Abstract
We examined the susceptibility of field strains (BO-1, BO-2, TO-1, and YH-1) and one laboratory strain (H-1) of the western flower thrip, Frankliniella occidentalis, to benzoylureas. LC50 values of novaluron were determined as 0.64 ppm against laboratory strain and 2.1–130 ppm against field strains. In the presence of piperonyl butoxide, a cytochrome P450 inhibitor, the insecticidal activity of novaluron tended to be enhanced. To examine whether point mutations in chitin synthase 1 (CHS1) discovered in an etoxazole-resistant strain of Tetranychus urticae and a benzoylurea-resistant strain of Plutella xylostella exist in F. occidentalis, the nucleotide sequence of CHS1 was analyzed. We found a nonsynonymous substitution that corresponded to the location of the mutations found in T. urticae and P. xylostella in the field strains of F. occidentalis but not in the laboratory strain, indicating that this point mutation might be associated with the benzoylurea resistance exhibited by the field strains.
Keywords: Frankliniella occidentalis, chitin synthesis inhibitors, benzoylurea, chitin synthase 1
Introduction
The western flower thrip, Frankliniella occidentalis, is a polyphagous pest of a variety of crops. In various species of thrips, including F. occidentalis, resistance against major insecticides has been reported, and metabolic detoxification is often involved in enhanced insecticide resistance.1) For example, cytochrome P450 monooxygenases are reported to be involved in resistance against organophosphates,2,3) carbamates,2,4) pyrethroids,2,3) and neonicotinoids5) in F. occidentalis [reviewed in1,6)]. In addition, point mutations at target sites have been reported: for example, a nonsynonymous substitution in the nicotinic acetylcholine receptor (nAChR) has been identified in spinosad-resistant strains of F. occidentalis7) and in the melon thrips, Thrips palmi.8) Thus, both metabolic detoxification and target site mutation are involved in insecticide resistance in thrips, as reported in other insect species.9)
Benzoylureas [Group 15 in the Insecticide Resistance Action Committee (IRAC) Mode of Action (MoA) classification] are insect growth regulators that show insecticidal activity by inhibiting chitin biosynthesis. Chitin is a polymer of N-acetyl-D-glucosamine (GlcNAc) and an essential component of the insect cuticular exoskeleton as well as of peritrophic matrices in the midgut.10) The key reaction in chitin biosynthesis is the incorporation of the GlcNAc moiety of UDP-GlcNAc into chitin polymers, and this step is catalyzed by chitin synthase (CHS).10,11) To date, two CHS genes have been identified in the genome of the fruit fly Drosophila melanogaster,12) the tobacco hornworm Manduca sexta,13) and the red flour beetle Tribolium castaneum.14) In T. castaneum, for example, CHSA (also called CHS1) is responsible for synthesizing epidermal cuticle, whereas CHSB (CHS2) is responsible for synthesis of the midgut peritrophic matrix.14)
Meanwhile, a mite growth inhibitor, etoxazole [4-(4-tert-butyl-2-ethoxyphenyl)-2-(2,6-difluorophenyl)-1,3-oxazoline], belonging to Group 10B in the IRAC MoA classification,15) has been reported to induce molting defects, reduce chitin content, and inhibit the incorporation of radiolabeled GlcNAc into chitinous material in the fall armyworm Spodoptera frugiperda, as was also observed after benzoylurea treatment.16)
Although both benzoylureas and etoxazole are known to inhibit chitin synthesis, their molecular mode of action has been an enigma. Based on biochemical studies, it has been believed that benzoylureas inhibit either the polymerization of GlcNAc into elongating chitin chains or the accumulation of chitin fibrils.11,17) A recent study revealed that etoxazole resistance in the two-spotted spider mite Tetranychus urticae was attributed to a point mutation, I1017F, in CHS1.18) Furthermore, a mutation, I1042M/F, in the CHS1 of the benzoylurea-resistant diamondback moth Plutella xylostella was discovered at the same position as the I1017F of T. urticae, and in vivo functional analysis using a genome-editing approach revealed that this mutation conferred resistance to benzoylureas in D. melanogaster.19) These recent studies have provided evidence that CHS1 could be the target of benzoylureas and etoxazole.
In this study, we evaluated the susceptibility of field and laboratory strains of F. occidentalis against benzoylureas. The field strains were found to be more resistant to benzoylureas than the laboratory strain. In the presence of piperonyl butoxide (PBO), a cytochrome P450 inhibitor, the insecticidal activity against novaluron tended to be enhanced. In field strains, the analysis of the nucleotide sequence of CHS1 cDNA revealed a nonsynonymous substitution from isoleucine to methionine at the same position reported in T. urticae18) and in P. xylostella,19) suggesting that this might account for benzoylurea resistance in the field strains.
Materials and Methods
1. Thrips rearing
The laboratory strain of F. occidentalis (H-1) was provided by Professor T. Murai (Utsunomiya University). This strain was originally collected from gerbera flowers in Shimane Prefecture in Japan in 1994, and it has been reared in the laboratory without any exposure to insecticide. Other strains (BO-1, BO-2, TO-1, and YH-1) collected in Oita Prefecture were provided by Dr. S. Okazaki (Oita Prefectural Agriculture, Forestry and Fisheries Research Center).20) Larvae and adults were fed germinated broad bean seeds (Kokusai Pet Food) in plastic containers in accordance with a previously reported method,21) at 23±1°C with a long-day photoperiod (16L:8D).
2. Chemicals
Formulated lufenuron (Match 5% EC, Syngenta Japan), flufenoxuron (Cascade 10% EC, BASF Japan), and novaluron (Counter 8.5% EC, SDS Biotech) were diluted with distilled water. To calculate the median lethal concentration (LC50), novaluron (analytical standard, Sigma-Aldrich) was dissolved and diluted in acetone. PBO (Tokyo Chemical Industry, Co., Ltd.) was dissolved in acetone.
3. Bioassays
After anesthetization with carbon dioxide for 1 min, 10 propupae were gathered on a filter paper and treated twice with 4 µL of the test chemical solution or the solvent only, as a control, using a micropipette. Excessive solution was immediately removed by absorbent filter paper. After the treatment, propupae were transferred to a 1.5-mL microcentrifuge tube with a small filter paper loaded with 6 µL of distilled water to retain humidity. The few propupae that died right after treatment, probably due to either the toxicity of the solvent or physical damage (approximately less than 5% of all propupae tested), were excluded from the experiments. Mortality was assessed six days later while determining whether specimens had undergone pupa-to-adult molting. Insecticidal assays were conducted on at least three replicates so that at least 30 propupae were used for each compound. Corrected mortality was calculated according to Abbott’s formula.22)
To calculate the LC50, analytical-grade novaluron was diluted to at least four different concentrations and applied to propupae as described above. LC50 was calculated by a probit analysis with PriProbit ver. 1.63.23) To assess the synergistic effects of PBO, a solution containing varying concentrations of novaluron as well as 2000 ppm of PBO was applied to propupae as described above. Propupae were treated with 2000 ppm PBO as the control.
4. cDNA cloning of chitin synthase 1
For each strain, twenty pupae were collected in a microcentrifuge tube. Total RNA was extracted from the pooled pupae with TRIzol reagent (Thermo Fisher Scientific), and oligo-dT-primed reverse transcription was performed using PrimeScript II 1st strand cDNA Synthesis Kit (Takara Bio). Primers for CHS1 were designed based on the sequences identified from our unpublished transcriptome database (RNA-Seq) of F. occidentalis, as well as from transcriptome and genome database of F. occidentalis in i5k Workspace@NAL (https://i5k.nal.usda.gov/webapp/blast/), provided by Baylor College of Medicine Human Genome Sequencing Center. T. castaneum CHS1 sequence (accession number NP_001034491) was used as a query. Based on the obtained contig sequences, a pair of primers (forward primer, CAG CTC CTA TTT GCT CAG ATG C; reverse primer, GCT GTT CAC GTT CTG GAG GAA G) was designed to amplify a region of the CHS1 mRNA sequence that encompasses the point mutation site reported in T. urticae18) and P. xylostella.19) A partial CHS1 cDNA fragment was amplified based on an RT-PCR approach using TaKaRa Ex Taq Hot Start Version (Takara Bio). The PCR products were purified, cloned into pGEM-T Easy Vector (Promega), and sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific). Alternatively, purified PCR products were sequenced directly.
Results and Discussion
The mortalities of the five strains of F. occidentalis exposed to formulated lufenuron, flufenoxuron, and novaluron are listed in Table 1. Among the five strains, both H-1 and TO-1 were susceptible to 50 ppm lufenuron, whereas strain H-1 was the most susceptible to 50 ppm flufenoxuron and 43 ppm novaluron. Among the four field strains, BO-1, BO-2, and YH-1 were especially resistant to these formulated benzoylurea-type insecticides, whereas strain TO-1 was less resistant (Table 1). To calculate LC50 values, bioassays were performed using analytical-grade novaluron (Table 2). The LC50 was 0.64 ppm for strain H-1 and 2.1 ppm for strain TO-1. By contrast, the LC50 was 130 ppm for strain BO-1, 49 ppm for strain BO-2, and 71 ppm for strain YH-1. Thus, strains BO-1, BO-2, and YH-1 were less susceptible to novaluron, as compared with H-1 and TO-1. The resistance ratios against novaluron as compared to laboratory strain H-1 were 110 for YH-1 and 210 for BO-1 (Table 2). Although we attempted to enhance the resistance of these strains by the occasional application of novaluron, it was not very successful, due to the microbial infection of the thrips culture and other unknown reasons. In future studies, the establishment of a highly resistant population by treating thrips with benzoylureas will be needed.
Table 1. Mortality of F. occidentalis strains against formulated benzoylurea-type insecticides.
| Chemicals | Concentration (ppm) | Corrected mortality (%) | ||||
|---|---|---|---|---|---|---|
| H-1 | BO-1 | BO-2 | TO-1 | YH-1 | ||
| Lufenuron | 50 | 64 | 30 | 4.4 | 79 | 28 |
| Flufenoxuron | 50 | 47 | 5.2 | 16 | 8.9 | 19 |
| Novaluron | 43 | 54 | 4.8 | −10 | 34 | 0.83 |
Table 2. LC50 of novaluron and synergism by PBO.
| Strains | Chemicals | LC50 (ppm) | 95% CLa) | Synergism ratiob) | Resistance ratioc) |
|---|---|---|---|---|---|
| H-1 | novaluron | 0.64 | 0.099–3.3 | ||
| novaluron+PBO | 0.25 | 0.024–1.8 | 2.6 | ||
| BO-1 | novaluron | 130 | 23–800 | 210 | |
| novaluron+PBO | 110 | 14–920 | 1.2 | ||
| BO-2 | novaluron | 49 | 5.3–460 | 77 | |
| novaluron+PBO | 5.5 | 0.45–48 | 8.9 | ||
| TO-1 | novaluron | 2.1 | 0.19–15 | 3.2 | |
| novaluron+PBO | 6.9 | 0.66–63 | 0.30 | ||
| YH-1 | novaluron | 71 | 7.5–690 | 110 | |
| novaluron+PBO | 31 | 3.7–260 | 2.3 |
a) 95% confidence limitb) Synergism ratio was calculated as (LC50 of novaluron)/(LC50 of novaluron+PBO)c) Resistance ratio was calculated as (LC50 of tested strain)/(LC50 of H-1)
To evaluate the involvement of cytochrome P450 monooxygenase-mediated metabolism, bioassays were conducted with PBO. The synergism ratios were 8.9 for strain BO-2, 2.6 for strain H-1, 2.3 for strain YH-1, 1.2 for strain BO-1, and 0.30 for strain TO-1 (Table 2). Thus, the insecticidal activity of novaluron tended to be enhanced in strains H-1, BO-2, and YH-1 in the presence of PBO, indicating that cytochrome P450 monooxygenase-mediated metabolism was involved in novaluron resistance in these strains.
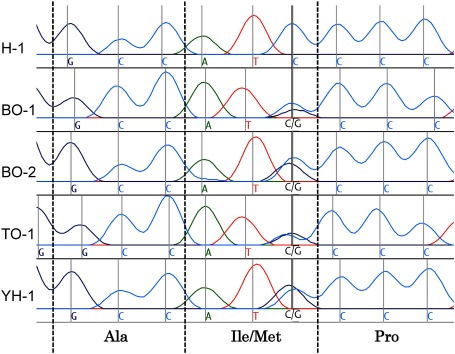
From the transcriptome database of F. occidentalis, two contigs homologous to T. castaneum CHS1 were identified. After assembling the sequences of the two contigs, a pair of RT-PCR primers was designed to amplify a portion of CHS1 cDNA surrounding the point mutation site that corresponded to I1017F in T. urticae18) and I1042M/F in P. xylostella.19) First, we subcloned and sequenced the PCR product derived from the cDNA pool of laboratory strain H-1. The determined nucleotide sequence of the subclone (399 bp, DDBJ/EMBL/GenBank accession number LC197806) was found to be 99% identical to the counterpart from the above-mentioned transcriptome. Thus, we confirmed that the RT-PCR procedure indeed amplified the specific product correctly. Then, we directly sequenced the RT-PCR products derived from cDNA of pooled pupae of the respective strains to determine whether a similar nucleotide sequence substitution in the case of T. urticae or P. xylostella had occurred. For laboratory strain H-1, the codon for this site was ATC, encoding isoleucine (Fig. 1). In contrast, the codon in the four field strains was ATC/G, encoding isoleucine or methionine. Thus, there was a nonsynonymous substitution-isoleucine to methionine-in field strains, but not in the laboratory strain. As shown in Fig. 2, this substitution was at the same position as I1042M/F of P. xylostella CHS119) and I1017F of T. urticae CHS1.18) Van Leeuwen and colleagues performed a functional analysis of chitin synthase using a yeast expression system and concluded that the isoleucine-to-phenylalanine point mutation found in T. urticae was associated with etoxazole resistance.18) Furthermore, functional analysis using a genome-editing approach revealed that isoleucine-to-phenylalanine or isoleucine-to-methionine substitution conferred resistance to chemicals, including etoxazole and benzoylureas, in D. melanogaster.19) Taken together, we assume that isoleucine-to-methionine substitution of CHS1 in the field strains of F. occidentalis was likely to be involved in benzoylurea resistance.
Fig. 1. Electropherograms of CHS1 of five strains of F. occidentalis corresponding to the I1017F site in T. urticae. A portion of CHS1 cDNA was amplified by RT-PCR using cDNA of pooled pupae, and the purified PCR product was sequenced directly.
Fig. 2. Aligned amino acid sequences of helix 5 in the C-terminal 5 transmembrane segments of CHS1. The positions of the I1017F substitution in etoxazole-resistant Tetranychus urticae18) and I1042M/F in benzoylurea-resistant Plutella xylostella19) are indicated in gray. Conserved amino acid residues are shown in bold letters. PxCHS1-S, P. xylostella CHS1 of the susceptible strain BCS-S (DDBJ/EMBL/GenBank accession number API61825); PxCHS1-Sud-TFM, P. xylostella CHS1 of the resistant strain Sud-TFM (API61827); PxCHS1-China, P. xylostella CHS1 of the resistant strain China19); TuCHS1-S, T. urticae CHS1 of the etoxazole-susceptible strain GSS (AFG28413); TuCHS1-R, T. urticae CHS1 of the etoxazole-resistant strain EtoxR (AFG28419); FoCHS1-S, Frankliniella occidentalis CHS1 of the susceptible strain H-1; FoCHS1-field, F. occidentalis CHS1 of field strains BO-1, BO-2, TO-1, and YH-1.
Since we analyzed the sequence of CHS1 from pooled individuals in this study, it was not clear whether CHS1 alleles exist as homozygous or heterozygous in each individual. Therefore, to precisely determine the genotype of many individuals for each strain, it would be necessary to amplify genomic DNA from individuals instead of sequencing cDNA from pooled individuals.
It is interesting that a point mutation in one amino acid in CHS1 exists at the same location among etoxazole-resistant mites,18) benzoylurea-resistant diamondback moths,19) and benzoylurea-resistant thrips (Fig. 2), although etoxazole and benzoylurea belong to different groups in the IRAC MoA classification. Using a high-resolution genetic mapping technique, Demaeght and colleagues demonstrated that resistance to mite growth inhibitors clofentezine (Group 10A of IRAC MoA) and hexythiazox (MoA Group 10A) is also attributed to CHS1.24) Very recently, it was reported using genome-modified D. melanogaster that isoleucine-to-phenylalanine or isoleucine-to-methionine substitution in CHS1 conferred resistance to etoxazole (MoA Group 10B), benzoylureas (MoA Group 15), and buprofezin (MoA Group 16).19) These studies indicated that clofentezine, hexythiazox, etoxazole, benzoylureas, and buprofezin share the same molecular mode of action. Douris and colleagues suggested the possibility that these chemicals could be classified in a single MoA group.19) Further analysis, such as X-ray crystallography of CHS1 that is bound to one of these insecticides, might help to elucidate the mode of action at the molecular level.
Acknowledgments
We thank Professor Tamotsu Murai (Utsunomiya University) and Dr. Shin-ichiro Okazaki (Oita Prefectural Agriculture, Forestry and Fisheries Research Center) for providing F. occidentalis, and Ms. Sayumi Tanaka (Nagoya University) for assistance with insect rearing.
References
- 1).P. Bielza: Pest Manag. Sci. 64, 1131–1138 (2008). [DOI] [PubMed] [Google Scholar]
- 2).P. J. Espinosa, J. Contreras, V. Quinto, C. Gravalos, E. Fernandez and P. Bielza: Pest Manag. Sci. 61, 1009–1015 (2005). [DOI] [PubMed] [Google Scholar]
- 3).G. Zhao, W. Liu, J. M. Brown and C. O. Knowles: J. Econ. Entomol. 88, 1164–1170 (1995). [Google Scholar]
- 4).S. E. Jensen: J. Econ. Entomol. 93, 464–471 (2000). [DOI] [PubMed] [Google Scholar]
- 5).C. Minakuchi, Y. Inano, X. Shi, D. Song, Y. Zhang, K. Miura, T. Miyata, X. Gao, T. Tanaka and S. Sonoda: Appl. Entomol. Zool. (Jpn.) 48, 507–513 (2013). [Google Scholar]
- 6).Y. Gao, Z. Lei and S. R. Reitz: Pest Manag. Sci. 68, 1111–1121 (2012). [DOI] [PubMed] [Google Scholar]
- 7).A. M. Puinean, S. J. Lansdell, T. Collins, P. Bielza and N. S. Millar: J. Neurochem. 124, 590–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).W. X. Bao, Y. Narai, A. Nakano, T. Kaneda, T. Murai and S. Sonoda: Pestic. Biochem. Physiol. 112, 51–55 (2014). [DOI] [PubMed] [Google Scholar]
- 9).R. Feyereisen, W. Dermauw and T. Van Leeuwen: Pestic. Biochem. Physiol. 121, 61–77 (2015). [DOI] [PubMed] [Google Scholar]
- 10).H. Merzendorfer: J. Comp. Physiol. B 176, 1–15 (2006). [DOI] [PubMed] [Google Scholar]
- 11).H. Merzendorfer: Insect Sci. 20, 121–138 (2013). [DOI] [PubMed] [Google Scholar]
- 12).M. E. Gagou, M. Kapsetaki, A. Turberg and D. Kafetzopoulos: Insect Biochem. Mol. Biol. 32, 141–146 (2002). [DOI] [PubMed] [Google Scholar]
- 13).D. G. Hogenkamp, Y. Arakane, L. Zimoch, H. Merzendorfer, K. J. Kramer, R. W. Beeman, M. R. Kanost, C. A. Specht and S. Muthukrishnan: Insect Biochem. Mol. Biol. 35, 529–540 (2005). [DOI] [PubMed] [Google Scholar]
- 14).Y. Arakane, S. Muthukrishnan, K. J. Kramer, C. A. Specht, Y. Tomoyasu, M. D. Lorenzen, M. Kanost and R. W. Beeman: Insect Mol. Biol. 14, 453–463 (2005). [DOI] [PubMed] [Google Scholar]
- 15).T. Ishida, J. Suzuki and Y. Tsukidate: YI-5301, a novel oxazoline acaricide. in Proceedings of British Crop Protection Conference on Pests and Diseases 1994. Brighton, UK.
- 16).R. Nauen and G. Smagghe: Pest Manag. Sci. 62, 379–382 (2006). [DOI] [PubMed] [Google Scholar]
- 17).Y. Nakagawa: J. Pestic. Sci. 21, 460–467 (1996) (In Japanese). [Google Scholar]
- 18).T. Van Leeuwen, P. Demaeght, E. J. Osborne, W. Dermauw, S. Gohlke, R. Nauen, M. Grbic, L. Tirry, H. Merzendorfer and R. M. Clark: Proc. Natl. Acad. Sci. U.S.A. 109, 4407–4412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).V. Douris, D. Steinbach, R. Panteleri, I. Livadaras, J. A. Pickett, T. Van Leeuwen, R. Nauen and J. Vontas: Proc. Natl. Acad. Sci. U.S.A. 113, 14692–14697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).S. Okazaki, R. Ohtsubo and K. Kato: Kyushu Plant Protection Research 60, 79–83 (2014) (In Japanese). [Google Scholar]
- 21).T. Murai and A. J. M. Loomans: Entomol. Exp. Appl. 101, 281–289 (2001). [Google Scholar]
- 22).W. S. Abbott: J. Econ. Entomol. 18, 265–267 (1925). [Google Scholar]
- 23).M. Sakuma: Appl. Entomol. Zool. (Jpn.) 33, 339–347 (1998). [Google Scholar]
- 24).P. Demaeght, E. J. Osborne, J. Odman-Naresh, M. Grbic, R. Nauen, H. Merzendorfer, R. M. Clark and T. Van Leeuwen: Insect Biochem. Mol. Biol. 51, 52–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]