Abstract
Viscosity of protein solution is one of the most troublesome issues for the high-concentration formulation of protein drugs. In this review, we summarize the practical methods that suppress the viscosi-ty of protein solution using small molecular additives. The small amount of salts decreases the viscosity that results from electrostatic repulsion and attraction. The chaotrope suppresses the hydrophobic attraction and cluster formation, which can lower the solution viscosity. Arginine hydrochloride (ArgHCl) also sup-presses the solution viscosity due to the hydrophobic and aromatic interactions between protein molecules. The small molecular additives are the simplest resolution of the high viscosity of protein solution as well as understanding of the primary cause in complex phenomena of protein interactions.
Keywords: Viscosity, arginine, antibody, immunoglobulin G, protein aggregation, serum albumin, molecular interaction
1. Introduction
Biopharmaceutical proteins have been widely developed for the last decade [1]. Recent advances in the protein drugs have resulted in a new type of problems related to the difficulty in handling the protein solutions, typically aggregation [2] and viscosity [3]. To deliver a therapeutic protein, a highly concentrated protein solution is needed for intramuscular and subcutaneous injections. With increasing concentration of the protein, the viscosity of the protein solution also increases.
The viscosity of protein solutions results from several types of interactions between protein molecules [4, 5], as listed in Table 1 [6]. The electrostatic repulsion governed by Coulomb’s law is a major cause that determines the overall solution property because of the long-range interaction. The other repulsive potential is the exclusion volume effect by steric repulsion, which is a very short interaction with a contact distance between atoms. By contrast, attractive interactions are varied, though charge–dipole and charge-induced dipole are major causes due to the electrostatic interactions that affect the protein solution viscosity in a low ionic strength solution. Van der Waal interaction is classified into a short-range interaction that is proportional to the inverse of the sixth power of distances between regions. The other types of attractive interactions, hydrogen bond and hydrophobic contact, are another face of van der Waals interaction. The interactions between proteins are simply explained by the fact that the long-range repulsive electrostatic interaction plays a dominant role in solutions and that several types of short-range attractions characterize the individual solution property [3, 7]. The aggregation also alters the solution property [8]. Taken together, the cause of protein solution viscosity is too complex to reconstruct from individual interaction types among thousands of atoms constructed in protein molecules.
Table 1.
Major interaction depended on the distance.
| Interaction | Distance | Interaction Type |
|---|---|---|
| Charge–dipole | Long | Attraction |
| Charge–charge | Medium | Repulsion |
| Charge–charge | Medium | Attraction |
| van der Waals | Short | Attraction |
| Exclude volume effect | Short | Repulsion |
| Hydrophobic (in water) | Short | Attraction |
This review article is intended to focus on the control of solution viscosity using small molecular additives. The control of solution viscosity is quite easier than understanding the cause from the physical forces. Fig. (1) summarizes the overview of the approach that controls the protein solution viscosity, which can be classified into four types of factors.
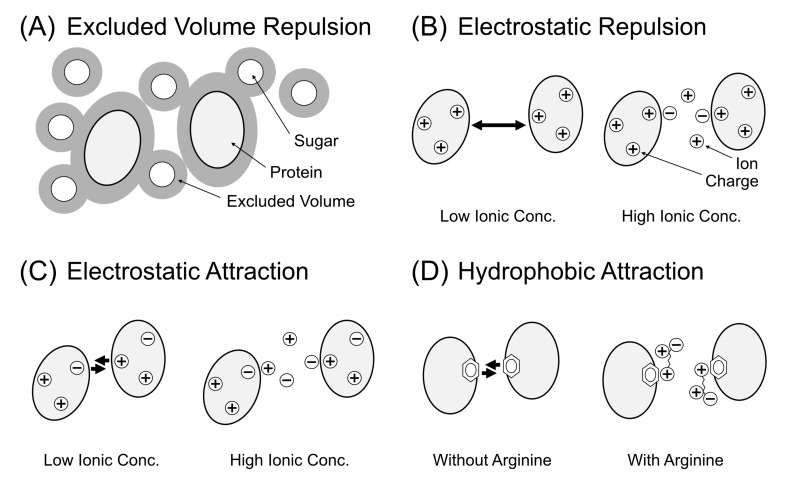
Fig. (1).
Summary of the strategy to decrease the viscosity of protein solution using small molecular additives. (A) The case of exclusion volume effect by steric repulsion. Small solutes (sugar molecules) hamper the mobility of proteins. Thus, the removal of the solutes can decrease the solution viscosity. (B) The case of electrostatic repulsion between proteins. Bold arrows show higher repulsive force between proteins. In the high ionic concentration, the repulsive force decreases, leading to a decrease in the viscosity. (C) The case of electrostatic attraction by charge–charge and charge–dipole interactions. Ions can decrease the viscosity by electrostatic shield by the prevention of the local interactions. (D) The case of hydrophobic interaction between nonpolar and aromatic surfaces of proteins. Arginine molecules weaken the hydrophobic interaction.
Briefly, the reduced concentration of protein and solute can decrease the solution viscosity by lowering the probability of the steric repulsion between solutes (Fig. 1A). The addition of small amount of salts, typically 50 mM NaCl, creates an electrostatic shield between protein molecules, leading to a decrease in the electrostatic repulsion (Fig. 1B) or attraction (Fig. 1C). On the other hand, the addition of chaotrope and Arginine hydrochloride (Arg) weakens the hydrophobic and aromatic interactions between protein molecules (Fig. 1D), which also decreases the solution viscosity.
2. Case A: Exclusion volume effect
First, we consider the condition that contains only protein molecules in aqueous solution. Let the protein molecules be a hard sphere without any specific interaction. Even under the simple condition, the solution viscosity increases depending on the volume fraction [9]. The relationship between the solution viscosity and the volume fraction is described by the following Einstein’s equation (2):
| (2) |
where η, η0, and φ represent the solution viscosity, the water viscosity, and the volume fraction of the solute, respectively [10]. The value 2.5 is a parameter for solid sphere depending on the shape of the solute. Accordingly, with increasing concentration of the solute, the solution viscosity increases without specific interactions.
At high protein concentrations, the Einstein’s equation cannot describe the solution viscosity of protein due to the steric repulsive interactions [11, 12]. Thus, the solution viscosity deviates the ideal solution with increasing concentration of solutes [13]. Living cells usually contain ~300 mg/mL macromolecules [14]. Under the crowding condition, the solute results in the increase of solution viscosity by the steric repulsion between molecules [15]. The colligative property was reported in 1928 using an experimental approach [16]. In that article, Adair showed that the osmotic pressure of the solution of sheep hemoglobin increased with increasing protein concentration [17].
Mooney described the relationship between concentration and viscosity under the high protein concentration according to the following empirical equation [18]:
| (3) |
where φ and λ indicate the volume fraction of the protein and a constant, respectively. η and η0 are the viscosities of the solution and the extrapolated solution to the concentration at zero, respectively. As described in equation (3), the solution viscosity increases with increasing concentration of protein with exponential growth. This equation is an empirical derivation, which reflects the experimental protein solution under the high concentration. The following equation is a derivative of equation (3):
| (4) |
where C is the protein concentration, and Cmax is the maximum concentration of the protein in solution. Mooney’s equation (4) explains hemoglobin [18, 19] and IgG [20] under the high concentration of proteins.
Mooney’s equation (4) is a good approximation for the experimental solution viscosities of various proteins. However, some proteins with high concentration deviate from this equation. This is because high-concentration protein solutions contain some specific interactions between molecules as well as the exclusion volume effect as a neutral crowder. In 1950, Krieger and Dougherty reported the following equation that could be better fitted to the experimental raw data than Mooney’s equation (4) [9]:
| (5) |
where φ is the volume fraction of the protein molecule, φm is the maximum volume fraction of the protein molecule, and [η] is a constant 2.5 if the protein assumes a solid sphere. The φm values of a monodisperse sphere and loose random packing are theoretically the values of 0.74 and 0.6, respectively [21].
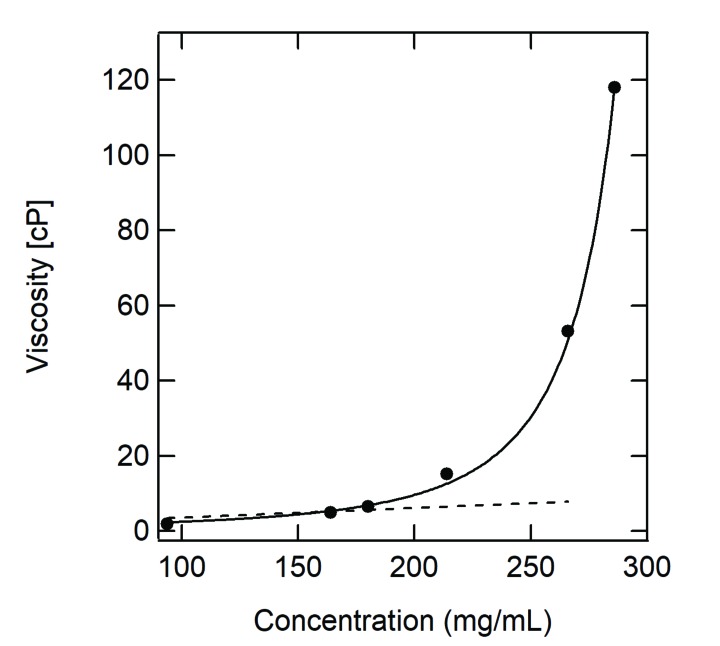
Fig. (2) shows the practical viscosity of bovine serum albumin (BSA) as a function of protein concentration. The data were fitted with Einstein’s equation (2) and Krieger–Dougherty’s equation (5). As shown in the figure, the linear extrapolation of the data by equation (2) is also deviated with increasing concentration of protein. By contrast, equation (5) is in good agreement with the actual viscosity data.
Fig. (2).
Solution viscosity as a function of protein concentration. The data were obtained by bovine serum albumin in 50 mM Na phosphate (pH 7.4) at 25°C. The dotted line and solid curve represent Einstein’s equation (2) and Krieger–Dougherty’s equation (5), respectively. The dotted line was fitted to the data below 180 mg/mL.
Connolly described the solution viscosity of protein as the following simple equation:
| (6) |
where k and C are a constant and the protein concentration, respectively [22]. Equation (6) is much simpler than other equations, but this can only describe high concentration of antibody solution below 300 mg/mL [23]. Although the various types of equations have been proposed, the important point is that all equations are exponential, rather than linear. Thus, the protein viscosity increases steeply above a threshold of the concentration.
The solid sphere model can, in part, predict the viscosity of protein solution. Schere et al. investigated the viscosity of two types of IgG1 monoclonal antibodies at concentrations from 0.5 mg/mL to 275 mg/mL by static light scattering [24]. They analyzed the data by Rayleigh scattering intensity and revealed that the solid sphere model represents the viscosity of monoclonal antibody when the solution contains high concentration of NaCl. To observe the data more clearly, the viscosity of IgG1 monoclonal antibodies results primarily from the electrostatic attractions at neutral pHs under the low ionic strength. When the high ionic strength weakens the electrostatic attractive interactions by the electrostatic shield, the solution viscosity would be increased by the predominant steric repulsion.
Here, we consider the examples of the solution viscosity in the presence of small additives. Protein solutions usually contain buffer and salt as well as protein and water molecules. The important point is that the third component causes the increase in the solution viscosity by the exclusion volume effect. Actually, sugars (typically trehalose and glucose) and polyols (typically polyethylene glycol and glycerol) increase the viscosity of protein solution, though these additives function as a protein stabilizer [25]. For example, it was reported that trehalose, sucrose, sorbitol, glucose, fructose, xylose, and galactose increased the viscosity of monoclonal antibody solution, which results from the exclusion volume effect [26]. It is interesting to note that the presence of 150 mM salt reduced the solution viscosity of antibody by half [25]. These data are related to two ambivalent interactions; sugars increase the probability of the interaction between molecules, followed by increase in the solution viscosity, while ions weaken the electrostatic affinity between protein molecules. Such electrostatic repulsive and attractive interactions are discussed later in Case B and Case C.
In summary, the viscosity of protein solution increases by the exclusion volume effect even in the absence of specific interactions between protein molecules. To suppress the viscosity, decreasing the solute concentration is one of the simple strategies. Sugars and polyols have been used as solution additives that can stabilize the tertiary protein structure [27], and hence, we need to remember that high-concentration solutes increase the solution viscosity when utilizing such protein stabilizers. If we use sugars as the protein stabilizer, decreasing the concentration of the additive is one of the good choices to control viscosity.
3. Case B: Electrostatic repulsion
In Case A, we described the solution viscosity without specific interactions. In Cases B, C, and D, we will describe the solution viscosity with specific interactions between protein molecules.
Electrostatic repulsion and attraction are the most dominant factors in an overall protein solution. Several articles have reported about the high concentration of an antibody solution, indicating the same conclusion that the solution viscosity of a monoclonal antibody deviates from the ideal solution under low ionic strength [24]. To explain the deviation from the ideal solution, the following Taylor expansion has been used:
| (7) |
where Π is the osmotic pressure of solution, R is the gas constant, T is the absolute temperature, c is the protein concentration in mass unit, M is the molecular mass of protein, and B2 and B3 are the constants. Protein has a large M value, so that the second and third correction factors are generally not negligible in a protein solution. The factor B2 is the most important factor to understand the behavior of polymers, originally described by Zimm in 1940 by the analysis of dynamic light scattering (DLS) [28, 29]. The factor B2 is named as the osmotic second virial coefficient. When analyzed by static light scattering (SLS), the parameter is sometimes named as interaction parameter, kD [30].
The subscripts are generally 1 for water, 2 for protein, and 3 for additive. Thus, the second virial coefficient is often noted as B22. The positive value of B22 indicates the repulsive interaction between protein molecules, while the negative value indicates the attractive interaction. The experimental B22 of the protein solution can be determined by DLS and SLS [6, 24, 31-33], neutron scattering [30], ultracentrifugation sedimentation equilibrium [31], molecular dynamics simulation of potential of mean force [34] based on McMillan and Mayer [35], and simulation by Coarse -grained model [36].
Here, we discuss the electrostatic repulsion between protein molecules (Fig. 1B). The electrostatic interaction includes the repulsion between same electric charges and the attraction between opposite electric charges, in addition to the hydrogen bond between O and H atoms, dipole–dipole interaction, and dipole–charge interaction (Table 1). The magnitudes of interaction depend on the distance. In short, the most strong interaction is electrostatics with long range proportional to r-1, followed by the hydrogen bond, which is one order of magnitude lower than the electrostatics. When one type of protein is dissolved into aqueous solution, the protein has a positive or negative net charge in the solution. For example, a protein that has an isoelectric point of pH 5 possesses a negative net charge in the solution of pH 7. As a result, there is the repulsive interaction between molecules in the solution (Fig. 1B). The electrostatic repulsion suppresses the mobility of the protein molecule, which leads to an increase in the solution viscosity. In particular, the electrostatic repulsion is generally the most dominant factor at low concentration of protein solution, because the electrostatic repulsion is the long-range interaction even in the diluted protein solution. When the protein concentration increases, the dominant factor changes from the electrostatic repulsion to the other attractive interactions classified into short range (Table 1) [32].
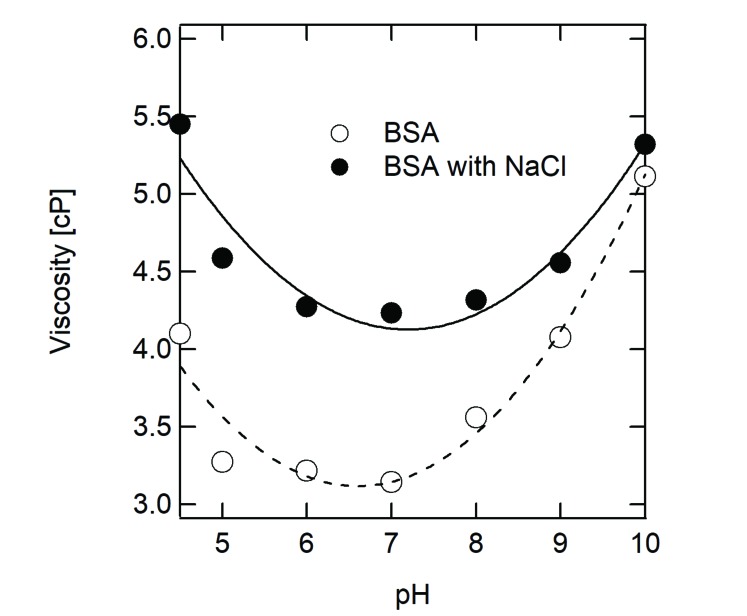
We examined a simple example shown in Fig. (3). BSA has an isoelectric point of about pH 5.1, which is a representative protein for which the viscosity increases in acidic or alkaline condition. The U-shaped viscosity as a function of pH results from the fact that the dominant factor of the solution viscosity is electrostatic repulsion. Thus, to decrease the viscosity of this type of protein solution, pH shift toward neutral pH is one of the simple strategies. When 150 mM NaCl was added into the protein solution, the viscosity curve was an apparently flat form with upper shift (Fig. 3). The change in the viscosity curve by 150 mM NaCl results from the combination of two facts as follows: (i) the addition of solute (NaCl, in this case) increases the viscosity across the whole range of pH by steric repulsion and (ii) the electrostatic shield by NaCl decreases the solution viscosity at alkaline and acidic pH, rather than at neutral pH. In other words, the difference in the viscosity curves between protein alone and protein with 150 mM NaCl represents the combination of both Case A and Case B.
Fig. (3).
Viscosity of bovine serum albumin (BSA) in the presence or absence of NaCl. The viscosities of 180 mg/mL BSA alone (open circles) and 180 mg/mL albumin with 150 mM NaCl (closed circles) were measured at 25°C using an oscillation viscometer VM-10A-L (CBC Materials Co. Ltd., Japan).
Inoue et al. have reported the viscosity of BSA and human serum albumin (HSA) at high concentration (~300 mg/mL) in the presence of various additives at pH 7.4 [37]. The addition of 200 mM NaCl and GdnHCl decreases the viscosities of BSA and HSA by two-third. The decrease in the viscosity was observed for LysHCl, ArgHCl, and NaCl, while no decrease was observed for Gly, Ala, and other non-salt amino acids. These data indicate that the viscosities of BSA and HSA result from monopole-monopole repulsive force, and hence they can be controlled by the addition of salt by the electrostatic shield.
Wang et al. investigated the viscosity of BSA solution at pH 6.0 that affects the amino acids and salts as additives [38]. The addition of 150 mM ArgHCl rather increased the viscosity of BSA (~10%), which was apparently different from that described by Inoue [37]. The different behavior of the additive in BSA solution gives a consistent explanation regarding the electrostatic repulsion with solution viscosity. First, the viscosity of BSA solution in Wang’s study is ~12 cP, which is 5-fold lower viscosity than the condition mentioned in Inoue’s study. Under the condition, the electrostatic repulsion has a smaller influence on the solution viscosity due to the low protein concentration. In addition, the condition at pH 6.0 in Wang’s study is close to the isoelectric point of BSA. As shown in (Fig. 3), the electrostatic repulsion is not a dominant factor when the solution is at neutral pH. Accordingly, the solution viscosity increased resulting from the increasing amino acid concentration. In reality, amino acids show positive values of second virial coefficient [39].
The protein is also not a solid sphere but an anisotropic macromolecule with a flexible structure. Therefore, it is naturally thought that the actual isoelectric point of a protein in an aqueous solution is different from the calculated isoelectric point from the primary structure of the protein. In reality, the isoelectric point of protein depends on the measuring methods. A monoclonal antibody of IgG1 has different isoelectric points at pH 7.8 by the isoelectric focusing phoresis and at pH 6.7 by the zeta potential [40]. Such structural complexity due to the inhomogeneity affects the solution viscosity that deviates from the simple monotonic behavior [33, 41]. For example, an antibody of IgG1 has a larger heterogeneity of the surface charge than the other, which results in the high viscosity based on the electrostatic repulsion between the protein molecules [42].
In summary, the viscosity of an antibody solution is too difficult to understand the theoretical approach due to the nonhomogeneous charges of the surface on the complex structure. However, the control of the solution viscosity using solution additives is comparatively easy. As discussed above, the addition of salt can weaken the electrostatic interaction between molecules, leading to decrease in the viscosity. In particular, the electrostatic shield by the ion is more effective under the pH that is different from the protein isoelectric point. Actually, the solution viscosity of BSA can be described only under the alkaline and acidic conditions [43]. Under the extreme pHs, the electrostatic repulsion plays a crucial role in the overall solution viscosity. By contrast, when the solution pH is close to the isoelectric point of the protein, the theoretical calculation is much more difficult due to the various types of short-range interactions as well as long-range dominant electrostatic repulsions.
4. Case C: Electrostatic attraction
Case A and Case B have discussed the repulsive interactions. The following cases describe the attractive interactions. The attractive interactions are also the crucial factors that determine the protein solution viscosity, because the intermolecular attraction hampers the mobility of protein molecules in the solution (Fig. 1C). The interactions concerned about dipoles are described inversely with the fourth or sixth powers of the distance between particles. The dipole–dipole and dipole-induced dipole interactions are short-range interactions, which increase the attraction with increasing protein concentration [44].
Immunoglobulin is classified into five classes, and the most abundant is IgG. Among the four types of IgG isotype, IgG1 is the most used monoclonal antibody in pharmaceutical drugs. IgG has a common structure with two heavy chains and two light chains crosslinked by disulfide bonds, but the solubility and stability of IgG exhibit diversity by the slight difference in the amino acid sequence [45]. The solution viscosity of IgG1 can be calculated when the solution pH is different from the isoelectric points [46]. Under the pH, the electrostatic repulsion plays a crucial role in the solution viscosity, similar to BSA as shown in Case B. However, when the solution pH is close to the isoelectric points of protein, the theoretical calculation cannot describe the experimental data. Under the low electrostatic repulsive condition, various types of attracted interactions occur between the protein molecules.
Chari et al. investigated the pH dependence of the viscosity of IgG2 monoclonal antibody solution [6]. In the presence of 0.3 M ionic strength, 20 mg/mL and 120 mg/mL antibody solutions showed a constant viscosity independence on pH. By contrast, under the low ionic strength of 4 mM, the viscosity of the antibody solution shows a bell-shaped curve at a maximum pH of 8–9, which corresponds to the isoelectric point of pH 9.0. Similarly, Binabaji et al. [23] and Saito et al. [31] have shown the bell-shaped viscosity curves as a function of pH for antibody. These data indicate that the solution viscosity of high-concentration antibody results from the short-range electrostatic and/or dipole attractions.
Accordingly, the molecular mechanism of the antibody solution at around neutral pH is complex compared to the serum albumin as follows. Yadav et al. investigated four types of IgG1 under the 15 mM ionic strength [47]. Under the condition, the electrostatic interactions are a predominant factor in the protein solution. However, the viscosity is not correlated to the positive charge of IgG1. This is because the viscosity results from not only the amount of charges but also its heterogeneity of the surface charges on the nonglobular structure. Shire et al. investigated the dependence of the monoclonal antibody IgG1 on the ionic strength [4]. Under the low ionic condition, 130 mg/mL antibody solution showed 120 cP, while the viscosity of this antibody solution decreased to 20 cP in the presence of 150 mM NaCl, implying that the extremely high viscosity results from the electrostatic multipoint attractions between molecules. Other studies have reported that the electrostatic interaction plays a crucial role in the high viscosity of antibody solution in low ionic strength conditions when the solution pH is close to the isoelectric point of protein [40, 48].
The charged location on the protein surface is not negligible for solution viscosity. Shire et al. prepared IgG1 mutants that exchange the charged residues and investigated the relationship between structure and function by rheometer and DLS [49]. The results indicated that the exposed charge groups on the Fab region play an important role in increasing the solution viscosity at neutral pH. Thus, the deletion mutant of complementarity-determining regions has low propensity to interact with each other, leading to a decrease in the viscosity even with high-concentration antibody. Indeed, it is interesting that the identical amino acid composition without slight difference in sequence changes the solution viscosity of protein.
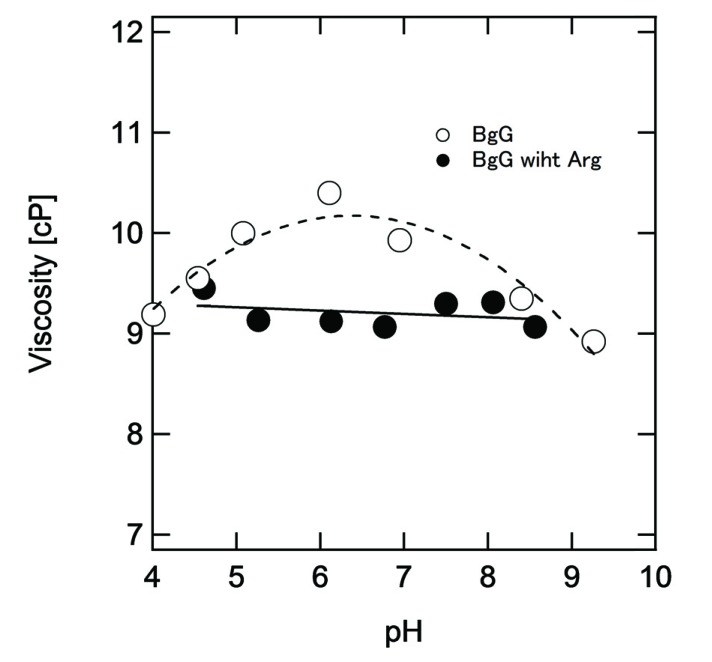
The antibody mixture like γ-globulin shows higher viscosity at around the isoelectric point [45]. Fig. (4) reconfirms the viscosity of bovine γ-globulin as a function of pH. As discussed above, the maximum viscosity was observed at around pH 7, which is a similar value to the isoelectric point of γ-globulin. ArgHCl as the additive changed the viscosity curve of γ-globulin as follows: (i) the viscosity at neutral pH decreased in the presence of Arg sample because of the weakening of the electrostatic interaction such as dipole attraction and hydrophobic interaction between protein molecules by cation–π interaction [50] and (ii) the pH dependence diminished in the presence of Arg because Arg can bind on the aromatic surface at around neutral pH [51], which will be discussed in detail in Case D.
Fig. (4).
Viscosity of bovine γ-globulin (BgG). The protein solution of 180 mg/mL was prepared at the respective pH in the presence (closed circles) or absence (open circles) of 250 mM ArgHCl. At pH above 9.0, the protein solution with ArgHCl cannot be prepared because of the decreasing pH by ArgHCl.
In summary, the antibody solution has a maximum viscosity at around the isoelectric point of the protein. To decrease the viscosity of this type of protein, the addition of small amount of salts, typically 50 mM ionic strength, is one of the simplest approaches. Any types of salts can act to suppress the electrostatic attraction by the electrostatic shield, including NaCl, ArgHCl, and LysHCl [38].
5. Case D: Hydrophobic interaction
Hydrophobic effect is an important phenomenon that drives the interaction between nonpolar solutes in aqueous solution [52]. Frank and Evans represented the behavior of nonpolar solute in aqueous solution as “the water forms frozen patches or microscopic icebergs around such solute molecules” [53]. This is a historical important description of nonpolar solutes in aqueous solution by an entropic point of views. In 1950, Kauzmann described the importance of the hydrophobic bond for stabilizing the native conformation of proteins [54]. Later, the solubility of the respective amino acid residues [55] and the folding theory with thermodynamics [56] revealed the protein folding phenomenon in aqueous solutions. Privalov is one of the most important researchers in protein folding by a calorimeter [57]. The thermodynamic analysis of the solute in aqueous solution evidently showed the accurate picture of the protein in solution; Chandler described as “oil and water molecules actually attract each other, but not nearly as strongly as water attracts itself” [58]. Thus, the protein tertiary structure shows high stability in vacuum than in water [54]. In other words, the protein structure is stabilized by the intramolecular interaction of protein, rather than the entropic factor with water.
Based on recent advances in ultrafast spectroscopy, we can observe the dynamic interaction between ion and water [59]. A classical view of the function of ion was that water affects the bulk structure, the so-called structure-making and structure-breaking context. However, today, it is believed that ion does not interact with water molecule beyond the hydration shell [60]. Accordingly, ion molecules interact directly with solutes and proteins [61, 62]. In other words, ions and nonpolar solutes pertain only in the local water molecule around the protein surface [63]. The water molecules around the hydrophobic surface do not have a static structure like iceberg but show slow orientational dynamics than the bulk-free-water molecules [64].
When protein concentration increases, a protein molecule interacts with another molecule, leading to an increase in the solution viscosity (Fig. 1D). This is because the intermolecular attraction slows the mobility of protein molecules. Here the hydrophobic interaction is also controlled by the addition of small molecules. The strategy is quite simple; an additive that increases the solubility of a hydrophobic solute weakens the hydrophobic interaction between protein molecules. Typically, the additives are denaturant and chaotrope, such as urea, guanidine hydrochloride, thiocyanate, and iodide, which are highly soluble themselves in water [65]. These denaturants lack hydration shell, as observed by neutron scattering [66], which is prone to bind on a protein surface and unfolds the tertiary structure of the protein. Similarly, urea binds to a protein by hydrogen bonds and unfolds the tertiary structure of the protein [67].
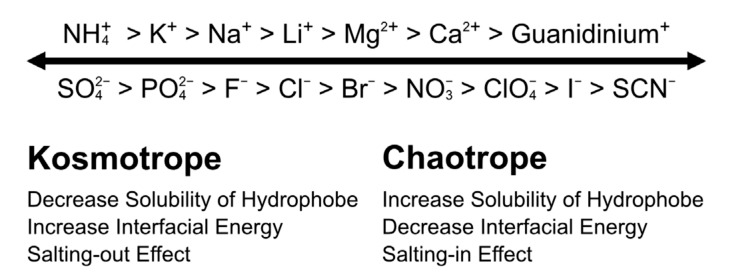
As shown in (Fig. 1C), low-concentration salts (typically 50 mM) suppress the electrostatic interaction by the electrostatic shield. Further increase in the salt concentration (500 mM, for example) causes a specific interaction between the ion and solute [68-71]. This salt-concentration-defending effect has been historically known as Hofmeister effect. As shown in Fig. 5 of the brief summary, chaotropic ions, such as thiocyanate and iodide, bind preferentially on the protein surface [72, 73]. Such specific binding increases the solubility of a nonpolar solute and decreases the interfacial energy. The opposite side of the ions shows the reverse effects of chaotrope on the solute, the so-called kosmotrope that is named from Greek chaos and kosmos meaning disorder and order, respectively [74]. Recent studies on the Hofmeister effect by molecular dynamics (MD) simulation and ultrafast spectroscopy have shown that ions do not change the bulk water structure but bind specifically to the solute surface [73, 75-77].
Fig. (5).
Hofmeister series and effects. Right side is chaotrope that decreases the interfacial energy between solutes, so that the chaotrope increases the solubility of the solute by the salting-in effect. Left side is kosmotrope, which has the opposite effects on the chaotrope.
Shire et al. have investigated the viscosity of solution of high-concentration monoclonal antibody in various ions [78]. In the absence of ions, the solution viscosity of an IgG1 antibody was 20 cP at pH 6.0 at 125 mg/mL. The solution viscosity steeply decreased with increasing concentration of the ions. The ionic strength of 0.1 M showed almost half of the viscosity compared to that in the absence of the ions. The iodide and thiocyanate ions were more effective in suppressing the solution viscosity than did sodium and chloride ions. Thus, chaotrope is more favorable for decreasing the solution viscosity of the antibody solution, resulting from the chaotrope, and is prone to bind on theprotein surface that neutralizes the surface charge. The neutralization of the surface charge, as well as the increasing solubility with chaotropic ions, weakens the protein–protein interaction.
Klibanov et al. have shown that hydrophobic solutes decrease the solution viscosity of BSA and γ-globulin [79]. The solution viscosity of serum albumin at pH 7.4 at 50 cP decreased with increasing hydrophobicity of the hydrophobic solutes. In addition, when chaotrope was selected as the counterion, the viscosity was effectively decreased. Accordingly, the hydrophobic salt decreased the hydrophobic interaction between the protein molecules. Both negatively or positively charged hydrophobic salts showed similar reduction in the viscosity of the high-concentration γ-globulin solution. The same groups have shown that hydrophobic salts successfully decreased the viscosity of humanized monoclonal antibodies [80]. Thus, hydrophobic salts decreased the solution viscosity of high-concentration proteins weakened by both hydrophobic and electrostatic interactions. However, hydrophobic salts are usually toxic in living cells.
Arginine (Arg) is one of the most useful additives in aqueous protein solution [81]. Arg prevents protein–protein interaction without unfolding, which is a favorable property for various situations [82]. The first report of Arg as the solution additive was in 1991 for the refolding of the antibody Fab fragment reported by Buchner and Rudolph [83]. Later, in 2002, it was reported that Arg prevents heat-induced aggregation of proteins [84]. The molecular mechanism of Arg as the solution additive is mainly caused by the cation–π interaction between the planner guanidinium group in Arg and the aromatic groups in a protein [85, 86]. A X-ray crystallographic analysis showed that three Arg molecules bind on the surface of hen egg-white lysozyme [87]. In addition, guanidine has a hydrophobic property as analyzed by MD simulation and neutron diffraction analysis with isotopic substitution experiment [88]. Accordingly, it is also thought that the hydrophobic region of Arg interacts with the hydrophobic aromatic residues [89].
The binding energy of Arg with an aromatic ring is weak from 0.5 to 2.0 kJ/mol estimated by a transfer free energy of the aromatic compounds from water to 1 M Arg solution [86, 90, 91]. Arg does not exclude on the protein surface similar to sugar and polyol but stabilizes the native-state protein [92]. The weak interaction of Arg with protein results in apparently controversial data. For example, preferential interaction of Arg on the protein surface depends on the concentration [93-95]. As another example, the binding of Arg on the protein depends on the solution pH; the solution at pH 4.5 was found with three Arg molecules on the surface of hen egg-white lysozyme [87], while the solution at pH 7.5 was not directly found with the Arg molecule but differed from the hydration water structure on the lysozyme surface [96]. This is because lysozyme is a basic protein with an isoelectric point as pH 11, so that Arg molecules may bind with the protein more strongly under the acidic condition. In addition, Arg forms a cluster by head-to-tail hydrogen bonding, which acts as an aggregation suppressor [97]. Although the molecular mechanism of the solution condition remains controversial due to the weakness of interaction and the so many types of interactions between Arg and protein, Arg is not harmful to the protein structure and solution state with that property.
As shown in (Fig. 4), the solution viscosity of high-concentration γ-globulin had a maximum value at around neutral pH. This is because the primary force of the viscosity is from the electrostatic attraction and/or hydrophobic attraction under the neutral pH. Inoue et al. have shown that the addition of 0.5 M Arg decreased the viscosity of 250 mg/mL bovine γ-globulin solution from 60 to 35 cP [98]. By contrast, at pH 9.4, the addition of 0.5 M Arg slightly decreased the viscosity of the same concentration of γ-globulin solution from 41 to 39 cP [98]. The comparison of the two results showed the following findings: (i) the pH shift apart from the isoelectric point of protein decreases the viscosity from 60 to 41 cP; (ii) Arg can decrease the viscosity of γ-globulin solution under the neutral pH that is close to the isoelectric point of protein; and (iii) Arg cannot decrease the viscosity of γ-globulin solution at alkaline pH. These facts suggest that the hydrophobic attraction plays a crucial role in the high viscosity of high-concentration γ-globulin solution. Under the conditions, Arg can weakly bind to the protein, which suppresses the protein–protein interaction [51], leading to a decrease in the solution viscosity compared to that with Lys, Gly, NaCl, and other additives.
Arg has been used as a hydrochloride salt, that is, ArgHCl. However, counterion plays an important role in the suppression of protein–protein interactions. In reality, it was shown that Arg-Glu and Arg-Asp mixtures give higher resistance for thermal aggregation of IgG1 monoclonal antibodies than does ArgHCl alone [99, 100]. As another example, Asp-Glu mixture decreases the solution viscosity of monoclonal antibodies of IgG1, IgG2, and IgG4 solutions [101]. The melting temperature of the protein is affected by the type of counterion on Arg; Arg-Glu and Arg-acetate increased the melting temperature of IgG1, while ArgHCl and Arg2(SO4) increased [102]. Arg-chaotrope salt is one of the good combinations to suppress the hydrophobic interaction [103]. The buffer itself has an effect on protein solubility and stability [104, 105].
Histidine is another amino acid for the additive that suppresses the solution viscosity of high concentration of protein [106]. Histidine is a well-used buffer from pH 5.5 to 6.5 in antibody solution because of its stability and nontoxicity. Histidine has a multiface, which has a cation with aromatic property as well as connects with protein by hydrogen bonds [107]. Actually, 40 mM histidine decreased the viscosity by one-half of 150 mg/mL human interleukin 8 [106]. Accordingly, histine buffer is a good selection for decreasing the solution viscosity of high-concentration protein at weakly acidic condition.
6. Cluster and aggregates
We discussed in Case A to Case D about monodispersed molecules in solution. Finally, we will describe reversible aggregation with the solution viscosity.
Protein is generally an aggregated-prone molecule in aqueous solution that also hampers the application of protein from industries to academics. For example, the viscosity of IgG1 monoclonal antibody steeply increased during heat treatment at 70°C for 20 min for 60 mg/mL protein, due to the aggregate formation [108]. The increase in viscosity depended on the protein concentration. If protein molecule forms association transiently, the solution viscosity is also increase, as shown in equation (2). Recent studies have revealed this mechanism [46, 108, 109].
The association and growth processes of protein aggregation are too complex to describe by the classical DLVO theory. The practical models to describe protein aggregation have been proposed in various manners, though the empirical models were classified into 19 types in a review reported in 2009 [110]. The complexity of the protein aggregation results from the various possible tertiary structures in solution as well as the complex composition of the primary sequence. In addition, the term “protein aggregation” is not in agreement with a general consensus by scientists and researchers due to the various fields of the protein solution problem, as pointed out by Murphy and Roberts in 2013 [111].
The report about dynamic cluster in protein solution was first reported in 2004 [112, 113]. After the high impact articles, the dynamic and transient protein clusters became a subject of interest in pharmaceutical applications. The molecular mechanism of the formation of the cluster results from a short-ranged attraction and a long-ranged repulsion, which are the same as the principle of the solution viscosity of protein [114, 115]. That is, the particle size is determined by the balance between growth of short-range attraction and dispersion of long-range repulsion.
Equilibrium cluster formation of hen egg-white lysozyme was first described by small-angle scattering and confocal microscopy [112]. The equilibrium dimer formation was observed under the low protein concentration [116]. In particular, the cluster was grown under the high protein concentration of 17.5%–22.5% [117]. Such dynamic cluster formation also changes the solution viscosity [118]. The cluster is stabilized by the balance between short-range attraction and long-range repulsion; small-angle neutron scattering (SANS) and small-angle X-ray scattering (SAXS) analyses showed that the intermediate-range order plays a crucial role in the stabilization [119, 120]. Thus, the molecular mechanism of the formation of dynamic equilibrium cluster is still controversial.
Lilyestrom et al. investigated the viscosity of IgG1 antibody solution in the presence of sodium sulfate as a function of protein and additive concentrations by DLS and SAXS [121]. The data indicated that the dynamic clusters are formed with increasing concentration of protein. The authors concluded that transient and equilibrium clusters from two to nine are formed dynamically at a high protein concentration above 100 mg/mL, rather than solid sphere dispersed in solution. Godfrin et al. also reported the cluster formation of IgG1 in the presence of 50 mM sodium sulfate [122]. Sodium sulfate is also a type of kosmotrope, which may lead to cluster formation by the excluded effect. Taken together, it is plausible that the cluster is formed in high protein concentration solution; further addition of neutral crowder also accelerates the cluster formation.
Schere investigated the cluster formation of 275 mg/mL IgG1 monoclonal antibody and found that the formed cluster was dissolved by salts and ArgHCl [95]. The addition of NaSCN and ArgHCl dissolved more effectively than NaCl, indicating that chaotrope is favorable for the cluster dissolution. On the contrary, Borwankar et al. showed that a disaccharide of trehalose is an inert crowder that stabilizes the clustered-state protein [123]. Similar experiments were reported under the extreme high concentration of protein by Johnston et al. showing that trehalose stabilizes the cluster state at 700 mg/mL monoclonal antibodies [124]. The nanoclusters have diameters of 50–300 nm, which are dispersed by the colloidal balance between electrostatic repulsion and short-range attractions. Such a stabilized clustered state may be used for a new type formulation of high-concentration protein, if the cluster is fully reversible without irreversible aggregate formation.
Similarly, an emulsion state of protein–polyelectrolyte complex (PPC) has been developed for the formulation of high-concentration protein drugs [125]. The molecular mechanism of PPC formation is only from electrostatic interaction. Briefly, if only one type of protein disperses into a solution, protein molecules disperse by the electrostatic repulsion between molecules. Under the condition, when the oppositely charged polyelectrolyte is added into the protein solution, the polyelectrolyte neutralizes the protein charge by the complex formation. The formed PPC is also a neutral net charge that diminishes the electrostatic repulsion between protein molecules, leading to form large and soft emulsions [126]. That study showed that the PPC formation is used for 10 types of antibodies, hormones, and pharmaceutical enzymes. The important point is that the PPC is highly reversible when 150 mM or more salts redissolve the protein into solution without protein denaturation [127]. The precipitated-state PPC is stable for heat, oxidation, and agitation [128] and is nontoxic [129]. Thus, the emulsion-state PPC will be a new candidate for the formulation of high-concentration protein to suppress the viscosity of the solution [130].
In this review, the viscosity caused by the interaction between solute molecules has been described only regarding to a folded state of globular proteins, without consideration of protein unfolding or intrinsically disordered proteins. The actual globular proteins might change their structures depending on the conditions of the solution, such as the presence of co-solvent and the concentration of protein. However, the globular proteins, structures of which are (partially) unfolded in aqueous media, are usually lost due to their irreversible aggregation (which might cause significant increase in solution viscosity). Accordingly, it is noted that the control of aggregation is more effective than the control of the protein structure for lowering the viscosity of the solution.
Conclusion
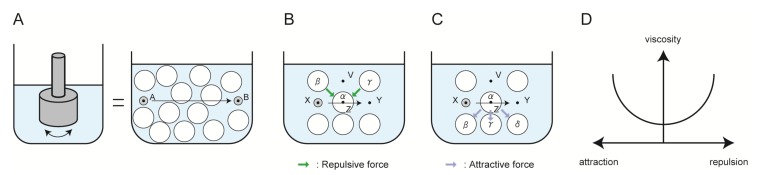
Solution viscosity increases by the attraction and repulsion between the protein molecules, as shown in the above discussion. Therefore, the protein structure plays an important role for the solution viscosity. However, the protein structure is too complex to be understood in details. Accordingly, the proposes of this review was to describe the use of small additives to decrease the viscosity of protein solutions by hampering these interactions between protein molecules. Figure 6 summarizes a simple scheme of the origin of viscosity. The viscosity of solution is typically measured by how easily a particle moves from position A to B (Fig. 6A), which can be detected by a conventional viscometer. When the particle moves from position X to Y, it requires movement of the solute α from position Z to position V. However, it becomes difficult for solute α to move from position Z to position V due to the presence of the repulsive force originating from particles β and γ (Fig. 6B) or the attractive force from particles β, γ, and δ (Fig. 6C). Therefore, viscosity of solution increases by both attraction and repulsion between protein molecules (Fig. 6D).
Fig. (6).
Origin of viscosity. (A) Typical viscometer and the mechanism of viscosity. (B) A case of the repulsive force between α, β, and γ. (C) A case of the attractive force between α, β, γ, and δ. (D) The relationship between molecular interaction and solution viscosity.
In this review, we described the small molecules that control the viscosity of protein solution. This approach provides a view to consider the solution viscosity using solution additives. For example, if the addition of 50 mM NaCl decreases the solution viscosity of a protein, the cause of the viscosity is determined as the electrostatic attraction or electrostatic repulsion. If the addition of 50 mM NaSCN or NaCl does not affect the solution viscosity of a protein, but if the addition of 500 mM NaSCN decreases the viscosity, then the primary cause of the solution viscosity is the hydrophobic attraction between protein molecules, rather than the electrostatic interactions.
The solution viscosity of the high-concentration protein results in various types of interactions (Table 1), as well as the structural change and transient cluster formation. Owing to this, the protein solution is quite complex to understand even today. By contrast, the application possibility described in this review provides practical problem solution that controls the experimental behavior of protein solution viscosity (Table 2). Finally, we conclude that ArgHCl has the best additives for both (i) the electrostatic shield under the acidic or alkaline conditions for albumin-type protein [98] and (ii) the weakening of the hydrophobic interaction at neutral pH like antibodies [37]. In addition, Arg is a naturally occurring amino acid with safety and stability and being inexpensive.
Table 2.
Practical solution for lowering viscosity of protein solution.
| Primary cause of viscosity | Primary approach |
|---|---|
| Steric repulsion | Decrease the concentration of additive |
| Electrostatic repulsion | Add 50 mM salts and/or approximate pI |
| Electrostatic attraction | Add 50 mM salts and/or separate from pI |
| Hydrophobic attraction | Add 500 mM chaotrope and/or 500 mM Arg |
AcknowledgEments
Declared none.
LIST OF ABBREVIATIONS
- Arg
Arginine
- HCl
Hydrochloride
- IgG
Immunoglobulin G
- NaCl
Sodium chloride
- BSA
Bovine serum albumin
- DLS
Dynamic light scattering
- SLS
Static light scattering
- HAS
Human serum albumin
- Gdn
Guanidinium
- Lys
Lysine
- Gly
Glycine
- Ala
Alanine
- Fab
Fragment, antigen-binding
- MD
Molecular dynamics
- Glu
Glutamic acid
- Asp
Aspartic acid
- SO4
Sulfate
- DLVO
Derjaguin-Landau-Verwey-Overbeek
- SANS
Small-angle neutron scattering
- SAXS
Small-angle X-ray scattering
- SCN
Thiocyanate
- PPC
Protein–polyelectrolyte complex
Consent for Publication
Not applicable.
conflict of interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Mitragotri S., Burke P.A., Langer R. Overcoming the challenges in administering biopharmaceuticals: Formulation and delivery strategies. Nat. Rev. Drug Discov. 2014;13:655–672. doi: 10.1038/nrd4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li W., Prabakaran P., Chen W., Zhu Z., Feng Y., Dimitrov D.S. Antibody aggregation: Insights from sequence and structure. Antibodies (Basel) 2016;5:19. doi: 10.3390/antib5030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jezek J., Rides M., Derham B., Moore J., Cerasoli E., Simler R., Perez-Ramirez B. Viscosity of concentrated therapeutic protein compositions. Adv. Drug Deliv. Rev. 2011;63:1107–1117. doi: 10.1016/j.addr.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Nguyen M.D., Andya J.D., Shire S.J. Reversible self-association increases the viscosity of a concentrated monoclonal antibody in aqueous solution. J. Pharm. Sci. 2005;94:1928–1940. doi: 10.1002/jps.20347. [DOI] [PubMed] [Google Scholar]
- 5.Burckbuchler V., Mekhloufi G., Giteau A.P., Grossiord J.L., Huille S., Agnely F. Rheological and syringeability properties of highly concentrated human polyclonal immunoglobulin solutions. Eur. J. Pharm. Biopharm. 2010;76:351–356. doi: 10.1016/j.ejpb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Chari R., Jerath K., Badkar A.V., Kalonia D.S. Long- and short-range electrostatic interactions affect the rheology of highly concentrated antibody solutions. Pharm. Res. 2009;26:2607–2618. doi: 10.1007/s11095-009-9975-2. [DOI] [PubMed] [Google Scholar]
- 7.Roger H., Adrian V.P., Rudolf P., Rick F.R., Anand J., Jian L., Dilip A., Manoj K.C., Yet-ming C., Steve G., Sergei K., Mehran K., Roland K., David C.L., Jennifer L., Steve L., David W., John S.W., Wai-Yim C., Mike F., Frank H., O. Anatole L., Carel Jan O., Thomas Z. Long range interactions in nanoscale science. Rev. Mod. Phys. 2010;82:1887–1944. [Google Scholar]
- 8.Wang W., Nema S., Teagarden D. Protein aggregation - pathways and influencing factors. Int. J. Pharm. 2010;390:89–99. doi: 10.1016/j.ijpharm.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 9.Genovese D.B. Shear rheology of hard-sphere, dispersed, and aggregated suspensions, and filler-matrix composites. Adv. Colloid Interface Sci. 2012;171:1–16. doi: 10.1016/j.cis.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Goldsack D.E., Franchetto R. The viscosity of concentrated electrolyte solutions. I. Concentration dependence at fixed temperature. Can. J. Chem. 1977;55:1062–1072. [Google Scholar]
- 11.Mooney M. The viscosity of a concentrated suspension of spherical particles. J. Colloid Sci. 1951;6:162–170. [Google Scholar]
- 12.Zimmerman S.B., Minton A.P. Macromolecular crowding: Biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 13.Ross P.D., Minton A.P. Hard quasispherical model for the viscosity of hemoglobin solutions. Biochem. Biophys. Res. Commun. 1977;76:971–976. doi: 10.1016/0006-291x(77)90950-0. [DOI] [PubMed] [Google Scholar]
- 14.Zimmerman S.B., Trach S.O. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 15.Zhou H.X., Rivas G., Minton A.P. Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adair G.S. A theory of partial osmotic pressures and membrane equilibria, with special reference to the application of Dalton’s law to haemoglobin solutions in the presence of salts. Proc. R. Soc. Lond. 1928;120:573–603. [Google Scholar]
- 17.Minton A.P. The influence of macromolecular crowding and macromolecular confinement on biochemical reactions in physiological media. J. Biol. Chem. 2001;276:10577–10580. doi: 10.1074/jbc.R100005200. [DOI] [PubMed] [Google Scholar]
- 18.Mooney M. The viscosity of a concentrated suspension of spherical particles. J. Colloid Sci. 1951;6:162–170. [Google Scholar]
- 19.McClain B.L., Finkelstein I.J., Fayer M.D. Dynamics of hemoglobin in human erythrocytes and in solution: Influence of viscosity studied by ultrafast vibrational echo experiments. J. Am. Chem. Soc. 2004;126:15702–15710. doi: 10.1021/ja0454790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monkos K., Turczynski B. A comparative study on viscosity of human, bovine and pig IgG immunoglobulins in aqueous solutions. Int. J. Biol. Macromol. 1999;26:155–159. doi: 10.1016/s0141-8130(99)00080-x. [DOI] [PubMed] [Google Scholar]
- 21.Quemada D., Berli C. Energy of interaction in colloids and its implications in rheological modeling. Adv. Colloid Interface Sci. 2002;98:51–85. doi: 10.1016/s0001-8686(01)00093-8. [DOI] [PubMed] [Google Scholar]
- 22.Connolly B.D., Petry C., Yadav S., Demeule B., Ciaccio N., Moore J.M., Shire S.J., Gokarn Y.R. Weak interactions govern the viscosity of concentrated antibody solutions: High-throughput analysis using the diffusion interaction parameter. Biophys. J. 2012;103:69–78. doi: 10.1016/j.bpj.2012.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binabaji E., Ma J., Zydney A.L. Intermolecular interactions and the viscosity of highly concentrated monoclonal antibody solutions. Pharm. Res. 2015;32:3102–3109. doi: 10.1007/s11095-015-1690-6. [DOI] [PubMed] [Google Scholar]
- 24.Scherer T.M., Liu J., Shire S.J., Minton A.P. Intermolecular interactions of IgG1 monoclonal antibodies at high concentrations characterized by light scattering. J. Phys. Chem. B. 2010;114:12948–12957. doi: 10.1021/jp1028646. [DOI] [PubMed] [Google Scholar]
- 25.Minton A.P. Influence of macromolecular crowding upon the stability and state of association of proteins: Predictions and observations. J. Pharm. Sci. 2005;94:1668–1675. doi: 10.1002/jps.20417. [DOI] [PubMed] [Google Scholar]
- 26.He F., Woods C.E., Litowski J.R., Roschen L.A., Gadgil H.S., Razinkov V.I., Kerwin B.A. Effect of sugar molecules on the viscosity of high concentration monoclonal antibody solutions. Pharm. Res. 2011;28:1552–1560. doi: 10.1007/s11095-011-0388-7. [DOI] [PubMed] [Google Scholar]
- 27.Arakawa T., Timasheff S.N. Stabilization of protein structure by sugars. Biochemistry. 1982;21:6536–6544. doi: 10.1021/bi00268a033. [DOI] [PubMed] [Google Scholar]
- 28.Zimm B.H. Application of the methods of molecular distribution to solutions of large molecules. J. Chem. Phys. 1946;14:164–179. [Google Scholar]
- 29.Zimm B.H. The scattering of light and the radial distribution function of high polymer solutions. J. Chem. Phys. 1948;16:1093–1099. [Google Scholar]
- 30.Velev O.D., Kaler E.W., Lenhoff A.M. Protein interactions in solution characterized by light and neutron scattering: Comparison of lysozyme and chymotrypsinogen. Biophys. J. 1998;75:2682–2697. doi: 10.1016/S0006-3495(98)77713-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito S., Hasegawa J., Kobayashi N., Kishi N., Uchiyama S., Fukui K. Behavior of monoclonal antibodies: Relation between the second virial coefficient B2 at low concentrations and aggregation propensity and viscosity at high concentrations. Pharm. Res. 2012;29:397–410. doi: 10.1007/s11095-011-0563-x. [DOI] [PubMed] [Google Scholar]
- 32.Saluja A., Badkar A.V., Zeng D.L., Nema S., Kalonia D.S. Ultrasonic storage modulus as a novel parameter for analyzing protein-protein interactions in high protein concentration solutions: Correlation with static and dynamic light scattering measurements. Biophys. J. 2007;92:234–244. doi: 10.1529/biophysj.106.095174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arzenšek D., Kuzman D., Podgornik R.J. Hofmeister effects in monoclonal antibody solution interactions. Phys. Chem. B. 2015;119:10375–10389. doi: 10.1021/acs.jpcb.5b02459. [DOI] [PubMed] [Google Scholar]
- 34.Elcock A.H., McCammon J.A. Calculation of weak protein-protein interactions: The pH dependence of the second virial coefficient. Biophys. J. 2001;80:613–625. doi: 10.1016/S0006-3495(01)76042-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMillan W.G., Jr, Mayer J.E. The statistical thermodynamics of multicomponent systems. J. Chem. Phys. 1945;13:276–305. [Google Scholar]
- 36.Calero-Rubio C., Saluja A., Roberts C.J. Coarse-grained antibody models for “weak” protein-protein interactions from low to high concentrations. J. Phys. Chem. B. 2016;120:6592–6605. doi: 10.1021/acs.jpcb.6b04907. [DOI] [PubMed] [Google Scholar]
- 37.Inoue N., Takai E., Arakawa T., Shiraki K. Arginine and lysine reduce the high viscosity of serum albumin solutions for pharmaceutical injection. J. Biosci. Bioeng. 2014;117:539–543. doi: 10.1016/j.jbiosc.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 38.Wang S., Zhang N., Hu T., Dai W., Feng X., Zhang X., Qian F. Viscosity-lowering effect of amino acids and salts on highly concentrated solutions of two IgG1 monoclonal antibodies. Mol. Pharm. 2015;12:4478–4487. doi: 10.1021/acs.molpharmaceut.5b00643. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H. Viscosity B-coefficients and standard partial molar volumes of amino acids, and their roles in interpreting the protein (enzyme) stabilization. Biophys. Chem. 2006;122:157–183. doi: 10.1016/j.bpc.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Yadav S., Liu J., Shire S.J., Kalonia D.S. Specific interactions in high concentration antibody solutions resulting in high viscosity. J. Pharm. Sci. 2010;99:1152–1168. doi: 10.1002/jps.21898. [DOI] [PubMed] [Google Scholar]
- 41.Sarangapani P.S., Hudson S.D., Jones R.L., Douglas J.F., Pathak J.A. Critical examination of the colloidal particle model of globular proteins. Biophys. J. 2015;108(3):724–737. doi: 10.1016/j.bpj.2014.11.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yadav S., Laue T.M., Kalonia D.S., Singh S.N., Shire S.J. The influence of charge distribution on self-association and viscosity behavior of monoclonal antibody solutions. Mol. Pharm. 2012;9:791–802. doi: 10.1021/mp200566k. [DOI] [PubMed] [Google Scholar]
- 43.Sarangapani P.S., Hudson S.D., Migler K.B., Pathak J.A. The limitations of an exclusively colloidal view of protein solution hydrodynamics and rheology. Biophys. J. 2013;105:2418–2426. doi: 10.1016/j.bpj.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh S.N., Yadav S., Shire S.J., Kalonia D.S. Dipole-dipole interaction in antibody solutions: Correlation with viscosity behavior at high concentration. Pharm. Res. 2014;31:2549–2558. doi: 10.1007/s11095-014-1352-0. [DOI] [PubMed] [Google Scholar]
- 45.Correia I.R. Stability of IgG isotypes in serum. MAbs. 2010;2:221–232. doi: 10.4161/mabs.2.3.11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicoud L., Jagielski J., Pfister D., Lazzari S., Massant J., Lattuada M., Morbidelli M. Kinetics of monoclonal antibody aggregation from dilute toward concentrated conditions. J. Phys. Chem. B. 2016;120:3267–3280. doi: 10.1021/acs.jpcb.5b11791. [DOI] [PubMed] [Google Scholar]
- 47.Yadav S., Shire S.J., Kalonia D.S. Factors affecting the viscosity in high concentration solutions of different monoclonal antibodies. J. Pharm. Sci. 2010;99:4812–4829. doi: 10.1002/jps.22190. [DOI] [PubMed] [Google Scholar]
- 48.Roberts D., Keeling R., Tracka M., van der Walle C.F., Uddin S., Warwicker J., Curtis R. The role of electrostatics in protein-protein interactions of a monoclonal antibody. Mol. Pharm. 2014;11:2475–2489. doi: 10.1021/mp5002334. [DOI] [PubMed] [Google Scholar]
- 49.Yadav S., Sreedhara A., Kanai S., Liu J., Lien S., Lowman H., Kalonia D.S., Shire S.J. Establishing a link between amino acid sequences and self-associating and viscoelastic behavior of two closely related monoclonal antibodies. Pharm. Res. 2011;28:1750–1764. doi: 10.1007/s11095-011-0410-0. [DOI] [PubMed] [Google Scholar]
- 50.Kyne C., Ruhle B., Gautier V.W., Crowley P.B. Specific ion effects on macromolecular interactions in Escherichia coli extracts. Protein Sci. 2015;24:310–318. doi: 10.1002/pro.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miyatake T., Yoshizawa S., Arakawa T., Shiraki K. Charge state of arginine as an additive on heat-induced protein aggregation. Int. J. Biol. Macromol. 2016;87:563–569. doi: 10.1016/j.ijbiomac.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 52.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 53.Frack H.S., Evans M.W. Free volume and entropy in condensed systems III. Entropy in binary liquid mixtures; partial molal entropy in dilute solutions; structure and thermodynamics in aqueous electrolytes. J. Chem. Phys. 1945;13:507–532. [Google Scholar]
- 54.Kauzmann W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 55.Nozaki Y., Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J. Biol. Chem. 1971;246:2211–2217. [PubMed] [Google Scholar]
- 56.Baldwin R.L. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Privalov P.L., Gill S.J. Stability of protein structure and hydrophobic interaction. Adv. Protein Chem. 1988;39:191–234. doi: 10.1016/s0065-3233(08)60377-0. [DOI] [PubMed] [Google Scholar]
- 58.Chandler D. Hydrophobicity: Two faces of water. Nature. 2002;417:491. doi: 10.1038/417491a. [DOI] [PubMed] [Google Scholar]
- 59.Rezus Y.L., Bakker H.J. Observation of immobilized water molecules around hydrophobic groups. Phys. Rev. Lett. 2007;99:148301. doi: 10.1103/PhysRevLett.99.148301. [DOI] [PubMed] [Google Scholar]
- 60.Omta A.W., Kropman M.F., Woutersen S., Bakker H.J. Negligible effect of ions on the hydrogen-bond structure in liquid water. Science. 2003;301:347–349. doi: 10.1126/science.1084801. [DOI] [PubMed] [Google Scholar]
- 61.Marcus Y. Effect of ions on the structure of water: Structure making and breaking. Chem. Rev. 2009;109:1346–1370. doi: 10.1021/cr8003828. [DOI] [PubMed] [Google Scholar]
- 62.Ball P., Hallsworth J.E. Water structure and chaotropicity: Their uses, abuses and biological implications. Phys. Chem. Chem. Phys. 2015;17:8297–8305. doi: 10.1039/c4cp04564e. [DOI] [PubMed] [Google Scholar]
- 63.Chaplin M. Do we underestimate the importance of water in cell biology? Nat. Rev. Mol. Cell Biol. 2006;7:861–866. doi: 10.1038/nrm2021. [DOI] [PubMed] [Google Scholar]
- 64.Laage D., Stirnemann G., Hynes J.T. Why water reorientation slows without iceberg formation around hydrophobic solutes. J. Phys. Chem. B. 2009;113:2428–2435. doi: 10.1021/jp809521t. [DOI] [PubMed] [Google Scholar]
- 65.Greene R.F., Jr, Pace C.N. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, alpha-chymotrypsin, and beta-lactoglobulin. J. Biol. Chem. 1974;249:5388–5393. [PubMed] [Google Scholar]
- 66.Mason P.E., Neilson G.W., Dempsey C.E., Barnes A.C., Cruickshank J.M. The hydration structure of guanidinium and thiocyanate ions: Implications for protein stability in aqueous solution. Proc. Natl. Acad. Sci. USA. 2003;100:4557–4561. doi: 10.1073/pnas.0735920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zou Q., Habermann-Rottinghaus S.M., Murphy K.P. Urea effects on protein stability: Hydrogen bonding and the hydrophobic effect. Proteins. 1998;31:107–115. [PubMed] [Google Scholar]
- 68.Kunza W., Lo Nostrob P., Ninham B.W. The present state of affairs with Hofmeister effects. Curr. Opin. Colloid Interface Sci. 2004;9:1–18. [Google Scholar]
- 69.Kunz W., Henle J., Ninham B.W. ‘Zur Lehre von der Wirkung der Salze’ (about the science of the effect of salts): Franz Hofmeister’s historical papers. Curr. Opin. Colloid Interface Sci. 2004;9:19–37. [Google Scholar]
- 70.Collins K.D. Ions from the Hofmeister series and osmolytes: Effects on proteins in solution and in the crystallization process. Methods. 2004;34:300–311. doi: 10.1016/j.ymeth.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y., Cremer P.S. Chemistry of Hofmeister anions and osmolytes. Annu. Rev. Phys. Chem. 2010;61:63–83. doi: 10.1146/annurev.physchem.59.032607.093635. [DOI] [PubMed] [Google Scholar]
- 72.Arakawa T., Timasheff S.N. Preferential interactions of proteins with salts in concentrated solutions. Biochemistry. 1982;21:6545–6552. doi: 10.1021/bi00268a034. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y., Cremer P.S. Interactions between macromolecules and ions: The Hofmeister series. Curr. Opin. Chem. Biol. 2006;10:658–663. doi: 10.1016/j.cbpa.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 74.Kozo H., Geiduschek E.P. The effect of electrolytes on the stability of the deoxyribonucleate helix. J. Am. Chem. Soc. 1962;84:1329–1338. [Google Scholar]
- 75.Zangi R. Can salting-in/salting-out ions be classified as chaotropes/kosmotropes? J. Phys. Chem. B. 2010;114:643–650. doi: 10.1021/jp909034c. [DOI] [PubMed] [Google Scholar]
- 76.Zangi R., Berne B.J. Aggregation and dispersion of small hydrophobic particles in aqueous electrolyte solutions. J. Phys. Chem. B. 2006;110:22736–22741. doi: 10.1021/jp064475+. [DOI] [PubMed] [Google Scholar]
- 77.Corridoni T., Mancinelli R., Ricci M.A., Bruni F. Viscosity of aqueous solutions and local microscopic structure. J. Phys. Chem. B. 2011;115:14008–14013. doi: 10.1021/jp202755u. [DOI] [PubMed] [Google Scholar]
- 78.Kanai S., Liu J., Patapoff T.W., Shire S.J. Reversible self-association of a concentrated monoclonal antibody solution mediated by Fab-Fab interaction that impacts solution viscosity. J. Pharm. Sci. 2008;97:4219–4227. doi: 10.1002/jps.21322. [DOI] [PubMed] [Google Scholar]
- 79.Du W., Klibanov A.M. Hydrophobic salts markedly diminish viscosity of concentrated protein solutions. Biotechnol. Bioeng. 2011;108:632–636. doi: 10.1002/bit.22983. [DOI] [PubMed] [Google Scholar]
- 80.Guo Z., Chen A., Nassar R.A., Helk B., Mueller C., Tang Y., Gupta K., Klibanov A.M. Structure-activity relationship for hydrophobic salts as viscosity-lowering excipients for concentrated solutions of monoclonal antibodies. Pharm. Res. 2012;29:3102–3109. doi: 10.1007/s11095-012-0802-9. [DOI] [PubMed] [Google Scholar]
- 81.Hamada H., Arakawa T., Shiraki K. Effect of additives on protein aggregation. Curr. Pharm. Biotechnol. 2009;10:400–407. doi: 10.2174/138920109788488941. [DOI] [PubMed] [Google Scholar]
- 82.Arakawa T., Kita Y. Multi-faceted arginine: Mechanism of the effects of arginine on protein. Curr. Protein Pept. Sci. 2014;15:608–620. doi: 10.2174/138920371506140818113015. [DOI] [PubMed] [Google Scholar]
- 83.Buchner J., Rudolph R. Renaturation, purification and characterization of recombinant Fab-fragments produced in Escherichia coli. Biotechnology (N. Y.) 1991;9:157–162. doi: 10.1038/nbt0291-157. [DOI] [PubMed] [Google Scholar]
- 84.Shiraki K., Kudou M., Fujiwara S., Imanaka T., Takagi M. Biophysical effect of amino acids on the prevention of protein aggregation. J. Biochem. 2002;132:591–595. doi: 10.1093/oxfordjournals.jbchem.a003261. [DOI] [PubMed] [Google Scholar]
- 85.Hirano A., Arakawa T., Shiraki K. Arginine increases the solubility of coumarin: Comparison with salting-in and salting-out additives. J. Biochem. 2008;144:363–369. doi: 10.1093/jb/mvn078. [DOI] [PubMed] [Google Scholar]
- 86.Hirano A., Kameda T., Arakawa T., Shiraki K. Arginine-assisted solubilization system for drug substances: Solubility experiment and simulation. J. Phys. Chem. B. 2010;114:13455–13462. doi: 10.1021/jp101909a. [DOI] [PubMed] [Google Scholar]
- 87.Ito L., Shiraki K., Matsuura T., Okumura M., Hasegawa K., Baba S., Yamaguchi H., Kumasaka T. High-resolution X-ray analysis reveals binding of arginine to aromatic residues of lysozyme surface: Implication of suppression of protein aggregation by arginine. Protein Eng. Des. Sel. 2011;24:269–274. doi: 10.1093/protein/gzq101. [DOI] [PubMed] [Google Scholar]
- 88.Mason P.E., Neilson G.W., Enderby J.E., Saboungi M.L., Dempsey C.E., MacKerell A.D., Jr, Brady J.W. The structure of aqueous guanidinium chloride solutions. J. Am. Chem. Soc. 2004;126:11462–11470. doi: 10.1021/ja040034x. [DOI] [PubMed] [Google Scholar]
- 89.Li J., Garg M., Shah D., Rajagopalan R. Solubilization of aromatic and hydrophobic moieties by arginine in aqueous solutions. J. Chem. Phys. 2010;133:054902. doi: 10.1063/1.3469790. [DOI] [PubMed] [Google Scholar]
- 90.Hirano A., Arakawa T., Shiraki K. Arginine increases the solubility of coumarin: Comparison with salting-in and salting-out additives. J. Biochem. 2008;27:253–257. doi: 10.1093/jb/mvn078. [DOI] [PubMed] [Google Scholar]
- 91.Ariki R., Hirano A., Arakawa T., Shiraki K. Arginine increases solubility of alkyl gallates through interaction with the aromatic ring. J. Biochem. 2011;149:389–394. doi: 10.1093/jb/mvr004. [DOI] [PubMed] [Google Scholar]
- 92.Arakawa T., Ejima D., Tsumoto K., Obeyama N., Tanaka Y., Kita Y., Timasheff S.N. Suppression of protein interactions by arginine: A proposed mechanism of the arginine effects. Biophys. Chem. 2007;127:1–8. doi: 10.1016/j.bpc.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 93.Schneider C.P., Trout B.L. Investigation of cosolute-protein preferential interaction coefficients: New insight into the mechanism by which arginine inhibits aggregation. J. Phys. Chem. B. 2009;113:2050–2058. doi: 10.1021/jp808042w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shukla D., Trout B.L. Preferential interaction coefficients of proteins in aqueous arginine solutions and their molecular origins. J. Phys. Chem. B. 2011;115:1243–1253. doi: 10.1021/jp108586b. [DOI] [PubMed] [Google Scholar]
- 95.Scherer T.M. Role of cosolute-protein interactions in the dissociation of monoclonal antibody clusters. J. Phys. Chem. B. 2015;119:13027–13038. doi: 10.1021/acs.jpcb.5b07568. [DOI] [PubMed] [Google Scholar]
- 96.Nakakido M., Tanaka Y., Mitsuhori M., Kudou M., Ejima D., Arakawa T., Tsumoto K. Structure-based analysis reveals hydration changes induced by arginine hydrochloride. Biophys. Chem. 2008;137:105–109. doi: 10.1016/j.bpc.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 97.Shukla D., Trout B.L. Interaction of arginine with proteins and the mechanism by which it inhibits aggregation. J. Phys. Chem. B. 2010;114:13426–13438. doi: 10.1021/jp108399g. [DOI] [PubMed] [Google Scholar]
- 98.Inoue N., Takai E., Arakawa T., Shiraki K. Specific decrease in solution viscosity of antibodies by arginine for therapeutic formulations. Mol. Pharm. 2014;11:1889–1896. doi: 10.1021/mp5000218. [DOI] [PubMed] [Google Scholar]
- 99.Kheddo P., Tracka M., Armer J., Dearman R.J., Uddin S., van der Walle C.F., Golovanov A.P. The effect of arginine glutamate on the stability of monoclonal antibodies in solution. Int. J. Pharm. 2014;473:126–133. doi: 10.1016/j.ijpharm.2014.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fukuda M., Kameoka D., Torizawa T., Saitoh S., Yasutake M., Imaeda Y., Koga A., Mizutani A. Thermodynamic and fluorescence analyses to determine mechanisms of IgG1 stabilization and destabilization by arginine. Pharm. Res. 2013;31:992–1001. doi: 10.1007/s11095-013-1221-2. [DOI] [PubMed] [Google Scholar]
- 101.Fukuda M., Moriyama C., Yamazaki T., Imaeda Y., Koga A. Quantitative correlation between viscosity of concentrated MAb solutions and particle size parameters obtained from small-angle X-ray scattering. Pharm. Res. 2015;32:3804–3812. doi: 10.1007/s11095-015-1739-6. [DOI] [PubMed] [Google Scholar]
- 102.Zhang J., Frey V., Corcoran M., Zhang-van Enk J., Subramony J.A. Influence of arginine salts on the thermal stability and aggregation kinetics of monoclonal antibody: Dominant role of anions. Mol. Pharm. 2016;13:3362–3369. doi: 10.1021/acs.molpharmaceut.6b00255. [DOI] [PubMed] [Google Scholar]
- 103.Yoshizawa S., Arakawa T., Shiraki K. Effect of counter ions of arginine as an additive for the solubilization of protein and aromatic compounds. Int. J. Biol. Macromol. 2016;91:471–476. doi: 10.1016/j.ijbiomac.2016.05.085. [DOI] [PubMed] [Google Scholar]
- 104.Salis A., Monduzzi M. Not only pH. Specific buffer effects in biological systems. Curr. Opin. Colloid Interface Sci. 2016;23:1–9. [Google Scholar]
- 105.Roberts D., Keeling R., Tracka M., van der Walle C.F., Uddin S., Warwicker J., Curtis R. Specific ion and buffer effects on protein-protein interactions of a monoclonal antibody. Mol. Pharm. 2015;12:179–193. doi: 10.1021/mp500533c. [DOI] [PubMed] [Google Scholar]
- 106.Chen B., Bautista R., Yu K., Zapata G.A., Mulkerrin M.G., Chamow S.M. Influence of histidine on the stability and physical properties of a fully human antibody in aqueous and solid forms. Pharm. Res. 2003;20:1952–1960. doi: 10.1023/b:pham.0000008042.15988.c0. [DOI] [PubMed] [Google Scholar]
- 107.Liao S.M., Du Q.S., Meng J.Z., Pang Z.W., Huang R.B. The multiple roles of histidine in protein interactions. Chem. Cent. J. 2013;7:44. doi: 10.1186/1752-153X-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nicoud L., Lattuada M., Yates A., Morbidelli M. Impact of aggregate formation on the viscosity of protein solutions. Soft Matter. 2015;11:5513–5522. doi: 10.1039/c5sm00513b. [DOI] [PubMed] [Google Scholar]
- 109.Imamura H., Honda S. Kinetics of antibody aggregation at neutral pH and ambient temperatures triggered by temporal exposure to acid. J. Phys. Chem. B. 2016;120:9581–9589. doi: 10.1021/acs.jpcb.6b05473. [DOI] [PubMed] [Google Scholar]
- 110.Morris A.M., Watzky M.A., Finke R.G. Protein aggregation kinetics, mechanism, and curve-fitting: A review of the literature. Biochim. Biophys. Acta. 2009;1794:375–397. doi: 10.1016/j.bbapap.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 111.Murphy R.M., Roberts C.J. Protein misfolding and aggregation research: Some thoughts on improving quality and utility. Biotechnol. Prog. 2013;29:1109–1115. doi: 10.1002/btpr.1812. [DOI] [PubMed] [Google Scholar]
- 112.Stradner A., Sedgwick H., Cardinaux F., Poon W.C., Egelhaaf S.U., Schurtenberger P. Equilibrium cluster formation in concentrated protein solutions and colloids. Nature. 2004;432:492–495. doi: 10.1038/nature03109. [DOI] [PubMed] [Google Scholar]
- 113.Sciortino F., Mossa S., Zaccarelli E., Tartaglia P. Equilibrium cluster phases and low-density arrested disordered states: The role of short-range attraction and long-range repulsion. Phys. Rev. Lett. 2004;93:055701. doi: 10.1103/PhysRevLett.93.055701. [DOI] [PubMed] [Google Scholar]
- 114.Mossa S., Sciortino F., Tartaglia P., Zaccarelli E. Ground-state clusters for short-range attractive and long-range repulsive potentials. Langmuir. 2004;20:10756–10763. doi: 10.1021/la048554t. [DOI] [PubMed] [Google Scholar]
- 115.Campbell A.I., Anderson V.J., van Duijneveldt J.S., Bartlett P. Dynamical arrest in attractive colloids: The effect of long-range repulsion. Phys. Rev. Lett. 2005;94:208301. doi: 10.1103/PhysRevLett.94.208301. [DOI] [PubMed] [Google Scholar]
- 116.Falus P., Porcar L., Fratini E., Chen W.R., Faraone A., Hong K., Baglioni P., Liu Y. Distinguishing the monomer to cluster phase transition in concentrated lysozyme solutions by studying the temperature dependence of the short-time dynamics. J. Phys. Condens. Matter. 2012;24:064114. doi: 10.1088/0953-8984/24/6/064114. [DOI] [PubMed] [Google Scholar]
- 117.Porcar L., Falus P., Chen W.R., Faraone A., Fratini E., Hong K., Baglioni P., Liu Y. Formation of the dynamic clusters in concentrated lysozyme protein solutions. J. Phys. Chem. Lett. 2010;1:126–129. [Google Scholar]
- 118.Cardinaux F., Zaccarelli E., Stradner A., Bucciarelli S., Farago B., Egelhaaf S.U., Sciortino F., Schurtenberger P. Cluster-driven dynamical arrest in concentrated lysozyme solutions. J. Phys. Chem. B. 2011;115:7227–7237. doi: 10.1021/jp112180p. [DOI] [PubMed] [Google Scholar]
- 119.Liu Y., Porcar L., Chen J., Chen W.R., Falus P., Faraone A., Fratini E., Hong K., Baglioni P. Lysozyme protein solution with an intermediate range order structure. J. Phys. Chem. B. 2011;115:7238–7247. doi: 10.1021/jp109333c. [DOI] [PubMed] [Google Scholar]
- 120.Godfrin P.D., Hudson S.D., Hong K., Porcar L., Falus P., Wagner N.J., Liu Y. Short-time glassy dynamics in viscous protein solutions with competing interactions. Phys. Rev. Lett. 2015;115:228302. doi: 10.1103/PhysRevLett.115.228302. [DOI] [PubMed] [Google Scholar]
- 121.Lilyestrom W.G., Yadav S., Shire S.J., Scherer T.M. Monoclonal antibody self-association, cluster formation, and rheology at high concentrations. J. Phys. Chem. B. 2013;117:6373–6384. doi: 10.1021/jp4008152. [DOI] [PubMed] [Google Scholar]
- 122.Godfrin P.D., Zarraga I.E., Zarzar J., Porcar L., Falus P., Wagner N.J., Liu Y. Effect of hierarchical cluster formation on the viscosity of concentrated monoclonal antibody formulations studied by neutron scattering. J. Phys. Chem. B. 2016;120:278–291. doi: 10.1021/acs.jpcb.5b07260. [DOI] [PubMed] [Google Scholar]
- 123.Borwankar A.M., Dinin A.K., Laber J.R., Twu A., Wilson B.K., Maynard J.A., Truskett T.M., Johnston K.P. Tunable equilibrium nanocluster dispersions at high protein concentrations. Soft Matter. 2013;9:1766–1771. [Google Scholar]
- 124.Johnston K.P., Maynard J.A., Truskett T.M., Borwankar A.U., Miller M.A., Wilson B.K., Dinin A.K., Khan T.A. Kaczorowski, K.J. Concentrated dispersions of equilibrium protein nanoclusters that reversibly dissociate into active monomers. ACS Nano. 2012;28:1357–1369. doi: 10.1021/nn204166z. [DOI] [PubMed] [Google Scholar]
- 125.Kurinomaru T., Shiraki K. Aggregative protein-polyelectrolyte complex for high-concentration formulation of protein drugs. Int. J. Biol. Macromol. 2017;100:11–17. doi: 10.1016/j.ijbiomac.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 126.Kurinomaru T., Maruyama T., Izaki S., Handa K., Kimoto T., Shiraki K. Protein-poly(amino acid) complex precipitation for high-concentration protein formulation. J. Pharm. Sci. 2014;103:2248–2254. doi: 10.1002/jps.24025. [DOI] [PubMed] [Google Scholar]
- 127.Izaki S., Kurinomaru T., Maruyama T., Uchida T., Handa K., Kimoto T., Shiraki K. Feasibility of antibody-poly(glutamic acid) complexes: Preparation of high-concentration antibody formulations and their pharmaceutical properties. J. Pharm. Sci. 2015;104:1929–1937. doi: 10.1002/jps.24422. [DOI] [PubMed] [Google Scholar]
- 128.Maruyama T., Izaki S., Kurinomaru T., Handa K., Kimoto T., Shiraki K. Protein-poly(amino acid) precipitation stabilizes a therapeutic protein l-asparaginase against physicochemical stress. J. Biosci. Bioeng. 2015;120:720–724. doi: 10.1016/j.jbiosc.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 129.Izaki S., Kurinomaru T., Handa K., Kimoto T., Shiraki K. Stress tolerance of antibody-poly(Amino Acid) complexes for improving the stability of high concentration antibody formulations. J. Pharm. Sci. 2015;104:2457–2463. doi: 10.1002/jps.24515. [DOI] [PubMed] [Google Scholar]
- 130.Shiraki K., Kurinomaru T., Tomita S. Wrap-and-strip technology of protein-polyelectrolyte complex for biomedical application. Curr. Med. Chem. 2016;23:276–289. doi: 10.2174/0929867323666151127201126. [DOI] [PubMed] [Google Scholar]