH, Wiedmann M. 2015. Spore populations among bulk tank raw milk and dairy powders are significantly different. J Dairy Sci 98:8492–8504. doi: 10.3168/jds.2015-9943. [DOI] [PubMed] [Google Scholar]
- 18.Watterson MJ, Kent DJ, Boor KJ, Wiedmann M, Martin NH. 2014. Evaluation of dairy powder products implicates thermophilic sporeformers as the primary organisms of interest. J Dairy Sci 97:2487–2497. doi: 10.3168/jds.2013-7363. [DOI] [PubMed] [Google Scholar]
- 19.Weenk GH, van den Brink JA, Struijk CB, Mossel DAA. 1995. Modified methods for the enumeration of spores of mesophilic Clostridium species in dried foods. Int J Food Microbiol 27:185–200. doi: 10.1016/0168-1605(94)00164-2. [DOI] [PubMed] [Google Scholar]
- 20.Tallent SM, Kotewicz KM, Strain EA, Bennett RW. 2012. Efficient isolation and identification of Bacillus cereus group. J AOAC Int 95:446–451. doi: 10.5740/jaoacint.11-251. [DOI] [PubMed] [Google Scholar]
- 21.Wehrle E, Moravek M, Dietrich R, Burk C, Didier A, Martlbauer E. 2009. Comparison of multiplex PCR, enzyme immunoassay and cell culture methods for the detection of enterotoxinogenic Bacillus cereus. J Microbiol Methods 78:265–270. doi: 10.1016/j.mimet.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Sharpton TJ. 2014. An introduction to the analysis of shotgun metagenomic data. Front Plant Sci 5:209. doi: 10.3389/fpls.2014.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh AM, Crispie F, Daari K, O'Sullivan O, Martin JC, Arthur CT, Claesson MJ, Scott KP, Cotter PD. 2017. Strain-level metagenomic analysis of the fermented dairy beverage nunu highlights potential food safety risks. Appl Environ Microbiol 83:e01144-. doi: 10.1128/AEM.01144-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quigley L, O'Sullivan DJ, Daly D, O'Sullivan O, Burdikova Z, Vana R, Beresford TP, Ross RP, Fitzgerald GF, McSweeney PL, Giblin L, Sheehan JJ, Cotter PD. 2016. Thermus and the pink discoloration defect in cheese. mSystems 1:e00023-. doi: 10.1128/mSystems.00023-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DG. 2014. Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front Microbiol 5:648. doi: 10.3389/fmicb.2014.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leonard SR, Mammel MK, Lacher DW, Elkins CA. 2015. Application of metagenomic sequencing to food safety: detection of Shiga toxin-producing Escherichia coli on fresh bagged spinach. Appl Environ Microbiol 81:8183–8191. doi: 10.1128/AEM.02601-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piro VC, Matschkowski M, Renard BY. 2017. MetaMeta: integrating metagenome analysis tools to improve taxonomic profiling. Microbiome 1:101. doi: 10.1186/s40168-017-0318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 29.Wielinga PR, de Heer L, de Groot A, Hamidjaja RA, Bruggeman G, Jordan K, van Rotterdam BJ. 2011. Evaluation of DNA extraction methods for Bacillus anthracis spores spiked to food and feed matrices at biosafety level 3 conditions. Int J Food Microbiol 150:122–127. doi: 10.1016/j.ijfoodmicro.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 30.Rudi K, Moen B, Dromtorp SM, Holck AL. 2005. Use of ethidium monoazide and PCR in combination for quantification of viable and dead cells in complex samples. Appl Environ Microbiol 71:1018–1024. doi: 10.1128/AEM.71.2.1018-1024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forghani F, Langaee T, Eskandari M, Seo KH, Chung MJ, Oh DH. 2015. Rapid detection of viable Bacillus cereus emetic and enterotoxic strains in food by coupling propidium monoazide and multiplex PCR (PMA-mPCR). Food Control 55:151–157. doi: 10.1016/j.foodcont.2015.02.049. [DOI] [Google Scholar]
- 32.Feehery GR, Yigit E, Oyola SO, Langhorst BW, Schmidt VT, Stewart FJ, Dimalanta ET, Amaral-Zettler LA, Davis T, Quail MA, Pradhan S. 2013. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLoS One 8:e76096. doi: 10.1371/journal.pone.0076096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokouchi H, Fukuoka Y, Mukoyama D, Calugay R, Takeyama H, Matsunaga T. 2006. Whole-metagenome amplification of a microbial community associated with scleractinian coral by multiple displacement amplification using phi29 polymerase. Environ Microbiol 8:1155–1163. doi: 10.1111/j.1462-2920.2006.01005.x. [DOI] [PubMed] [Google Scholar]
- 34.Binga EK, Lasken RS, Neufeld JD. 2008. Something from (almost) nothing: the impact of multiple displacement amplification on microbial ecology. ISME J 2:233–241. doi: 10.1038/ismej.2008.10. [DOI] [PubMed] [Google Scholar]
- 35.Ahsanuddin S, Afshinnekoo E, Gandara J, Hakyemezoglu M, Bezdan D, Minot S, Greenfield N, Mason CE. 2017. Assessment of REPLI-g multiple displacement whole genome amplification (WGA) techniques for metagenomic applications. J Biomol Tech 28:46–55. doi: 10.7171/jbt.17-2801-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thoendel M, Jeraldo P, Greenwood-Quaintance KE, Yao J, Chia N, Hanssen AD, Abdel MP, Patel R. 2017. Impact of contaminating DNA in whole genome amplification kits used for metagenomic shotgun sequencing for infection diagnosis. J Clin Microbiol 55:1789–1801. doi: 10.1128/JCM.02402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postollec F, Mathot AG, Bernard M, Divanac'h ML, Pavan S, Sohier D. 2012. Tracking spore-forming bacteria in food: from natural biodiversity to selection by processes. Int J Food Microbiol 158:1–8. doi: 10.1016/j.ijfoodmicro.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigues MX, Lima SF, Canniatti-Brazaca SG, Bicalho RC. 2017. The microbiome of bulk tank milk: characterization and associations with somatic cell count and bacterial count. J Dairy Sci 100:2536–2552. doi: 10.3168/jds.2016-11540. [DOI] [PubMed] [Google Scholar]
- 39.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 40.André S, Zuber F, Remize F. 2013. Thermophilic spore-forming bacteria isolated from spoiled canned food and their heat resistance. Results of a French ten-year survey. Int J Food Microbiol 165:134–143. doi: 10.1016/j.ijfoodmicro.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 41.O-Thong S, Prasertsan P, Karakashev D, Angelidaki I. 2008. Thermophilic fermentative hydrogen production by the newly isolated Thermoanaerobacterium thermosaccharolyticum PSU-2. Int J Hydrogen Energy 33:1204–1214. doi: 10.1016/j.ijhydene.2007.12.015. [DOI] [Google Scholar]
- 42.Mtimet N, Guegan S, Durand L, Mathot AG, Venaille L, Leguerinel I, Coroller L, Couvert O. 2016. Effect of pH on Thermoanaerobacterium thermosaccharolyticum DSM 571 growth, spore heat resistance and recovery. Food Microbiol 55:64–72. doi: 10.1016/j.fm.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Lai Q, Goker M, Meier-Kolthoff JP, Wang M, Sun Y, Wang L, Shao Z. 2015. Genomic insights into the taxonomic status of the Bacillus cereus group. Sci Rep 5:14082. doi: 10.1038/srep14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Begley M, Cotter PD, Hill C, Ross RP. 2009. Identification of a novel two-peptide lantibiotic, lichenicidin, following rational genome mining for LanM proteins. Appl Environ Microbiol 75:5451–5460. doi: 10.1128/AEM.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown BL, Watson M, Minot SS, Rivera MC, Franklin RB. 2017. MinION™ nanopore sequencing of environmental metagenomes: a synthetic approach. Gigascience 6(3):1–10. doi: 10.1093/gigascience/gix007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva GG, Green KT, Dutilh BE, Edwards RA. 2016. SUPER-FOCUS: a tool for agile functional analysis of shotgun metagenomic data. Bioinformatics 32:354–361. doi: 10.1093/bioinformatics/btv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menzel P, Ng KL, Krogh A. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 52.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, Asnicar F, Truong DT, Tett A, Morrow AL, Segata N. 2016. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods 13:435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 55.Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. 2015. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. PeerJ 3:e1029. doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Heel AJ, de Jong A, Montalbán-López M, Kok J, Kuipers OP. 2013. BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res 41:W448–W453. doi: 10.1093/nar/gkt391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.