Abstract
Tafa is a family of small secreted proteins with conserved cysteine residues and restricted expression in the brain. It is composed of five highly homologous genes referred to as Tafa-1 to -5. Among them, Tafa-2 is identified as one of the potential genes responsible for intellectual deficiency in a patient with mild mental retardation. To investigate the biological function of Tafa-2 in vivo, Tafa-2 knockout mice were generated. The mutant mice grew and developed normally but exhibited impairments in spatial learning and memory in Morris water maze test and impairments in short- and long-term memory in novel object recognition test, accompanied with increased level of anxiety-like behaviors in open-field test and elevated plus maze test, and decreased level of depression-like behaviors in forced-swim test and tail-suspension test. Further examinations revealed that Tafa-2 deficiency causes severe neuronal reduction and increased apoptosis in the brain of Tafa-2−/− mice via downregulation of PI3K/Akt and MAPK/Erk pathways. Conformably, the expression levels of CREB target genes including BDNF, c-fos and NF1, and CBP were found to be reduced in the brain of Tafa-2−/− mice. Taken together, our data indicate that Tafa-2 may function as a neurotrophic factor essential for neuronal survival and neurobiological functions.
Keywords: Tafa-2, knockout mice, learning and memory deficiency, neuronal survival, PI3K/Akt, MAPK/Erk
Introduction
Tafa-2, also known as Tafa2, 6330575M02, and Fam19a2, is one of the five members of the TAFA family discovered by using a novel database searching strategy [1]. The highly homologous Tafa genes encode the proteins with common features, such as approximately 100-amino acid (aa) in length, conserved cysteine residues at fixed positions, predominantly expressed in specific brain regions, and produce secreted polypeptides when transfected into mammalian cells. A few tentative hypotheses about the biological functions of Tafa genes have been proposed, including: (1) may act as brain-specific chemokines to modulate immune responses in the central nervous system (CNS), (2) may represent a novel class of neurokines to regulate immune and nervous cells, and (3) may control axonal sprouting following brain injury [1]. In fact, Tafa-3 has been suggested as a promising candidate for a pars tuberalis messenger involved in transmitting the photoperiodically controlled melatonin signal to the hypothalamo-hypophysial circuitry [2].
Mouse Tafa-2 is located on chromosome 10D2 and composed of nine exons spanning 477 kb. Several isoforms for mouse Tafa-2 have been identified, encoding proteins varied from 131 to 70 aa. In contrast, human Tafa-2 is located on chromosome 12q14.1 and composed of 10 exons spanning approximately 485 kb. Although several transcripts have been detected or predicted, all of them encode the same protein of 131 aa. The percentage identity of TAFA-2 proteins between mouse and human reaches up to 97%, indicating an important evolutionarily conserved role for this protein. The relative mRNA expression of Tafa-2 in human CNS is 50- to 1000-folds higher than that in other tissues, such as colon, heart, lung, spleen, kidney, and thymus, consistent with our results on mouse Tafa-2 mRNA tissue expression pattern (Supplementary Fig. S1A). Moreover, the transcription of mouse Tafa-2 mRNA in the brain coincides with the neuronal development, as indicated by NeuN (a well-recognized marker for mature neurons) expression at different stages of individual growth (Supplementary Fig. S1B). Tafa-2 protein exists in different brain regions, including cortex, amygdala and hippocampus, and it colocalizes with NeuN-positive neuron cells, as examined by both immunohistochemistry and immunofluorescence on sections of different brain regions. Tafa-2 appears to be present in the cytoplasm and the nucleus of neurons and it also seems to be located in the nucleus of GFAP-positive astrocytes (Supplementary Fig. S1C,D). Considering the conservation of protein sequences and the specific expression pattern of Tafa-2, it would be reasonable to hypothesize that Tafa-2 may play a crucial role in neurobiological functions. A previous report, suggesting that Tafa-2 might be a potential gene disrupted as a result of chromosomal rearrangement in a patient with mild mental retardation [3], reinforced our presumption.
In the present study, Tafa-2 knockout mice were generated and used to investigate the consequences of Tafa-2 gene deletion on behaviors and neurobiological functions in mice at the whole animal level. Tafa-2−/− mice exhibited impairments not only in learning and memory but also in emotional regulation examined by a variety of behavioral tests. Further studies revealed that Tafa-2 plays an essential role in neuronal survival via regulating PI3K/Akt and MAPK/Erk pathways, suggesting that Tafa-2 may function as a neurotrophic factor in brain development and function.
Materials and Methods
Generation of Tafa-2 knockout mice
Tafa-2 knockout mice were generated by routine homologous recombination. The targeting vector containing homology arms of 4528 bp (5′ arm, sequences in intron 5) and 3350 bp (3′ arm, sequences in intron 7) was electroporated into ES cells, aiming to delete mouse Tafa-2 genomic 2172-bp fragment covering exons 6 and 7, which are common for all Tafa-2 transcripts. The deleted region was replaced by phosphoglycerate kinase-neomycin resistance cassette (PGK-Neo) for positive selection. Targeted ES cell clones and offsprings were genotyped by PCR using primers P1 and P2 for 5′ arm evaluation and P3 and P4 for 3′ arm evaluation. Products of 5380-bp and 4200-bp were obtained respectively and verified by DNA sequencing. Later, triple primer PCR strategy (using primers P5, P6, and P7) was designed for routine genotyping using the following amplification conditions: 94°C for 3 min and 32 cycles of 94°C for 10 s, 72°C for 5 min 30 s, and a 5-min incubation at 72°C at the end of the run. PCR products were resolved on 1% agarose gels. Product lengths of wild-type (wt) and targeted alleles were 3092-bp and 1602-bp, respectively. The primer sequences of P1–P7 are listed in Supplementary Table S1. All mice were housed under specific pathogen free (SPF) conditions at a constant room temperature of 22–24°C with a 12/12-h light/dark cycle, with free access to a diet of regular chow and water. The F1 and F2 generations were backcrossed with C57BL/6 mice. Age- and sex-matched littermates with different genotypes from generations F4–F6 were used for phenotypic analyses. Animal protocols and experiments were approved by Institutional Animal Care and Use Committee of Shanghai Research Center for Model Organisms (Permit number: 2012-0019, 2016-0003).
mRNA expression analyses
Total RNAs from mouse tissues were isolated using Trizol reagent (Life Technologies Inc., Gaithersburg, USA) according to the manufacturer’s instructions. The first-strand cDNAs were synthesized from 2-μg of total RNAs with oligo (dT) primer and random six mer primer using PrimerScript RTase (TaKaRa, Dalian, China) at 37°C for 15 min. cDNAs were amplified using specific sets of primers listed in Supplementary Table S1. PCR products were separated by electrophoresis on 2.0% agarose gels and visualized by ethidium bromide staining. Real-time PCR was performed with a Mastercycler ep realplex (Eppendorf) using SYBR Premix Ex Taq kit (TaKaRa). Resolution of the product of interest from nonspecific product amplification was achieved by melting curve analysis. Gene expression levels were normalized to GAPDH content using the 2−ΔΔCt or 2−ΔCt method [4].
Histological analysis
Mice were euthanized by CO2 inhalation and were transcardially perfused with 1 × PBS followed by 4% paraformaldehyde (PFA). Fixed brains were dehydrated through a graded series of ethanol and embedded in paraffin. For Nissl staining, coronal serial sections (10 μm) were deparaffinized, rehydrated, rinsed in 1 × PBS for 5 min and then stained with 0.1% cresyl violet/0.5% acetic acid for 12 min. The number of Nissl positive neurons per specific surface area (around 220 μm × 164 μm) of section was detected in both cortex and hippocampus by image analysis using Image-Pro Plus software. For immunohistochemistry, brain sections were dewaxed, rehydrated and subject to microwave antigen retrieval. After pretreatment with 3% H2O2 in methanol at room temperature for 10 min to block the endogenous peroxidase activity, tissue sections were blocked for 1 h in 1 × PBS containing 5% BSA (w/v) or normal goat serum (v/v) at room temperature and incubated overnight at 4°C with diluted antibodies against TAFA-2 and CBP. After three washes with PBS (5 min each time), sections were detected using a VECTASTAIN® ABC kit (Vector Laboratories, Burlingame, USA) according to the manufacturer’s instructions. For immunofluorescence, tissue sections were incubated for 10 min in PBS containing 0.1% Triton X-100 and blocked for 1 h in 1 × PBS containing 5% BSA (w/v) or normal goat serum (v/v) at room temperature after antigen retrieval. Primary antibodies used were antibodies against Tafa-2, NeuN, GFAP, microtubule associated protein 2 (MAP2) and synaptophysin (Syn). After extensive washing in PBS, brain sections were incubated with either Alexa Fluor 488- or 594-conjugated secondary antibodies (Invitrogen, Carlsbad, USA) and nuclei were visualized by DAPI (Invitrogen) counterstain. Antibodies used are listed in Supplementary Table S2. Slides were mounted in fluorescence mounting medium (DAKO), coverslipped and examined under a Nikon Eclipse 90i microscope (Tokyo, Japan). For quantitative determinations, images were analyzed by Image-Pro Plus software.
Western blot analysis
For protein lysate preparation, hemi-brains (four samples per group) were homogenized in ice-cold, high-detergent lysis buffer (10 × w/v, 50 mM Tris, pH 7.4, 150 mM NaCl, 2% NP-40, 0.5% sodium deoxycholate, 4% SDS, and protease and phosphatase inhibitor cocktails) and centrifuged at 12,000 g for 30 min. The supernatant was harvested, and total protein concentration was determined using a bicinchoninic acid protein assay kit (Thermo Scientific, Waltham, USA). Samples were supplemented with 6-fold concentrated SDS-PAGE sample buffer containing 2-mercaptoethanol and boiled at 100°C for 5 min. Equal amounts of proteins (80–100 μg) were separated by 10–12% SDS-PAGE and transferred onto nitrocellulose membranes (catalog no. 162-0112; Bio-Rad, Hercules, USA). Membranes were blocked with 5% nonfat milk for 1 h and incubated with the primary antibodies as indicated overnight at 4°C. Antibodies used are listed in Supplementary Table S2. GAPDH was used as a loading control. To visualize specific protein signals, the secondary antibodies conjugated with IRdye800CW (LI-COR, Lincoln, USA) were used and the membranes were scanned by Odyssey Infrared Imager (LI-COR).
Antibodies
In total, 15 primary antibodies and six secondary antibodies were used in this study. Except for the anti-Tafa-2 antibody, the remaining antibodies were all typical ones that had been used in published papers. The catalog number, supplier, dilution as well as the references are shown in Supplementary Table S2. The references cited used the primary antibodies with the same catalog numbers as those used by us. And the applications (western blotting and IF/IHC) of the primary antibodies in the references were the same as the technique used in our study. The specificity of the primary antibodies used in western blot analysis could also be evaluated by the molecular weight of the protein band. The specificity of the primary antibodies used in IF/IHC was evaluated by performing negative controls without adding primary antibodies in parallel. The specificity of the goat anti-Tafa-2 (A-14) polyclonal antibody (sc-244255) is illustrated by our results in Supplementary Fig. S1C, Fig. 1E and Fig. 5D, where negative controls without adding primary antibodies and tissue sections from Tafa-2−/− mice were used in parallel.
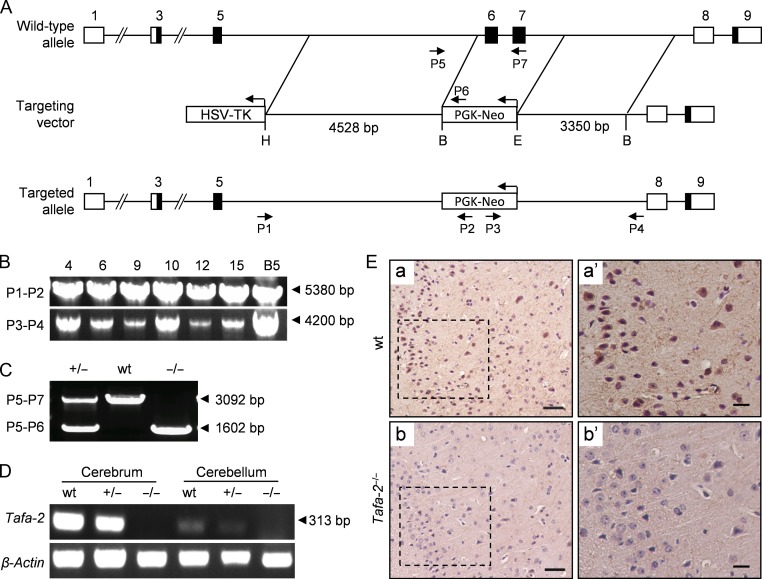
Figure 1.
Generation of Tafa-2 knockout mice (A) Graphic representation of the Tafa-2 targeting strategy. Boxes represent exons with the coding region in black. The targeting vector contained 4528 bp of 5′ homology and 3350 bp of 3′ homology. PGK-Neo and HSV-TK were used for positive and negative selection, respectively. P1–P7, primers for genotyping and their relative positions are indicated. H, HindIII; B, BamHI; E, EcoRI. (B) Representative PCR genotyping of the ES cell clones and F1 hybrid generation obtained by crossing male chimeras with C57BL/6J females, using primers P1 and P2 to the 5′ arm and P3 and P4 to the 3′ arm. Products of 5380 bp and 4200 bp were obtained, respectively, and verified by DNA sequencing. (C) Triple primer PCR strategy using primers P5-P7 was designed for routine genotyping. The size of PCR products (arrowhead) was shown on the right of the images. (D) RT-PCR with total RNAs from cerebrum and cerebellum of mice with different Tafa-2 genotypes. Primers amplifying a specific region of the Tafa-2 coding sequence were used. β-Actin was used as an internal control. (E) Immunohistochemistry analysis of cortex sections from wt and Tafa-2−/− mice. Boxed areas in a and b were magnified in a’ and b’. Scale bar = 50 μm in a and b. Scale bar = 20 μm in a′ and b′.
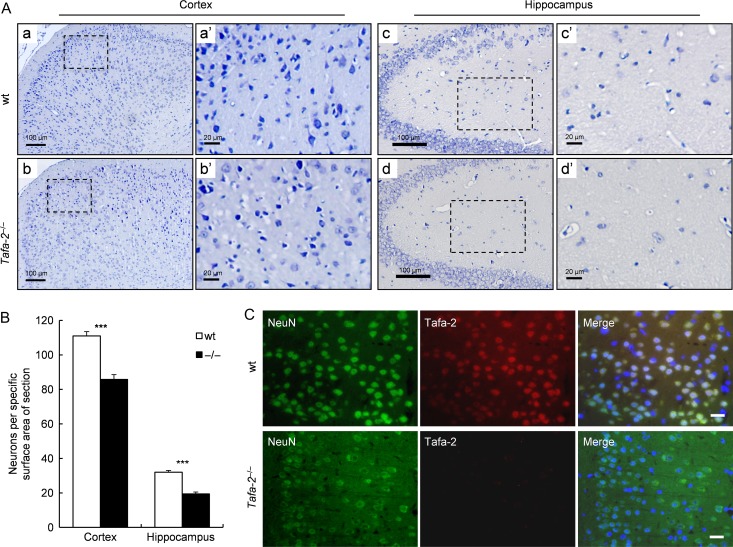
Figure 5.
Severe neuronal loss in the brain of Tafa-2−/− mice (A) Representative images of Nissl staining on sections of cortex and hippocampus from wt and Tafa-2−/− mice. Boxed areas in a, b, c, and d were magnified in a′, b′, c′, and d′. Scale bar = 100 μm in a, b, c, and d. Scale bar = 20 μm in a′, b′, c′, and d′. (B) The number of Nissl positive neurons per specific surface area (around 220 μm × 164 μm) of section from cortex and hippocampus was quantified by image analysis using Image-Pro Plus software and compared between wt and Tafa-2−/− mice. The data were average results of 12 sections from four mice each group (three sections per mouse). Data are shown as the mean ± SE. Student’s t test: ***P < 0.001. (C) Representative images of immunofluorescence of NeuN, a neuron marker, and Tafa-2 on sections of cortex from wt and Tafa-2−/− mice. Scale bar = 20 μm.
Behavioral testing
All behavioral tests were conducted during the animals’ light cycle, between 9:00 a.m. and 6:00 p.m. The apparatus and analysis software used in behavioral tests were supplied by Shanghai Mobile Datum Information Technology Co., Ltd (Shanghai, China). Behavioral analyses of wt and Tafa-2−/− male mice were conducted at the age of 10–18 weeks (adult). In order to facilitate adaptation to the experimental environment, mice were housed in the testing room for at least 30 min before the experiments. The animals were naive to the test situation and were used only once. At the time interval between two successive measurements, the apparatus was cleaned with 75% ethanol and dried using clean paper. Each behavioral test was conducted repeatedly with two different batches of animals. All behavioral experiments were conducted and analyzed in a double-blind manner with respect to mouse genotype.
Morris water maze test
The Morris water maze test was performed according to previous reports [5–7] with minor modifications. A circular pool (120 cm in diameter and 45 cm in height) was filled with water maintained at 21–23°C, and a circular escape platform with a diameter of 8 cm was submerged 1 cm below the water surface. The water was colored white by addition of non-toxic opaque paint to hide the escape platform. The test procedure consisted of a training period (five consecutive days) and a test period (on the sixth day). During the training period, mice were given four trials per day with a 15 min intertrial interval and 60 s per trial to find the hidden platform, which was always located in the center of the same quadrant (target quadrant) for all animals. Mice were guided to the hidden platform by hand if they were unable to find it after 60 s. After reaching the platform, each mouse was allowed to remain in it for 30 s, and then it was quickly dried with a towel and put under a heating lamp set at 37°C to avoid hypothermia. Mice were put in different quadrants during the four trials each day and the order of quadrants was changed on different training days. Mice were subject to the test on the sixth day when the platform was removed and mice were allowed to search for it for 60 s. The swimming behavior of each mouse was recorded with a video camera, and data were analyzed individually using custom-written software. The latency time during training, time spent in the target quadrant and the number of times crossing the original platform position were determined as a measure of spatial learning and memory and compared between wt and Tafa-2−/− mice.
Novel object recognition test
The novel object recognition test was performed as described previously [7–10] with minor modifications. The experimental apparatus was an open-field chamber (40 cm × 40 cm × 30 cm). Mice were individually habituated in the empty chamber for 2 days (10 min per day) before testing. A full experiment consisted of a sample phase (training session) and a choice phase (retention session). During the sample phase, two identical objects were placed into the chamber at fixed locations and the mice were allowed to explore freely for 10 min. An animal was considered to be exploring the object when its head was facing the object or it was touching or sniffing the object. The time spent in exploring each object was recorded. After training, mice were immediately returned to their home cages. During the choice phase (1 or 24 h after the sample phase), mice were placed back into the same chamber, with one of the familiar objects used during the sample phase replaced by a novel object. The animals were then allowed to explore freely for 10 min and the time spent in exploring each object was recorded. A preference index, a ratio of the amount of time spent in exploring any one of the two objects (training session) or the novel object (retention session) over the total time spent in exploring both objects, was determined as a measure of recognition memory.
Open-field test and elevated plus maze test
The open-field and elevated plus maze tests were performed as previously published [5]. Mice were placed in the 24 × 24 × 38 cm3 open-field chambers which were equipped with video cameras. A square region of 12 × 12 cm2 in the center of the chamber was defined as the ‘central zone’. During the test, mice were allowed to ambulate in it for 15 min. Horizontal locomotion (total distance traveled), average movement speed, and time spent in the central zone were recorded by a video/computer system and analyzed by a custom-written computer software. The elevated plus maze was constructed of white chipboards, elevated 50 cm above ground, and consisted of two opposite closed arms with 30 cm high opaque walls and two opposite open arms of the same size (36 cm × 6 cm). The animals were placed onto the central platform of the maze facing the open-arm. During a 5-min test, exploratory activity was recorded by a video/computer system, and a custom-written computer software was used to calculate the time spent in the open area and the distance traveled in the open and closed arms.
Forced-swimming test and tail-suspension test
In the forced-swimming test (FST), mice were placed into a Plexiglas cylinder (45 cm high and 19 cm internal diameter) filled with water (23 ± 1°C) to a depth of 22 cm. Mice were forced to swim for 6 min and then dried and put back to their cages. The amount of time spent immobile, defined as floating motionless in the water and making only small movements necessary to keep its head above the water, was recorded during the last 4 min of the test. The tail-suspension test (TST) was performed as previously described [5,11]. Each mouse was suspended individually by its tail from a metal rod, which was fixed 50 cm above the surface of a table covered with soft cloth in a sound-proof room. The tip of the mouse tail was fixed on the rod using adhesive scotch tape. The duration of the test was 6 min and the immobility time of the tail-suspended mice was measured during the last 4 min of the test.
TUNEL labeling and quantification
Apoptotic cells on sections were detected using an In Situ Cell Death Detection kit (11684795910; Roche, Mannheim, Germany) according to the manufacturer’s protocol. Briefly, the paraffin-embedded brain sections were blocked using 5% of goat serum for 30 min followed by the treatment of fluorescein at 37°C for 1 h. After being washed with PBS for three times, the slides were counterstained by DAPI, mounted in DAKO, coverslipped and examined under a Nikon Eclipse 90i microscope. All positive cells were counted per section in the cortex and hippocampus by an experimenter blind to genotype. The percentage of apoptotic cells per 20-nm-thick section was calculated as the ratio between TUNEL-positive cells and DAPI positive cells, and then averaged per genotype.
Caspase-3 activity assay
The activity of caspase-3 was determined using the Caspase-3 Activity kit (Beyotime, Haimen, China). Tissues were homogenized in lysis buffer and incubated on ice for 10 min before they were centrifugated at 18,000 g for 15 min at 4°C. Protein concentration of supernatants was measured by Bradford’s method and equal amounts of proteins (50 μl) were incubated in a total volume of 100 μl comprised of 40 μl detection buffer and 10 μl caspase-3 substrate Ac-DEVD-pNA (2 mM). After incubation for 2 h at 37°C, cleavage of the substrate was detected using BioTek Synergy 2 (Gene Company Limited, Shanghai, China) with the absorbance at 405 nm.
PET-CT
The animals were fasted for 12 h, and each animal was weighed before the experiment. After a short isoflurane (2% in 100% oxygen for 5 min) inhalation anesthesia period, the mice were intravenously injected with 18F-FDG. Mice were scanned on an Inveon microPET/CT scanner (Siemens Medical Solutions; Siemens Healthcare Molecular Imaging, Boston, USA) 60 min after 18F-FDG injection. Image acquisition and analysis were performed with the Siemens Inveon Research Acquisition Workplace Software (IRW, version 3.0). According to the PET/CT fusion images generated by the computer, the accurate calculation of standardized uptake values (SUVs) in the mouse brain was quantitatively analyzed.
Statistical analysis
All data are expressed as the mean± standard error (SE) (n ≥ 3), unless otherwise stated. Two-way ANOVA followed by Bonferroni post hoc tests and two-tailed Student’s t test were used for comparisons between groups. Time distribution in the four quadrants during the Morris water maze test was compared using Chi-square test. For all of the statistical tests, P values less than 0.05 were considered statistically significant.
Results
Tafa-2 −/− mice are born alive and appear grossly normal
To explore the physiological function of Tafa-2 in vivo, we constructed a targeting vector using ET cloning method [12,13] and used homologous recombination in ES cells to disrupt the Tafa-2 gene. Graphic representation of the Tafa-2 targeting strategy is shown in Fig. 1A. F1 hybrid generation obtained by crossing male chimeras with C57BL/6J females was genotyped by PCR analysis of genomic DNA, with a targeted ES cell clone B5 as a positive control. Primers used and the product length are indicated in Fig. 1B. Interbreeding of Tafa-2+/− mice produced the offspring with three genotypes, as illustrated by triple primer PCR strategy (Fig. 1C). Tafa-2 mRNA was not detected by RT-PCR in either cerebrum or cerebellum of homozygotes (Fig. 1D) and Tafa-2 protein was not detected by immunohistochemistry on cortex sections of Tafa-2−/− mice (Fig. 1E), demonstrating the targeted deletion of Tafa-2 in mice. We examined the basic phenotypes of Tafa-2 knockout mice, and found that Tafa-2 deletion in mice had no significant effect on embryonic development and postnatal growth, as illustrated by genotype distribution, sex ratio, and body weight of the offspring (Supplementary Fig. S2). Thus, Tafa-2−/− mice were born alive and appeared grossly normal.
Tafa-2 −/− mice exhibit significant impairment in spatial learning and memory, as well as short- and long-term memory
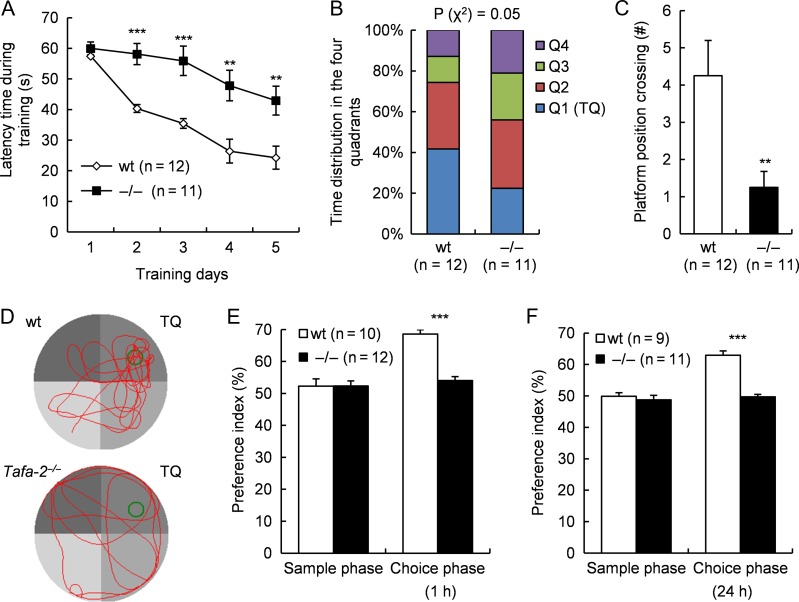
The prominent and brain-restricted expression of Tafa-2 impelled us to investigate the intellectual ability and emotional regulation of Tafa-2−/− mice. Morris water maze test and novel object recognition test were applied to assess spatial learning and memory and recognition memory, respectively. In the Morris water maze test, Tafa-2−/− mice displayed significantly longer latency to find the hidden platform during the training period as compared with wt littermates (two-way ANOVA: interaction F (4) = 2.42, P = 0.0534, genotype F (1) = 52.38, P < 0.0001, training days F (4) = 15.98, P < 0.0001, Bonferroni post-test: P < 0.001, n = 12 for wt, n = 11 for Tafa-2−/−; Fig. 2A), although mice of both genotypes exhibited progressive improvement in finding the hidden platform, as indicated by the decrease in escape latency time during the training period (Fig. 2A). In the test period when the platform was removed, time distribution in the four quadrants was significantly different between wt and Tafa-2−/− mice (Chi-square test, P = 0.05) (Fig. 2B). Tafa-2−/− mice spent significantly less time in the target quadrant (wt, 24.7 ± 3.0; Tafa-2−/−, 12.2 ± 1.9; Student’s t test: t (21) = 3.48, P < 0.01) and significantly fewer number of times crossing the original platform position (wt, 4.3 ± 0.9; Tafa-2−/−, 1.0 ± 0.4; Student’s t test: t (21) = 3.08, P < 0.01; Fig. 2C) when compared with wt littermates. Representative swimming paths of wt and Tafa-2−/− mice during the test period are shown in Fig. 2D. The ability of nonspatial memory was analyzed by the novel object recognition test. In the sample phase, comparable exploratory preference index existed between wt and Tafa-2−/− mice (wt, 52.3% ± 2.3% and Tafa-2−/−, 52.4% ± 1.6%; Fig. 2E; wt, 49.9% ± 1.1% and Tafa-2−/−, 48.8% ± 1.4%; Fig. 2F). However, in the choice phase performed 1 h or 24 h after sample phase using two different batches of mice, a marked decrease in the exploratory preference for novel objects was evident in Tafa-2−/− mice compared with that in wt controls (wt, 68.6% ± 1.4% and Tafa-2−/−, 53.8%± 1.1%; Student’s t test: t (20) = 8.38, P < 0.001; Fig. 2E; wt, 62.9% ± 1.4%; and Tafa-2−/−, 49.7% ± 0.7%; Student’s t test: t (18) = 8.97, P < 0.001; Fig. 2F). There was no significant difference in the total time taken to explore two objects during sample phase and choice phase (data not shown). Taken together, Tafa-2−/− mice exhibited significant impairment in both spatial learning and memory, and nonspatial short- and long-term recognition memory.
Figure 2.
Tafa-2−/− mice exhibited significant impairment in spatial learning and memory, as well as short- and long-term memory (A–D) Morris water maze test. (A) The latency time during training period (learning curves) (two-way ANOVA: interaction F (4) = 2.42, P = 0.0534, genotype F (1) = 52.38, P < 0.0001, training days F (4) = 15.98, P < 0.0001, Bonferroni post-tests: **P < 0.01, ***P < 0.001, n = 12 for wt, n = 11 for Tafa-2−/−). (B) Time distribution in the four quadrants during the test period (Chi-square test: P = 0.05). (C) Number of times crossing the original platform position during the test period (Student’s t test: **P < 0.01). (D) Representative swimming paths during the test period. TQ, target quadrant. (E,F) Novel object recognition test. (E) Exploratory preference index during the sample phase and the choice Phase 1 h after the sample phase was determined as a measure of short-term recognition memory (Student’s t test: ***P < 0.001, n = 10 for wt, n = 12 for Tafa-2−/−). (F) Exploratory preference index during the sample phase and the choice Phase 24 h after the sample phase was determined as a measure of long-term recognition memory (Student’s t test: ***P < 0.001, n = 9 for wt, n = 11 for Tafa-2−/−).
Tafa-2 −/− mice display increased level of anxiety-like behaviors and decreased level of depression-like behaviors
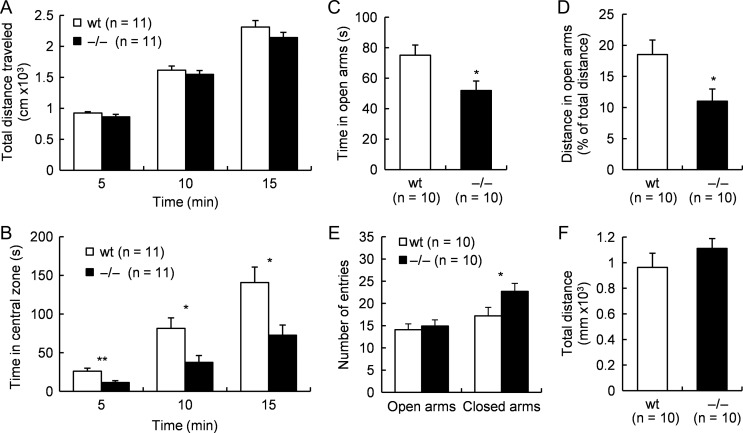
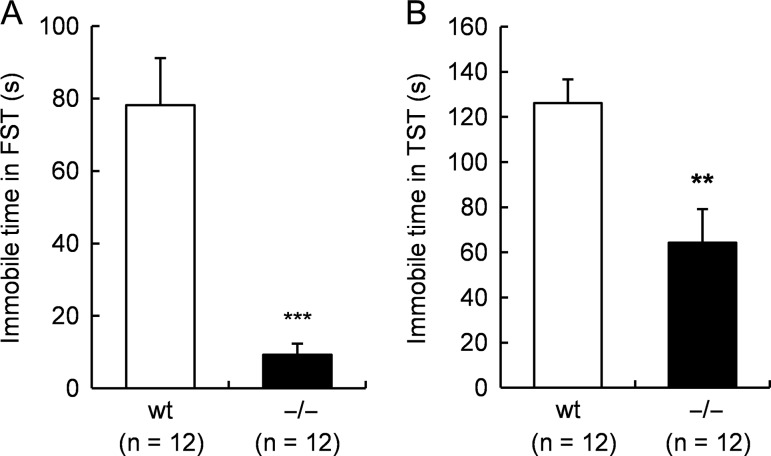
Next, we explored whether Tafa-2−/− mice had any emotional regulation deficits by additional behavioral tests. First, open-field test and elevated plus maze test were conducted to evaluate anxiety-like behaviors. Tafa-2−/− mice spent significantly less time in the central zone of the open-field, analyzed at 5, 10, and 15 min during the test, as compared with their wt littermates (Student’s t tests: 5 min, t (20) = 3.37, P < 0.01; 10 min, t (20) = 2.69, P < 0.05; 15 min, t (20) = 2.83, P < 0.05; Fig. 3B), although mice of both genotypes displayed similar locomotion activity as revealed by total distance traveled (P > 0.05; Fig. 3A) during the test. Furthermore, elevated plus maze test showed that Tafa-2−/− mice had reduced open-arm exploration in terms of both time spent in open arms (wt, 75.1 ± 6.6 and Tafa-2−/−, 51.9 ± 6.2, Student’s t test: t (18) = 2.56, P < 0.05; Fig. 3C) and distance traveled in open arms (wt, 18.5% ± 2.3% and Tafa-2−/−, 11.0% ± 2.0%, Student’s t test: t (18) = 2.46, P < 0.05; Fig. 3D) as compared with wt controls. However, the number of entries to the open arms were comparable between wt and Tafa-2−/− mice (wt, 14.1 ± 1.3 and Tafa-2−/−, 14.9 ± 1.9, Student’s t test: t (18) = −0.34, P > 0.05; Fig. 3E). On the other hand, the number of entries to the closed arms was significantly more in Tafa-2−/− mice than that in wt mice (wt, 17.2 ± 1.4 and Tafa-2−/−, 22.7 ± 1.8, Student’s t test: t (18) = −2.39, P < 0.05; Fig. 3E). No significant difference existed in the total number of entries to both the open and the closed arms (wt, 31.3 ± 2.4 and Tafa-2−/−, 37.6 ± 2.3, Student’s t test: t (18) =−1.88, P > 0.05), although the Tafa-2−/− mice showed relatively more entries. Consistent with the results from the open-field test, all mice exhibited similar total distance traveled (P > 0.05; Fig. 3F). Taken together, these data suggested that Tafa-2 deficiency in mice caused a tendency towards anxiety-like behaviors which seemed irrelevant to the locomotor activity of mice. Second, we conducted FST and TST to evaluate depression-like behaviors. As shown in Fig. 4A, Tafa-2−/− mice spent significantly less immotile time in FST, compared with that of wt controls (wt, 78.2 ± 13.0 and Tafa-2−/−, 9.3 ± 2.8, Student’s t test: t (22) = 5.19, P < 0.001). Conformably, there was a significant genotypic effect on the immobility time in the TST (wt, 126.1 ± 9.6 and Tafa-2−/−, 64.3 ± 14.8, Student’s t test: t (22) = 3.51, P < 0.01; Fig. 4B). Collectively, these results indicated that Tafa-2 deficiency in mice caused decreased level of depression-like behaviors.
Figure 3.
Tafa-2−/− mice displayed increased level of anxiety-like behaviors (A,B) Open-field test (n = 11 for each genotype). Total distance traveled (A) and time spent in the central zone (B) were compared between wt and Tafa-2−/− mice. (C–F) Elevated plus maze test (n = 10 for each genotype). Time spent in open arms (C), distance traveled in open arms expressed as percentage values of total distance (D), number of entries to the open and closed arms (E) and total distance traveled (F) were compared between wt and Tafa-2−/− mice. Data are shown as the mean ± SE. Student’s t test: *P < 0.05, **P < 0.01.
Figure 4.
Tafa-2−/− mice displayed decreased level of depression-like behaviors (A) Forced-swimming test. (B) Tail-suspension test. Data are shown as the mean ± SE (n = 12 for each genotype). Student’s t test: **P < 0.01, ***P < 0.001.
Tafa-2 −/− mice show neuronal loss, defects in dendritic and synaptic morphology, and apoptosis in the brain
To elucidate the underlying mechanisms by which Tafa-2 deficiency in mice caused the behavioral phenotypes mentioned above, we carried out a number of histological analyses. Nissl staining on sections of cortex and hippocampus from wt and Tafa-2−/− mice demonstrated a striking loss of Nissl positive neurons in cerebral cortex and hippocampus (Fig. 5A,B). Significant reductions in the number of Nissl positive neurons per specific surface area (around 220 μm × 164 μm) of section was detected in both cortex and hippocampus (P < 0.001; Fig. 5B) by image analysis using Image-Pro Plus software, although Nissl-stained brain sections of mutant mice showed no evidence of gross changes in brain organization or connectivity when compared with those of wt mice (data not shown). Moreover, immunofluorescence of NeuN showed a dramatic decrease of NeuN-positive neurons in the brain of Tafa-2−/− mice, where Tafa-2 protein could not be detected, in contrast to that of wt controls (Fig. 5C).
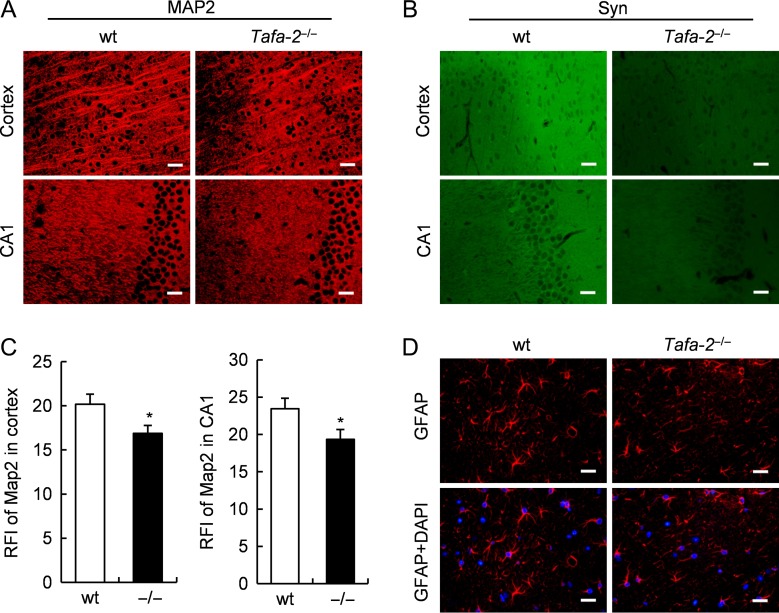
To examine whether Tafa-2 deficiency affects neuronal and synaptic morphology, we performed immunofluorescence on brain sections to analyze markers for dendrites and presynaptic terminals. Immunostaining of MAP2, a dendritic marker, was slightly weaker in the cortex and the hippocampal CA1 area of Tafa-2−/− mice than those of wt controls (Fig. 6A). Quantitative analysis showed that the relative fluorescence intensity (RFI) of MAP2 staining was significantly lower in the cortex and the hippocampal CA1 area of Tafa-2−/− mice than those of wt controls (n = 4 for each genotype, P < 0.05; Fig. 6C). Immunostaining of synaptophysin, a presynaptic marker, was also markedly changed in the cortex and the hippocampal CA1 area between the two genotypic groups (Fig. 6B). To assess the potential effects of Tafa-2 deletion on astrocytes, we conducted immunofluorescence of GFAP, a marker for reactive astrocytes, and found no difference on GFAP immunostaining (Fig. 6D). Collectively, these results showed neuronal loss and defects in dendritic and synaptic morphology in the brain of mice lacking Tafa-2, which may constitute the structural basis for the impaired learning and memory ability and the emotional regulation abnormality in Tafa-2−/− mice.
Figure 6.
Defects in dendritic and synaptic morphology in the brain of Tafa2−/− mice (A) Immunofluorescence of MAP2, on sections of cortex and hippocampal CA1 area of wt and Tafa-2−/− mice. Scale bar = 20 μm. (B) Immunofluorescence of synaptophysin (Syn), on sections of cortex and hippocampal CA1 area of wt and Tafa-2−/− mice. Scale bar = 20 μm. (C) Quantitative analysis of the relative fluorescence intensity (RFI) of MAP2 staining in A. n = 4 for each genotype. Student’s t test: *P < 0.05. (D) Immunofluorescence of GFAP. Nuclei were counterstained by DAPI. Scale bar = 20 μm.
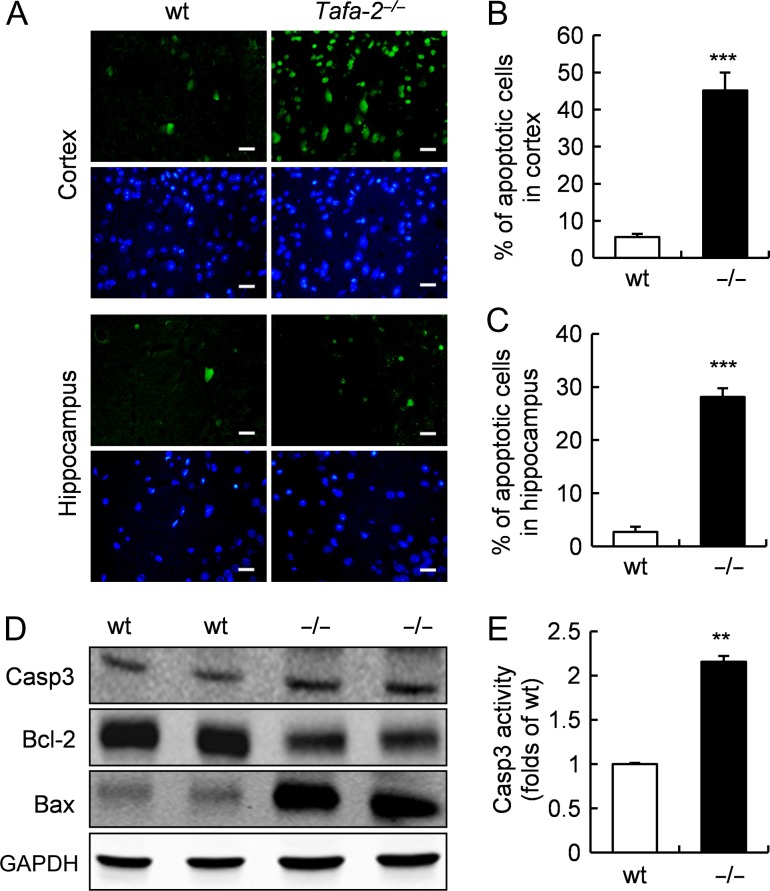
To determine whether the neuronal loss in Tafa-2−/− mice is due to apoptosis, we performed TUNEL assay. As shown in Fig. 7A-C, TUNEL-positive (TUNEL+) apoptotic cells were significantly elevated in both cortex (wt, 5.6% ± 0.9%; Tafa-2−/−, 45.2% ± 4.8%, P < 0.001; Fig. 7A,B) and hippocampus (wt, 2.7% ± 1.0%; Tafa-2−/−, 28.2% ± 1.6%, P < 0.001; Fig. 7A,C) of Tafa-2−/− mice, compared with wt controls. Western blot analysis of brain lysates revealed that Bcl-2 was dramatically decreased, whereas Bax was markedly increased in the brain of Tafa-2−/− mice (Fig. 7D). To determine whether caspase-3 is activated in the brain of Tafa-2−/− mice, caspase-3 protein level was measured by western blot analysis and caspase-3 activity was determined. Although no significant difference was detected at caspase-3 protein level between wt and Tafa-2−/− mice (Fig. 7D), its activity in Tafa-2−/− mice was significantly higher than that in wt controls (folds of wt: 2.16 ± 0.06, P < 0.01; Fig. 7E). Collectively, these data suggested that apoptosis was significantly increased in the brain of Tafa-2−/− mice.
Figure 7.
Increased apoptosis in the brain of Tafa-2−/− mice (A) Representative images of TUNEL staining (green) on sections of cortex and hippocampus from wt and Tafa-2−/− mice. The nuclei were visualized by DAPI counterstain (blue). Scale bar = 20 μm. (B,C) The number of apoptotic cells in cortex and hippocampus was quantified by image analysis and compared between wt and Tafa-2−/− mice. n = 4 for each genotype. Student’s t test: ***P < 0.001. (D) Western blot analysis of caspase-3, Bcl-2 and Bax in the brain lysates of wt and Tafa-2−/− mice. GAPDH was used as a protein loading control. (E) Relative caspase-3 activity in the brain of wt and Tafa-2−/− mice. Data are shown as the mean ± SE. Student’s t test: **P < 0.01, n = 6 for wt and n = 5 for Tafa-2−/− mice.
It has been proposed that impairment of energy metabolism in the brain is among the main reasons thought to underlie the pathogenesis of the neurodegenerative disorders and mental illnesses, such as Alzheimer’s, Huntington’s, and depression [14–16]. Since glucose is the main substrate for energy metabolism of the brain, we performed PET/CT to test whether glucose metabolism was changed due to Tafa-2 disruption. As shown in Supplementary Fig. S3, the global uptake of glucose in the brain of Tafa-2−/− mice was significantly increased (P < 0.05).
Downregulaton of PI3K/Akt and MAPK/Erk signaling pathways and reduced expression of CREB-dependent genes and CBP in the brain of Tafa-2−/− mice
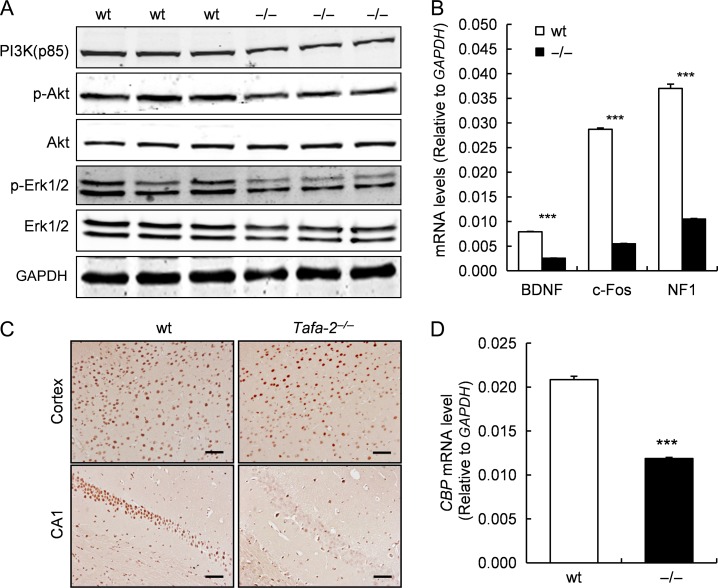
To examine whether Tafa-2 deficiency affects PI3K/Akt and MAPK/Erk signaling pathways in the brain, western blot analysis was conducted to measure the expression levels of PI3K (p85), p-Akt, Akt, p-Erk1/2, Erk1/2 in the brain lysates of wt and Tafa-2−/− mice. Results showed that protein levels of PI3K (p85), p-Akt, and p-Erk1/2 were remarkably downregulated in the brain lysates of Tafa-2−/− mice, compared with those of wt controls, although the levels of total Akt and Erk1/2 proteins were comparable between the two genotypic groups (Fig. 8A). In addition, quantitative real-time RT-PCR results demonstrated significant reduction of BDNF, c-fos and neurofibromin 1 (NF1) in the brain of Tafa-2−/− mice, compared with those of wt mice (P < 0.001; Fig. 8B). Immunohistochemistry of CBP on sections of cortex and hippocampus CA1 from wt and Tafa-2−/− mice displayed moderate and robust reduction of CBP staining in the cortex and the hippocampal CA1 area, respectively (Fig. 8C). The reduced protein level of CBP in the brain of Tafa-2−/− mice was further supported by significantly lower transcription of CBP, as assayed by quantitative real-time RT-PCR (P < 0.001; Fig. 8D).
Figure 8.
Downregulaton of PI3K/Akt and MAPK/Erk signaling pathways and reduced expressions of CREB-dependent genes and CBP in the brain of Tafa-2−/− mice (A) Western blot analysis of PI3K (p85), p-Akt, Akt, p-Erk1/2, Erk1/2 in the brain lysates of wt and Tafa-2−/− mice. GAPDH was used as a protein loading control. (B) Real-time PCR analysis of BDNF, c-Fos, and NF1 mRNA levels in the brain of wt and Tafa-2−/− mice. GAPDH was used as an internal control. (C) Representative images of immunohistochemistry of CBP on sections of cortex and hippocampus CA1 from wt and Tafa-2−/− mice. Scale bar = 50 μm. (D) Real-time PCR analysis of CBP mRNA level in the brain of wt and Tafa-2−/− mice. GAPDH was used as an internal control. Data are shown as the mean ± SE. Student’s t test: ***P < 0.001, n = 3 for each genotype.
Discussion
In the present study, we explored the physiological function of Tafa-2 in vivo by generating a global Tafa-2 knockout mouse model. Tafa-2 is predominantly expressed in the CNS, demonstrated by previous work and our current study. The appearance of Tafa-2 mRNA coincides with the neuronal development and Tafa-2 protein appears to be present in the cytoplasm and the nucleus of neurons and it also seems to be located in the nucleus of GFAP-positive astrocytes, presuming a potential role in neuronal function. Indeed, Tafa-2−/− mice exhibited impairments not only in learning and memory but also in emotional regulation examined by a variety of behavioral tests. Considering the conservation of Tafa-2 between human and mouse, a better understanding of the role of this endogenous peptide in health may pave the way to exploit Tafa-2 as a novel therapeutic agent for detection and prevention of neurological disorders.
The seemingly different subcellular localization of Tafa-2 in neurons and astrocytes sounds fascinating and has not been reported by others. As a member of the Tafa family which produces secreted polypeptides, the detection of Tafa-2 in the cytoplasm of neurons is in line with our expectations. But why is Tafa-2 nuclear localization in neurons and/or astrocytes? Astrocytes are the most abundant glial cells in the CNS and are closely associated with neuronal synapses. They play a number of active roles in the brain, including biochemical support of endothelial cells that form the blood-brain barrier, provision of nutrients to the nervous tissue, maintenance of extracellular ion balance, and a role in the repair and scarring process of the brain. The nuclear localization of Tafa-2 in the astrocytes may play an important role of Tafa-2 in the function of astrocytes, which is not evident in the present study. Nevertheless, as far as we know, there is no nuclear localization signal in the amino acid sequence of Tafa-2 protein. Whether Tafa-2 is truly localized in the nucleus of neurons and/or astrocytes and how it is positioned in the nucleus need to be further studied.
Tafa-2 deficiency in mice caused increased level of anxiety-like behaviors and decreased level of depression-like behaviors, which is particularly interesting because anxiety disorders and depression are often co-morbid condition, and seems that these measures are discordant in these mice. As far as we know, these seemingly contradictory behaviors occur not only in the present Tafa-2−/− mice, but also in other mouse models. It was reported that CLOCK knockdown in the ventral tegmental area (VTA) leads to less anxiety-related behavior and increased depression-like behavior [17]. Moreover, prolonged corticotrophin-releasing factor (CRF) over-expression at the central nucleus of the amygdala (CeA) attenuated stress-induced anxiety-like behaviors, whereas prolonged CRF over-expression at the dorsolateral subdivision of the bed nucleus of the stria terminalis (BNSTdl) increased depressive-like behaviors without affecting anxiety levels [18]. In addition, PACAP−/− mice showed increased depression-like behavior in FST accompanied with reduced anxiety [19]. Although the phenotype of mood regulation is opposite to what we found in our study, it provides evidence supporting that anxiety and depression may have different regulatory mechanisms. Nevertheless, the mechanisms by which Tafa-2 affects these behavioral phenotypes remain illusive.
Histologically, Tafa-2−/− mice showed neuronal loss and defects in dendritic and synaptic morphology in the brain, which might constitute the structural basis for the impaired learning and memory ability and the emotional regulation abnormality in Tafa-2−/− mice, even though no evidence of gross changes in brain organization or connectivity was found. The brain weight and the brain/body weight ratio were comparable between wt and Tafa-2−/− mice (data not shown). Furthermore, Tafa-2 deficiency caused dramatic neuronal apoptosis without affecting the survival of GFAP-positive astrocytes. One would expect that the reduced neuronal cell density should be associated with lower glucose uptake, which is also our presumption. However, PET-CT results showed increased basal glucose metabolism in the brain of Tafa-2−/− mice (Supplementary Fig. S3). Glucose is the main energy source for the adult brain and it is metabolized primarily via glycolysis. The brain’s functional connectivity is complex, has high energetic cost, and requires efficient use of glucose. It has been proposed that regions with a high degree of functional connectivity are energy efficient and can minimize consumption of glucose [20]. However, due to the reduced neuronal cells in the brain of Tafa-2−/− mice, the functional connectivity is impaired, which may result in increased glucose uptake. The higher energy demands in the brain of Tafa-2−/− mice could render it more vulnerable to deficits in energy delivery or utilization, and help explain their sensitivity to neurobiological disorders. However, further research is necessary to draw conclusions about the specificity and significance of the observed changes in brain glucose metabolism.
In the brain of Tafa-2−/− mice, we observed a remarkably reduction in the protein levels of PI3K (p85), p-Akt, and p-Erk1/2. PI3K/Akt and MAPK/Erk signaling pathways are well-recognized to be involved in the regulation of neuronal survival [21–32]. Downregulation of PI3K/Akt and MAPK/Erk signaling pathways induce pro-apoptotic members of Bcl-2 family, inhibit anti-apoptotic Bcl-2 proteins and activate caspase(s), consistent with the increased level of Bax, decreased level of Bcl-2 and higher activity of caspase-3 in the brain of Tafa-2−/− mice (Fig. 7D,E). The pro-apoptotic protein Bax and the anti-apoptotic protein Bcl-2 play crucial roles in regulating apoptotic cell death and the balance of them is very important in modulating cellular survival/death [33,34]. Caspase proteins, which function on the downstream of the Bcl-2 family during apoptosis, play key roles in execution of cellular breakdown [35]. Among the effector caspases, caspase-3 is the most frequently involved caspase protein in neuronal apoptosis. Thus, downregulation of these signaling pathways probably contributes to the neuronal apoptosis in the brain of Tafa-2−/− mice.
Moreover, PI3K/Akt and MAPK/Erk signaling pathways can activate the transcription factor, cAMP response-element binding protein (CREB), and the transcriptional coactivator, CREB-binding protein (CBP), both of which modulate CRE-dependent gene expressions. CREB is activated by a phosphorylation at Ser-133 [36] and disruption of its function in brain leads to neurodegeneration [37]. CBP is a transcriptional coactivator with histone acetyltransferase activity and is required for both short-term and long-term memory formation [38]. Ablation of Tafa-2 in mice significantly reduced the expressions of CREB-dependent genes and CBP in the brain. The expressions of CREB-dependent genes have been demonstrated to play essential roles in the consolidation of long-term forms of synaptic plasticity and memory, including BDNF, c-Fos, and NF1. BDNF is one of the neurotrophic factors that play important roles in neuronal survival and growth. It binds to its high-affinity receptor TrkB (tyrosine kinase B) and results in activation of different intracellular pathways, leading to neural plasticity, neurogenesis, stress resistance and cell survival [39]. Clinically, low BDNF is associated with cognitive impairment in chronic patients with schizophrenia [40]. c-Fos belongs to a bigger Fos family of transcription factors. Mice lacking c-Fos in the CNS have impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity [41]. And a recent report demonstrated a role of c-Fos in experience-dependent plasticity and learning [42]. NF1 is found within the mammalian postsynapse, where it is known to bind to NMDA receptor complex [43]. Mutations in the NF1 gene are associated with neurofibromatosis type I (also known as von Recklinghausen disease) and 30%-60% of these patients have cognitive deficits and learning disabilities [44–46]. Thus, the downregulation of PI3K/Akt and MAPK/Erk signaling pathways, the decreased expressions of CREB-dependent genes, especially BDNF, c-Fos, and NF1, and the lower level of CBP in the brain of Tafa-2−/− mice all contribute to the neuronal loss and the defects in dendritic and synaptic morphology, synergistically or independently, leading to impaired learning and memory ability and the emotional regulation abnormality in Tafa-2−/− mice.
Upon interpretation of our results, we have to note the following limitations: (1) Tafa-2 is one of the five members of the Tafa family, and the expression of Tafa mRNAs in different brain regions is distinct. The developmental knockout of Tafa-2 in mice may cause compensations of the other family members, which may also influence the phenotypes of Tafa-2−/− mice. (2) We observed defects not only in learning and memory but also in emotional regulations in Tafa-2−/− mice. We do not know whether the emotional dysregulation, such as anxiety and depression, aggravates the impairments in learning and memory, or vise versa. Since these neurobiological functions are controlled by different brain regions, the relationship between them and how they communicate to affect each other remain illusive. (3) As a member of the Tafa family which produces secreted polypeptides, Tafa-2 is probably functioning as a cytoprotective molecule and exerting its function through downstream signaling pathways by binding to its high-affinity receptor, which has not been identified yet. (4) The expression of Tafa-2 in astrocytes and its physiological significance for the function of astrocytes need to be further clarified.
In summary, our data demonstrated that Tafa-2 plays crucial roles in neuronal survival and neurobiological functions. Tafa-2 is predominantly expressed by NeuN-positive neurons and its disruption in mice causes a significant neuronal reduction and apoptosis, leading to disorders not only in learning and memory but also in emotional regulation. PI3K/Akt and MAPK/Erk signaling pathways are involved in the regulation of these physiological changes initiated by Tafa-2 deficiency. Given the high conservation of Tafa-2 protein, it might be a potential drug target for the treatment of neurobiological disorders.
Supplementary Material
Funding
This work was supported by the grants from National Natural Science Foundation of China (No. 81430028 to Z.W.), and Science and Technology Commission of Shanghai Municipality (No. 16DZ2280800 to Z.W., and No. 18ZR1423500 to C.S.).
References
- 1. Tom Tang Y, Emtage P, Funk WD, Hu T, Arterburn M, Park EE, Rupp F. TAFA: a novel secreted family with conserved cysteine residues and restricted expression in the brain. Genomics 2004, 83: 727–734. [DOI] [PubMed] [Google Scholar]
- 2. Fischer C, Christ E, Korf HW, von Gall C. Tafa-3 encoding for a secretory peptide is expressed in the mouse pars tuberalis and is affected by melatonin 1 receptor deficiency. Gen Comp Endocrinol 2012, 177: 98–103. [DOI] [PubMed] [Google Scholar]
- 3. Borg K, Stankiewicz P, Bocian E, Kruczek A, Obersztyn E, Lupski JR, Mazurczak T. Molecular analysis of a constitutional complex genome rearrangement with 11 breakpoints involving chromosomes 3, 11, 12, and 21 and a approximately 0.5-Mb submicroscopic deletion in a patient with mild mental retardation. Hum Genet 2005, 118: 267–275. [DOI] [PubMed] [Google Scholar]
- 4. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 5. Zhang LL, Wang JJ, Liu Y, Lu XB, Kuang Y, Wan YH, Chen Y, et al. GPR26-deficient mice display increased anxiety- and depression-like behaviors accompanied by reduced phosphorylated cyclic AMP responsive element-binding protein level in central amygdala. Neuroscience 2011, 196: 203–214. [DOI] [PubMed] [Google Scholar]
- 6. Balu DT, Carlson GC, Talbot K, Kazi H, Hill-Smith TE, Easton RM, Birnbaum MJ, et al. Akt1 deficiency in schizophrenia and impairment of hippocampal plasticity and function. Hippocampus 2012, 22: 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakai T, Nagai T, Tanaka M, Itoh N, Asai N, Enomoto A, Asai M, et al. Girdin phosphorylation is crucial for synaptic plasticity and memory: a potential role in the interaction of BDNF/TrkB/Akt signaling with NMDA receptor. J Neurosci 2014, 34: 14995–15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tang YP, Shimizu E, Dube GR, Rampon C, Kerchner GA, Zhuo M, Liu G, et al. Genetic enhancement of learning and memory in mice. Nature 1999, 401: 63–69. [DOI] [PubMed] [Google Scholar]
- 9. Nagai T, Takuma K, Kamei H, Ito Y, Nakamichi N, Ibi D, Nakanishi Y, et al. Dopamine D1 receptors regulate protein synthesis-dependent long-term recognition memory via extracellular signal-regulated kinase 1/2 in the prefrontal cortex. Learn Mem 2007, 14: 117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen G, Chen KS, Knox J, Inglis J, Bernard A, Martin SJ, Justice A, et al. A learning deficit related to age and beta-amyloid plaques in a mouse model of Alzheimer’s disease. Nature 2000, 408: 975–979. [DOI] [PubMed] [Google Scholar]
- 11. Bilkei-Gorzo A, Racz I, Michel K, Zimmer A. Diminished anxiety- and depression-related behaviors in mice with selective deletion of the Tac1 gene. J Neurosci 2002, 22: 10046–10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet 1998, 20: 123–128. [DOI] [PubMed] [Google Scholar]
- 13. Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 2003, 13: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bohnen NI, Djang DS, Herholz K, Anzai Y, Minoshima S. Effectiveness and safety of 18F-FDG PET in the evaluation of dementia: a review of the recent literature. J Nucl Med 2012, 53: 59–71. [DOI] [PubMed] [Google Scholar]
- 15. Olah J, Klivenyi P, Gardian G, Vecsei L, Orosz F, Kovacs GG, Westerhoff HV, et al. Increased glucose metabolism and ATP level in brain tissue of Huntington’s disease transgenic mice. FEBS J 2008, 275: 4740–4755. [DOI] [PubMed] [Google Scholar]
- 16. Detka J, Kurek A, Kucharczyk M, Glombik K, Basta-Kaim A, Kubera M, Lason W, et al. Brain glucose metabolism in an animal model of depression. Neuroscience 2015, 295: 198–208. [DOI] [PubMed] [Google Scholar]
- 17. Mukherjee S, Coque L, Cao JL, Kumar J, Chakravarty S, Asaithamby A, Graham A, et al. Knockdown of Clock in the ventral tegmental area through RNA interference results in a mixed state of mania and depression-like behavior. Biol Psychiatry 2010, 68: 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Regev L, Neufeld-Cohen A, Tsoory M, Kuperman Y, Getselter D, Gil S, Chen A. Prolonged and site-specific over-expression of corticotropin-releasing factor reveals differential roles for extended amygdala nuclei in emotional regulation. Mol Psychiatry 2011, 16: 714–728. [DOI] [PubMed] [Google Scholar]
- 19. Gaszner B, Kormos V, Kozicz T, Hashimoto H, Reglodi D, Helyes Z. The behavioral phenotype of pituitary adenylate-cyclase activating polypeptide-deficient mice in anxiety and depression tests is accompanied by blunted c-Fos expression in the bed nucleus of the stria terminalis, central projecting Edinger-Westphal nucleus, ventral lateral septum, and dorsal raphe nucleus. Neuroscience 2012, 202: 283–299. [DOI] [PubMed] [Google Scholar]
- 20. Tomasi D, Wang GJ, Volkow ND. Energetic cost of brain functional connectivity. Proc Natl Acad Sci USA 2013, 110: 13642–13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yao R, Cooper GM. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science 1995, 267: 2003–2006. [DOI] [PubMed] [Google Scholar]
- 22. Crowder RJ, Freeman RS. Phosphatidylinositol 3-kinase and Akt protein kinase are necessary and sufficient for the survival of nerve growth factor-dependent sympathetic neurons. J Neurosci 1998, 18: 2933–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96: 857–868. [DOI] [PubMed] [Google Scholar]
- 24. Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 1999, 286: 1358–1362. [DOI] [PubMed] [Google Scholar]
- 25. Xu Y, Zhang Q, Yu S, Yang Y, Ding F. The protective effects of chitooligosaccharides against glucose deprivation-induced cell apoptosis in cultured cortical neurons through activation of PI3K/Akt and MEK/ERK1/2 pathways. Brain Res 2011, 1375: 49–58. [DOI] [PubMed] [Google Scholar]
- 26. Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, et al. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 1997, 275: 661–665. [DOI] [PubMed] [Google Scholar]
- 27. Brunet A, Datta SR, Greenberg ME. Transcription-dependent. and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr Opin Neurobiol 2001, 11: 297–305. [DOI] [PubMed] [Google Scholar]
- 28. Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell 2007, 129: 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barnabe-Heider F, Miller FD. Endogenously produced neurotrophins regulate survival and differentiation of cortical progenitors via distinct signaling pathways. J Neurosci 2003, 23: 5149–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaillant AR, Mazzoni I, Tudan C, Boudreau M, Kaplan DR, Miller FD. Depolarization and neurotrophins converge on the phosphatidylinositol 3-kinase-Akt pathway to synergistically regulate neuronal survival. J Cell Biol 1999, 146: 955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma Y, Lu C, Li C, Li R, Zhang Y, Ma H, Zhang X, et al. Overexpression of HSPA12B protects against cerebral ischemia/reperfusion injury via a PI3K/Akt-dependent mechanism. Biochim Biophys Acta 2013, 1832: 57–66. [DOI] [PubMed] [Google Scholar]
- 32. Chen L, Wei X, Hou Y, Liu X, Li S, Sun B, Liu H. Tetramethylpyrazine analogue CXC195 protects against cerebral ischemia/reperfusion-induced apoptosis through PI3K/Akt/GSK3beta pathway in rats. Neurochem Int 2014, 66: 27–32. [DOI] [PubMed] [Google Scholar]
- 33. Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 2007, 26: 1324–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kitagawa K, Matsumoto M, Tsujimoto Y, Ohtsuki T, Kuwabara K, Matsushita K, Yang G, et al. Amelioration of hippocampal neuronal damage after global ischemia by neuronal overexpression of BCL-2 in transgenic mice. Stroke 1998, 29: 2616–2621. [DOI] [PubMed] [Google Scholar]
- 35. Salakou S, Kardamakis D, Tsamandas AC, Zolota V, Apostolakis E, Tzelepi V, Papathanasopoulos P, et al. Increased Bax/Bcl-2 ratio up-regulates caspase-3 and increases apoptosis in the thymus of patients with myasthenia gravis. In Vivo 2007, 21: 123–132. [PubMed] [Google Scholar]
- 36. Gonzalez GA, Yamamoto KK, Fischer WH, Karr D, Menzel P, Biggs W 3rd, Vale WW, et al. A cluster of phosphorylation sites on the cyclic AMP-regulated nuclear factor CREB predicted by its sequence. Nature 1989, 337: 749–752. [DOI] [PubMed] [Google Scholar]
- 37. Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, et al. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet 2002, 31: 47–54. [DOI] [PubMed] [Google Scholar]
- 38. Chen G, Zou X, Watanabe H, van Deursen JM, Shen J. CREB binding protein is required for both short-term and long-term memory formation. J Neurosci 2010, 30: 13066–13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bathina S, Das UN. Brain-derived neurotrophic factor and its clinical implications. Arch Med Sci 2015, 11: 1164–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang XY, Liang J, Chen DC, Xiu MH, Yang FD, Kosten TA, Kosten TR. Low BDNF is associated with cognitive impairment in chronic patients with schizophrenia. Psychopharmacology (Berl) 2012, 222: 277–284. [DOI] [PubMed] [Google Scholar]
- 41. Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, Layer LE, Kvello A, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci 2003, 23: 9116–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Hoz L, Gierej D, Lioudyno V, Jaworski J, Blazejczyk M, Cruces-Solis H, Beroun A, et al. Blocking c-Fos expression reveals the role of auditory cortex plasticity in sound frequency discrimination learning. Cereb Cortex 2018, 28: 1645–1655. [DOI] [PubMed] [Google Scholar]
- 43. Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci 2000, 3: 661–669. [DOI] [PubMed] [Google Scholar]
- 44. North KN, Riccardi V, Samango-Sprouse C, Ferner R, Moore B, Legius E, Ratner N, et al. Cognitive function and academic performance in neurofibromatosis. 1: consensus statement from the NF1 cognitive disorders task force. Neurology 1997, 48: 1121–1127. [DOI] [PubMed] [Google Scholar]
- 45. Rasmussen SA, Friedman JM. NF1 gene and neurofibromatosis 1. Am J Epidemiol 2000, 151: 33–40. [DOI] [PubMed] [Google Scholar]
- 46. Trovo-Marqui AB, Tajara EH. Neurofibromin: a general outlook. Clin Genet 2006, 70: 1–13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.