ABSTRACT
Immune responses in human populations are highly variable, with this variability presenting challenges for vaccine design. As such a better understanding of the factors that determine this variability will help in the development of precision vaccination strategies. The Milieu Interieur consortium was established to address this challenge through a definition of the normal boundaries of a healthy immune response, and the characterization of their genetic and environmental determinants. To do this we have implemented standardized tools for monitoring functional immune responses at the proteomic and transcriptional level, which have been applied to a 1,000 healthy donor cohort. This approach has recently allowed us to quantify the extent of genetic control of cellular variability and transcriptional responses to infection. Initial findings on the influence of age, sex, and genetics may already be included in considerations for improved vaccine development, and ongoing analysis will further define the factors behind inter-individual variability in diverse immune responses. This approach will help to guide the development of the next generation of vaccines that will take into account differences in populations and eventually individuals.
KEYWORDS: Immunity, Healthy donors, Immunomonitoring, Systems Immunology, Genetic associations, adjuvants, DNA, immune modulators, influenza, molecular biology, vaccinology
Dissecting immune response variability
Human immune responses show high levels of inter-individual variability across all populations.1 Part of this variability is a result of the flexible and dynamic nature of immune responses, which also makes possible vaccination strategies for a variety of diseases ranging from infection to cancer. However, this inherent variability can also present challenges for vaccine design as not all recipients will respond in the same manner or to the same degree. Well known examples include the Hepatitis B vaccine from which 5–10% of healthy recipients fail to seroconvert.2 While personalized vaccines may be applicable in conditions such as cancer, they are not yet a feasible approach for preventative population-based interventions. Nevertheless, precision vaccination strategies may take into account more general population differences in order to improve vaccine efficacy and minimize potential adverse reactions. However, to achieve this we first need to better understand and define what factors contribute to and define inter-individual immune response heterogeneity. It was within this context that the Milieu Interieur consortium was established in 2011 to define the normal boundaries of a healthy immune response in a European population, and to define their genetic and environmental determinants.3 Additional complementary healthy human cohorts have been established in parallel to address similar questions that include the Human Functional Genomics Project,4 the Human Immunology Project Consortium (HIPC),5" and the 10K Immunomes.6 From these studies we are now beginning to obtain a better definition of immune response variability and the key factors that drive it.
Variability in immune responses can be due to multiple factors that may be biological (age, sex), genetic (SNPs, gene methylation), environmental (microbiome, latent or chronic infections), or lifestyle (diet, smoking, medical history) factors. While many studies have examined how each of these components individually impact immune responses, fewer studies have successfully combined these co-factors in an integrative analysis. From analysis of the Milieu Interieur cohort we recently identified both genetic and non-genetic determinants that contribute to variance within circulating immune cell populations.7 As immune cells are the major targets for most vaccines it is important to understand how and why they may differ between individuals. Leveraging the 1,000 well-defined healthy donors and the associated metadata, we identified a significant impact of age, sex and latent cytomegalovirus (CMV) infection on numbers of adaptive immune cells such as T and B cells.7 These are the immune cells whose differentiation into long-lived memory cells is the classical hallmark of successful vaccination. Taking advantage of our age-balanced cohort we showed that naïve CD8+ T cells decrease more than twice as rapidly with age as compared to naïve CD4+ T cells (3.5% and 1.5% per year, respectively) showing how age may significantly impact vaccines that target these cellular populations.7 CMV latent infection was also revealed to have a major impact on the differentiation status of T cell subsets and was associated with a 12- and 4.5-times higher number of CD4+ and CD8+ T effector memory (TEM) and T effector memory RA (TEMRA) cells.7 Another strong environmental or lifestyle effect was active smoking, which showed a significant positive effect on the number of circulating leukocytes, and particularly in the number of circulating regulatory T cells. Interestingly, genetic effects on circulating leukocytes were more preferentially identified in innate immune cell populations as compared to adaptive cells. In total 25% of the measured cellular immunophenotypes showed a genome-wide significant association (p < 1 × 10−10), and of those phenotypes, 80% were innate immune cell-specific.7 This may reflect the shorter half-life of innate cells as compared to adaptive cells, resulting in environmental effects having a stronger relative influence on these cells as compared to potential genetic determinants. Extrapolating from these results for precision vaccination strategies, we could propose that adjuvant selection should consider genetic variability as it targets innate immune cells, and antigen selection, which is more relevant for adaptive responses should include factors such as age, CMV serostatus and smoking. However, prospective in vivo human vaccination clinical studies, such as those planned by the Human Vaccine Project, are required to test and validate such hypotheses.8
The need for standardized immunomonitoring
While the phenotyping of cellular populations can reveal important determinants of immune variability, immune responses are highly dynamic and often require stimulation or perturbation to reveal their full depth and function. For example, vaccine efficacy studies often require ex vivo antigen stimulation of cells from the recipient to assess induction of T cell-mediated immunity by intra-cellular flow cytometry or ELISPOT assays, with many efforts recently made to standardize the reporting of such approaches.9 These techniques usually rely upon the isolation of peripheral blood mononuclear cells (PBMCs) by density gradient separation. However, PBMC isolation introduces technical variability and remains challenging to standardize within and across clinical laboratories. It also requires trained laboratory staff and sterile conditions, with the blood often transported from clinical sites to central labs, potentially introducing pre-analytic variability. In addition, such experimental manipulation of innate immune cells can also induce non-specific activation or cell death. To overcome these challenges within Milieu Interieur we have implemented whole blood stimulation assays using TruCulture syringe-based devices. Working with whole blood also includes granulocytes, platelets and plasmatic components, which can make important contributions to immune responses. Recently, we described a multi-center clinical study that directly compared TruCulture with conventional PBMCs, utilizing lipopolysaccharide (LPS) and combined anti-CD3/CD28 antibodies as the stimulants and Luminex multi-analyte proteins as the readout.10 The major finding was the improved reproducibility of TruCulture as compared to PBMC stimulation, in particular, for inter-center differences highlighting the suitability of this approach for multi-center immunomonitoring studies.10 Other groups have utilized similar standardized whole blood plate-based stimulations that require lower blood volumes, an advantage for pediatric studies.11
Dissecting signatures of induced immune responses for correlates of protection
An additional advantage of TruCulture is its adaptability to diverse immune stimuli and that the biological readout can be modified to monitor alterations in cellular phenotype,12 induced protein secretion,12 RNA expression changes,13 and metabolic or lipidomic functional changes. Transcriptional analysis of stimulated whole blood can be challenging due to multiple technical reasons, but we have developed a high-throughput single-step RNA extraction method from stimulated TruCulture samples allied to a Nanostring hybridization array readout.13 Utilizing the diverse stimulation conditions applied to the Milieu Interieur cohort, we tested the hypothesis that the transcriptional signature of key effector cytokines can capture the responses to Toll-like Receptor (TLR) ligands or microbes. To do this we analyzed the expression of 572 genes in 25 healthy donors following stimulation with 4 key innate cytokines (IFN-β, IFN-γ, IL-1β, and TNF-α) applying unsupervised principal component analysis (PCA) and a linear support vector machine (SVM) algorithm.13 This allowed the identification of a 44-gene signature that captured the diversity of complex innate immune responses with improved segregation between distinct TLR and microbial stimuli. Furthermore, we can now use this approach to assess how potential vaccine adjuvants such as TLR ligands induce specific cytokine associated gene expression (Fig. 1).
Figure 1.
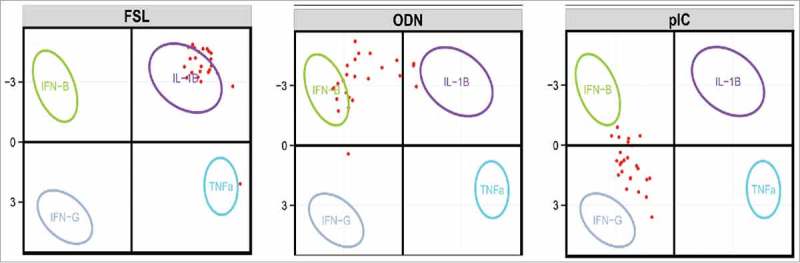
PCA defined by the eigenvectors and eigenvalues as based on the four-cytokine (IFN-β, IFN-γ, IL-1β, and TNF-α) induced mRNA expression data of the 44 genes that were defined to capture the optimal signature. Ellipses representing 95% confidence interval (CI) were constructed and replaced the individual samples. Projected sample vectors of TLR stimuli (shown in red) for each of the 25 donors (FSL, ODN, pIC), individually projected onto the first 2 PC vectors, using the 44 selected genes. PC1 is on the X axis, and PC2 on the Y axis. (From Urrutia et al, Cell Reports 201613 using the paper R Shiny application at https://www.synapse.org/ – !Synapse:syn7059574).
From analysis of 25 healthy donors we observed differential levels of inter-individual variability in gene expression depending on the TLR targeted. For example, the responses to fibroblast-stimulating lipopeptide (FSL-1) stimulation, which targets TLR2, were associated with IL-1β-induced gene expression with relatively little inter-donor variability (with the exception of a single outlier donor who revealed a TNF-α response). In contrast, for CpG oligodeoxynucleotides (ODN) that activate TLR9, the majority of donors mapped to IFN-β transcriptomic responses as expected, but certain individuals showed more IL-1β-driven responses. Similarly, the responses to polyinosinic:polycytidylic acid (poly I:C), which activates TLR3, were spread between the IFN-γ and IFN-β-induced gene signatures. Collectively, these results illustrate how inter-individual differences to TLR stimulation driven by vaccination may result in differing immune responses and variable efficacy and protection. As such, a better understanding of the factors behind such variability could be integrated into vaccine development and eventually included in precision vaccination strategies.
Integrating non-genetic differences in vaccine responses
Given such observed high levels of inter-individuality in induced immune responses, large sample sizes are required for sufficient statistical power to dissect which factors drive this variability. We recently utilized the 1,000 donor cohort of Milieu Interieur to dissect the relative contributions of age, sex, cellular composition and genetics to induced immune responses to microbial stimulation.14 Of particular interest for vaccine responses was the age-specific association with IFN-γ, a commonly used correlate of protection in vaccine clinical studies given its key role in adaptive immunity.15 Notably, IFN-γ gene expression showed a strong and significant decline with age for bacterial, fungal, and viral responses (Fig. 2). Gene expression data sets were regressed on the numbers of major circulating immune cells, indicating that this was not merely due to a decline in CD4+ or CD8+ T cells as observed by flow cytometric analysis. In contrast, IFN-γ gene expression following stimulation with a superantigen, staphylococcal enterotoxin B (SEB), did not decline with age (Fig. 2). Therefore while immune responses are known to generally weaken with age,16 these results show that it is both stimulation and the specific context that have important consequences for vaccination strategies in elderly populations.
Figure 2.
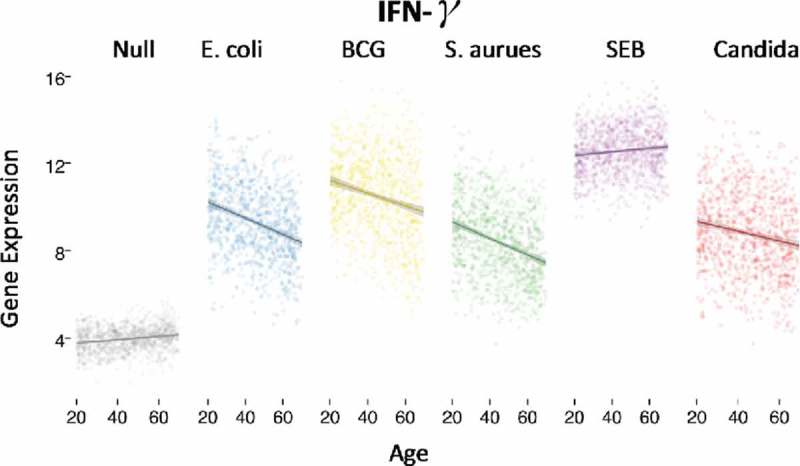
Age specific decline of IFNγ gene expression following stimulation with E. coli, BCG, S. aurues, and C. albicans, but not SEB. Each dot represents an individual of the 1,000 donor MI cohort as stratified by age, and IFNγ gene expression measured by Nanostring. (Adapted from Piasecka et al, PNAS 201816 using the R Shiny application at http://www.misage.pasteur.fr).
Another interesting age-specific association that we observed was related to the influenza-induced response. We observed some striking gene expression differences between donors aged 20–29 and 30–39 years old. These differences were also observed between the youngest donors (20-29) and the older decades (40-69 years) but the largest differences were seen between these two specific groups. We suggest that this may reflect pre-existing immunity in the older cohort to the strain used in the TruCulture tube (H1N1) that may not be present in the youngest group. This lack of immunity in the younger group could possibly be explained by antigenic drift of circulating influenza strains that occurred since 1982 (when 29 year old donors were born) resulting in the absence of exposure to this specific strain. This hypothesis is supported by the relatively higher induction of anti-viral response genes (e.g., type I interferon responses), and the lower induction of antibody associated genes (e.g., FCGRT, CR1) in younger donors. Our gene expression results correspond with a very similar age-specific association that was observed in a recent H1NI vaccination study, where most gene expression divergence was also observed in 30 year old donors17 and older participants. However, no age-associated differences were observed in their influenza-specific antibodies, as measured by hemagglutination inhibition (HAI) or microneutrualization (MN) assays, suggesting that differential gene expression between the younger and older groups was not due to preexisting influenza exposure or B cell memory responses. immunity. The alternative suggestion is that it reflects a real decline in influenza vaccine responses from the age of 30 years onward, which suggests a decline in immune responsiveness much earlier than previously believed. A recent multi-cohort analysis from the HIPC revealed a baseline transcriptional signature that was predictive of influenza vaccination responses.18 Intriguingly, an inflammatory gene signature was associated with better antibody responses to vaccination in young individuals (<35 years), and with worse responses in older individuals (>60 years).18 Additional transcriptional analysis will be required to see whether this signature can also be identified in an ex vivo-like stimulation and therefore used to potentially stratify vaccine recipients.
Integrating genetic differences in vaccine responses
Due to rapid advances in technology and subsequent decreases in the cost of analysis, the integration of genetic information is an increasingly attractive approach for tailoring vaccines to distinct populations.19 In addition, many vaccines have been initially developed in Western Caucasian populations but then show decreased efficacy in other ethnic backgrounds. Understanding how host genetic differences may contribute to differential immune responses may therefore help to guide vaccine development and implementation, however a better basic understanding of genetic regulation of immunity is still required. Towards this goal, we recently described within Milieu Interieur a master genetic regulator of bacteria-induced immune responses. Variability in the TLR 1/6/10 gene locus regulated 105 genes after stimulation with E. coli, 80 genes after stimulation with BCG, 7 genes after stimulation with S. aureus, and 13 genes after stimulation with SEB14 (measurement of 560 immune related genes). Individuals with the homozygous dominant, TT genotype, of the most differential single nucleotide polymorphism (SNP) (rs4833095, T-allele frequency = 0.79) displayed lower expression for many inflammatory response genes (e.g., IL1B, IL6, IL12B) and higher expression for regulatory response genes (e.g., TGFB1, TGFBI, IL1RAP). This genetic variant may be highly relevant for explaining differences among ethnic populations as work from our group previously showed that this specific genetic regulation is more prevalent in Europeans as compared to Africans.20 Therefore, this approach may explain not only immune response heterogeneity present in Europeans, but perhaps some of the variability observed between individuals from different racial and ethnic backgrounds populations. Additional studies will continue to further explore how these differences may be regulated at the agonist and ligand level and in more diverse immune phenotypes.
Conclusion
In summary, recent population-based studies that focused on healthy populations and integrated genetic and immunological phenotyping have begun to define the factors behind variable immune responses.1 Depending on the specific immune phenotype studied, up to 50% of the observed variance is estimated to be due to environmental factors,21 with an estimated 20–40% due to genetics.22,23 However, a striking amount of the specific individual factors behind this variation remains to be defined, and in particular, the specific environmental factors that act to influence immunity. The large percentage of unexplained variation is likely due to complex gene-environment interactions that will require large scale and longitudinal population based studies to decipher more fully. Nevertheless, such an understanding is crucial to achieve the promise of precision vaccination strategies that will take into account an individual's specific immune response to optimize efficacy and reduce adverse reactions. This will ensure that vaccination strategies capitalize on recent scientific progress and are adapted to address 21st century public health challenges for a biologically, genetically and environmentally diverse global population.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The author acknowledges support from the Milieu Interieur consortium, part of the French government's “Invest in the Future program,” managed by the Agence Nationale de la Recherche (ANR, reference 10-LABX-69-01), and thanks Dr Stacey Wooden for critical review of the manuscript.
References
- 1.Liston A, Carr EJ, Linterman MA. Shaping variation in the human immune system. Trends Immunol. 2016;37:637–46. doi: 10.1016/j.it.2016.08.002. PMID:27692231. [DOI] [PubMed] [Google Scholar]
- 2.McDermott AB, Cohen SB, Zuckerman JN, Madrigal JA. Hepatitis B third-generation vaccines: improved response and conventional vaccine non-response–evidence for genetic basis in humans. J Viral Hepat. 1998;5 (Suppl 2:9–11. doi: 10.1046/j.1365-2893.1998.0050s2009.x. PMID:9857354. [DOI] [PubMed] [Google Scholar]
- 3.Thomas S, Rouilly V, Patin E, Alanio C, Dubois A, Delval C, Marquier LG, Fauchoux N, Sayegrih S, Vray M, et al. . The milieu intérieur study – an integrative approach for study of human immunological variance. Clin Immunol Orlando Fla. 2015;157:277–93. doi: 10.1016/j.clim.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Pappalardo JL, Hafler DA. The Human Functional Genomics Project: Understanding generation of diversity. Cell. 2016;167:894–6. doi: 10.1016/j.cell.2016.10.040. PMID:27814519. [DOI] [PubMed] [Google Scholar]
- 5.Brusic V, Gottardo R, Kleinstein SH, Davis MM, HIPC steering committee Computational resources for high-dimensional immune analysis from the Human Immunology Project Consortium. Nat Biotechnol. 2014;32:146–8. doi: 10.1038/nbt.2777. PMID:24441472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zalocusky KA. et al. . The 10,000 Immunomes Project: A resource for human immunology. bioRxiv. 2017. [Google Scholar]
- 7.Patin E, Hasan M, Bergstedt J, Rouilly V, Libri V, Urrutia A, Alanio C, Scepanovic P, Hammer C, Jönsson F, et al. . Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat Immunol. 2018. doi: 10.1038/s41590-018-0049-7. PMID:29476184. [DOI] [PubMed] [Google Scholar]
- 8.Koff WC, Gust ID, Plotkin SA. Toward a human vaccines project. Nat Immunol. 2014;15:589–92. doi: 10.1038/ni.2871. PMID:24940943. [DOI] [PubMed] [Google Scholar]
- 9.Janetzki S. et al. . ‘MIATA’-minimal information about T cell assays. Immunity. 2009;31:527–8. doi: 10.1016/j.immuni.2009.09.007. PMID:19833080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy D, Rouilly V, Braudeau C, Corbière V, Djebali R, Ungeheuer MN, Josien R, LaBrie ST, Lantz O, Louis D, et al. . Standardized whole blood stimulation improves immunomonitoring of induced immune responses in multi-center study. Clin Immunol Orlando Fla 2017;183:325–35. doi: 10.1016/j.clim.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Smolen KK, Ruck CE, Fortuno ES, Ho K, Dimitriu P, Mohn WW, Speert DP, Cooper PJ, Esser M, Goetghebuer T, et al. . Pattern recognition receptor-mediated cytokine response in infants across 4 continents. J Allergy Clin Immunol. 2014;133:818–26.e4. doi: 10.1016/j.jaci.2013.09.038. PMID:24290283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duffy D, Rouilly V, Libri V, Hasan M, Beitz B, David M, Urrutia A, Bisiaux A, Labrie ST, Dubois A, et al. . Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity. 2014;40:436–50. doi: 10.1016/j.immuni.2014.03.002. PMID:24656047. [DOI] [PubMed] [Google Scholar]
- 13.Urrutia A, Duffy D, Rouilly V, Posseme C, Djebali R, Illanes G, Libri V, Albaud B, Gentien D, Piasecka B, et al. . Standardized whole-blood transcriptional profiling enables the deconvolution of complex induced immune responses. Cell Rep. 2016;16:2777–91. doi: 10.1016/j.celrep.2016.08.011. PMID:27568558. [DOI] [PubMed] [Google Scholar]
- 14.Piasecka B, Duffy D, Urrutia A, Quach H, Patin E, Posseme C, Bergstedt J, Charbit B, Rouilly V, MacPherson CR, et al. . Distinctive roles of age, sex, and genetics in shaping transcriptional variation of human immune responses to microbial challenges. Proc Natl Acad Sci USA. 2018;115:E488–97. doi: 10.1073/pnas.1714765115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thakur A, Pedersen LE, Jungersen G. Immune markers and correlates of protection for vaccine induced immune responses. Vaccine. 2012;30:4907–20. doi: 10.1016/j.vaccine.2012.05.049. PMID:22658928. [DOI] [PubMed] [Google Scholar]
- 16.Nikolich-Žugich J. The twilight of immunity: Emerging concepts in aging of the immune system. Nat Immunol. 2018;19:10–19. doi: 10.1038/s41590-017-0006-x. PMID:29242543. [DOI] [PubMed] [Google Scholar]
- 17.Sobolev O, Binda E, O'Farrell S, Lorenc A, Pradines J, Huang Y, Duffner J, Schulz R, Cason J, Zambon M, et al. . Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat Immunol. 2016;17:204–13. doi: 10.1038/ni.3328. PMID:26726811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. HIPC-CHI Signatures Project Team & HIPC-I Consortium Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci Immunol. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mentzer AJ, O'Connor D, Pollard AJ, Hill AVS. Searching for the human genetic factors standing in the way of universally effective vaccines. Philos Trans R Soc Lond B Biol Sci. 2015;370. doi: 10.1098/rstb.2014.0341. PMID:25964463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quach H, Rotival M, Pothlichet J, Loh YE, Dannemann M, Zidane N, Laval G, Patin E, Harmant C, Lopez M, et al. . Genetic adaptation and neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–56.e17. doi: 10.1016/j.cell.2016.09.024. PMID:27768888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carr EJ, Dooley J, Garcia-Perez JE, Lagou V, Lee JC, Wouters C, Meyts I, Goris A, Boeckxstaens G, Linterman MA, et al. . The cellular composition of the human immune system is shaped by age and cohabitation. Nat Immunol. 2016;17:461–8. doi: 10.1038/ni.3371. PMID:26878114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olaru ID, Lange C, Heyckendorf J. Personalized medicine for patients with MDR-TB. J Antimicrob Chemother. 2016;71:852–5. doi: 10.1093/jac/dkv354. PMID:26507429. [DOI] [PubMed] [Google Scholar]
- 23.Mangino M, Roederer M, Beddall MH, Nestle FO, Spector TD. Innate and adaptive immune traits are differentially affected by genetic and environmental factors. Nat Commun. 2017;8:13850. doi: 10.1038/ncomms13850. PMID:28054551. [DOI] [PMC free article] [PubMed] [Google Scholar]


