Abstract
Research on the carcinogenesis process is currently focused primarily on understanding its genetic basis and molecular abnormalities that may be predictive factors and therapeutic targets. It was clearly confirmed recently that microRNAs are involved in the mechanisms of leukocyte development, differentiation, and apoptosis, as well as in the pathogenesis of proliferative diseases of the hematopoietic system. Currently, research strategies allow determination of the deregulation of microRNA profiles in relation to other cytogenetic aberrations, as well as prognostic factors and primary end points. The problem of the possibility of their use as therapeutic targets is also increasingly discussed. In this article, we analyze literature data on abnormalities in microRNA expression in proliferative diseases of the hematopoietic system in the context of classic cytogenetic and molecular aberrations.
Keywords: hematological malignancies, microRNA, cytogenetic abnormalities
Introduction
Hematological malignancies belong to a group of clonal diseases of stem and progenitor cells. They result from genetic and epigenetic changes that cause disorders of signal transduction and gene expression, leading to dysregulation of self-renewal, proliferation, and differentiation of cells. These processes are multifactorial and multistage.1
Recently published studies on carcinogenesis have focused mainly on genetic and molecular disorders that can serve as prognostic factors and become potential therapeutic targets.1,2 Three large groups of genes associated with etiopathogenesis of cancer have been identified so far: oncogenes, tumor-suppressor genes, and genes that provide genome stability. Their dysfunction does not, however, fully represent molecular changes in carcinogenesis.2,3 Many genetic abnormalities have been described in hematological malignancies. They contribute not only to identifying new histoclinical entities and prognostic factors but are also used to follow up effects of the therapy.3
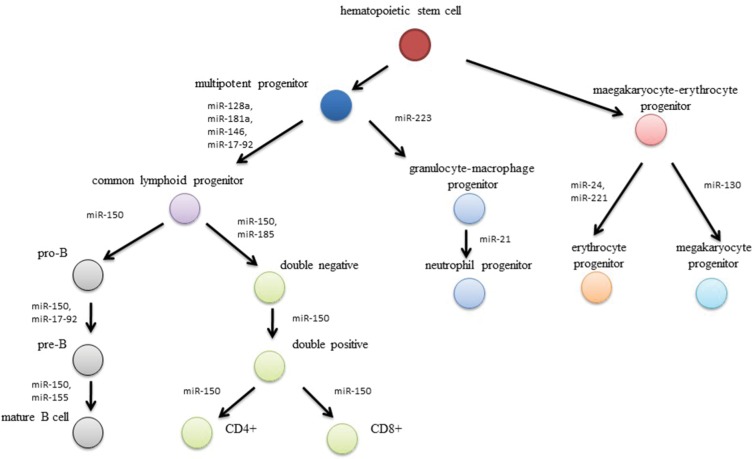
It has been revealed that small RNA molecules, known as microRNA, are involved in the leukemogenesis (Figure 1).4 MicroRNA are family of short (20–22 nucleotides), non-coding RNA molecules that posttranscriptionally mediate gene expression.5 They were first described in the 1990s by Ambros et al during studies of Caenorhabditis elegans. Shortly after this, microRNAs were identified in many species of animals, plants, viruses, and fungi. More than 5,000 genes for microRNA have been described so far.6,7
Figure 1.
Effect of microRNA-expression disturbances on leukemogenesis processes.
MicroRNAs play a role in regulation of cell proliferation, differentiation, apoptosis, hemostasis, oncogenesis, and angiogenesis. It is estimated that genes coding microRNAs account for 1%–5% of the human genome. DNA sequences coding microRNAs are highly conserved, and about 70% of them are found in introns and/or exons of other genes, whereas 30% are situated between genes coding proteins and noncoding RNA.5,6 In most cases, microRNAs are negative regulators of gene expression, but their functions may be much more extensive, as there have been reports about their ability to activate translation. Single microRNAs may influence expression of a few genes, whereas genes are complementary to different microRNAs.6,7 Over 50% of described microRNAs are located within the regions of the genome that undergo malignant transformation, ie, fragile sites, loss-of-heterozygosity regions, minimal regions of amplification, hot spots, or common breakpoint regions. This is associated with such aberrations as deletion and amplification, point mutation in a gene’s DNA sequence, and single-nucleotide polymorphism, which may lead not only to changes in the expression of microRNA but also may influence formation of complexes between target transcript and microRNA.3,6,8
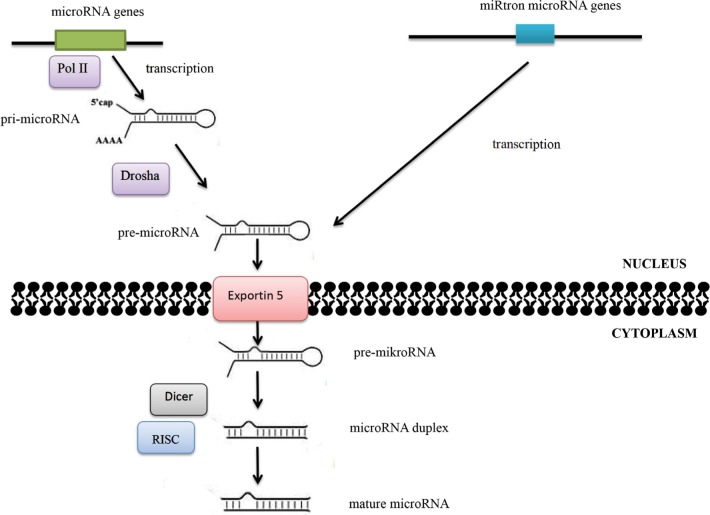
Alterations in microRNA expression in carcinogenesis may also result from changes in the regulation of transcription. Some microRNAs are controlled by transcription factors encoded by oncogenes or suppressor genes, while the expression of other microRNAs is influenced by changes in the methylation status of promoters. Abnormalities in microRNA functions may also be associated with damages incurred to the complementary sequence in the target gene in the process of neoplastic transformation. Quantitative changes in mature-microRNA levels may result from abnormalities in Dicer1, DGCR8, Drosha, and other proteins associated with the process of their biogenesis (Figure 2).5,6
Figure 2.
MicroRNA-biogenesis process.
Notes: The maturation process of microRNAs takes place in two main stages. The RNA II polymerase (Pol II) is mainly responsible for transfer of microRNA genes. As a result of transcription, primary precursors are formed, ie, pri-microRNA, in which maturation enzymes from the family of ribonuclease III – Drosha and Dicer – are involved. Double-stranded pri-microRNA structures are recognized by the DGCR8 protein, which is associated with Drosha ribonuclease and together with other cofactors of the reaction forms a microprocessor complex involved in the processing of pri-microRNA transcripts in the cell nucleus. The pre-microRNA formed as a result of maturation is then transported to the cytoplasm using the exportin 5–Ran-GTP protein complex. Within the cytoplasm, the pre-microRNA joins the Dicer ribonuclease. As a result of Dicer ribonuclease activity in cooperation with cofactors (argonaute protein, TAR binding protein, or PKR protein activator), double-stranded microRNA duplexes are formed. The active form of duplex microRNA is incorporated into the RISC protein, in which it obtains its biological maturity.
MicroRNA expression in chronic lymphocytic leukemia
The first clinical data about dysregulation of microRNAs were associated with the pathogenesis of chronic lymphocytic leukemia (CLL).6 A lymphoproliferative disorder, CLL is the most commonly diagnosed leukemia in adults. It is characterized by a highly variable clinical course. Some patients live long even without treatment. In other patients, the disease is more aggressive, and despite intensive treatment, leads to death within 2–3 years. Throughout the last few years, many new prognostic factors have been recognized and has become a basis for new therapeutic strategies.9,10 These factors include immunophenotypic antigens of leukemic cells, such as the expression of CD38 antigen and ZAP70 assessed by flow cytometry, somatic hypermutation of immunoglobulin heavy-chain variable-region genes and cytogenetic aberrations found in >80% of cases. Deletions of 13q14.3, 11q23, and 6q12 and trisomy 12 are common abnormalities associated with CLL.9–11 It has been proven that in patients with deletion of 13q14.3, miR15a and miR16–1 expression is inhibited or at very low levels. MiR15a expression reduced in 25% of patients with CLL, and low expression of miR16-1 reported in 45% of cases.12 Monoallelic deletion may result in increased expression of Bcl2, which plays a role as an apoptosis antagonist. It also leads to the increased expression of cell-cycle-related genes, including CCND2, CCND3, CDK4, and CDK6, whose activity is required for cell-cycle G1–S transition.13,14 Studies have shown that miR15a and miR16-1 expression is inversely correlated with Bcl2 expression in CLL samples and that these microRNAs negatively regulate Bcl2 at a posttranscriptional level.13,15 It has also been reported that Bcl2 repression by these microRNAs induces apoptosis in a leukemic cell-line model. Abnormal expression of miR15a/16-1 resulting from 13q14.3 deletion is found in about 66% of patients with CLL.16
MiR34a, a member of the miR34 family, is regulated by the tumor-suppressor gene TP53 and contributes to its downstream effects on proliferation arrest and induction of apoptosis. Low miR34a expression is found in patients with chemotherapy-refractory CLL associated with deletion of p53, which results in decreased DNA damage response and apoptosis resistance, leading in turn to cell-cycle arrest.17,18 SIRT1 and TLC1 transcription factors, which play an important role in cell-death inhibition and overexpressed in most cases of CLL, have been identified as a relevant miR34 target.19 Decreased expression of miR34a has been observed in aggressive subtypes of CLL with TP53 gene abnormali ties, leading to the development of a fludarabine-refractory phenotype.20
The decreased expression of miR29c and miR17-5p, which is part of the miR17-92 cluster, were also observed in patients with refractory CLL. It was demonstrated that miR17-5p targets E2F1, p21, and cyclin D, thus suggesting its role as a tumor suppressor.20 It is hypothesized, but not proven, that high expression of miR331 in CLL patients targets SOCS1, whose reduced levels have been previously found in many cancers and might allow cell survival, proliferation, and angiogenesis.21
MiR181 is also involved in the development of CLL. Decreased miR181 expression increases antiapoptotic protein levels, including MCL1 and Bcl2. Studies have proved that miR181b regulates the expression of TCL1, and its low expression has been regarded as an unfavorable prognostic factor.22,23 Overexpression of TCL1, which is an important lymphocytic survival factor associated with CLL, might also be explained by miR29b downregulation.24
MiR155-dependent downregulation of SHIP1 and CEBPβ proteins has been observed in leukemic pre-B lymphocytes. This results in a block of B-cell differentiation, and might induce reactive proliferation of the myeloid lineage.25 MiR155 suppresses the expression of HDAC4 and Bcl6 during miR155-dependent leukemogenesis. All these processes result in derepression of some of the known Bcl6 targets, which may block B-cell development and induce uncontrolled cell proliferation.26
Recent studies have shown that high expression of miR221/222 is associated with resistance to fludarabine and may serve as a prognostic marker. It also correlates with shorter overall survival (OS) and time to first treatment.27 MiR21 and miR92 are also considered prognostic factors. MiR21 is involved in downregulation of PTEN, CCND2, and DPH1, which might act as regulators of the cell cycle.16
MicroRNA expression in Hodgkin’s lymphoma
Hodgkin’s lymphoma (HL) is characterized by the presence of giant Reed–Sternberg cells and Hodgkin mononuclear cells. Although the etiology of the disease has not been elucidated, epidemiological and serological studies indicate a connection between Epstein–Barr virus infection and HL development.28,29 There are two basic types of disease: classical HL, which covers about 95% of all cases, and nodular lymphocyte-predominant HL much less often, affecting 5% of patients.29 Data on cytogenetic changes in patients with HL are scarce: there have been no large studies that would confirm specific gene rearrangements. This is mainly due to the small number of malignant cells characterized by a low mitotic index, complex karyotype, and poor chromosome morphology.30 In the HL population, however, microRNA expression has been reported to be abnormal. In classical HL, decreased expression of miR520a, miR200a, and miR614 is observed. Overexpression, demonstrated for miR20a, miR21, miR9, miR155, miR16, miR140, miR18a, miR30b, miR30a-5p, miR196a, miR374, and miR186, may involve activation of the JAK–STAT pathway.31–33 This pathway is also regulated by miR135a. Reduction in its expression leads to impaired apoptosis and proliferation, which translates into a worse prognosis in this group of patients.34 In contrast, the increased expression of miR21 and miR30D is associated with resistance to chemotherapy. Additionally, low miR9 expression is associated with impairment of cytokine production.35
MicroRNA expression in non-Hodgkin’s lymphomas
Non-HLs (NHLs) are a heterogeneous group of hematological malignancies that originate from lymphatic hematopoietic tissue. They are classified into two groups: B-cell lymphomas, which constitute about 90% of cases, and T-cell lymphomas, which account for about 10% of them. Strong experimental data support a critical role of microRNAs in NHL lymphomagenesis and pathogenesis. It has been demonstrated that microRNAs can function either as tumor-suppressive, inhibiting tumor proliferation, or oncogenes, which promote tumor escape from apoptosis induced by chemotherapy.36
Diffuse large B-cell lymphoma
Diffuse large B-cell lymphoma (DLBCL) is the most common type of NHL, representing ~30%–40% of all NHLs. Gene-expression profiling has been used to classify these lymphomas into three subgroups: germinal center B-cell-like DLBCL, activated B-cell-like DLBCL, and primary mediastinal DLBCL.37 These subtypes can be distinguished not only by gene-expression profiles but also by distinct microRNA-expression profiles. For example, the miR17-92 cluster of microRNAs is significantly upregulated in germinal center B-cell-like DLBCL tumors. Additionally, miR155, miR221, and miR21 are expressed more highly in activated B-cell-like DLBCL.36 Recently, it was shown that increased expression of miR125a and miR125b activates one of the most deregulated pathways in the pathogenesis of DLBCL: nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB).38 It has been observed that miR155 expression is ~20–30 times higher in DLBCL cells than in normal cells.39 There are two negative regulators of the PI3K–Akt pathway: SHIP1 and PIK3R1, which are both downregulated by miR155. In addition, miR155 downregulates BMP.36,40
Current data show that the expression of miR222 is associated with shorter progression-free survival (PFS) in DLBCL patients treated with R-CHOP (rituximab, vincristine, doxorubicin, cyclophosphamide, and prednisone) immunochemotherapy via inducing downregulation of p27 that might facilitate cell proliferation and survival.39,41 Furthermore, increased expression of miR142, miR146-5p, and miR223 and increased expression of miR146a and miR200c correlate with longer relapse and PFS in DLBCL patients treated with R-CHOP.36
MicroRNA expression is tissue-specific in DLBCL cells and microRNA-expression profiles differ, depending on the primary tumor site. MiR17-5p is upregulated in DLBCL tumors arising from the central nervous system, while miR127 is upregulated in DLBCL tumors arising from the testes.42 In the diagnosis of primary central nervous system lymphoma (PCNSL), microRNAs play an important role as disease markers in cerebrospinal fluid. MiR21, miR19, and miR92a show a significant presence in the cerebrospinal fluid of patients with PCNSL. The role of these microRNAs in the diagnosis of PCNSL is high, with 95.7% sensitivity and 96.7% specificity.43
Follicular lymphoma
Follicular lymphoma (FL) is a lymphoproliferative disease that is derived from B lymphocytes of lymph-gland media. A characteristic genetic aberration, described in 70%–95% of patients with FL, is the acquired t(14;18), which results in the transfer of a chromosome fragment containing the BCL2 gene (18q21.3) to the region of the gene encoding the chain severe immunoglobulins (14q32.33). The effect of this is excessive production of the antiapoptotic Bcl2 protein.44 However, the exact mechanisms underlying lymphomagenesis have not yet been identified, and t(14;18) fragments have also been described in healthy people, which may indicate that these processes are more complex. Studies on gene-expression profiles in patients with FL confirm that the lymph-node microenvironment can play a significant role in FL biology as well.44–46
Diagnostic difficulties are caused by cases in which t(14;18) is not found. At the same time, it has been shown that the presence of t(14;18) and/or morphologically high-grade (grade 3A or B) FL is very often not present in CD10-negative patients, and these patients require intensive treatment. In the context of these data, it is important to look for new prognostic factors that would facilitate the diagnosis.45,46 In t(14;18)-negative patients, disorders in microRNA profiles, such decreased expression of miR16, miR26a, miR101, miR29c, and miR138, have also been described.47
It has also been shown that patients with FL have low miR17-5p expression, while increased levels of this microRNA have been observed in patients with disease transformation in DLBCL. A reverse relationship was found in the case of miR31: the expression of this microRNA was high in patients in the early stages of the disease and decreased as it progressed, while in transforming patients, it may be not expressed at all.48 In FL patients, elevated miR20a/20b levels have also been reported, which were associated with decreased expression of CDRN1A/p21 and overexpression of miR194, resulting in a decrease in SOCS2 expression. These changes led to disturbances in the lymphomagenesis process and affect cell proliferation and differentiation.49
Mantle-cell lymphoma
Mantle-cell lymphoma (MCL) is an aggressive lymphoma that is characterized by t(11;14)(q13;q32) resulting in over-expression of the CCND1 gene. This leads to overproduction of cyclin D1, which binds to cyclins dependent on CDK4 and CDK6, induces phosphorylation of RB1 and thus inhibits its suppressor role, promoting cell migration into the S phase of the cell cycle. However, other genetic changes (ATM, CDKN2A, TP53, MYC, SYK, and BCL2) are necessary for the development of the disease, which affect the deregulation of DNA-repair mechanisms and conditioning the cell’s survival. Secondary genetic aberrations regarding 3q and 9q are associated with an unfavorable prognosis.50–52
Literature data indicate a link between the loss of the coding region miR15/16 and members of the cluster in the 3’UTR of CCND1 and the overexpression of cyclin D1.53 In addition, reduced expression of the miR17-92 cluster correlates with overexpression of Myc and is associated with aggressive disease course. It was also shown that the PHLPP2, a key regulator of the PI3K–Akt pathway, is a target for this microRNA.54 A potential prognostic value predicting the course of MCL is also reduction in miR29 expression, which leads to activation of CDK4/DK6.55 In turn, low miR34a expression results in overexpression of CDK6 and high expression of the MYC oncogene. Overexpression of miR181a leads to a reduction in expression of the gene encoding the proapoptotic Bim protein.56
Husby et al57 confirmed the prognostic and predictive value of miR18b-expression disorders and proposed introducing into daily clinical practice the new biological MCL International Prognostic Index (MIPI-B)-miR prognosticator, combining expression levels of miR18b with MIPI-B data. Arakava et al,58 in turn, confirmed that the relationship between miR15b overexpression and classical MCL transformation is an aggressive form of MCL. It was also indicated that overexpression of miR10a, miR20b, and miR363 translates into OS reduction. Also, miR127-3p, miR615-3p, and miR146a expression may have a potential prognostic value, but further studies are required.56
MicroRNA expression in multiple myeloma
Multiple myeloma (MM) is a multistage disease characterized by proliferation and accumulation of monoclonal plasmacytes that produce monoclonal immunoglobulin or their immunoglobulin fragments. The first stage in disease development is immortalization of B-cells following chromosome trans-locations within immunoglobulin heavy-chain genes during class switching and somatic hypermutations that result in the assembly of immunoglobulin-gene-enhancer sequences with oncogenes.59,60 The primary translocation of immunoglobulin is associated with ectopic expression of CCND1, CCND3, and FGFR3/MMSET/MMMAF oncogenes, which induces the proliferation of a long-lived plasmablast/plasmacyte population.61 Relations between karyotype dysfunctions and translocations within the immunoglobulin heavy chain have not been fully understood. Literature data indicate that karyotype instability may initiate carcinogenesis. Secondary translocations often include the MYC gene, and TP53 mutations/deletions are associated with disease progression.59,61
Reduced expression of miR15a has been described in relapsed and/or refractory MM.62 At the same time, higher expression of miR15a and miR16 has been observed in patients with newly diagnosed MM compared to the healthy population.63 Literature data also indicate a relationship between miRNA17-92 cluster expression abnormalities and increased antiapoptotic activity of the Bcl2 protein. MCL2 is the molecular target for miRNA29b. Decreased MCL2 expression is associated with induction of apoptotic processes.64
In MM patients, the miRNA193b-365 cluster is also over-expressed.65 Expression of miR720, miR1308, and miR1246 in this group of patients is statistically significantly higher than in the healthy population.66 MiR146b, miR140, miR145, miR125a, miR151, miR223, miR155, and Let7f regulate the expression of Hox9, c-Myc, Bcl2, and Shp1/2, and expression changes in these microRNAs may be involved in myelomagenesis and be a prognostic factor.67 Also, miR17-92 clusters are activated by Myc, and disorders in their expression correlate with MM progression.68 Literature data also indicate a relationship between miR15a, miR16-1, miR17, miR20, miR92, miR148a, and shorter PFS and miR548d, miR373, miR554, and miR888 translate into shorter OS. High expression of miR153, miR490, miR455, miR642, miR500, and miR296 in MM patients may be treated as a favorable prognostic factor.67
Neri et al69 indicated for the first time the relationship between disturbances in microRNA-expression profiles and bortezomib resistance. Zhang et al70 showed that miR16-5 p, miR15a-5p, miR20a-5p, and miR17-5 p were significantly downregulated in patients with resistance. Literature data indicate that miR21 overexpression may induce resistance to dexamethasone, bortezomib, and doxorubicin-induced apoptosis.71 In turn, miR27a expression is associated with bortezomib resistance.72 In melphalan-resistant patients, miR221/222-expression levels are significantly higher and the percentage of apoptotic cells lower.73
MicroRNA expression in acute myeloid leukemia
Acute myeloid leukemia (AML) belongs to a group of clonal, proliferative diseases of the hematopoietic system that originate from cells at early stages of differentiation. The course of these diseases is characterized by inhibition of apoptosis and disorders of proliferation, which lead to proliferation of immature hematopoietic stem cells and infiltration of other tissues and organs of the body. AML is a heterogeneous group of diseases, with ~200 recurrent chromosomal abnormalities, including molecular abnormalities, such as mutations in FLT3 internal tandem duplication, WT1, NPM1, and CEBPA, MLL gene duplications, RUNX1 mutations, and chromosomal changes, such as t(11q23), t(8;21), t(8,21), inv(16), and trisomy 8.74 In addition to these abnormalities, disruptive microRNA expression and epigenetic dysregulation have also been reported. Many microRNAs are hyper-methylated, including miR124a, which targets EVI1, CEBPA, and CDK6,75 miR193a, which targets KIT and WT1,76,77 and miR34b associated with the regulation of HGM1.78 It has been demonstrated that the deletion of the MIR145 and MIR146A genes on chromosome 5q, often observed in patients with myeloid disorders, results in neutropenia and megakaryocytic dysplasia.79
MicroRNAs associated with the proliferation and differentiation of AML cells include miR155, miR221, miR126, Let7, and miR196b. Their expressions differ depending on the subtype of the disease.80 Overexpression of miR126 results in the inhibition of programmed cell death caused by decreased activity of the tumor-suppressor gene PLK2,81,82 whereas increased expression of miR1-2 and miR133a-1 is associated with aberrant hematopoietic stem-cell proliferation and correlates with the incidence of highly aggressive forms of AML.83 Repression of miR193 family members leads to the increased expression of the CKIT oncogene, which induces aberrant cell division and is associated with therapy failure.84 Overexpression of miR34a and miR221/222 and repression of miR23b are also typical of AML. AML cells also exhibit significantly increased expression of miR181 family members (miR181a and miR181b).6
It has also been proven that microRNA-expression level influences the prognosis in patients with AML. Overexpression of miR199a, miR191, miR20a, miR25, and miR199b correlates with shorter survival PFS, while increased expression of miRlet7b and miR9 is observed only in patients of intermediate and high cytogenetic risk.6,85
MicroRNA expression in acute lymphoblastic leukemia
Acute lymphoblastic leukemia (ALL) is the most common hematological malignancy in children, with peak incidence at 1–4 years of age. It accounts for 76% of all leukemia cases in children, whereas it is responsible for only 20% of cases in adults.86,87 ALL is a malignancy of lymphoid cells that originates from the early stages of B-cell (B-ALL) or T-cell (T-ALL) development. This is a heterogeneous disease of variable clinical course. The current classification system, introduced by WHO, is based on morphology, immunology, cytogenetics, and molecular biology. However, patients from the same risk group differ in terms of response to treatment and prognosis. Chromosomal abnormalities and aberrant microRNA expression are seen as likely culprits for discrepancies.88
ALL patients present very high expression levels of miR128b, miR204, miR218, miR332, and miR181b-1.89 High expression of miR128 is typical of ALL, and for this reason it is considered a diagnostic tool.88 Numerous microRNAs that have been studied have prognostic value. Reduced expression of miR124a is an adverse prognostic factor associated with higher mortality rate. Hypermethylation of the miR124a promoter leads to overexpression of CDK6 and RB1 protein phosphorylation, which in turn results in abnormal and uncontrolled division of ALL cells. Inhibition of miR9 results in overexpression of FGFR1 and CDK6, which leads to the increased proliferation rate and inhibition of apoptosis of ALL cells.6,88
MIR19B, MIR20A, MIR26A, MIR92, and MIR223 are regulatory oncogenes. Their increased expression leads to the repression of suppressor genes: PTEN, BCL2L11, PHF6, NF1, and FBXW7, whereas overexpression of miR142-3p leads to the reduced PKA levels.90 Research has also shown that microRNAs of the miR9 family (miR9-1, miR9-2, miR9-3) miR451 and miR709 play a suppression role in the development of ALL.6,90 It has also been proven that expression of miR196 is significantly reduced in patients with ALL.91 Let7b is downregulated in B-ALL patients with MLL trans-locations, while the miR17-92 cluster is overexpressed.88,92
MicroRNA expression in chronic myelogenous leukemia
Chronic myelogenous leukemia (CML) is a clonal myelo-proliferative disorder characterized by translocation between the long arms of chromosomes 9 and 22, called the Philadelphia chromosome. This aberration is found in 95% of newly diagnosed CML patients, and results in the formation of a hybrid gene, the BCR–ABL fusion gene, which encodes the p210 hybrid protein with tyrosine-kinase activity. High BCR–ABL p210 oncoprotein levels result in uncontrolled cell proliferation, apoptosis deregulation, and inhibition of hematopoietic stem-cell adherence to matrix molecules.93,94 A number of microRNAs regulate the expression of BCR-ABL: miR23a, miR30a, miR30e, miR203, miR320, and miR424.95 Overexpression of miR203 reduces ABL1 and BCR–ABL1 fusion-protein levels.96 MiR199b expression is significantly reduced in CML patients.97 C-Myc-dependent aberrant expression of the miR17-92 cluster (onco-miR1) has also been observed in CD34+ cells.98
Recently, miR191, miR29a, miR422b, miR100, miR326, and miR26a were identified as possible predictors for clinical resistance to tyrosine-kinase inhibitors. A rapid increase in miR146a and miR150 expression and decrease in miR142-3p and miR199b-5p were found in patients treated with imatinib. Loss of miR217 and miR199b expression has been correlated with resistance to tyrosine-kinase inhibitors. Imatinib has been demonstrated to induce demethylation of the miR203-promoter region, resulting in low BCR–ABL1 expression.97
Lower expression of miR130a is associated with poor prognosis, indicated by shorter OS, whereas miR370 reduces leukemogenesis by targeting the oncogene FOXM1.99,100 Literature data report that miR17 and miR20a inhibit pro-apoptotic Bim and subsequent mitochondria-dependent apop-tosis.101 MiR27a is an important regulator of hematopoietic development, as it targets the 3′UTR of RUNX1 transcription factor, a master regulator in hematopoietic development and important mediator in the pathogenesis of CML.102,103
MicroRNA expression in myelodysplastic syndromes
Myelodysplastic syndrome (MDS) is a group of malignant clonal hematological disorders characterized by unilineage or multilineage dysplasia, cytopenias, and risk of progression to AML. In addition to the cytogenetic aberrations observed in MDS, current studies have also revealed aberrant expression of microRNAs.104 In MDS with deletion of the long arm of chromosome 5 [del(5q)] alleles of miR143, miR145, and miR146a are deleted within the commonly deleted region located on chromosome 5q32–5q33.105 Lenalidomide therapy, indicated for this group of patients, leads to normalization of expression of these microRNA.105,106 Significant differences in microRNA-expression levels have also been found between high- and low-risk groups. Patients with high miR150 levels are more often classified in the good-cytogenetic-risk group.106 High expression of MCL1, caused by the downregulation of miR29a/miR29b plays a role in the leukemic evolution of MDS as well.107 It has been found that miR181c, miR181a, miR181b, and miR181d are selectively overexpressed in high-risk cases of MDS.108 The literature also reports upregulation of miR720 and miR21, whereas levels of miR210 and miR155 are increased in CD34+ MDS cells.109–111 At the same time, decreased expression of the Let7 family of microRNAs was shown.112
MicroRNA expression in Philadelphia-negative myeloproliferative neoplasms
Myeloproliferative neoplasms (MPNs) are a group of clonal hematopoietic disorders in which uncontrolled proliferation of hematopoietic bone-marrow stem cells plays a key role in the pathogenesis of apoptosis, genomic instability, and abnormal expression of adhesion molecules. The discovery of the JAK2V617F mutation, the structure of exons 12 and 14 of the JAK2 gene, and the role of mutations in the MPL gene have significantly contributed to the understanding of the pathogenesis of this group of diseases. The phenotypic diversity of individual MPN forms appears to be the result of the presence of additional, different molecular disturbances that significantly alter the way the intracellular signal is transmitted. Recent studies have confirmed the role of microRNA expression in the pathogenesis of this group of diseases.113,114
It has so far been confirmed that microRNAs can degrade their expression to MPN stem-cell-clone expansion and also underlie phenotypic diversity.113 Hypermethylation of miR203 is found in patients diagnosed with Philadelphia-negative MPN. Associations between miR28 overexpression and suppression of the region of thrombopoietin receptor Mpl and ectopic overexpression of miR433 have also been reported to result in abnormal hematopoietic bone-marrow stem-cell proliferation. Interestingly, miR134, miR214, and miR433 expression is not associated with changes in JAK2 activity, which may indicate other molecular mechanisms underlying the regulation expression of these microRNAs in patients with MPN.115,116
In the polycythemia vera population, reduced expression of erythropoiesis-related microRNAs (miR15a, miR150, miR221, miR223, miR24, and miR103) has been observed to result in increased erythropoiesis. Overexpression of miR320, miR451, miR144, however, leads to a decrease in the expression of inhibitors of erythropoiesis. In patients with the JAK2V617 mutation, overexpression of miR143, miR342, miR30c, miR150 and Let7a levels is observed. In patients with primary myelofibrosis, decreased miR31, miR34a, miR95, miR105, miR147, miR149, miR151, miR342, miR326, and miR4319 and overexpression of miR10a, miR146b and miR223 is observed. In the case of essential thrombocythemia, decreased levels of miR34a, miR105, miR147, miR149 and miR223 overexpression disorders have been demonstrated.113,115,116
Conclusion
Attempts to identify new prognostic and predictive factors in patients with hematopoietic systemic diseases have been undertaken for many years. Despite many reports on this subject, there is still a need to find new indicators. Further research is also needed on microRNA-expression disorders in patients with secondary hematological malignancies and rare MDS/MDN. MicroRNAs seem to be such new and useful biomarkers. They exhibit high durability during preparation, are highly informative, and their expression profiles can be evaluated in both body fluids and cells. These molecules are involved in the process of differentiation, development, and apoptosis, and abnormalities in their expression are associated with cytogenetic and molecular aberrations characteristic of hematopoietic proliferative diseases.
Current trends in experimental hematooncology have led to dynamic developments of research on microRNA expression and their introduction into clinical practice. The main assumption of using microRNA in the treatment of proliferative diseases of the hematopoietic system is influencing the control of the expression of target proteins involved in their pathogenesis. It has been demonstrated so far that inhibition of selected microRNAs by introducing knockdown mutations into the microRNA in vitro leads to a reduction in tumor-cell proliferation.117,118 It has also been proven that the role of endogenous microRNAs can be played by synthetic oligoribonucleotides or microRNA precursors. Their introduction into cells in specially designed vectors results in the silencing of gene expression that is important for the development or progression of cancer.119,120
The new therapeutic strategy is the use of anti-microRNA oligonucleotides, called antagomirs. Their use is related to the modification of protein activity by silencing genes for microRNAs that regulate their expression.119 Cell-transfection methods based on the use of microRNA vectors are currently developing extremely dynamically, and can bring progress in the treatment of proliferative diseases of the hematopoietic system.119,120
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Taylor J, Xiao W, Abdel-Wahab O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood. 2017;130(4):410–423. doi: 10.1182/blood-2017-02-734541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yao Y, Dai W. Genomic Instability and Cancer. J Carcinog Mutagen. 2014;5:1000165. doi: 10.4172/2157-2518.1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prakash G, Kaur A, Malhotra P, et al. Current role of genetics in hematologic malignancies. Indian J Hematol Blood Transfus. 2016;32(1):18–31. doi: 10.1007/s12288-015-0584-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Zhong H. The diagnosis, prognosis, and therapeutic application of MicroRNAs in haematological malignancies. Hematology. 2016;21(5):263–271. doi: 10.1080/10245332.2015.1114766. [DOI] [PubMed] [Google Scholar]
- 5.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microR-NAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17(3):189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grenda A, Budzyński M, Filip AA. Biogenesis of microRNAs and their role in the development and course of selected hematologic disorders. Postepy Hig Med Dosw. 2013;67:174–185. doi: 10.5604/17322693.1038361. [DOI] [PubMed] [Google Scholar]
- 7.Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 8.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585(13):2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Parker TL, Strout MP. Chronic lymphocytic leukemia: prognostic factors and impact on treatment. Discov Med. 2011;11(57):115–123. [PubMed] [Google Scholar]
- 10.Hallek M. Leukemia: Clymphocytic. Update on diagnosis, risk stratification, and treatment. Am J Hematol. 2015;2015(90):446–460. doi: 10.1002/ajh.23979. [DOI] [PubMed] [Google Scholar]
- 11.Hus I, Bojarska-Junak A, Dmoszyńska A, et al. ZAP-70 and CD38 expression are independent prognostic factors in patients with B-cell chronic lymphocytic leukaemia and combined analysis improves their predictive value. Folia Histochem Cytobiol. 2008;46(2):147–152. doi: 10.2478/v10042-008-0022-7. [DOI] [PubMed] [Google Scholar]
- 12.Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin Oncol. 2006;33(2):167–173. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Acunzo M, Croce CM. Downregulation of miR-15a and miR-16-1 at 13q14 in Chronic Lymphocytic Leukemia. Clin Chem. 2016;62(4):655–656. doi: 10.1373/clinchem.2015.240036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zanesi N, Pekarsky Y, Trapasso F, Calin G, Croce CM. MicroRNAs in mouse models of lymphoid malignancies. J Nucleic Acids Investig. 2010;1(1):8–40. doi: 10.4081/jnai.2010.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanlon K, Rudin CE, Harries LW. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL. PLoS One. 2009;4(9):e7169. doi: 10.1371/journal.pone.0007169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balatti V, Pekarky Y, Croce CM. Role of microRNA in chronic lymphocytic leukemia onset and progression. J Hematol Oncol. 2015;8:12. doi: 10.1186/s13045-015-0112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 18.Fatica A, Fazi F. MicroRNA-regulated pathways in hematological malignancies: how to avoid cells playing out of tune. Int J Mol Sci. 2013;14(10):20930–20953. doi: 10.3390/ijms141020930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Audrito V, Vaisitti T, Rossi D, et al. Nicotinamide blocks proliferation and induces apoptosis of chronic lymphocytic leukemia cells through activation of the p53/miR-34a/SIRT1 tumor suppressor network. Cancer Res. 2011;71(13):4473–4483. doi: 10.1158/0008-5472.CAN-10-4452. [DOI] [PubMed] [Google Scholar]
- 20.Shahjahani M, Mohammadiasl J, Noroozi F, et al. Molecular basis of chronic lymphocytic leukemia diagnosis and prognosis. Cell Oncol. 2015;38(2):93–109. doi: 10.1007/s13402-014-0215-3. [DOI] [PubMed] [Google Scholar]
- 21.Zanette DL, Rivadavia F, Molfetta GA, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40(11):1435–1440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 22.Bresin A, Callegari E, D’Abundo L, et al. miR-181b as a therapeutic agent for chronic lymphocytic leukemia in the Eµ-TCL1 mouse model. Oncotarget. 2015;6(23):19807–19818. doi: 10.18632/oncotarget.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Visone R, Veronese A, Rassenti LZ, et al. miR-181b is a biomarker of disease progression in chronic lymphocytic leukemia. Blood. 2011;118(11):3072–3079. doi: 10.1182/blood-2011-01-333484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pekarsky Y, Croce CM. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget. 2010;1(3):224–227. doi: 10.18632/oncotarget.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costinean S, Sandhu SK, Pedersen IM, et al. Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice. Blood. 2009;114(7):1374–1382. doi: 10.1182/blood-2009-05-220814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandhu SK, Volinia S, Costinean S, et al. miR-155 targets histone deacetylase 4 (HDAC4) and impairs transcriptional activity of B-cell lymphoma 6 (BCL6) in the Eµ-miR-155 transgenic mouse model. Proc Natl Acad Sci U S A. 2012;109(49):20047–20052. doi: 10.1073/pnas.1213764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferracin M, Zagatti B, Rizzotto L, et al. MicroRNAs involvement in fludarabine refractory chronic lymphocytic leukemia. Mol Cancer. 2010;9:123. doi: 10.1186/1476-4598-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vockerodt M, Cader FZ, Shannon-Lowe C, Murray P. Epstein-Barr virus and the origin of Hodgkin lymphoma. Chin J Cancer. 2014;33(12):591–597. doi: 10.5732/cjc.014.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai M, Papoudou-Bai A, Kitsoulis P, et al. Cell cycle and apoptosis deregulation in classical Hodgkin lymphomas. In Vivo. 2005;19(2):439–454. [PubMed] [Google Scholar]
- 30.Joos S, Menz CK, Wrobel G, et al. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood. 2002;99(4):1381–1387. doi: 10.1182/blood.v99.4.1381. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Mercado M, Manterola L, Lawrie CH. MicroRNAs in lymphoma: regulatory role and biomarker potential. Curr Genomics. 2015;16(5):349–358. doi: 10.2174/1389202916666150707160147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cordeiro A, Monzó M, Navarro A. Non-Coding RNAs in Hodgkin lymphoma. Int J Mol Sci. 2017;18(6):E1154. doi: 10.3390/ijms18061154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuccaro A, Bartolomei F, Cupelli E, Galli E, Giachelia M, Hohaus S. Prognostic factors in hodgkin lymphoma. Mediterr J Hematol Infect Dis. 2014;6(1):e2014053. doi: 10.4084/MJHID.2014.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navarro A, Diaz T, Martinez A, et al. Regulation of JAK2 by miR-135a: prognostic impact in classic Hodgkin lymphoma. Blood. 2009;114(14):2945–2951. doi: 10.1182/blood-2009-02-204842. [DOI] [PubMed] [Google Scholar]
- 35.van den Berg AV, Magalhães L, Ferreira Vidal A, Vidal AF. MicroR-NAs as biomarkers of the response to treatment with ABVD scheme in Hodgkin lymphoma. J Leuk. 2015;03(04):1–6. [Google Scholar]
- 36.Zheng RL, Jiang YJ, Wang X. Role of microRNAs on therapy resistance in Non-Hodgkin’s lymphoma. Int J Clin Exp Med. 2014;7(11):3818–3832. [PMC free article] [PubMed] [Google Scholar]
- 37.Nowakowski GS, Czuczman MS. ABC, GCB, and double-hit diffuse large B-cell lymphoma: does subtype make a difference in therapy selection? Am Soc Clin Oncol Educ Book. 2015;35:e449–e457. doi: 10.14694/EdBook_AM.2015.35.e449. [DOI] [PubMed] [Google Scholar]
- 38.Kim S-W, Ramasamy K, Bouamar H, Lin A-P, Jiang D, Aguiar RCT. MicroRNAs miR-125a and miR-125b constitutively activate the NF- B pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20. Proceedings of the National Academy of Sciences. 2012;109(20):7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni H, Tong R, Zou L, Song G, Cho WC. MicroRNAs in diffuse large B-cell lymphoma. Oncol Lett. 2016;11(2):1271–1280. doi: 10.3892/ol.2015.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Shen Y, Liu M, et al. Quantitative proteomics reveals that miR-155 regulates the PI3K-AKT pathway in diffuse large B-cell lymphoma. Am J Pathol. 2012;181(1):26–33. doi: 10.1016/j.ajpath.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alencar AJ, Malumbres R, Kozloski GA, et al. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clinical Cancer Research. 2011;17(12):4125–4135. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertus J-L, Harms G, Blokzijl T, et al. Specific expression of miR-17-5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Modern Pathology. 2009;22(4):547–555. doi: 10.1038/modpathol.2009.10. [DOI] [PubMed] [Google Scholar]
- 43.Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117(11):3140–3146. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 44.Sorigue M, Sancho J-M. Current prognostic and predictive factors in follicular lymphoma. Ann Hematol. 2018;97(2):209–227. doi: 10.1007/s00277-017-3154-z. [DOI] [PubMed] [Google Scholar]
- 45.Graham C, Lebrun DP. Tumor suppressors in follicular lymphoma. Leuk Lymphoma. 2015;56(7):1981–1988. doi: 10.3109/10428194.2014.982638. [DOI] [PubMed] [Google Scholar]
- 46.Leich E, Ott G, Rosenwald A, Pathology RA. Pathology, pathogenesis and molecular genetics of follicular NHL. Best Pract Res Clin Haematol. 2011;24(2):95–109. doi: 10.1016/j.beha.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Leich E, Zamo A, Horn H, et al. MicroRNA profiles of t(14;18)-negative follicular lymphoma support a late germinal center B-cell phenotype. Blood. 2011;118(20):5550–5558. doi: 10.1182/blood-2011-06-361972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson MA, Edmonds MD, Liang S, et al. miR-31 and miR-17-5p levels change during transformation of follicular lymphoma. Hum Pathol. 2016;50:118–126. doi: 10.1016/j.humpath.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, Corrigan-Cummins M, Hudson J, et al. MicroRNA profiling of follicular lymphoma identifies microRNAs related to cell proliferation and tumor response. Haematologica. 2012;97(4):586–594. doi: 10.3324/haematol.2011.048132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogt N, Dai B, Erdmann T, Berdel WE, Lenz G, et al. The molecular pathogenesis of mantle cell lymphoma. Leuk Lymphoma. 2017;58(7):1530–1537. doi: 10.1080/10428194.2016.1248965. [DOI] [PubMed] [Google Scholar]
- 51.Ahmed M, Zhang L, Nomie K, Lam L, Wang M, et al. Gene mutations and actionable genetic lesions in mantle cell lymphoma. Oncotarget. 2016;7(36):58638–58648. doi: 10.18632/oncotarget.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vose JM. Lymphoma: Mcell. Update on diagnosis, risk-stratification, and clinical management. Am J Hematol. 2013;2013(88):1082–1088. doi: 10.1002/ajh.23615. [DOI] [PubMed] [Google Scholar]
- 53.Masamha CP, Albrecht TR, Wagner EJ. Discovery and characterization of a novel CCND1/MRCK gene fusion in mantle cell lymphoma. J Hematol Oncol. 2016;9(1):30. doi: 10.1186/s13045-016-0260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rinaldi A, Poretti G, Kwee I, et al. Concomitant MYC and microRNA cluster miR-17-92 (C13orf25) amplification in human mantle cell lymphoma. Leuk Lymphoma. 2007;48(2):410–412. doi: 10.1080/10428190601059738. [DOI] [PubMed] [Google Scholar]
- 55.Zhao J-J, Lin J, Lwin T, et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115(13):2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: how a complex biology gets more complex. Leukemia. 2015;29(5):1004–1017. doi: 10.1038/leu.2014.351. [DOI] [PubMed] [Google Scholar]
- 57.Husby S, Ralfkiaer U, Garde C, et al. miR-18b overexpression identifies mantle cell lymphoma patients with poor outcome and improves the MIPI-B prognosticator. Blood. 2015;125(17):2669–2677. doi: 10.1182/blood-2014-06-584193. [DOI] [PubMed] [Google Scholar]
- 58.Arakawa F, Kimura Y, Yoshida N, et al. Identification of miR-15b as a transformation-related factor in mantle cell lymphoma. Int J Oncol. 2016;48(2):485–492. doi: 10.3892/ijo.2015.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weaver CJ, Tariman JD. Multiple myeloma genomics: a systematic review. Semin Oncol Nurs. 2017;33(3):237–253. doi: 10.1016/j.soncn.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 60.Alzrigat M, Párraga AA, Jernberg-Wiklund H. Epigenetics in multiple myeloma: From mechanisms to therapy. Semin Cancer Biol. 2017;579X:S104430104–104430109. doi: 10.1016/j.semcancer.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 61.Gabrea A, Leif Bergsagel P, Michael Kuehl W. Distinguishing primary and secondary translocations in multiple myeloma. DNA Repair. 2006;5(9–10):1225–1233. doi: 10.1016/j.dnarep.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 62.Roccaro AM, Sacco A, Thompson B, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113(26):6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Zhang B, Li W, et al. MiR-15a/16 regulates the growth of myeloma cells, angiogenesis and antitumor immunity by inhibiting Bcl-2, VEGF-A and IL-17 expression in multiple myeloma. Leuk Res. 2016;49:73–79. doi: 10.1016/j.leukres.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 64.Benetatos L, Vartholomatos G. Deregulated microRNAs in multiple myeloma. Cancer. 2012;118(4):878–887. doi: 10.1002/cncr.26297. [DOI] [PubMed] [Google Scholar]
- 65.Unno K, Zhou Y, Zimmerman T, Platanias LC, Wickrema A, et al. Identification of a novel microRNA cluster miR-193b-365 in multiple myeloma. Leuk Lymphoma. 2009;50(11):1865–1871. doi: 10.3109/10428190903221010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones CI, Zabolotskaya MV, King AJ, et al. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br J Cancer. 2012;107(12):1987–1996. doi: 10.1038/bjc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmad N, Haider S, Jagannathan S, Anaissie E, Driscoll JJ, et al. MicroRNA theragnostics for the clinical management of multiple myeloma. Leukemia. 2014;28(4):732–738. doi: 10.1038/leu.2013.262. [DOI] [PubMed] [Google Scholar]
- 68.Chen L, Li C, Zhang R, et al. miR-17-92 cluster microRNAs confers tumorigenicity in multiple myeloma. Cancer Lett. 2011;309(1):62–70. doi: 10.1016/j.canlet.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 69.Neri P, Gratton K, Ren L. Role of mir-30e in multiple myeloma mells resistance to lenalidomide and bortezomib. Blood. 2012;120:323. [Google Scholar]
- 70.Zhang L, Pan L, Xiang B, et al. Potential role of exosome-associated microRNA panels and in vivo environment to predict drug resistance for patients with multiple myeloma. Oncotarget. 2016;7(21):30876–30891. doi: 10.18632/oncotarget.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Li C, Ju S, et al. Myeloma cell adhesion to bone marrow stromal cells confers drug resistance by microRNA-21 up-regulation. Leuk Lymphoma. 2011;52(10):1991–1998. doi: 10.3109/10428194.2011.591004. [DOI] [PubMed] [Google Scholar]
- 72.Rastgoo N, Abdi J, Hou J, Chang H, et al. Role of epigenetics-microRNA axis in drug resistance of multiple myeloma. J Hematol Oncol. 2017;10(1):121. doi: 10.1186/s13045-017-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abdi J, Jian H, Chang H. Role of micro-RNAs in drug resistance of multiple myeloma. Oncotarget. 2016;7(37):60723–60735. doi: 10.18632/oncotarget.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar CC. Genetic abnormalities and challenges in the treatment of acute myeloid leukemia. Genes Cancer. 2011;2(2):95–107. doi: 10.1177/1947601911408076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.X-Xing Chen, Lin J, Qian J, et al. Dysregulation of miR-124-1 predicts favorable prognosis in acute myeloid leukemia. Clin Biochem. 2014;47(1–2):63–66. doi: 10.1016/j.clinbiochem.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 76.Li Y, Gao L, Luo X, et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121(3):499–509. doi: 10.1182/blood-2012-07-444729. [DOI] [PubMed] [Google Scholar]
- 77.Li H, Xing C, Zhou B, et al. A regulatory circuitry between miR-193a/miR-600 and WT1 enhances leukemogenesis in acute myeloid leukemia. Exp Hematol. 2018;61:59–68. doi: 10.1016/j.exphem.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 78.Liu L, Ren W, Chen K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cellular Physiology and Biochemistry. 2017;41(5):1981–1992. doi: 10.1159/000475277. [DOI] [PubMed] [Google Scholar]
- 79.Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q– syndrome phenotype. Nat Med. 2010;16(1):49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 80.Liao Q, Wang B, Li X, Jiang G, et al. miRNAs in acute myeloid leukemia. Oncotarget. 2017;8(2):3666–3682. doi: 10.18632/oncotarget.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lechman ER, Gentner B, Ng SWK, Sw N, et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29(2):214–228. doi: 10.1016/j.ccell.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li Z, Lu J, Sun M, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proceedings of the National Academy of Sciences. 2008;105(40):15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trino S, Lamorte D, Caivano A, et al. MicroRNAs as new biomarkers for diagnosis and prognosis, and as potential therapeutic targets in acute myeloid leukemia. Int J Mol Sci. 2018;19(2):E460. doi: 10.3390/ijms19020460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao X-N, Lin J, Li Y-H, Yh L, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011;30(31):3416–3428. doi: 10.1038/onc.2011.62. [DOI] [PubMed] [Google Scholar]
- 85.Garzon R, Volinia S, Liu C-G, et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111(6):3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hossain MJ, Xie L, Mccahan SM. Characterization of pediatric acute lymphoblastic leukemia survival patterns by age at diagnosis. J Cancer Epidemiol. 2014;2014(1):865979–9. doi: 10.1155/2014/865979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marinescu C, Vladareanu A-M, Mihai F. Acute lymphocytic leukemia in adults. Pathologic features and prognosis. Rom J Intern Med. 2015;53(1):33–38. doi: 10.1515/rjim-2015-0004. [DOI] [PubMed] [Google Scholar]
- 88.Luan C, Yang Z, Chen B. The functional role of microRNA in acute lymphoblastic leukemia: relevance for diagnosis, differential diagnosis, prognosis, and therapy. Onco Targets Ther. 2015;8:2903–2914. doi: 10.2147/OTT.S92470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zanette DL, Rivadavia F, Molfetta GA, et al. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40(11):1435–1440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 90.Yeh C-H, Moles R, Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol Cancer. 2016;15(1):37. doi: 10.1186/s12943-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bhatia S, Kaul D, Varma N. Potential tumor suppressive function of miR-196b in B-cell lineage acute lymphoblastic leukemia. Mol Cell Biochem. 2010;340(1–2):97–106. doi: 10.1007/s11010-010-0406-9. [DOI] [PubMed] [Google Scholar]
- 92.Fuziwara CS, Kimura ET. Insights into regulation of the miR-17-92 cluster of miRNAs in cancer. Front Med. 2015;2(34):64. doi: 10.3389/fmed.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Litwińska Z, Machaliński B. miRNAs in chronic myeloid leukemia: small molecules, essential function. Leuk Lymphoma. 2017;58(6):1297–1305. doi: 10.1080/10428194.2016.1243676. [DOI] [PubMed] [Google Scholar]
- 94.Tertian G, Avalos MR, Léonard C, et al. Additional translocation (9;12) (p13;q24.1) in newly diagnosed chronic myeloid leukemia: complete cytogenetic remission after interferon therapy. Hematol Cell Ther. 1996;38(3):269–273. doi: 10.1007/s00282-996-0269-4. [DOI] [PubMed] [Google Scholar]
- 95.Yeh C-H, Moles R, Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol Cancer. 2016;15(1):37. doi: 10.1186/s12943-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shibuta T, Honda E, Shiotsu H, et al. Imatinib induces demethylation of miR-203 gene: An epigenetic mechanism of anti-tumor effect of imatinib. Leuk Res. 2013;37(10):1278–1286. doi: 10.1016/j.leukres.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 97.di Stefano C, Mirone G, Perna S, Marfe G, et al. The roles of microR-NAs in the pathogenesis and drug resistance of chronic myelogenous leukemia (Review. Oncol Rep. 2016;35(2):614–624. doi: 10.3892/or.2015.4456. [DOI] [PubMed] [Google Scholar]
- 98.Venturini L, Battmer K, Castoldi M, et al. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109(10):4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 99.Zhu X, Zhao H, Lin Z, Zhang G, et al. Functional studies of miR-130a on the inhibitory pathways of apoptosis in patients with chronic myeloid leukemia. Cancer Gene Ther. 2015;22(12):573–580. doi: 10.1038/cgt.2015.50. [DOI] [PubMed] [Google Scholar]
- 100.Zhou M, Zeng J, Wang X, et al. MiR-370 sensitizes chronic myeloid leukemia K562 cells to homoharringtonine by targeting Forkhead box M1. J Transl Med. 2013;11(1):265. doi: 10.1186/1479-5876-11-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, He D, Zeng J, et al. The effects of antisense miRNA-20a alone or in combination with imatinib on K562 cell proliferation. Front Pharmacol. 2017;8:127. doi: 10.3389/fphar.2017.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci U S A. 2009;106(1):238–243. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.San José-Enériz E, Román-Gómez J, Jiménez-Velasco A, et al. MicroRNA expression profiling in Imatinib-resistant Chronic Myeloid Leukemia patients without clinically significant ABL1-mutations. Mol Cancer. 2009;8:69. doi: 10.1186/1476-4598-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Setijono SR, Kwon HY, Song SJ, Microrna SSJ. MicroRNA, an antisense RNA, in sensing myeloid malignancies. Front Oncol. 2017;7:331. doi: 10.3389/fonc.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Merkerova MD, Krejcik Z, Belickova M, et al. Genome-wide miRNA profiling in myelodysplastic syndrome with del(5q) treated with lenalidomide. Eur J Haematol. 2015;95(1):35–43. doi: 10.1111/ejh.12458. [DOI] [PubMed] [Google Scholar]
- 106.Rhyasen GW, Starczynowski DT. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia. 2012;26(1):13–22. doi: 10.1038/leu.2011.221. [DOI] [PubMed] [Google Scholar]
- 107.Kirimura S, Kurata M, Nakagawa Y, et al. Role of microRNA-29b in myelodysplastic syndromes during transformation to overt leukaemia. Pathology. 2016;48(3):233–241. doi: 10.1016/j.pathol.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 108.Kuang X, Chi J, Wang L. Deregulated microRNA expression and its pathogenetic implications for myelodysplastic syndromes. Hematology. 2016;21(10):593–602. doi: 10.1080/10245332.2016.1193962. [DOI] [PubMed] [Google Scholar]
- 109.Borze I, Scheinin I, Siitonen S, et al. miRNA expression profiles in myelodysplastic syndromes reveal Epstein-Barr virus miR-BART13 dysregulation. Leuk Lymphoma. 2011;52(8):1567–1573. doi: 10.3109/10428194.2011.568652. [DOI] [PubMed] [Google Scholar]
- 110.Kim Y, Cheong JW, Kim YK, et al. Serum microRNA-21 as a potential biomarker for response to hypomethylating agents in myelodysplastic syndromes. PLoS One. 2014;9(2):e86933. doi: 10.1371/journal.pone.0086933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shikama Y, Cao M, Ono T, et al. Reduction of c-Fos via overexpression of miR-34a results in enhancement of TNF- production by LPS in neutrophils from myelodysplastic syndrome patients. PLoS One. 2016;11(8):e0158527. doi: 10.1371/journal.pone.0158527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ikeda K, Ogawa K, Takeishi Y. The role of HMGA2 in the proliferation and expansion of a hematopoietic cell in myeloproliferative neoplasms. Fukushima J Med Sci. 2012;58(2):91–100. doi: 10.5387/fms.58.91. [DOI] [PubMed] [Google Scholar]
- 113.Zhan H, Cardozo C, Raza A. MicroRNAs in myeloproliferative neoplasms. Br J Haematol. 2013;161(4):471–483. doi: 10.1111/bjh.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Padrnos L, Mesa RA. Myeloproliferative neoplasms: translating new discoveries into better outcomes, better quality of life. Oncology. 2017;31(7):521–529. [PMC free article] [PubMed] [Google Scholar]
- 115.Alizadeh S, Azizi SG, Soleimani M, Farshi Y, Kashani Khatib Z. The role of microRNAs in myeloproliferative neoplasia. Int J Hematol Oncol Stem Cell Res. 2016;10(3):172–185. [PMC free article] [PubMed] [Google Scholar]
- 116.Fatica A, Fazi F. MicroRNA-regulated pathways in hematological malignancies: how to avoid cells playing out of tune. Int J Mol Sci. 2013;14(10):20930–20953. doi: 10.3390/ijms141020930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee YS, Dutta A. MicroRNAs: small but potent oncogenes or tumor suppressors. Curr Opin Investig Drugs. 2006;7(6):560–564. [PubMed] [Google Scholar]
- 118.Fan Z, Cui H, Yu H, et al. MiR-125a promotes paclitaxel sensitivity in cervical cancer through altering STAT3 expression. Oncogenesis. 2016;5:e223. doi: 10.1038/oncsis.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ciepłucha A, Jamroziak K, Robak T. Perspektywy zastosowania mikroRNA w leczeniu nowotworów. Acta Haematol Pol. 2007;38:425–435. [Google Scholar]
- 120.Li Z, Zhan W, Wang Z, et al. Inhibition of PRL-3 gene expression in gastric cancer cell line SGC7901 via microRNA suppressed reduces peritoneal metastasis. Biochem Biophys Res Commun. 2006;348(1):229–237. doi: 10.1016/j.bbrc.2006.07.043. [DOI] [PubMed] [Google Scholar]