Three nuclear encoded multi-functional pentatricopeptide repeat proteins localizing in the mitochondrion are involved in splicing of a cohort of mitochondrial group II introns and thereby required for complex I biogenesis.
Keywords: Arabidopsis thaliana, complex I biogenesis, mitochondria, NADH dehydrogenase, PPR, splicing
Abstract
Group II introns are common features of most angiosperm mitochondrial genomes. Intron splicing is thus essential for the expression of mitochondrial genes and is facilitated by numerous nuclear-encoded proteins. However, the molecular mechanism and the protein cofactors involved in this complex process have not been fully elucidated. In this study, we characterized three new pentatricopeptide repeat (PPR) genes, called MISF26, MISF68, and MISF74, of Arabidopsis and showed they all function in group II intron splicing and plant development. The three PPR genes encode P-type PPR proteins that localize in the mitochondrion. Transcript analysis revealed that the splicing of a single intron is altered in misf26 mutants, while several mitochondrial intron splicing defects were detected in misf68 and misf74 mutants. To our knowledge, MISF68 and MISF74 are the first two PPR proteins implicated in the splicing of more than one intron in plant mitochondria, suggesting that they may facilitate splicing differently from other previously identified PPR splicing factors. The splicing defects in the misf mutants induce a significant decrease in complex I assembly and activity, and an overexpression of mRNAs of the alternative respiratory pathway. These results therefore reveal that nuclear encoded proteins MISF26, MISF68, and MISF74 are involved in splicing of a cohort of mitochondrial group II introns and thereby required for complex I biogenesis.
Introduction
Mitochondria are essential eukaryotic organelles that are responsible for energy production via aerobic respiration and are thought to have originated from the endosymbiotic association of an ancient α-proteobacterium (Gray et al., 1999; Andersson et al., 2003; Gray, 2012). Over the course of evolution, most genes that were present in the original endosymbiont have been transferred to the nucleus of the host cell, resulting in a significant reduction of the mitochondrial gene content (Martin and Herrmann, 1998). Nonetheless, a small proportion of the ancestral genes have been retained in modern mitochondria. In plants, mitochondrial genomes exhibit considerable variations in size, structure and organization (Kubo and Newton, 2008; Gualberto and Newton, 2017). Plant mitochondrial genomes are highly recombinogenic and complex RNA expression processes have likely evolved in response (Hammani and Giegé, 2014). Among these, the splicing of numerous introns, which interrupt protein-coding genes, is required to generate translatable mature mRNAs and is accomplished for the most part by nuclear-encoded protein trans-factors (Bonen, 2008; Colas des Francs-Small and Small, 2014; Hammani and Giegé, 2014). As observed in mutants affected in the genes encoding such trans-factors, dysfunction in the removal of plant mitochondrial introns often has severe consequences for plant fitness (Brown et al., 2014; Colas des Francs-Small and Small, 2014; Hammani and Giegé, 2014).
Based on RNA structures and splicing mechanisms, mitochondrial introns can be classified into two families and in seed plants most of them are categorized as group II. A total of 23 group II introns have been identified in Arabidopsis mitochondria, requiring either cis-splicing or trans-splicing reactions for their removal (Bonen, 2008). Classical group II introns share a conserved structure consisting of six helical domains (DI–DVI) that radiate from a central hub (Bonen, 2008; Glanz and Kuck, 2009). The DV and DVI domains are especially important for the splicing reactions. DV is a highly conserved domain comprising the intron catalytic core, and DVI contains an internal bulged adenosine that acts as the attacking nucleophile to initiate the splicing reaction (Zimmerly and Semper, 2015). At least two splicing pathways for group II introns have been identified in seed plants (Li-Pook-Than and Bonen, 2006; Gualberto et al., 2015; Massel et al., 2016). The ‘branching’ pathway occurs via a two-step trans-esterification reaction. In the first step, the 2′-OH group of the branch point adenosine attacks the phosphate at the 5′-splice site. The 5′-exon is released and the intron, still attached to the 3′-exon, adopts a branched structure giving the intron a lariat form. In the second step, the 3′-OH of the 5′-exon attacks the 3′-splice site, resulting in ligated exons and the release of the intron as a lariat with a tiny tail (Brown et al., 2014). An alternative splicing pathway, known as the ‘hydrolytic pathway’, generates a linear intron and was adopted by some group II introns whose certain conserved features have been lost during the course of evolution (Li-Pook-Than and Bonen, 2006; Gualberto et al., 2015; Massel et al., 2016).
While some group II introns can undergo autocatalytic (i.e. self-) splicing in vitro, all are thought to require one or more proteins in vivo, such as intron-encoded maturases. Plant organellar group II introns are highly degenerate and lack regions that are essential for self-splicing. MatR is the only intron-encoded maturase that has been maintained in seed plant mitochondria (Wahleithner et al., 1990), and a recent study indicated that it is associated with splicing of multiple mitochondrial introns in vivo (Sultan et al., 2016). Four matR homologs (nMAT1–nMAT4) reside in the nuclear genome of angiosperms and their participation in the splicing of mitochondrial group II introns has been established (Keren et al., 2009, 2012; Cohen et al., 2014; Zmudjak et al., 2017). Other nuclear-encoded cofactors facilitating mitochondrial intron splicing include RUG3 (RCC1/UVR8/GEF-like 3), mTERF15 (Mitochondrial Transcription Termination Factor 15), WTF9 (What’s This Factor? 9), mCSF1 (Mitochondrial CAF-like Splicing Factor 1), ABO6 (ABA Overly Sensitive 6) and PMH2 (Putative Mitochondrial RNA Helicase 2) (Köhler et al., 2010; Kuhn et al., 2011; Francs-Small et al., 2012; He et al., 2012; Zmudjak et al., 2013; Hsu et al., 2014; Zmudjak et al., 2017). Recently, the RAD52-like protein ODB1, which was originally identified as a mitochondrial DNA repair component, has also been shown to be required for the efficient splicing of two group II introns (Gualberto et al., 2015). Aside from these splicing factors, the pentatricopeptide repeat (PPR) proteins are by far the most represented class of proteins playing roles in the splicing of mitochondrial introns (Barkan and Small, 2014). PPR proteins are composed of 2–30 tandem repeats of highly degenerate 35-amino-acid motifs that fold into a pair of antiparallel α-helices (Ke et al., 2013; Yin et al., 2013; Barkan and Small, 2014; Shen et al., 2016). Based on the nature of their repeats, PPR proteins can be divided into two major subfamilies (Lurin et al., 2004). The PLS-type PPR proteins contain triplets of P, L (Long), and S (Short) motifs and are almost exclusively implicated in RNA editing in plant organelles (Takenaka et al., 2013b; Sun et al., 2016). P-type PPR proteins contain only canonical 35-amino-acid repeats and take part in a wide range of post-transcriptional processes ranging from RNA maturation to translation in organelles (reviewed in Barkan and Small, 2014). Several PPR proteins implicated in RNA splicing have been characterized in Arabidopsis, such as OTP43 (Organelle Transcript Processing defect 43), ABO5 (ABA Overly Sensitive 5), ABO8 (ABA Overly Sensitive 8), BIR6 (Buthionine Sulfomixine-Insensitive Roots 6), OTP439 (Organelle Transcript Processing 439), and TANG2 or SLO3 (Slow Growth 3). Each of these proteins is highly specific and required for the splicing of a single mitochondrial intron in most cases (Colas des Francs-Small et al., 2014; de Longevialle et al., 2007; Koprivova et al., 2010; Liu et al., 2010; Yang et al., 2014; Hsieh et al., 2015). Taken together, these studies demonstrated the important role of PPR proteins in RNA splicing of plant mitochondria, but the role they play in this process is currently unknown.
In this study, we report the function of three newly characterized mitochondria-targeted PPR proteins, named Mitochondrial Intron Splicing Factor 26 (MISF26), Mitochondrial Intron Splicing Factor 68 (MISF68) and Mitochondrial Intron Splicing Factor 74 (MISF74), which are essential for the splicing of either one or several mitochondrial introns in Arabidopsis. The loss-of-function of these nuclear-encoded splicing factors affects complex I assembly, increases the expression of alternative oxidase genes, and results in retarded plant growth.
Materials and methods
Plant material
Arabidopsis Col-0 plants were obtained from the Arabidopsis Stock Centre of the Institut National de la Recherche Agronomique in Versailles (http://publiclines.versailles.inra.fr/). Arabidopsis T-DNA mutants N508252 (misf26-1), N816920 (misf26-2), N562109 (misf68-1), N510785 (misf68-2), and N554210 (misf74-1) were obtained from the European Arabidopsis Stock Centre (http://arabidopsis.info/), and N191C05 (misf74-2) was acquired from the GABI-Kat mutant collection (https://www.gabi-kat.de/). Plants homozygous for the insertions were identified by PCR genotyping. Specific primers used for genotyping are listed in Supplementary Table S1 at JXB online. Plants were grown on soil in a greenhouse under long-day conditions (16 h of light and 8 h of dark).
Subcellular localization of MISF26, MISF68, and MISF74
DNA regions encoding the N-terminal targeting sequence of each MISF protein were PCR amplified, cloned into pDONR207 by GatewayTM BP reaction (Invitrogen) and sequenced to check PCR accuracy. The obtained entry clones were subsequently transferred into pGWB5 (Nakagawa et al., 2007) by the GatewayTM LR reaction (Invitrogen) to create a translational fusion between the targeting sequences and the coding region of green fluorescent protein (GFP). The fusion constructs were transformed into Agrobacterium tumefaciens C58C51 and used for floral dip transformation of Arabidopsis Col-0 plants. Transgenic plants were selected on hygromycin, and GFP fluorescence was visualized in root cells by Leica TCS SP2 confocal microscopy. Prior to observation, roots were soaked in a solution of MitotrackerTM Red (0.1 μM) to label mitochondria.
RNA extraction and analysis
Total RNA was extracted from 8-week-old Arabidopsis flower buds using TRIzol reagent (Life Technologies) following the manufacturer’s instructions and treated with RTS DNase (Mobio) only when RNAs were prepared for reverse transcription (RT)-PCR or quantitative RT-PCR (qRT-PCR). RT-PCR analysis was performed to check the expression of MISF genes using primer pairs listed in Supplementary Table S1. The BIO2 gene was used as control to check cDNA integrity. RNA gel blot analyses were performed as described in (Haïli et al., 2016). Specific probes were amplified by PCR using primers indicated in Supplementary Table S1. To quantify the splicing efficiency of mitochondrial introns, quantitative RT-PCR was performed using primers designed across intron–exon regions (unspliced forms) and exon–exon junctions (spliced forms) as previously described (Koprivova et al., 2010; Haïli et al., 2013). Two biological and three technical repeats were performed and the nuclear 18S ribosomal RNA gene was used for data normalization.
Blue Native gel and in-gel activity assays
Eight-week-old Arabidopsis flower buds were used to prepare crude mitochondrial extracts as previously described (Dahan et al., 2014). One hundred micrograms of total proteins from purified organelles was loaded and separated on 4–16% (w/v) polyacrylamide NativePAGETM Bis/Tris gels (Invitrogen). Following electrophoresis, Blue Native (BN)-PAGE gels were transferred to polyvinylidene difluoride (PVDF) membranes or subjected to a complex I in-gel activity assay (Wang et al., 2017a). Succinctly, complex I activity was revealed by incubating gels in the presence of 0.2% nitroblue tetrazolium, 0.2 mM NADH and 0.1 M Tris–HCl (pH 7.4). When sufficient coloration was obtained, gels were soaked in fixing solution containing 30% (v/v) methanol and 10% (v/v) acetic acid to stop the reactions. For immuno-detection, BN-PAGE gels were transferred overnight to PVDF membranes in transfer buffer (50 mM Bis/Tris and 50 mM Tricine) at 30 V and at 4 °C. Membranes were then hybridized with antibodies to carbonic anhydrase (Cohen et al., 2014). Hybridization signals were revealed using enhanced chemiluminescence reagents (Western Lightning Plus ECL, Perkin Elmer).
Results
Three ppr mutants displaying slow-growth phenotypes
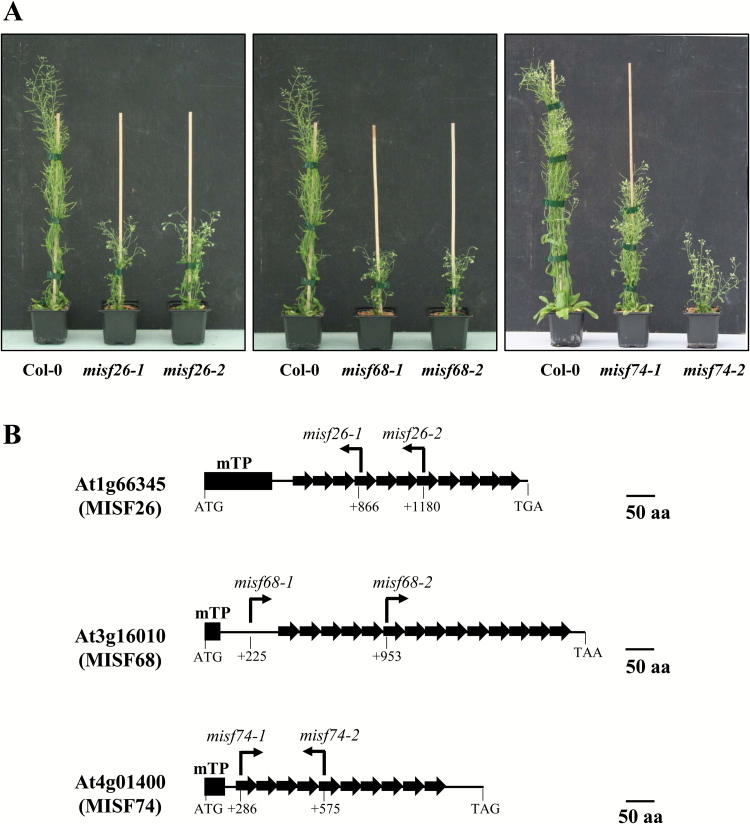
In an effort to better understand mitochondrial gene expression in plants, we assembled a large collection of Arabidopsis T-DNA insertion mutants disrupted in nuclear genes encoding putative mitochondria-targeted PPR proteins. Among them, we found three mutants showing global growth retardation phenotypes compared with wild-type plants, and that resemble previously identified Arabidopsis complex I mutants (Fig. 1A). The implicated PPR genes corresponded to AT1G66345, AT3G16010, and AT4G01400. After molecular characterization of the corresponding mutants (see below), we named these genes Mitochondrial Intron Splicing Factor 26 (MISF26), Mitochondrial Intron Splicing Factor 68 (MISF68), and Mitochondrial Intron Splicing Factor 74 (MISF74), respectively. Two independent T-DNA lines were identified for each PPR gene. Each mutant pair exhibited very similar phenotypes, supporting that the observed growth alterations were due to the inactivation of the corresponding PPR genes. The lines misf26-1 (SALK_008252), misf26-2 (SAIL_363_F11), misf68-2 (SALK_010785), misf74-1 (SALK_054210), and misf74-2 (GABI_191C05) contained T-DNA insertions within the PPR repeat coding regions of each corresponding gene, while the T-DNA of the misf68-1 (SALK_062109) mutant is located upstream of the first PPR repeat coding sequence of MISF68. The genes MISF26, MISF68, and MISF74 encode proteins that are predicted to comprise 11, 14, and 10 P-type PPR repeats, respectively, according to TPRpred (https://toolkit.tuebingen.mpg.de/#/tools/tprpred) and the PlantPPR database (http://ppr.plantenergy.uwa.edu.au/) (Fig. 1B). The accumulation of full-length transcripts derived from each PPR genes in the mutants could not be detected by RT-PCR analysis, strongly suggesting that the identified mutant lines corresponded to null mutants (see Supplementary Fig. S1). When grown under long day conditions (16 h light/8 h dark) in the greenhouse, misf74-2, misf68, and misf26 showed severely delayed growth compared with Col-0 reference plants, and reached out about half the size of wild-type plants after 3 months of culture. Intriguingly, misf74-1 plants exhibited a milder growth phenotype compared with misf74-2 mutants (Fig. 1A).
Fig. 1.
The three identified Arabidopsis misf mutants display strongly delayed developmental phenotypes. (A) Eight-week-old plants showing the global vegetative phenotype of the misf mutants compared with the wild-type (Col-0). (B) Schematic diagram of MISF26, MISF68, and MISF74 proteins depicting the 11, 14, and 10 predicted PPR-P repeats that they contain, respectively. The locations of corresponding T-DNA insertion sites are indicated. The putative mitochondrial targeting sequences (mTP) are shown as black boxes and were predicted using TargetP.
The AT4G01400, AT3G16010, and AT1G66345 PPR proteins are targeted to mitochondria in vivo
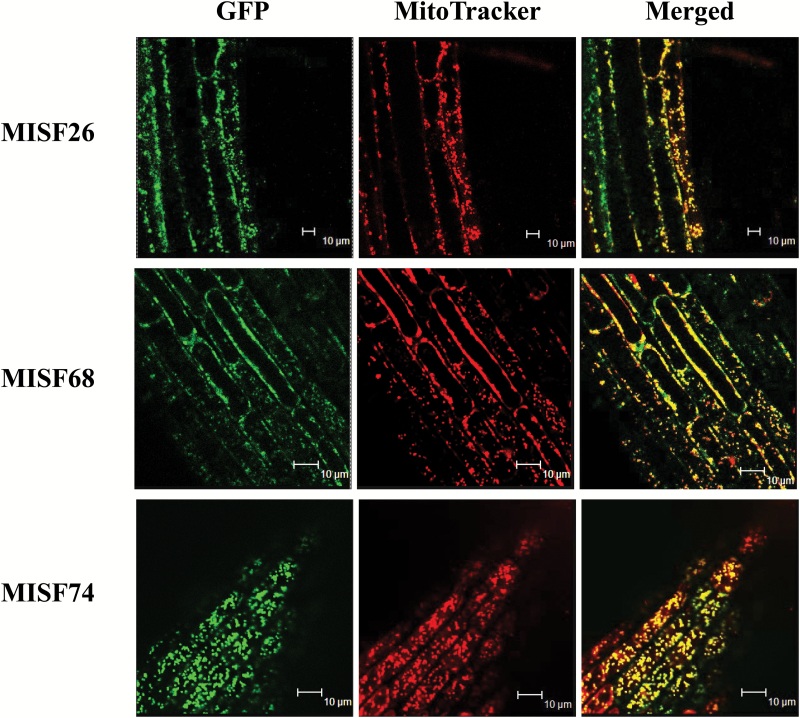
The three identified PPR proteins were predicted to reside in mitochondria according to the Arabidopsis subcellular database SUBA (http://suba3.plantenergy.uwa.edu.au/). To verify their subcellular distribution, we generated GFP translational fusion constructs comprising the region encoding the putative mitochondrial transit peptide of each PPR protein and then transformed them into Arabidopsis Col-0 plants. Confocal microscopy was used to detect the GFP fluorescence in the roots of the generated transgenic plants. In all cases, we observed GFP signals that co-localized with MitoTracker red signals in mitochondria (Fig. 2), which confirmed the mitochondrial localization of the three PPR proteins in vivo.
Fig. 2.
The MISF26, MISF68, and MISF74 genes encode mitochondria-targeted PPR proteins. Confocal microscope images showing the subcellular distribution of MISF: GFP translational fusions in root cells of young Arabidopsis transgenic plants. Prior to observation, roots were briefly soaked in MitoTracker Red to label mitochondria. The left panels show GFP fluorescence, the center show mitochondria labeled with the MitoTracker Red dye and the right panels present the merged signals.
The three identified PPR genes are involved in intron splicing of distinct mitochondrially encoded transcripts
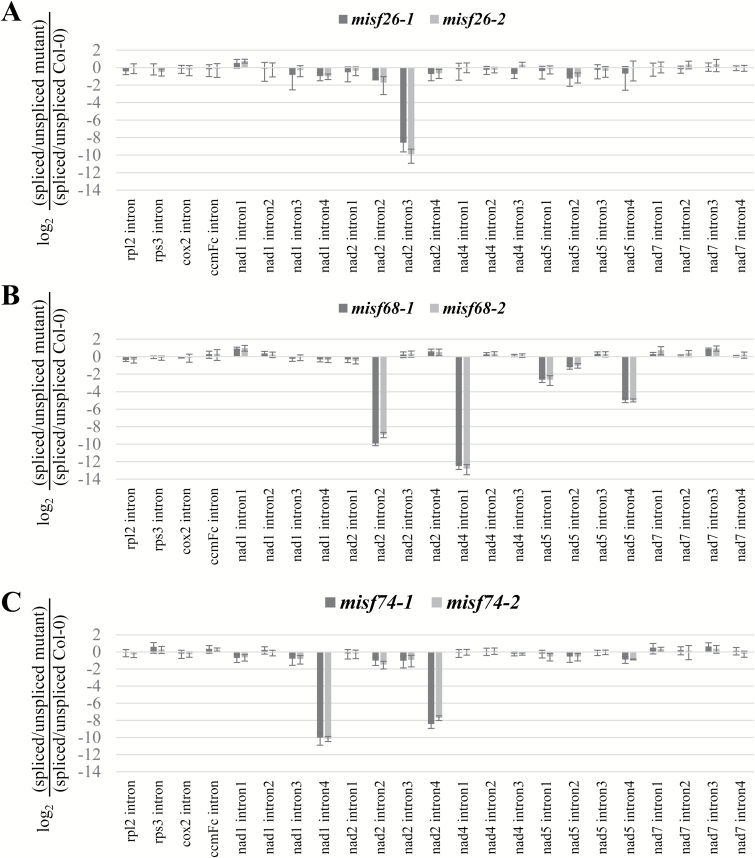
Previous studies have shown that PPR proteins are RNA-binding proteins and function in a wide range of mitochondrial and plastid RNA processing events (Barkan and Small, 2014; Hammani and Giegé, 2014). Our results indicated that the identified PPR proteins resided in mitochondria, which encouraged us to test whether they could play roles in mitochondrial RNA metabolism. To test this hypothesis, we first performed qRT-PCR to measure the steady state levels of mature mitochondrially encoded transcripts and determine the splicing efficiencies of all mitochondrial introns in each mutant line. No significant differences for most mature mitochondrial transcripts were found between the mutants and the wild-type (see Supplementary Fig. S2). However, a few specific regions of mRNAs encoding complex I respiratory subunits fail to accumulate to normal levels in the mutants. These concerned the nad2 mRNA in misf26, nad2 and nad4 mRNAs in misf68, and nad1 and nad2 transcripts in misf74 (Supplementary Fig. S2). To further specify the origin of these differences, we calculated the splicing efficiencies of all 23 introns in Arabidopsis mitochondria in the wild-type and mutant lines. As shown in Fig. 3, a single reduction in splicing efficiency was observed for nad2 intron 3 (cis-intron) in misf26 mutants (Fig. 3A). In contrast, several splicing defects were detected in misf68 and misf74 mutants (Fig. 3B, C). These concerned the nad5 intron 4 (cis-intron), the nad2 intron 2 (trans-intron) and the nad4 intron 1 (cis-intron) in misf68 plants (Fig. 3B). Regarding misf74 plants, large reductions in the splicing of nad1 intron 4 and nad2 intron 4 (both are cis-introns) were detected (Fig. 3C).
Fig. 3.
Splicing efficiencies of all mitochondrial introns were compared between the wild-type (Col-0) and the misf mutants. The bars depict the log2 ratio of spliced to unspliced forms for each intron in mutants as compared with the wild-type. Three technical replicates and two independent biological repeats were used for each genotype; standard errors are indicated. (A–C) show the results for the misf26, misf68, and misf74 mutants respectively.
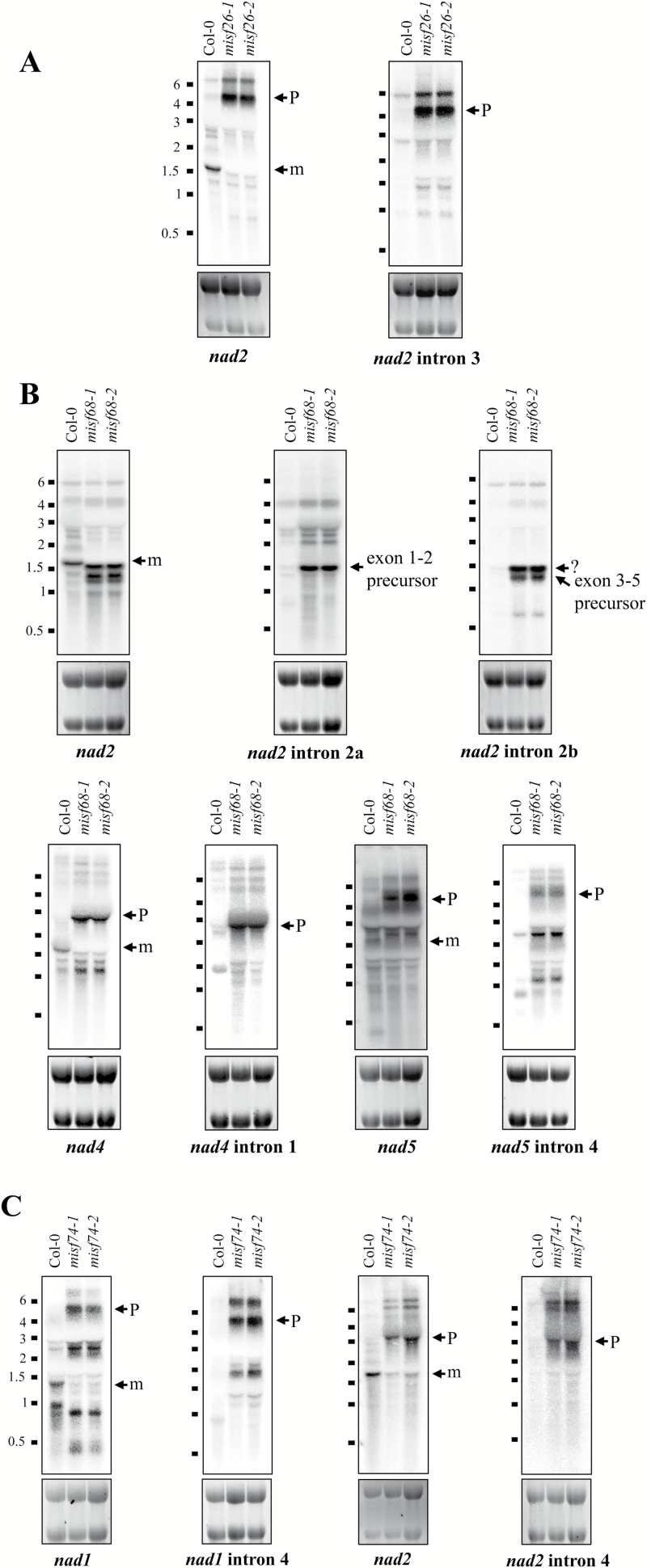
RNA gel blots were then carried out to confirm the results obtained from qRT-PCR analysis. Two kinds of probe were used for each affected transcript in this assay, the first one detecting mRNA exons of genes for which splicing defects were revealed and the second one specific to the impacted introns. The results showed that no mature transcripts for the impacted genes could be detected in any of the misf mutants, except for nad2 in misf74 plants in which trace amount of mature transcripts could be found. Accordingly, precursor transcripts comprising the introns whose splicing is reduced overaccumulated in all mutant lines (Fig. 4).
Fig. 4.
RNA gel blot analysis showing the accumulation pattern of affected complex I (nad) transcripts in the three misf mutants. Two probe sets were used in this assay: full-length cDNA probes to detect both mature and precursor transcripts and intron probes to specifically detect the affected precursor transcripts in each mutant. The blots were performed on total RNA isolated from ppr mutants and wild-type (Col-0) flowers and hybridized with the indicated probes. (A–C) show the corresponding results for misf26, misf68, and misf74 as compared with the wild-type. nad2 intron 2a and nad2 intron 2b are specific probes detecting the 5′- and 3′-portions of nad2 intron 2 (trans-intron). Ethidium bromide staining of ribosomal RNAs is shown below the blots and serves as a loading control. RNA marker sizes are indicated. m, mature mRNAs; p, precursor transcript.
Altogether, these results allowed us to conclude that the three newly identified Arabidopsis PPR genes are involved in splicing of different mitochondrial group II introns. MISF74 and MISF68 are required for splicing of two and three introns each, whereas MSIF26 is essential for a single one.
Complex I biogenesis is strongly affected in misf26, misf68, and misf74 mutants
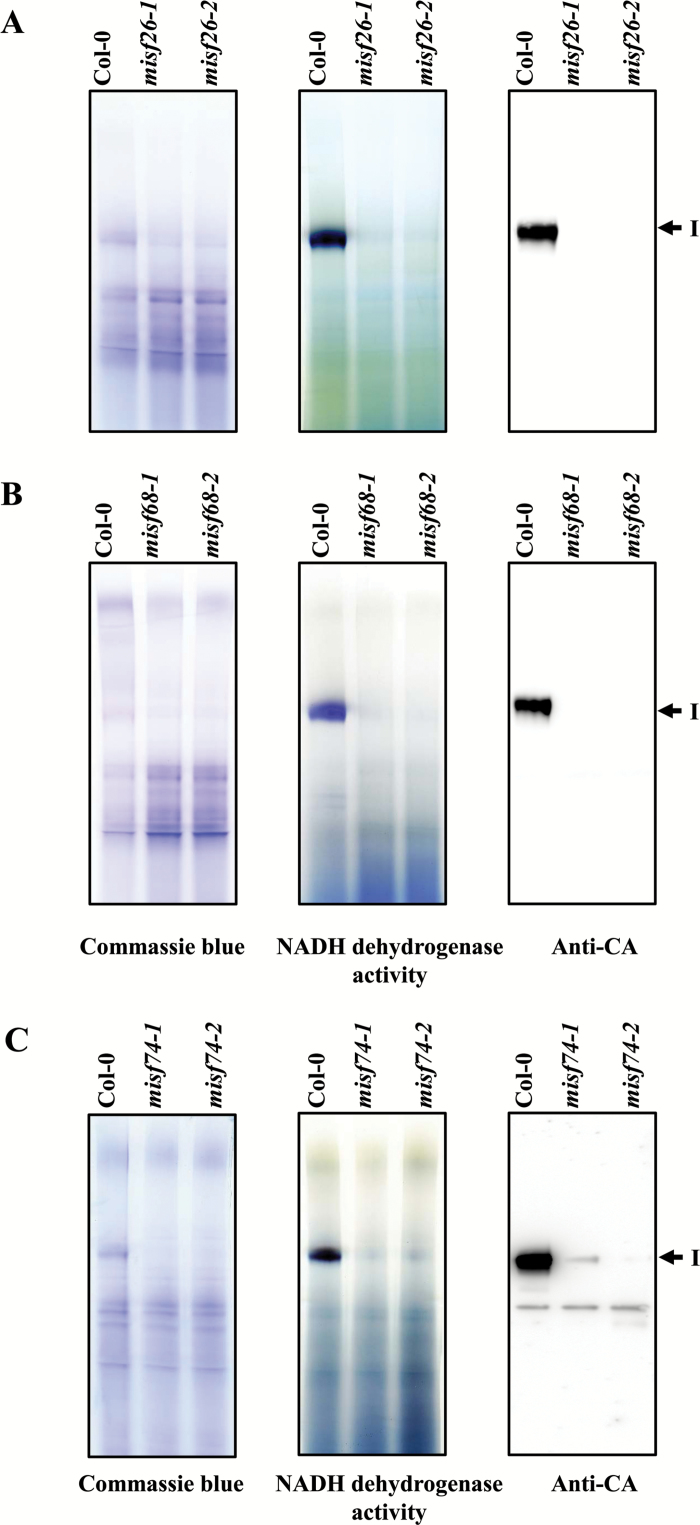
The mitochondrial nad genes encode essential subunits of respiratory complex I (NADH: ubiquinone oxidoreductase). The various splicing defects detected in the different misf mutants led us to consider that the corresponding plants may be affected in the production of complex I. We thus used BN-PAGE to separate the different mitochondrial respiratory complexes and estimate the abundance of complex I in wild-type plants and the three misf mutants. Crude mitochondrial extracts were prepared from both types of plants and solubilized in 1% n-dodecyl β-D-maltoside. Following gel migration, the abundance of respiratory complex I was visualized either by gel activity staining or by western blot analysis using antibodies recognizing the γ-carbonic anhydrases (CA) associated with complex I. These analyses revealed that the accumulation levels of complex I were significantly reduced in all misf mutants compared with the wild-type (Fig. 5). Intriguingly, a slight amount of complex I could be detected in the misf74-1 mutant but not in the misf74-2 mutant (Fig. 5C), which correlated with the milder growth alteration of misf74-1 compared with misf74-2 plants (Fig. 1).
Fig. 5.
Complex I biogenesis is strongly affected in misf mutants. Crude mitochondrial extracts were prepared from wild-type and mutant plants, separated on BN-PAGE gels and stained with Coomassie blue (left panel). In-gel staining revealing the NADH dehydrogenase activity of complex I is also present in the center. Following migration, BN-PAGE gels were transferred to membranes and blots were hybridized with antibodies to mitochondrial CA subunit (right panel). The holocomplex I is indicated by an arrowhead. (A–C) show the corresponding results for misf26, misf68, and misf74 mutants, respectively.
To further dissect the respiratory re-orientations resulting from the lack of complex I in the misf mutants, we examined the expression levels of the alternative oxidase (AOX) and NADH dehydrogenase (NDA, NDB, and NDC) genes by qRT-PCR. This analysis revealed a strong overaccumulation of AOX1A, NDA1, and NDB4 transcripts in most misf mutants compared with the wild-type. In contrast, the abundance of AOX2 mRNA was largely reduced in the misf mutants (see Supplementary Fig. S3).
Overall, our observations indicated that misf26, misf68, and misf74 mutants represent complex I deficient plants and that the alternative respiratory pathway is consequently activated in these lines. The loss of proper splicing for distinct nad transcripts in these mutants strongly suggests that these are responsible for the complex I deficiencies detected in these plants.
Discussion
MISF26, MISF68, and MISF74 are required for the splicing of various mitochondrial introns and affect complex I assembly and activity in Arabidopsis
The expression of mitochondrial transcripts in plants requires the activity of a plethora of nuclear-encoded trans-factors among which PPR proteins are by far the most represented class of proteins. This study reveals the function of three new mitochondria-targeted PPR proteins and shows that they are essential for the splicing of six separate mitochondrial introns. To reach such conclusions, we first observed that GFP translational fusions comprising the N-terminal regions of each of these PPR proteins localized in Arabidopsis mitochondria, strongly supporting the mitochondrial targeting of these proteins. The analysis of mitochondrial mRNAs in T-DNA insertion mutants clearly indicated that MISF26, MISF68, and MISF74 are indispensable for the splicing of one, three, and two mitochondrial introns, respectively (Figs 3, 4). The MISF26 PPR protein is crucial for the splicing of the third intron of the nad2 pre-mRNA. MISF68 was found to be essential for the removal of nad2 intron 2, nad4 intron 1, and nad5 intron 4, although the importance of MISF68 for this last intron appears to be weaker than for the other two. Regarding MISF74, we found that this PPR protein facilitates the splicing of both nad1 intron 4 and nad2 intron 4. These different splicing defects therefore impact the production of various mitochondrially encoded complex I subunits and accordingly all misf mutants show slow growth phenotypes that are typical of complex I mutants and that correlate with a very strong decrease of this respiratory complex (Figs 1A, 5). To compensate for the strong reduction in complex I accumulation, we found a strong increase in the steady state levels of mRNAs encoding alternative NADH dehydrogenases, notably NDA1 and NDB4, in the different misf mutants (see Supplementary Fig. S3).
MISF68 and MISF74 are multi-functional PPR proteins
Interestingly, no overlap was found among the intron targets of the three MISF proteins. The cis- and trans- nature of mitochondrial introns does not explain this lack of overlap, as the MISF68 protein facilitates the splicing of both types of introns. Two functional categories of trans-factors seem to play roles in intron splicing in plant mitochondria. Factors like maturases, RNA helicases or CRM (chloroplast RNA splicing and ribosome maturation) proteins are necessary for the removal of a large number of group II mitochondrial introns and can be considered as general splicing factors. They may be part of the core of a mitochondrial spliceosomal complex (if such complex exists) and are thereby required for the elimination of, virtually, all mitochondrial introns. The second category concerns factors like the PPR proteins that, in most cases, are necessary for the splicing of a single intron (see Supplementary Fig. S4). This second class of trans-factors seem to act as accessory factors aiding the recruitment of the mitochondrial splicing machinery on specific introns and/or by somehow improving the activity of this machinery on specific intron targets. The incorporation of intron-specific factors in the mitochondrial splicing machinery could result from sequence or structural changes in certain introns, creating mono-specific splicing complexes. Interestingly, our results indicate that some of these accessory factors can facilitate the splicing of more than one intron, as MISF74 is required for the elimination of two introns and three introns necessitate MISF68 for their splicing. The multi-functionality of PPR proteins in organellar RNA metabolism has been described in a few cases, notably for PPR engaged in RNA editing, but was rarely found for PPR proteins involved in splicing (de Longevialle et al., 2008; Khrouchtchova et al., 2012; Barkan and Small, 2014; Hammani and Giegé, 2014; Wang et al., 2017b). MISF68 and MISF74 represent two of the first counterexamples found in plant mitochondrion. The molecular functions of PPR proteins in splicing are still unclear but it has been suggested that they may help intron folding and stabilize their structure in a catalytically active form (Beick et al., 2008; Williams-Carrier et al., 2008; Solem et al., 2009; Barkan and Small, 2014; Aryamanesh et al., 2017). The multi-functionality of MISF68 and MISF74 may result from the recognition of the same functional domain shared by their intron targets, implying that they could play the same role in the splicing of the introns they facilitate. Some functional differences can be anticipated, though, as the lack of MISF68 does not impact the splicing efficiency of its different intron targets to the same extent (Fig. 3B). The multiplicity of roles that PPR proteins could play in splicing is also suggested by the fact that several of them are necessary for the removal of the same mitochondrial intron, such as ABO5 and MISF26 in Arabidopsis or Dek37 and EMP10 in maize (Liu et al., 2010; Cai et al., 2017; Dai et al., 2018). Finding the RNA biding sites of these PPR within their intron targets is certainly an important prerequisite for understanding the function of these proteins in splicing. It has been recently established that PPR proteins may recognize their RNA targets via a one PPR motif—one nucleotide rule, and that combinations of amino acids at positions 5 and 35 of each repeat are determinants for RNA base selection (Barkan et al., 2012; Takenaka et al., 2013a; Yagi et al., 2013; Kindgren et al., 2015; Shen et al., 2016). To predict their potential RNA binding sites, we applied the PPR-RNA recognition code to the three MISF proteins. Several candidate sites within most intron targets could be identified, except for MISF68 in nad5 intron 4 (Supplementary Fig. S5). However, the identified sites ranked rather low in the prediction lists for each protein, likely because several amino acid combinations at positions 5 and 35 of MISF PPR repeats are not filled by the PPR code. Advanced biochemical analyses such as RIP-Seq or similar kinds of approaches are thus necessary to identify the binding sites of these newly identified PPR proteins.
The three MISF proteins show different conservation levels in plants
Recent analyses have indicated that most PPR genes in plants appeared before the divergence of monocot and dicot species and that potential orthologs of Arabidopsis PPR proteins can be quite easily identified in other plant species (O’Toole et al., 2008). We compared protein similarities and identities between flowering plant orthologs of the three MISF proteins and found, surprisingly, that the levels of sequence identity varied quite extensively between them. With more than 80% of sequence identity between othologs, MISF68 appeared to be the most conserved factor among the three splicing proteins. The level of sequence identity between MISF74 homologs is around 74% whereas it is only around 60% for MISF26 closely related homologs (see Supplementary Fig. S6). Specific functional constraints may have limited the sequence diversification of MISF68 and MISF74 compared with MISF26, which could result from the fact that MISF68 and MISF74 facilitate the splicing of multiple introns. Because of the necessity to interact with several RNA targets, PPR proteins carrying multifarious activities may be effectively less incline to sequence changes throughout evolution. The high sequence conservation between MISF potential orthologs (Supplementary Fig. S6) suggested similar conservation levels for their in vivo RNA binding sites. Multiple sequence alignments were thus performed to see if any of the predicted target sites (Supplementary Fig. S5) could show higher sequence conservation compared with others. Certain sites show perfect or almost perfect sequence conservation and thus represent serious target site candidates of MISF proteins (Supplementary Fig. S7). Major genetic and biochemical advances are required before we understand how PPR proteins assist group II intron splicing and how that influences their sequence diversification. Taken together, MISF68 and MISF74 might serve as novel models to understand the multiple functions of PPR proteins, and their molecular deciphering will provide new insight into the mechanisms of intron splicing in plant mitochondria.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. RT-PCR analysis of each MISF transcript in wild-type and misf mutants.
Fig. S2. Various mature mitochondrial transcripts underaccumulate in misf mutants.
Fig. S3. Quantitative RT-PCR measuring the steady-state levels of mRNAs encoding alternative oxidases (AOX) and NADH-dehydrogenases (NDA, NDB and NDC).
Fig. S4. Listing of known nucleus-encoded proteinaceous factors involved in the splicing of mitochondrial group II introns in Arabidopsis.
Fig. S5. Predictions of MISF protein binding sites according to the PPR code.
Fig. S6. Multiple sequence alignments of MISF protein homologs from a representative selection of dicot and monocot plant species.
Fig. S7. Multiple sequence alignments analyzing the conservation of the predicted RNA targets of each MISF protein.
Table S1. Oligonucleotides used in this study.
Acknowledgements
This work was supported by Institut National de la Recherche Agronomique (INRA UMR 1318), China Scholarship Council (to CW). Funding for open access charge: IJPB, INRA UMR1318. No conflict of interest declared.
Author contributions
HM conceived the project; CW performed most of the experiments; FA performed part of the experiments; MQ and CG provided technical assistance; CW, FA and HM analyzed the data; HM and CW wrote the article with contributions from all the authors.
References
- Andersson SG, Karlberg O, Canback B, Kurland CG. 2003. On the origin of mitochondria: a genomics perspective. Philosophical Transactions of the Royal Society B: Biological Sciences 358, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aryamanesh N, Ruwe H, Sanglard LV, Eshraghi L, Bussell JD, Howell KA, Small I, des Francs-Small CC. 2017. The pentatricopeptide repeat protein EMB2654 is essential for trans-splicing of a chloroplast small ribosomal subunit transcript. Plant Physiology 173, 1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. 2012. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genetics 8, e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology 65, 415–442. [DOI] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. 2008. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Molecular and Cellular Biology 28, 5337–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonen L. 2008. Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8, 26–34. [DOI] [PubMed] [Google Scholar]
- Brown GG, Colas des Francs-Small C, Ostersetzer-Biran O. 2014. Group II intron splicing factors in plant mitochondria. Frontiers in Plant Science 5, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F. 2017. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. The Plant Journal 91, 132–144. [DOI] [PubMed] [Google Scholar]
- Cohen S, Zmudjak M, Colas des Francs-Small C, et al. 2014. nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. The Plant Journal 78, 253–268. [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Falcon de Longevialle A, Li Y, Lowe E, Tanz SK, Smith C, Bevan MW, Small I. 2014. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiology 165, 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Small I. 2014. Surrogate mutants for studying mitochondrially encoded functions. Biochimie 100, 234–242. [DOI] [PubMed] [Google Scholar]
- Dahan J, Tcherkez G, Macherel D, Benamar A, Belcram K, Quadrado M, Arnal N, Mireau H. 2014. Disruption of the CYTOCHROME C OXIDASE DEFICIENT1 gene leads to cytochrome c oxidase depletion and reorchestrated respiratory metabolism in Arabidopsis. Plant Physiology 166, 1788–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Luan S, Chen X, Wang Q, Feng Y, Zhu C, Qi W, Song R. 2018. Maize Dek37 encodes a P-type PPR protein that affects cis-splicing of mitochondrial nad2 intron 1 and seed development. Genetics 208, 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I. 2008. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. The Plant Journal 56, 157–168. [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small ID. 2007. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. The Plant Cell 19, 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francs-Small CC, Kroeger T, Zmudjak M, Ostersetzer-Biran O, Rahimi N, Small I, Barkan A. 2012. A PORR domain protein required for rpl2 and ccmF(C) intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. The Plant Journal 69, 996–1005. [DOI] [PubMed] [Google Scholar]
- Glanz S, Kuck U. 2009. Trans-splicing of organelle introns – a detour to continuous RNAs. Bioessays 31, 921–934. [DOI] [PubMed] [Google Scholar]
- Gray MW. 2012. Mitochondrial evolution. Cold Spring Harbor Perspectives in Biology 4, a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. 1999. Mitochondrial evolution. Science 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- Gualberto JM, Le Ret M, Beator B, Kühn K. 2015. The RAD52-like protein ODB1 is required for the efficient excision of two mitochondrial introns spliced via first-step hydrolysis. Nucleic Acids Research 43, 6500–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberto JM, Newton KJ. 2017. Plant mitochondrial genomes: dynamics and mechanisms of mutation. Annual Review of Plant Biology 68, 225–252. [DOI] [PubMed] [Google Scholar]
- Haïli N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, Colas des Francs-Small C, Vrielynck N, Mireau H. 2013. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Research 41, 6650–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haïli N, Planchard N, Arnal N, Quadrado M, Vrielynck N, Dahan J, des Francs-Small CC, Mireau H. 2016. The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrial NADH DEHYDROGENASE SUBUNIT7 mRNA in Arabidopsis. Plant Physiology 170, 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Giegé P. 2014. RNA metabolism in plant mitochondria. Trends in Plant Science 19, 380–389. [DOI] [PubMed] [Google Scholar]
- He J, Duan Y, Hua D, Fan G, Wang L, Liu Y, Chen Z, Han L, Qu LJ, Gong Z. 2012. DEXH box RNA helicase-mediated mitochondrial reactive oxygen species production in Arabidopsis mediates crosstalk between abscisic acid and auxin signaling. The Plant Cell 24, 1815–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh WY, Liao JC, Chang CY, Harrison T, Boucher C, Hsieh MH. 2015. The SLOW GROWTH3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH dehydrogenase subunit7 intron 2 in Arabidopsis. Plant Physiology 168, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YW, Wang HJ, Hsieh MH, Hsieh HL, Jauh GY. 2014. Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS ONE 9, e112360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke J, Chen RZ, Ban T, et al. 2013. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nature Structural & Molecular Biology 20, 1377–1382. [DOI] [PubMed] [Google Scholar]
- Keren I, Bezawork-Geleta A, Kolton M, Maayan I, Belausov E, Levy M, Mett A, Gidoni D, Shaya F, Ostersetzer-Biran O. 2009. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15, 2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I, Tal L, des Francs-Small CC, Araujo WL, Shevtsov S, Shaya F, Fernie AR, Small I, Ostersetzer-Biran O. 2012. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. The Plant Journal 71, 413–426. [DOI] [PubMed] [Google Scholar]
- Khrouchtchova A, Monde RA, Barkan A. 2012. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 18, 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Yap A, Bond CS, Small I. 2015. Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. The Plant Cell 27, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler D, Schmidt-Gattung S, Binder S. 2010. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Molecular Biology 72, 459–467. [DOI] [PubMed] [Google Scholar]
- Koprivova A, des Francs-Small CC, Calder G, Mugford ST, Tanz S, Lee BR, Zechmann B, Small I, Kopriva S. 2010. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. The Journal of Biological Chemistry 285, 32192–32199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Newton KJ. 2008. Angiosperm mitochondrial genomes and mutations. Mitochondrion 8, 5–14. [DOI] [PubMed] [Google Scholar]
- Kuhn K, Carrie C, Giraud E, Wang Y, Meyer EH, Narsai R, des Francs-Small CC, Zhang B, Murcha MW, Whelan J. 2011. The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. The Plant Journal 67, 1067–1080. [DOI] [PubMed] [Google Scholar]
- Li-Pook-Than J, Bonen L. 2006. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Research 34, 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z. 2010. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. The Plant Journal 63, 749–765. [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, et al. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. The Plant Cell 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Herrmann RG. 1998. Gene transfer from organelles to the nucleus: how much, what happens, and why?Plant Physiology 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massel K, Silke JR, Bonen L. 2016. Multiple splicing pathways of group II trans-splicing introns in wheat mitochondria. Mitochondrion 28, 23–32. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering 104, 34–41. [DOI] [PubMed] [Google Scholar]
- O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I. 2008. On the expansion of the pentatricopeptide repeat gene family in plants. Molecular Biology and Evolution 25, 1120–1128. [DOI] [PubMed] [Google Scholar]
- Shen C, Zhang D, Guan Z, et al. 2016. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nature Communications 7, 11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem A, Zingler N, Pyle AM, Li-Pook-Than J. 2009. Group II introns and their protein collaborators. In: Walter NG, Woodson SA, Batey RT, eds. Non-protein coding RNAs. Springer Series in Biophysics, vol 13. Berlin, Heidelberg: Springer, 167–182. [Google Scholar]
- Sultan LD, Mileshina D, Grewe F, et al. 2016. The reverse transcriptase/RNA maturase protein MatR is required for the splicing of various group II introns in Brassicaceae mitochondria. The Plant Cell 28, 2805–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Bentolila S, Hanson MR. 2016. The unexpected diversity of plant organelle RNA editosomes. Trends in Plant Science 21, 962–973. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Brennicke A, Graichen K. 2013a Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS ONE 8, e65343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A. 2013b RNA editing in plants and its evolution. Annual Review of Genetics 47, 335–352. [DOI] [PubMed] [Google Scholar]
- Wahleithner JA, MacFarlane JL, Wolstenholme DR. 1990. A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proceedings of the National Academy of Sciences, USA 87, 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Aubé F, Planchard N, Quadrado M, Dargel-Graffin C, Nogué F, Mireau H. 2017a The pentatricopeptide repeat protein MTSF2 stabilizes a nad1 precursor transcript and defines the 3′ end of its 5′-half intron. Nucleic Acids Research 45, 6119–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ren Y, Zhou K, et al. 2017. b WHITE STRIPE LEAF4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Frontiers in Plant Science 8, 1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R, Kroeger T, Barkan A. 2008. Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14, 1930–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Hayashi S, Kobayashi K, Hirayama T, Nakamura T. 2013. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 8, e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhang J, He J, Qin Y, Hua D, Duan Y, Chen Z, Gong Z. 2014. ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genetics 10, e1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, et al. 2013. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171. [DOI] [PubMed] [Google Scholar]
- Zimmerly S, Semper C. 2015. Evolution of group II introns. Mobile DNA 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmudjak M, Colas des Francs-Small C, Keren I, Shaya F, Belausov E, Small I, Ostersetzer-Biran O. 2013. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytologist 199, 379–394. [DOI] [PubMed] [Google Scholar]
- Zmudjak M, Shevtsov S, Sultan LD, Keren I, Ostersetzer-Biran O. 2017. Analysis of the roles of the Arabidopsis nMAT2 and PMH2 proteins provided with new insights into the regulation of group II Intron splicing in land-plant mitochondria. International Journal of Molecular Sciences 18, 2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.