Abstract
Background
NUDT21 is a mammalian precursor mRNA(pre-mRNA) 3’ end processing factor and plays an important role in the selection of poly(A) sites in 3’-untranslated region (3’-UTR). NUDT21 links alternative polyadenylation with regulation of glioblastoma and osteosarcoma progression and is found to be related to drug resistance in childhood acute leukemia. However, the effect of NUDT21 on leukemia cells and the underlying mechanism are unknown.
Methods
We knocked down NUDT21 in K562 cells and applied qRT-PCR and western blotting to quantitate the mRNA and protein expression. Cell proliferating and apoptosis were investigated subsequently by flow cytometry, BrdU, Caspase3/7. RNA microarray and intracellular signaling array were used to determine the important cell signaling pathways.
Results
We clarified that the mRNA expression levels of NUDT21 are higher in primary chronic myelocytic leukemia patients and K562 leukemic cells compared with healthy controls and PBMCs. Downregulation of NUDT21 expression in K562 cells inhibits proliferation and promotes apoptosis. Screening by mRNA chip and intracellular signaling array, we found that MAPK/ERK pathway represented the main molecular mechanism underlying the effects of NUDT21 knockdown in K562 cells.
Conclusion
NUDT21 played an important role in promoting proliferation and inhibiting apoptosis in leukemia K562 cells. The underlying mechanisms involved the modulation of PTEN and a set of downstream molecules including ERK1/2.
Impact statement
The present work shows that the expression of NUDT21 was upregulated in chronic myelocytic leukemia and K562 cells. Silencing NUDT21 inhibited the proliferation and promoted the apoptosis of K562 cells. Subsequent experiments confirmed that NUDT21 promoted K562 proliferation through regulating the expression of p-ERK. Our findings may provide insights into the molecular mechanism underlying the effects of NUDT21 on leukemia cells and a novel strategy for the treatment of leukemia.
Keywords: NUDT21, MAPK/ERK, K562 cell line, leukemia, cell proliferation, cell apoptosis
Introduction
Leukemia is a heterogeneous group of disorders resulting from the acquisition of chromosomal rearrangements, multiple gene mutations, and abnormal epigenetic regulation. Epigenetics indicates heritable changes in gene expression without nucleotide sequence variation, including DNA methylation, histone deacetylation, and chromatin remodeling.1 With the noncoding RNAs in the field of epigenetics becoming a new hot topic of research on abnormal epigenetic regulation in leukemia genesis,2 more studies have focused on posttranscriptional regulation of genes. Recently, NUDT21, a protein capable of regulating precursor messenger RNA (pre-mRNA) 3′-end formation at the posttranscriptional level, has been revealed to be closely related to disease progression.
It has become increasingly recognized that pre-mRNA 3′-end formation is crucial for mRNA maturation in eukaryotic cells, promoting mRNA stability, efficient nuclear transport, and translation.3 CFIm is a cleavage and polyadenylation specificity factor, and it was first purified from Hela cell nuclear extracts as four polypeptides of 25, 59, 68, and 72 kDa (CFIm25, CFIm59, CFIm68, and CFIm72, respectively). CFIm is a heterodimer composed of the smallest CFIm25 subunit and any one of the three large subunits, while CFIm59, CFIm68, and CFIm72 are structurally related.4 Although CFIm68 depletion decreases 3′-untranslated region (3′-UTR) length, the most significant primer activation signal switching was found to occur after knockdown of CFIm25.5
CFIm25, also known as NUDT21 (nudix, nucleoside diphosphate linked moiety X-type, motif21), has a NUDIX hydrolase domain that acts like an authentic RNA-binding protein. 3′-UTRs are important for fine-tuning transcript and protein levels because they contain binding sites for regulatory molecules such as RNA-binding proteins and miRNAs.6 NUDT21 regulates 3′-UTR length by binding to the proximal cleavage and polyadenylation site and directing alternative polyadenylation (APA).7 The global shortening of mRNAs through APA that occurs during enhanced cellular proliferation represents an important mechanism of regulated gene expression. The 3′-UTR truncation of growth-promoting mRNA transcripts that relieves intrinsic microRNA- and AU-rich element (ARE)-mediated repression has been observed to correlate with cellular transformation. Like alternative splicing, usage of APA signals allows a single gene to encode various mRNA transcripts.5 For example, use of APA signals often removes large parts of the 3′-UTR, which contain the possible miRNA-targeting sites and AREs, thus influencing the fate of mRNAs. Specific APA events play important roles in cell growth and differentiation or disease, such as immunoglobulin M (IgM) switching,8 spermatogenesis,9 and tumorigenesis.10,11
It has been demonstrated that NUDT21 plays pivotal roles in the regulation of glioblastoma cell proliferation and tumorigenicity.12,13 Redis et al14 reported that lncRNA CCAT2 regulated tumor metabolism through binding different CFIm subunits (CFIm25 and CFIm68). NUDT21 was also identified as a gene associated with resistance to etoposide in children with acute leukemia.15 These findings identified the potential relevance of NUDT21 to tumor progression. However, the role of NUDT21 in leukemia cell growth and the underlying mechanism remain to be fully elucidated.
In the present study, it was first confirmed that the expression of NUDT21 was upregulated in chronic myelocytic leukemia (CML) and K562 cells. Silencing NUDT21 inhibited the proliferation and promoted the apoptosis of K562 cells. Subsequent experiments confirmed that NUDT21 promoted K562 proliferation through regulating the expression of p-ERK, which is the effector of the MAPK/ERK pathway.
Materials and methods
Patients and cell culture
The human leukemia cell lines, K562, Jurkat, and HL-60 cells were purchased from the Cell Resource Center, IBMS, CAMS/PUMC, and maintained in DMEM supplemented with 10% FBS. Cells were incubated at 37°C and maintained at 5% CO2. Peripheral blood mononuclear cells (PBMCs) were collected from the healthy people’s medical examination specimen at the First Hospital of Shanxi Medical University. The use of the primary cells was approved by the ethics committee of The First Hospital of Shanxi Medical University (Taiyuan, China).
The bone marrow samples of primary CML patients and healthy control subjects were obtained from The First Hospital of Shanxi Medical University, China. The median age of the CML patients was 58 years (range, 49–76 years). All samples of CML were positive for the BCR/ABL gene. The study was approved by the ethics committee of The First Hospital of Shanxi Medical University (Taiyuan, China). Written informed consent was obtained from all subjects. All the bone marrow samples were kept frozen at −70°C until use.
Quantitative reverse polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cultured cell lines and bone marrow samples using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The IQ5 detection system (Bio-Rad Laboratories Inc., Hercules, CA, USA) and SYBR Green Real time PCR Master Mix (Thermo Fisher Scientific) were used for qRT-PCR analysis. The conditions of qRT-PCR were as follows: predenaturation for 5 minutes at 95°C, denaturation for 30 seconds at 90°C, annealing for 40 seconds at 60°C, and extension for 40 seconds at 72°C, for a total of 40 cycles. The PCR primers are listed as follows: NUDT21 forward, 5′-CGGCTACCCCATGTGTTACTG-3′; NUDT21 reverse, 5′-TCACCACCAGGTAGTTTGAAGAAA-3′; PTEN forward, 5′-CGACGGGAAGACAAGTTCAT-3′; PTEN reverse, 5′-AGGTTTCCTCTGGTCCTGGT-3′; GAPDH forward, 5′-TTTGTCAAGCTCATTTCCTG-3′; and GAPDH reverse, 5′-TGGTCCAGGGTTTCTTACTC-3′. GAPDH was used as an internal control, and fold changes were calculated by relative quantification (2−DDCt). Each experiment was conducted three times.
Western blotting
Protein was extracted from cells using RIPA buffer. After centrifugation for 15 minutes at 4°C at 14,000 g, upper supernatant was collected, and protein concentration was measured with the bicinchoninic acid method. An amount of 30 µg protein was electrophoresed in SDS-polyacrylamide gels (Thermo Fisher Scientific) and transferred to polyvinylidene difluoride membranes (EMD Burlington, Billerica, MA, USA). After blocking with 5% nonfat milk, membranes were incubated with specific primary antibodies overnight. Then, the membrane was washed three times with tris-buffered saline with Tween 20 buffer, incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody for 1 hour, and then developed onto X-ray films (Denville Scientific, Swedesboro, NJ, USA) using chemiluminescent reagents. Images were captured by Bio-Rad ChemiDoc XRS+ (Bio-Rad Laboratories Inc.). The antibodies used in this study were as follows: anti-Flag (F1804, Sigma-Aldrich Co., St Louis, MO, USA); anti-Proliferating Cell Nuclear Antigen (PCNA) (sc-56; Santa Cruz Biotechnology Inc., Dallas, TX, USA); anti-mouse IgG (sc-2005; Santa Cruz Biotechnology Inc.); anti-NUDT21 (ab183660; Abcam, Cambridge, UK); anti-cyclin E (ab133266; Abcam); anti-Bax (ab53154; Abcam); anti-Bcl-2 (ab32124; Abcam); anti-phosphatase and tensin homolog deleted on chromosome 10 (PTEN) (ab32199; Abcam); anti-ERK1+ ERK2 (phosphor T202+ Y204) (ab223500; Abcam); and anti-GAPDH (sc-32233; Santa Cruz Biotechnology Inc.) was used as a control.
Gene knockdown
Specific shRNA plasmid against NUDT21 targeting 5′-ACCTCCTCAGTATCCATAT-3′ and a non-silencing shRNA control plasmid targeting 5′-TTCTCCGAACGT-GTCACGT-3′ were synthesized by Santa Cruz Biotechnology Inc. The Control shRNA was transfected as a negative control in comparison with NUDT21 shRNA. Cell transfections were conducted using Lipofectamine 3,000 reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Cell Counting Kit-8 (CCK8) cell proliferation assay
The proliferative ability of leukemia cells was assessed using the CCK-8 (Dojindo, Tokyo, Japan) according to the manufacturer’s protocol. Leukemia cells stably transfected with shRNA control or NUDT21 shRNA were seeded into 96-well plates (2×103 cells/well) and then treated with temozolomide. After each indicated time point, the medium was replaced with fresh medium with 10% CCK-8. Then, the cells were incubated at 37°C for an additional 2 hours, and the absorbance was measured at 450 nm.
Bromodeoxyuridine (BrdU) incorporation analysis
DNA synthesis of the cells was assessed by the BrdU incorporation assay (Roche Applied Science, Penzberg, Germany). After 48 hours of transfection of the indicated shRNAs, the cells of each group were reseeded in 96-well plates at a density of 5×103 cells/well. Cells were incubated with 10 µM BrdU for 4 hours and then were fixed with 4% paraformaldehyde. After being rinsed with PBS, cells were treated with 2N HCl for 30 minutes. Nonspecific binding was blocked in 5% BSA for 1 hour at room temperature. Cells were respectively incubated with mouse anti-BrdU (1:200, MAB3222; EMD Millipore) overnight at 4°C one after another. Then followed by three times washes in PBS, they were incubated with Goat Anti-Mouse IgG H&L (Cy5®) preadsorbed (ab6563) for 1 hour at room temperature. Immunofluorescence was observed using flow cytometry.
Cell cycle assay
The cell cycle phase distribution of K562 cells was analyzed by flow cytometry. First, the K562 cells were fixed in 75% ice ethanol (–20°C) and preserved in a refrigerator at 4°C overnight. Then, the cells were centrifuged. To remove the stationary liquid, the cells were washed twice with cold PBS after centrifugation. Thereafter, cells mixed with RNaseA were placed in a water-bath for 30 minutes and stained using propidium iodide (PI). Finally, the ratio of cells in G0/G1, S, and G2/M phases to total cells was analyzed by using flow cytometry based on the red fluorescence in cells. Each experiment was conducted three times.
Caspase3/7 activity assay
The irradiated K562 cells were incubated for 18 hours. Caspase-Glo™ 3/7 buffer and lyophilized Caspase-Glo™ substrate (Promega Corporation, Fitchburg, WI, USA) were stored in a freezer and equilibrated to room temperature (20°C) prior to use. The buffer and substrate solutions were combined. Caspase-Glo™ reagent (100 µL) was added to each well. The contents of the wells were mixed on a plate shaker for 30 seconds. The 96-well plate was incubated at 20°C for 3 hours. The luminescence of the samples was measured using a Wallac Victor2 1420 Multilabel counter (PerkinElmer Inc., Waltham, MA, USA). Each experiment was conducted three times.
Annexin V/PI double-staining
The irradiated K562 cells were incubated for 18 hours, stained with annexin V-FITC (0.2 mg/mL) and PI (0.05 mg/mL) for 20 minutes, and were examined by flow cytometry (FACS Calibur; BD Biosciences, San Jose, CA, USA) using CellQuest Pro software after excitation with 488 nm laser and emission at 530 nm. A minimum of 10,000 cells were analyzed per sample and illustrated as a dot plot using Flowjo software.
mRNA array
Total RNA was prepared by Trizol reagent (Thermo Fisher Scientific) and the RNeasy kit (Qiagen NV, Venlo, the Nether-lands) according to the manufacturers’ guidelines. The RNA concentration quality control was performed by 1% agarose electrophoresis and NanoDrop Spectrophotometer 2000 (Thermo Fisher Scientific) and Agilent Bioanalyzer 2100. GeneChip Human Clariom™ D (Affymetrix) analysis was performed on RNA samples. The cDNA was purified with GeneChip WT PLUS Reagent Kit. The GeneChip Hybridization Wash and Stain Kit was used for hybridization and washing of the microarray. GeneChip Scanner 3000 was used for microarray scanning.
Intracellular signaling arrays
To evaluate the intracellular signaling activities, the PathScan Intracellular Signaling Array Kit (Cell Signaling Technology, Danvers, MA, USA) was used to assess the activities of components of several key signaling pathways, including the insulin signaling pathway and the apoptosis pathway.
First, the whole protein lysates of K562 cells were prepared using lysis buffer in the kit and then placed onto the membrane window of the antibody array slide. The lysate-treated slide was incubated overnight at 4°C on an orbital shaker. Then, the whole slide was washed with wash buffer (1×) and incubated on the orbital shaker for 5 minutes (20°C). Then, each of the 18 wells was added to the Detection Antibody Cocktail (1×) and incubated for 1 hour (20°C) on an orbital shaker. Next, the slide was incubated for 30 minutes with HRP-linked Streptavidin (1×) after washing three times. Finally, the slide was treated with Lumi Glo and peroxide and delivered to a GEL-DOC 2000 Gel Documentation System (Bio-Rad Laboratories Inc.) for taking pictures. The Quantity One software (Bio-Rad Laboratories Inc.) was used for assessing the intensity of bands. The following day, the antibody membrane was incubated with 1× HRP-linked Streptavidin and visualized with Lumi Glo and peroxide. Images were taken using the Bio-Rad gel documentation system.
Statistical analysis
Each experiment was performed at least three times. Quantitative data are presented as mean ± SD. An unpaired 2-sided Student’s t-test was used to determine the significant differences of all the results. The statistical significance levels were set at *P<0.05, **P<0.01, and ***P<0.001. Biostatistical analysis was performed using GraphPad software (GraphPad Prism 5).
Results
NUDT21 is highly expressed in primary CML patients and human leukemia cells
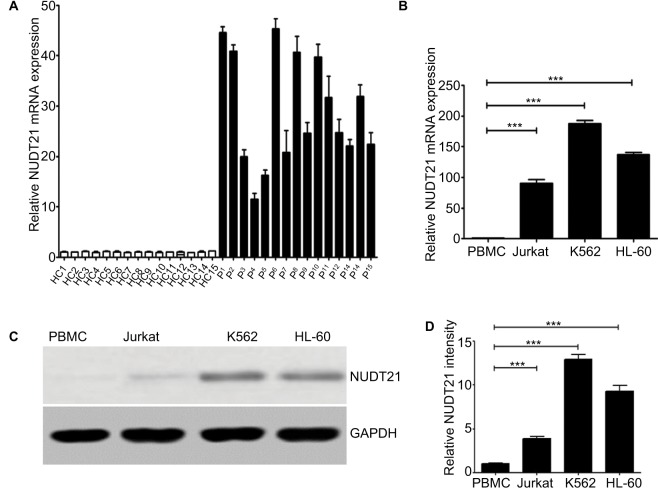
In order to identify and compare the expression levels of NUDT21 in primary CML patients and human leukemia cell lines, we conducted a preliminary exploration of NUDT21 expression in primary CML patients (n=15) and human leukemia cells (K562, Jurkat, and HL-60) by qRT-PCR and Western blotting. To evaluate the relative mRNA expression levels of NUDT21, we compared the Ct values of NUDT21 with those of the house-keeping gene GAPDH. The qRT-PCR analysis revealed that the expression levels of NUDT21 in bone marrow samples isolated from 15 primary CML patients were significantly higher than 15 healthy controls (Figure 1A). In human cell lines, the qRT-PCR and Western blotting analyses showed that both mRNA (Figure 1B) and protein (Figure 1C and D) expression levels of NUDT21 were significantly higher in human leukemia cells than normal cell PBMCs. These results indicated that NUDT21 may act as an oncogene in leukemia.
Figure 1.
Expression levels of NUDT21 in human leukemia.
Notes: (A) mRNA expression levels of NUDT21 in bone marrow samples from healthy control subjects (HC1–HC15) and CML patients (P1–P15). The mRNA (B) expression levels of NUDT21 were evaluated by qRT-PCR in human normal PBMCs, human leukemia cells K562, Jurkat, and hl-60 cells. The protein (C and D) expression levels of NUDT21 were evaluated by Western blotting in human normal PBMCs, human leukemia cells K562, Jurkat, and hl-60 cells. GAPDH was used as internal control; the data are the mean ± SD for duplicate experiments. ***P< 0.001.
Abbreviations: CML, chronic myelocytic leukemia; HC, healthy control; qRT-PCR, quantitative reverse polymerase chain reaction.
Knockdown of NUDT21 in leukemia cells
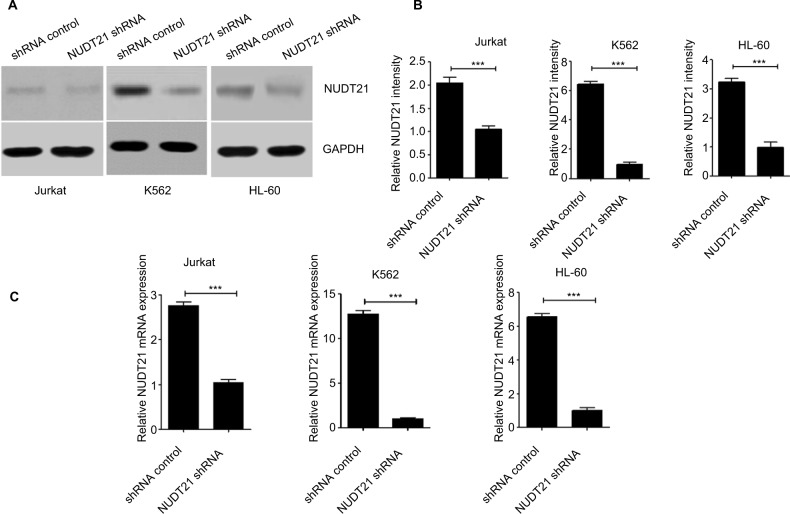
To assess the effect of NUDT21 on K562, Jurkat, and HL60 leukemia cells, shRNA targeting NUDT21 was transferred to leukemia cells with the negative shRNA (shRNA control) as a control. The knockdown effects of shRNAs were confirmed by Western blotting (Figure 2A and B) and qRT-PCR (Figure 2C) analyses.
Figure 2.
Knockdown of NUDT21 in leukemia cells.
Notes: Western blotting (A and B) and qPCR (C) were used to confirm the knockdown effects of shRNAs against NUDT21 in K562, Jurkat and HL-60 cells. The data are the mean ± SD for duplicate experiments; ***P<0.001.
Abbreviation: qRT-PCR, quantitative reverse polymerase chain reaction.
Knockdown of NUDT21 inhibits the growth and proliferation of leukemia cells
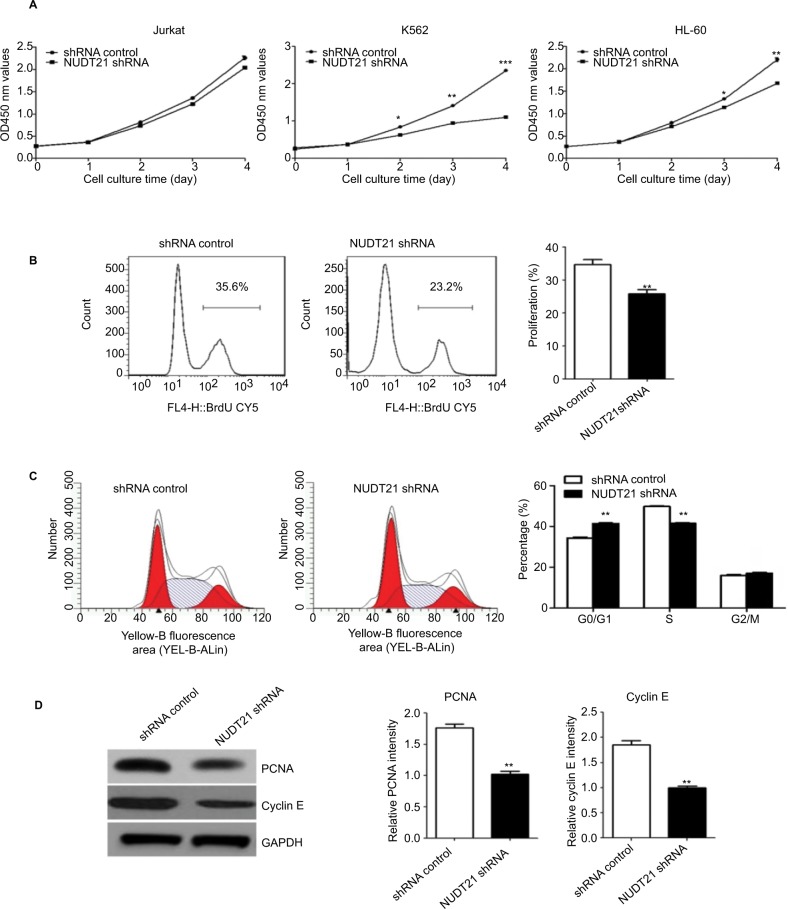
CCK-8 assays were then applied to determine the growth rate of K562, Jurkat, and HL60 leukemia cells over a specific time period. No significant difference was detected between the NUDT21 shRNA and the control shRNA group on the first day after incubation. Two to four days after incubation, the growth rate of K562, Jurkat, and HL60 leukemia cells in the NUDT21 shRNA group significantly decreased compared with the control shRNA group (P<0.05; Figure 3A). In NUDT21 shRNA-transfected K562 cells, there was a significant decrease in the BrdU incorporation compared with the control shRNA group (P<0.01; Figure 3B). We also compared the cell cycle phase distributions between the NUDT21 shRNA and the control shRNA groups by using flow cytometry. The observations revealed evidence relating to findings that compared with the control shRNA group, the number of cells in the G0/G1 phase in the NUDT21 shRNA group significantly increased (P<0.01; Figure 3C). In contrast, the number of cells in the S phase significantly reduced (P<0.01; Figure 3C), further indicating that inhibition of NUDT21 could suppress the cell transformation from the G0/G1 phase to the S phase. In line with this, we found that knockdown of NUDT21 led to a significant decrease in markers of cell proliferation capability, including PCNA (P<0.01) and cyclin E (P<0.01) (Figure 3D). Although the expression of NUDT21 in K562, Jurkat, and HL-60 was increased (the difference was statistically significant), the increase in K562 cells was significant, and the previous clinical study was based on clinical samples of chronic myelogenous leukemia. To study the mechanism of NUDT21 in CML, the cell line K562 of chronic myeloid leukemia was selected as the research object.
Figure 3.
K562 cell growth upon knockdown of NUDT21.
Notes: (A) Proliferation capabilities were detected by CCK-8 in K562, Jurkat, and HL-60 cells (2×103) for 5 days after transfection of shRNA control or NUDT21 shRNA. (B) K562 cells were transfected with shRNA control or NUDT21 shRNA for 48 hours and re-seeded in 96-well plate for BrdU assays. (C) Cell cycle phase distributions were detected by flow cytometry in K562 cells after transfection of shRNA control or NUDT21 shRNA. (D) Western blotting analysis of the protein expressions of PCNA and cyclin E after transfection of shRNA control or NUDT21 shRNA. The data are expressed as mean ± SD for duplicate experiments. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: CCK-8, Cell Counting Kit-8; BrdU, bromodeoxyuridine.
Knockdown of NDT21 induces apoptosis-promoting effect on K562 cells
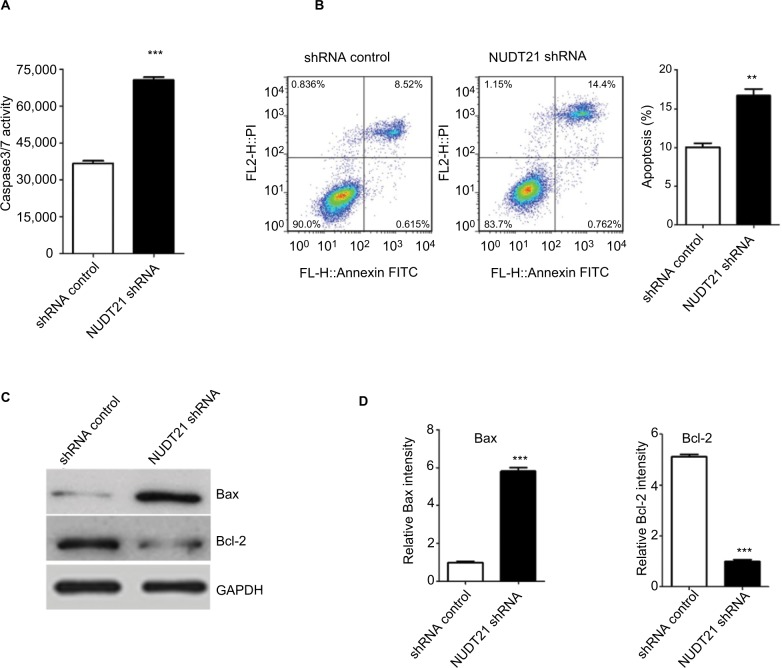
Thereafter, whether NUDT21 depletion inhibits cell growth through promoting the apoptosis rate of K562 cells was explored. The apoptosis rate of K562 cells was first determined by assessing the activities of the apoptosis markers Caspase3 and Caspase7. By using Caspase-Glo 3/7 assays in K562 cells (1.5×104), we demonstrated that knockdown of NUDT21 markedly induced the activation of Caspase3 and Caspase7 (P<0.05; Figure 4A). Annexin V/PI flow cytometry was also used to address whether NUDT21 depletion caused apoptosis. As shown in Figure 4B, NUDT21 depletion significantly induced apoptosis (defined as annexin V-positive) of K562 cells (P<0.01), especially late apoptosis (defined as annexin V-positive/PI-positive). To further determine whether NUDT21 depletion induces apoptosis by modulating the Bcl-2 family of proteins, the protein levels of Bax and Bcl-2 were estimated using Western blot analysis. We detected an increase in Bax and a decrease in Bcl-2 protein levels following NUDT21 depletion (Figure 4C and D). Taken together, these findings suggest that knockdown of NUDT21 induced apoptosis of K562 cells.
Figure 4.
K562 cell apoptosis upon knockdown of NUDT21.
Notes: (A) The activities of Caspase3/7 were determined by Caspase-Glo 3/7 assays in K562 cells (1.5×104) after transfection of shRNA control or NUDT21 shRNA. (B) The percentage of annexin V-positive revealed apoptosis. Percentages of cells undergoing apoptosis in different groups are shown. (C and D) Bax and Bcl-2 protein were determined by Western blot analysis in K562 cells after transfection of shRNA control or NUDT21 shRNA. Data are expressed as mean ± SD for duplicate experiments. **P<0.01; ***P<0.001.
Abbreviation: PI, propidium iodide.
Knockdown of NUDT21 results in inactivation of oncogenic signaling and reactivation of tumor-suppressive signaling
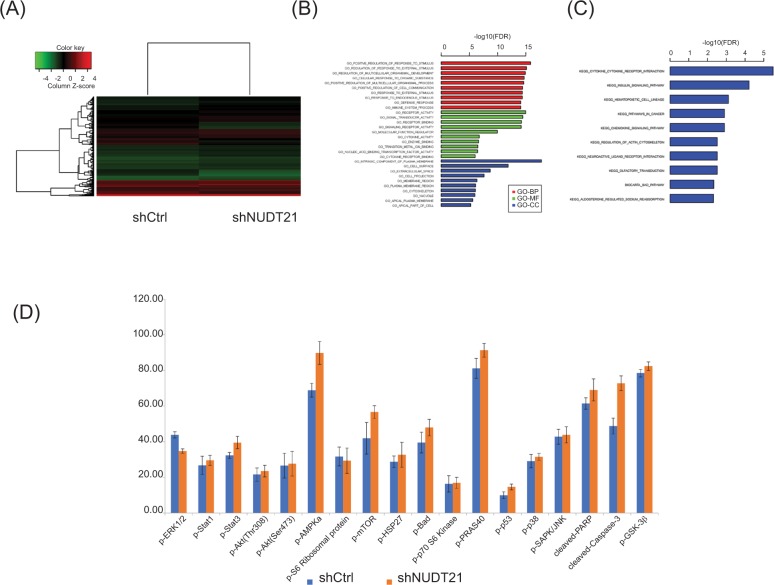
To dissect the mechanism underlying NUDT21’s oncogenic role in K562 cells, we further investigated cancer cellular response upon knockdown of NUDT21. To this end, we first performed microarray analysis in K562 cells upon NUDT21 depletion. There are 697 differentially expressed genes, with upregulation of 517 genes and downregulation of 180 genes after NUDT21 knockdown (Figure 5A). All the differentially expressed genes were further subjected to gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomics (KEGG) pathway enrichment analyses. The GO analysis showed that the differentially expressed genes were mainly involved in regulation of intracellular signal transduction, response to stimulus, multicellular organismal process, and cell communication (Figure 5B). The KEGG pathway analysis showed that the differentially expressed genes were mainly enriched in the insulin signaling pathway, cytokine receptor interaction, and hematopoietic cell lineage (Figure 5C).
Figure 5.
Deregulation of signaling pathways upon knockdown of NUDT21.
Notes: (A) Heatmap of 697 differentially expressed genes after knockdown of NUDT21 in K562 cells. (B) Enrichment analysis of differentially expressed genes in GO terms. (C) Enrichment analysis of differentially expressed genes in KEGG pathways. (D) The histogram showing the relative protein expressions of the key signaling components in K562 cells after transfection of shRNA control or NUDT21 shRNA. Data are expressed as mean ± SD for duplicate experiments.
Abbreviations: GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomics.
Additionally, we also used the human PathScan Intracellular Signaling Array Kit and performed the array analysis as described in the “Materials and methods” section. There are 20 targets on the arrays, including 18 key signaling components, a positive control, and a negative control (Table 1). The coordinates of the intracellular signaling array are also presented. First, we investigated the K562 cells transfected with shRNA control or NUDT21 shRNA and compared the signaling activation. Cell lysates were prepared and incubated onto the intracellular signaling array membrane overnight. The following day, antibody membrane was incubated with 1× HRP-linked Streptavidin and visualized with LumiGlo and peroxide. Images were taken using the Bio-Rad gel documentation system. As shown in Table 1 and Figure 5D, the phosphorylation of ERK1/2, which is the effector protein of the MPKA/ERK pathway, was reduced significantly. Meanwhile, the cleaved Caspase3 raised in the NUDT21 shRNA group compared with the shRNA control group. In addition, the phosphorylation levels of some insulin pathway-related proteins were elevated in the NUDT21 shRNA group compared with the shRNA control group, including Stat3, mTOR, BAD, PRAS40, GSK3β, and cleaved PARP. The levels of p-Akt (Thr308, Ser473), p-p38, and p-SAPK/JNK showed no statistically significant difference between the NUDT21 shRNA group and the shRNA control group.
Table 1.
Coordinates of human intracellular signaling array
| Target | Phosphorylation site | Modification | P-value | Up/down | |
|---|---|---|---|---|---|
| 1 | Positive control | N/A | N/A | \ | \ |
| 2 | Negative control | N/A | N/A | \ | \ |
| 3 | p-ERK1/2 | Thr202/Try204 | Phosphorylation | 0.000 | –0.207 |
| 4 | p-Stat1 | Tyr701 | Phosphorylation | 0.262 | 0.105 |
| 5 | p-Stat3 | Tyr705 | Phosphorylation | 0.001 | 0.223 |
| 6 | p-Akt(Thr308) | Thr308 | Phosphorylation | 0.357 | 0.090 |
| 7 | p-Akt(Ser473) | Ser473 | Phosphorylation | 0.790 | 0.041 |
| 8 | p-AMPKa | Thr172 | Phosphorylation | 0.000 | 0.304 |
| 9 | p-S6 Ribosomal Protein | Ser235/236 | Phosphorylation | 0.526 | –0.075 |
| 10 | p-mTOR | Ser2448 | Phosphorylation | 0.004 | 0.351 |
| 11 | p-HSP27 | Ser78 | Phosphorylation | 0.240 | 0.136 |
| 12 | p-Bad | Ser112 | Phosphorylation | 0.020 | 0.212 |
| 13 | p-p70 S6 Kinase | Thr389 | Phosphorylation | 0.799 | 0.036 |
| 14 | p-PRAS40 | Thr246 | Phosphorylation | 0.004 | 0.125 |
| 15 | p-p53 | Ser15 | Phosphorylation | 0.001 | 0.467 |
| 16 | p-p38 | Thr180/Tyr182 | Phosphorylation | 0.191 | 0.083 |
| 17 | p-SAPK/JNK | Thr183/Tyr185 | Phosphorylation | 0.692 | 0.024 |
| 18 | Cleaved PARP | Asp214 | Cleavage | 0.023 | 0.124 |
| 19 | Cleaved Caspase3 | Asp175 | Cleavage | 0.000 | 0.492 |
| 20 | p-GSK-3β | Ser9 | Phosphorylation | 0.015 | 0.050 |
Notes: The phosphorylation of ERK1/2, which is the effector protein of the MPKA/ERK pathway, was reduced significantly. Meanwhile, the cleaved Caspase3 raised in the shNUDT21 group compared with the shRNA control group (P<0.001).
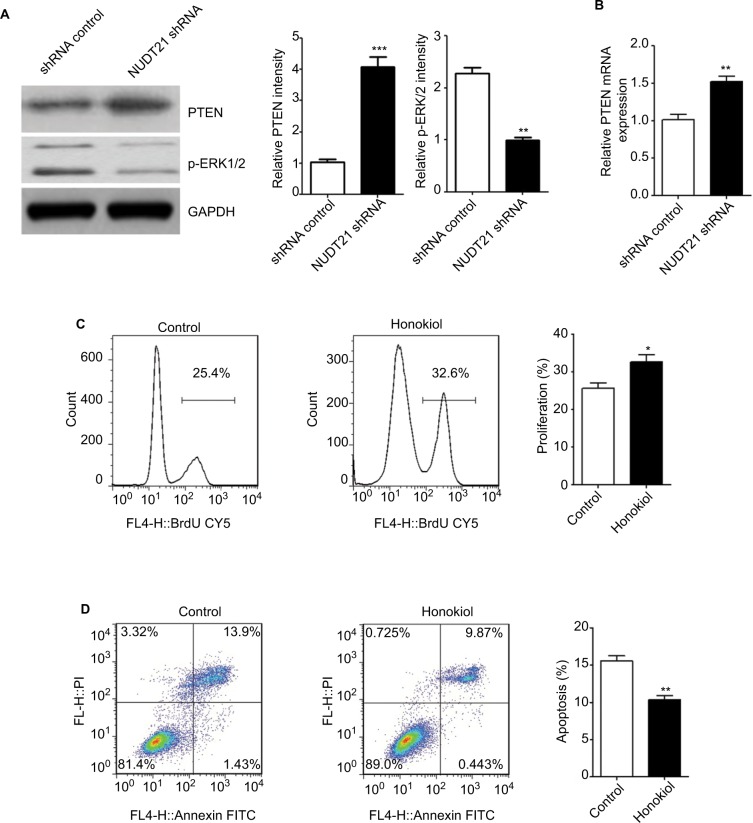
Knockdown of NUDT21 inhibits ERK pathway and promotes PTEN expression in K562 cells
To validate the variation of p-ERK levels following NUDT21 knockdown in K562 cells, we conducted Western blot analysis of K562 cells transfected with NUDT21 shRNA and shRNA control. We validated that the level of p-ERK1/2 was significantly inhibited and downregulated in the NUDT21 shRNA group (P<0.01; Figure 6A). Meanwhile, an obvious promotion of PTEN mRNA (P<0.01; Figure 6B) and protein (P<0.001; Figure 6A) expression was found in NUDT21-depleted K562 cells. We next evaluated whether the p-ERK level influences K562 cell growth. Honokiol has been used as an agonist of p-ERK. Our results showed that treatment with honokiol led to a promotion in cell proliferation (P<0.05; Figure 6C) and a reduction in cell apoptosis (P<0.01; Figure 6D).
Figure 6.
Potential mechanisms underlying the effects of NUDT21 on K562 cell growth.
Notes: (A) The protein levels of p-ERK1/2 and PTEN were shown in K562 cells transfected with shRNA control or NUDT21 shRNA using Western blot analysis. (B) The mRNA levels of PTEN were compared between K562 cells of shRNA control group and NUDT21 shRNA group using qRT-PCR analysis. (C) BrdU incorporation in K562 cells treated with honokiol or control. (D) Percentages of cells undergoing apoptosis (annexin V-positive) in different groups were shown. Data are expressed as mean ± SD for duplicate experiments. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: BrdU, bromodeoxyuridine; PI, propidium iodide; PTEN, phosphatase and tensin homolog deleted on chromosome 10; qRT-PCR, quantitative reverse polymerase chain reaction.
Discussion
It is now widely accepted that gene mutations and epigenetic modifications, including DNA methylation and histone modifications, are crucial determinants of leukemogenesis.16 Recent studies have also highlighted the important pathogenic roles of alternative splicing events in cancer, which participate in the posttranscriptional regulation of tumor suppressor genes and oncogenes.17 During alternative splicing events, particular exons of a gene may be included within or excluded from the final mRNA, enabling the generation of different proteins from one gene.
Leukemia is a common malignant hematologic disease. Cytogenetic aberrations and molecular genetic alterations are currently considered the most significant prognostic factors in determining the response to chemotherapy and survival outcome. Leukemogenesis is closely related to abnormal epigenetic regulation. However, the cellular events that initiate leukemia remain to be fully elucidated.
The generation of mRNA in eukaryotes is achieved by transcription from the DNA template and pre-mRNA processing reactions of capping, splicing, and polyadenylation. Eukaryotic pre-mRNA 3′-end processing (3′ processing) of mRNA precursors (pre-mRNAs) by polyadenylation is an essential step in gene expression.18 Defects in 3′ processing can have catastrophic consequences for the cell19 and have been associated with various human diseases.20 Moreover, 3′ processing can serve as a means of gene expression regulation through APA, which is widely utilized in modulating mRNA transcript levels in diverse cell types, developmental stages, and diseases.21,22
NUDT21 is an essential pre-mRNA 3′-end processing factor.23 Suppression of Nudt21 enhanced the generation of induced pluripotent stem cells, facilitated transdifferentiation into trophoblast stem cells, and impaired differentiation of myeloid precursors and embryonic stem cells, suggesting a broader role for Nudt21 in cell fate change.5 In Hela cells, dramatic changes in expression of several known oncogenes including cyclin D1 are observed because of NUDT21 depletion. NUDT21 knockdown increases MeCP2 in patient-derived lymphoblastoid cells, and reducing it to wild-type levels in NUDT21-duplication patients rescues MeCP2 protein levels to that of the healthy controls.24 NUDT21 also showed the strongest correlation with resistance to etoposide in childhood acute leukemia.15 However, the role of NUDT21 in the growth of leukemia cells and the underlying mechanisms remain to be fully explored. Therefore, we investigated the influence of NUDT21 knockdown on the cell proliferation and apoptosis in leukemia cells.
We showed that NUDT21 was highly expressed in CML patients as well as in multiple leukemia cell lines including K562, Jurkat, and HL-60 cells, indicating that NUDT21 might be a promising target for leukemia therapy. Future studies focusing on the expression levels of NUDT21 in samples from healthy controls and leukemia patients in different disease stages would provide more information about the association between NUDT21 expression levels and leukemia progression.
Our data suggested that knockdown of NUDT21 significantly inhibited cell proliferation, promoted cell apoptosis, and altered the distribution of the cell cycle in the K562 cell line. This is consistent with the previous study in which NUDT21 expression was identified to be associated with the resistance to etoposide in children with acute leukemia.15 This study proposed that NUDT21 may act as an oncogene in leukemia. However, a previous study showed that NUDT21 knockdown promoted glioblastoma tumor growth. The divergent effects of NUDT21 knockdown in leukemia and glioblastoma might result from different mechanisms. We therefore focused on the mechanism underlying the oncogenic role of NUDT21 in K562 cells in the further analyses. We assessed the specific intracellular signaling components that were modulated by NUDT21 knockdown in K562 cells. The intracellular signaling arrays showed that p-ERK1/2 was significantly inhibited by NUDT21 knockdown. The ERK pathway has been reported to be strongly associated with cell proliferation and apoptosis. We further observed that agonists of the ERK pathway promoted K562 cell proliferation and reduced K562 cell apoptosis in vitro. These results indicated that the effect of NUDT21 knockdown on K562 cells is associated with the ERK pathway.
PTEN is one of the classical inhibitors of the ERK pathways. The qRT-PCR and Western blot analyses demonstrated that the expression level of PTEN, which plays important roles in ERK1/2 phosphorylation and tumorigenesis, were significantly upregulated by NUDT21 knockdown. These data strongly suggested that NUDT21 knockdown regulated the proliferation and apoptosis of leukemia cells through upregulation of PTEN. However, in consideration of the complexity of cellular signaling in the regulation of cell growth, pathways beyond PTEN and ERK may also be involved. These results underscore the need for further research to elucidate the underlying mechanism of proliferation suppression and increase in apoptosis in K562 cells following NUDT21 knockdown.
Advanced sequencing and transcriptome analysis technologies have led to seminal discoveries uncovering the complexity of cancer. The global shortening of mRNAs through APA that occurs during enhanced cellular proliferation represents an important mechanism of posttranscriptional regulation.13,18 Notably, over 50% of the genes possess APA sites on their 3′-UTRs, leading to alternative 3′-UTR length and isoforms.19 APA has attracted increasing interest in cancer research as several studies have unraveled that dysregulation of APA represents a common feature among many oncological diseases.5 Nudt21 has been identified as a broad repressor of proximal poly(A) site usage. However, the mechanism of NUDT21 in K562 cell is unknown, and we will further study the APA regulatory mechanism of NUDT21 in our subsequent experiments.
Conclusion
This study indicated that NUDT21 played an important role in promoting cancer cell proliferation and inhibiting cell apoptosis in leukemia. The underlying mechanisms involved the modulation of PTEN and a set of downstream molecules including ERK1/2. This article clarifies that NUDT21 is involved in the malignant proliferation of leukemia cells as well as improves the understanding of the pathogenesis of leukemia. We will follow through the biological effects of NUDT21 on cells to find new therapeutic targets for leukemia.
Acknowledgments
This study was supported by grant from Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (2015116), Youth Project of Shanxi Science and Technology Department (201701D221274), and Innovation Fund of the First Affiliated Hospital of Shanxi Medical University (YG1501).
Footnotes
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Machova Polakova K, Koblihova J, Stopka T. Role of epigenetics in chronic myeloid leukemia. Curr Hematol Malig Rep. 2013;8(1):28–36. doi: 10.1007/s11899-012-0152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchese FP, Huarte M. Long non-coding RNAs and chromatin modifiers: their place in the epigenetic code. Epigenetics. 2014;9(1):21–26. doi: 10.4161/epi.27472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Dowell RD, Yi R. Genome-wide maps of polyadenylation reveal dynamic mRNA 3’-end formation in mammalian cell lineages. RNA. 2013;19(3):413–425. doi: 10.1261/rna.035360.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S, Yamamoto J, Chen Y, et al. Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes Cells. 2010;15(9):1003–1013. doi: 10.1111/j.1365-2443.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 5.Masamha CP, Xia Z, Yang J, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510(7505):412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayr C. Regulation by 3’-Untranslated Regions. Annu Rev Genet. 2017;51:171–194. doi: 10.1146/annurev-genet-120116-024704. [DOI] [PubMed] [Google Scholar]
- 7.Routh A, Ji P, Jaworski E, Xia Z, Li W, Wagner EJ, Poly WEJ. Poly(A)-ClickSeq: click-chemistry for next-generation 3′-end sequencing without RNA enrichment or fragmentation. Nucleic Acids Res. 2017;45(12):e112. doi: 10.1093/nar/gkx286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fusby B, Kim S, Erickson B, Kim H, Peterson ML, Bentley DL. Coordination of RNA Polymerase II Pausing and 3’ End Processing Factor Recruitment with Alternative Polyadenylation. Mol Cell Biol. 2016;36(2):295–303. doi: 10.1128/MCB.00898-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartini BL, Wang H, Wang W, Millette CF, Kilpatrick DL. Pre-messenger RNA cleavage factor I (CFIm): potential role in alternative polyadenylation during spermatogenesis. Biol Reprod. 2008;78(3):472–482. doi: 10.1095/biolreprod.107.064774. [DOI] [PubMed] [Google Scholar]
- 10.Gennarino VA, Alcott CE, Chen CA, et al. NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. eLife. 2015;4(2) doi: 10.7554/eLife.10782. eLife.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu ZJ, Huang P, Chong YX, et al. MicroRNA-181a promotes proliferation and inhibits apoptosis by suppressing CFIm25 in osteosarcoma. Mol Med Rep. 2016;14(5):4271–4278. doi: 10.3892/mmr.2016.5741. [DOI] [PubMed] [Google Scholar]
- 12.Lou JC, Lan YL, Gao JX, et al. Silencing NUDT21 Attenuates the Mesenchymal Identity of Glioblastoma Cells via the NF-κB Pathway. Front Mol Neurosci. 2017;10:420. doi: 10.3389/fnmol.2017.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erson-Bensan AE, Can T. Alternative Polyadenylation: Another Foe in Cancer. Mol Cancer Res. 2016;14(6):507–517. doi: 10.1158/1541-7786.MCR-15-0489. [DOI] [PubMed] [Google Scholar]
- 14.Redis RS, Vela LE, Lu W, et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol Cell. 2016;61(4):520–534. doi: 10.1016/j.molcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szczepanek J, Styczyński J, Tretyn A, Pogorzała M, Wysocki M. Identification of the genes expression profile associated with the ex vivo resistance to etoposide in childhood acute leukemias. Postepy Hig Med Dosw. 2012;66:401–408. doi: 10.5604/17322693.1000903. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010;10(1):23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22(9):976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang K, Tong L, Manley JL. Delineating the structural blueprint of the pre-mRNA 3’-end processing machinery. Mol Cell Biol. 2014;34(11):1894–1910. doi: 10.1128/MCB.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J, Hyman L, Moore C. Formation of mRNA 3’ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63(2):405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danckwardt S, Hentze MW, Kulozik AE. 3’ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO J. 2008;27(3):482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3’ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci U S A. 2009;106(17):7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brumbaugh J, di Stefano B, Wang X, et al. Nudt21 Controls Cell Fate by Connecting Alternative Polyadenylation to Chromatin Signaling. Cell. 2018;172(1–2):106–120. doi: 10.1016/j.cell.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gennarino VA, Alcott CE, Chen CA, et al. NUDT21-spanning CNVs lead to neuropsychiatric disease and altered MeCP2 abundance via alternative polyadenylation. Elife. 2015;4:4. doi: 10.7554/eLife.10782. [DOI] [PMC free article] [PubMed] [Google Scholar]