Abstract
Background
Novel therapeutics for inflammatory bowel disease (IBD) are under development, yet mechanistic readouts at the tissue level are lacking. Techniques to assess intestinal immune composition could represent a valuable tool for mechanism of action (MOA) studies of novel drugs. Mass cytometry enables analysis of intestinal inflammatory cell infiltrate and corresponding molecular fingerprints with unprecedented resolution. Here, we aimed to optimize the methodology for isolation and cryopreservation of cells from intestinal tissue to allow for the potential implementation of mass cytometry in MOA studies.
Methods
We investigated key technical issues, including minimal tissue requirements, cell isolation protocols, and cell storage, using intestinal biopsies and peripheral blood from healthy individuals. High-dimensional mass cytometry was employed for the analyses of biopsy-derived intestinal cellular subsets.
Results
Dithiothreitol and mechanical dissociation decreased epithelial cell contamination and allowed for isolation of adequate cell numbers from 2 to 4 colonic or ileal biopsies (6 × 104±2 × 104) after a 20-minute collagenase digestion, allowing for reliable detection of most major immune cell subsets. Biopsies and antibody-labeled mononuclear cells could be cryopreserved for later processing and acquisition (viability > 70%; P < 0.05).
Conclusions
Mass cytometry represents a unique tool for deep immunophenotyping intestinal cell composition. This technique has the potential to facilitate analysis of drug actions at the target tissue by identifying specific cellular subsets and their molecular signatures. Its widespread implementation may impact not only IBD research but also other gastrointestinal conditions where inflammatory cells play a role in pathogenesis.
Keywords: CyTOF, lamina propria, randomized control trials, Crohn’s disease, ulcerative colitis
INTRODUCTION
Inflammatory bowel disease (IBD) affect more than 1 million people in North America, and with an increasing worldwide incidence, it may soon become a global emergent disease.1 In response, a rapidly expanding range of therapeutics have been approved, or are currently under development, including several that specifically target the traffic of leukocytes (eg, vedolizumab, etrolizumab, ozanimod)2, 3. However, biologically based mechanistic readouts of treatment efficacy are limited. Currently, clinical trials rely mostly on subjective clinical end points, such as the Crohn’s Disease Activity Index (CDAI) or patient-reported outcomes.4, 5 These end points have undergone revision in recent years, incorporating objective evaluation of intestinal inflammation by endoscopy and histology to better understand the response to new therapies.6 Yet, objective mechanistic readouts to complement clinical parameters could be a valuable adjunct for both early proof-of-concept and late-stage clinical trials. In addition, a major limitation in the conduct of IBD trials is the lack of quantitative tools by which a drug’s efficacy can be assessed directly within the intestine, similar to measuring viral load for HIV and hepatitis C trials. Indeed, a number of recent IBD trials have failed (such as tofacitinib7, 8 and anti-MAdCAM-19 for Crohn’s disease [CD]), without a clear indication of why, or whether the drug achieved its intended effect in the target intestinal tissue. A technique by which the overall intestinal immune cell composition and its molecular fingerprint could be assessed would likely represent a significant advance, integrating progress in basic and clinical science in a comprehensive fashion.
High-dimensional mass cytometry (cytometry by time of flight [CyTOF]) combines flow cytometry with elemental mass spectrometry, allowing simultaneous quantification of up to 40 surface and/or intracellular markers on a single-cell basis.10–12 This emerging technology, together with computational tools capable of handling complex high-dimensional single-cell data sets,13–17 enables analyses of the inflammatory cell infiltrates and their molecular fingerprint in inflamed tissues with unprecedented resolution. The technique is particularly powerful for samples where cell numbers are limited,18 as up to 40 extracellular or intracellular markers can be examined simultaneously, instead of spread across multiple flow cytometry antibody panels.19 Whereas CyTOF has been utilized for analysis of cell subsets derived from peripheral blood, spleen, skin, liver, lung, and intestine,20–23 few studies have focused on the technical challenges and limitations of analyses of the gastrointestinal tract. One recent report utilized mass cytometry to examine the changes in immune cell composition occurring in duodenal and rectal lamina propria in patients with celiac disease and CD24 but did not explore a wider range of technical parameters such as yield and reproducibility.
To expand the use of mass cytometry to clinical IBD settings, whether for large-scale multicenter research studies involving human subjects or proof-of-concept mechanistic trials of new therapeutics, several logistical challenges exist. These include but are not limited to small cellular yields recoverable from limited numbers of intestinal pinch biopsies and whether these yields are sufficient for identification of cell populations; the length and complexity of cell isolation protocols; the limited access of clinical centers to wet laboratory facilities capable of on-site sample processing; and the storage of biopsy and cell samples for transport to adequately equipped wet laboratories, allowing for live cell isolation and centralized data acquisition by mass cytometry. We conceived this study as a pilot effort to assess mass cytometry as a potential tool for examining the cellular determinants and molecular fingerprints that are central to the pathogenesis of IBD, its potential role for assessing a drug’s mechanism of action, and its feasibility for incorporation into clinical trial protocols.
METHODS
Human Samples
Adult patients (age 48–69 years) without history of gastrointestinal disease undergoing ileocolonoscopy for evaluation of abdominal symptoms or colorectal cancer screening were recruited after written informed consent, as per San Diego Veterans Administration Medical Center (SDVAMC) and University of California, San Diego (UCSD) Institutional Review Board approved protocol numbers H130266 and 161756. Intestinal biopsies from “healthy” patients with normal colonoscopy exams were obtained from the terminal ileum and colon.
Isolation of Cells From Intestinal and Peripheral Blood Samples
Intestinal biopsies were collected using standard (2.8-mm) intestinal biopsy forceps into 50-mL conical tubes containing 15 mL Hank’s Balanced Salt Solution (HBSS; ThermoFisher Scientific, San Diego, CA, USA). All tissues were either processed or frozen within 2 hours of harvest. Biopsies were washed for 10 minutes in HBSS, transferred to 10 mL HBSS containing 5 mM dithiothreitol (DTT; Sigma Aldrich, Carlsbad, CA, USA), and incubated on a rocker for an additional 15 minutes at room temperature. Subsequently, a second HBSS/DTT wash was performed for 15 minutes, before a final wash in HBSS for 5 minutes. Where indicated, 1 mM EDTA (Hoefer, Holliston, MA, USA) was used alone or in combination with DTT. Biopsies were then transferred to weighing boats containing complete Roswell Park Memorial Institute (RPMI; RPMI supplemented with 10% fetal bovine serum [FBS], 50 mg/mL penicillin and streptomycin, and 2 mM sodium pyruvate [all ThermoFisher Scientific, San Diego, CA, USA]) and finely minced using surgical scissors. Minced tissue was next incubated with a digestion solution comprised of complete RPMI containing 1.5 mg/mL collagenase VIII (Sigma Aldrich, Carlsbad, CA, USA) and 50 µg/mL DNase I (Roche, Pleasanton, CA, USA) and incubated for 20 minutes at 37°C on a rocker at 200 RPM. Following digestion, the remaining tissue debris was removed by passing through a cell strainer. Cells were washed with HBSS and incubated with antibodies.
Peripheral blood mononuclear cells (PBMCs) were isolated from 5 mL of heparinized venous blood using Lymphoprep solution (Axis-Shield, Norton, MA, USA) and density-gradient centrifugation.
Tissue Preservation
Following isolation, PBMCs were placed in cryovials containing 1 mL freezing buffer (40% complete RPMI, 50% FBS, 10% dimethyl sulfoxide [DMSO]; Sigma Aldrich, Carlsbad, CA, USA) in a freezing container (Mr Frosty, ThermoFisher Scientific, San Diego, CA, USA) and placed at –80°C. Biopsies were preserved using 1 of 3 methods: (1) transfer to 10 mL complete RPMI and storage overnight at 4°C; (2) transfer to cryovials containing freezing buffer and slow-freezing in a Mr Frosty freezing container (as with PBMC); or (3) snap-freezing by incubation on dry ice for 5 minutes. All frozen samples were stored at –80°C for up to 2 months. Mr Frosty slow-freezing containers allow samples to be cooled at an approximate rate of 1°C per minute. Samples were recovered by placing cryovials into a 37°C water bath until thawed, then processed as with freshly harvested biopsies.
Antibody Staining and Poststaining Cryopreservation
Antibody staining was carried out as previously described.10 In brief, isolated cells were resuspended in Maxpar cell staining buffer (CSB; Fluidigm, San Francisco, CA, USA) and subsequently stained as follows: (1) viability staining using Cisplatin-195Pt (Fluidigm) for 5 minutes; (2) Fc receptor blockade using Human TruStain FcX (BioLegend, San Diego, CA, USA) for 10 minutes; (3) surface antibody staining (Fluidigm; BioLegend, San Diego, CA, USA) (a full antibody list is available in Supplementary Table 1) for 30 minutes; (4) fixation using Maxpar Fix I buffer (Fluidigm) overnight; (5) permeabilization using Maxpar Permeabilization buffer (Fluidigm) for 45 minutes; and (6) DNA-Intercalator labeling (Fluidigm) using a cationic nucleic acid intercalator containing 191Ir and 193Ir for detection of nucleated cells (20-minute incubation). Where indicated, purified antibodies were conjugated with metal isotopes in-house using antibody labeling kits (Fluidigm). Stained cells were kept in CSB for up to 1 week before acquisition on the mass cytometer, or alternatively cryopreserved for later acquisition. Stained samples were frozen in 90% FBS, 10% DMSO for up to 2 months.25
Sample Acquisition and Data Processing
Before acquisition, cells were washed twice with Milli-Q-purified water and resuspended in a 1:10 dilution of EQ Four Element Calibration Beads (Fluidigm) to a concentration of 0.5 × 106 cells/mL. Samples were acquired using a CyTOF Helios (Fluidigm) with super sampler addition (Scientific Apparatus LLC, Alamo, CA, USA) according to the manufacturer’s directions. Data were normalized to mass bead signal using Matlab software, as described previously.26
Data Analysis
Mass cytometry data were uploaded to Cytobank (Cytobank Inc, Santa Clara, CA, USA) for analyses. Before analysis, mass cytometry data were gated on nucleated, live CD45+ events, then gated on the populations of interest, followed by biaxial gating and t-distributed stochastic neighbor embedding (t-SNE, vi-SNE) analysis.16 Summary graphs were produced using GraphPad Prism, version 7, software (GraphPad Software Inc, La Jolla, CA, USA). t-SNE analysis was conducted using 100,000 total events proportionally drawn from samples, with 1000 iterations and a perplexity value of 30. t-SNE plot lineage overlays were produced using Inkscape software. For automated cluster identification, the R statistical programming environment was used, with the Bioconductor package Cytofkit27 loaded for t-SNE mapping, followed by automated cluster identification (ClusterX)27 for identification and characterization of immune cell subsets, using 25,000 events per sample. t-SNE analysis and ClusterX were performed using the markers shown in Supplementary Table 1. ClusterX subset data were exported, and heatmaps were generated using Heatmapper online software.28
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 6 software (GraphPad Software Inc). Column statistics tests were used to assess parametric distribution of data sets. For comparison of 2 groups, the Wilcoxon matched-pairs signed rank test was utilized, or the Mann-Whitney test for unpaired samples. For comparison of multiple groups, the Kruskal-Wallis test was used, followed by Dunn’s multiple comparison test. Descriptive statistics are displayed as mean ± standard deviation in all figures.
RESULTS
Rapid Leukocyte Isolation From Intestinal Pinch Biopsies
A major logistical challenge for the implementation of mass cytometry for mechanism of action studies in IBD is the low cell numbers that may be isolated from a limited number of intestinal biopsies, obtainable from patients undergoing ileocolonoscopy. To maximize recovery of live immune cells, we reviewed key elements of several published cell isolation protocols,29–31 with the goals of shortening isolation time, maximizing viable cell yields, and obtaining a valid and reproducible representation of intestinal lamina propria cell composition reflecting that of the native ileum and colon.
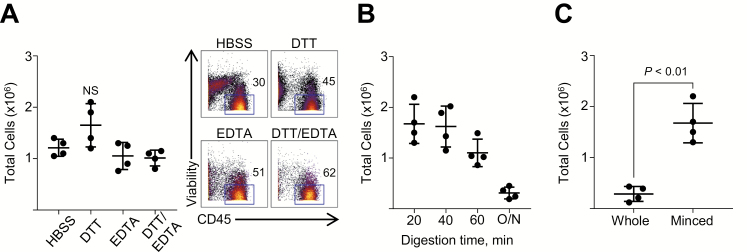
In the initial stage of cell isolation from intestinal biopsies, mucus and epithelial cells are typically removed, which facilitates subsequent tissue digestion and reduces clogging of the cytometer’s fluidic system. DTT and EDTA have been previously employed and were examined alone or in combination (Fig. 1A). Treatment of biopsies with DTT alone resulted in the highest total cell yield when compared with incubation with HBSS alone, EDTA alone, or a combination of DTT and EDTA. Furthermore, despite more efficient removal of CD45neg cells (such as epithelial cells and fibroblasts) with the combination of DTT and EDTA, treatment with DTT alone also resulted in the highest yield of CD45-expressing leukocytes.
FIGURE 1.
Optimization of tissue digestion protocols for isolation of viable immune cells from intestinal biopsies. A, Epithelial cells were removed from biopsies before digestion by washing with either DTT, EDTA, or a combination, compared with HBSS alone. Both total cell counts and mass cytometry plots displaying viability and CD45 expression are shown. B, Total live cell yields derived from biopsies after 20, 40, and 60 minutes or overnight (O/N), digestion at 37°C with Collagenase VIII and DNase I. C, Live cell yields derived from biopsies digested with or without prior mincing. Data are shown as means ± SD of 4 individual donors, and significance was determined using the Kruskal-Wallis test followed by Dunn’s multiple comparisons test (A), comparing data with HBSS, or the Wilcoxon test (B, C).
Biopsies were subsequently digested with a mixture of Collagenase and DNase, the combination of which has been widely utilized for human intestinal tissue digestion.29 Testing digestion times revealed that a short 20-minute incubation was sufficient, compared with longer incubation times (Fig. 1B). Increasing digestion times beyond 20 minutes diminished the total viable cell yield. The optimal concentration of Collagenase was determined to be 1.5 mg/mL, a high concentration that aids with digesting biopsy tissue in a short time period (Supplementary Fig. 1A). In addition, we tested the effect of mechanical dissociation before digestion.32 We found that this step had the greatest impact on cell yield (Fig. 1C). Lastly, placing biopsy tissue to digest while on a rocker at 200 RPM led to a slightly increased cell yield over samples that were stationary during digestion (Supplementary Fig. 1B). In summary, DTT washes followed by a 20-minute Collagenase/DNase digestion of minced colonic pinch biopsies resulted in the highest cell yield.
Required Number of Biopsies for Adequate Detection of Major Immune Cell Subsets
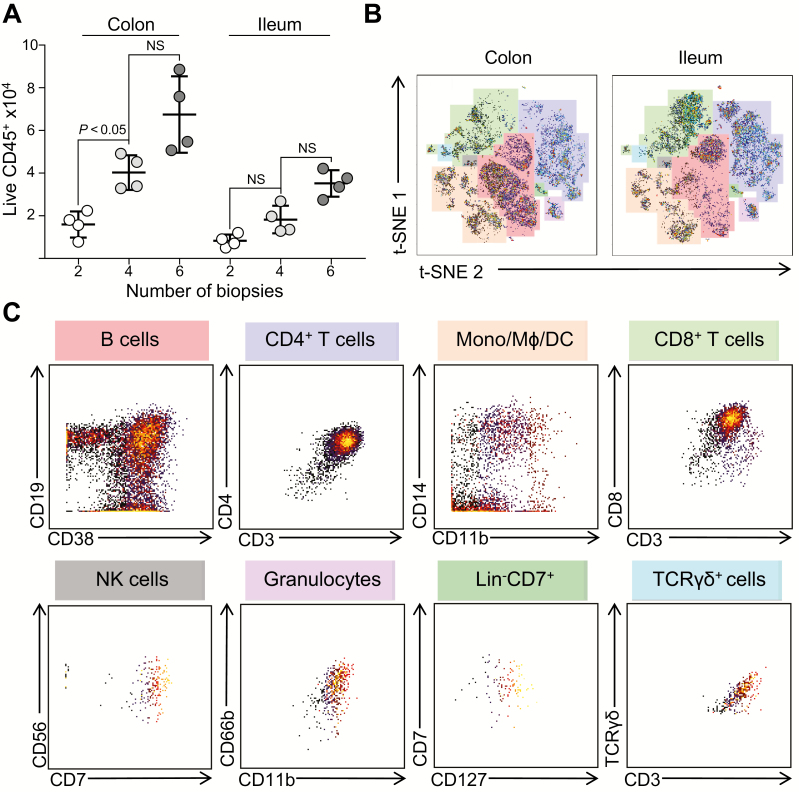
Tissue collection represents a logistical challenge during clinical studies, as it prolongs procedure duration. Thus, it was critical to assess the minimal number of biopsies necessary for adequate analysis of cell composition by mass cytometry. To this end, we compared the number of cells of all major immune subsets among live CD45-expressing cells using different numbers of colonic or ileal biopsies (Table 1, Fig. 2A). As expected, more biopsies led to isolation of greater cell numbers. Terminal ileal biopsies yielded approximately half of the live CD45positive events, compared with colonic biopsies. To determine whether the acquired events were sufficient for robust data analysis and visualization, t-SNE analyses were conducted on CD45positive events acquired from 4 colonic or ileal biopsies (Fig. 2B). t-SNE analysis is a dimensionality reduction analytical tool that allows visualization of multiparameter data sets on a single-cell basis,14, 15 generating 2-dimensional plots of single-cell data and grouping of cells with similar phenotypes, based on the overall expression of each marker. By analyzing cellular events recovered from either 4 colonic or ileal biopsies, the 8 major immune populations described in Table 1 were visualized as distinct islands on the t-SNE map, indicating effective identification of all major mononuclear cell subsets in the lamina propria. The identities of these 8 major population islands can be demonstrated by biaxial plots (Fig. 2C). These findings indicate that as few as 2 colonic or 4 ileal biopsies are sufficient for identifying the major immune cell subsets by mass cytometry. However, events derived from 2 ileal biopsies prevented analysis of low abundance subsets, such as TCRγδ+ cells, due to limited cell numbers.
Table 1.
Yields of Cells of Each Major Lineage Obtained From Differing Numbers of Colonic and Ileal Pinch Biopsies
| Lineage | Colonic Biopsies, No. | Ileal Biopsies, No. | ||||
|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 2 | 4 | 6 | |
| M +/- SD | M +/- SD | M +/- SD | M +/- SD | M +/- SD | M +/- SD | |
| CD45+ | 15,879 +/- 6237 | 40,273 +/- 8116 | 67,451 +/- 17,906 | 10,019 +/- 4321 | 18,224 +/- 6473 | 35,190 +/- 6221 |
| B cell | 5698 +/- 1189 | 14,246 +/- 2973 | 24,376 +/- 5087 | 1703 +/- 543 | 3157 +/- 658 | 6040 +/- 1260 |
| CD4+ T cell | 3566 +/- 744 | 8976 +/- 1873 | 15,210 +/- 3174 | 3005 +/- 806 | 5866 +/- 1224 | 11,605 +/- 2422 |
| Mono/Mϕ/DC | 2335 +/- 487 | 5574 +/- 1163 | 9757 +/- 2036 | 2204 +/- 352 | 2447 +/- 510 | 4676 +/- 976 |
| CD8+ T cell | 1750 +/- 365 | 4159 +/- 868 | 7320 +/- 1528 | 2604 +/- 753 | 4791 +/- 1000 | 9033 +/- 1885 |
| NK cell | 444 +/- 92 | 1163 +/- 242 | 2030 +/- 424 | 102 +/- 54 | 194 +/- 41 | 337 +/- 70 |
| Granulocyte | 406 +/- 84 | 993 +/- 207 | 1765 +/- 368 | 278 +/- 98 | 460 +/- 96 | 864 +/- 180 |
| Lin-CD7+ | 350 +/- 73 | 798 +/- 166 | 1412 +/- 295 | 148 +/- 44 | 322 +/- 67 | 538 +/- 112 |
| TCR γδ+ | 199 +/- 41 | 409 +/- 85 | 930 +/- 194 | 5 +/- 2 | 27 +/- 6 | 39 +/- 8 |
Live CD45positive events derived from either 2, 4, or 6 colonic or ileal biopsies, identified by gating on nucleated cells, followed by viability staining and CD45 expression. Lineages were identified as clusters using t-SNE analysis of live CD45positive events, and phenotype was confirmed using major immune markers for each subset. Numbers display means ± SD of 4 individual donors.
FIGURE 2.
Four to 6 colonic and ileal biopsies are sufficient for robust analyses of immune cell subsets using mass cytometry. A, Yields of live CD45positive events, derived from the indicated number of colon and ileal biopsies, as acquired by CyTOF. Live CD45positive events were identified by gating on nucleated cells, followed by viability staining and CD45 expression. Data points represent paired individual donors, shown as mean ± SD. B, t-SNE analysis of live CD45positive events derived from 4 colonic (left) or ileal (right) biopsies. Color overlays highlight each immune lineage. t-SNE plots display data representative of 4 individual donors. C, Biaxial plots displaying the phenotype of each major population identified by t-SNE analysis in (B). Plots are representative of 4 individual donors.
Preservation and Storage of Intestinal Biopsies for Later Mass Cytometry Analyses
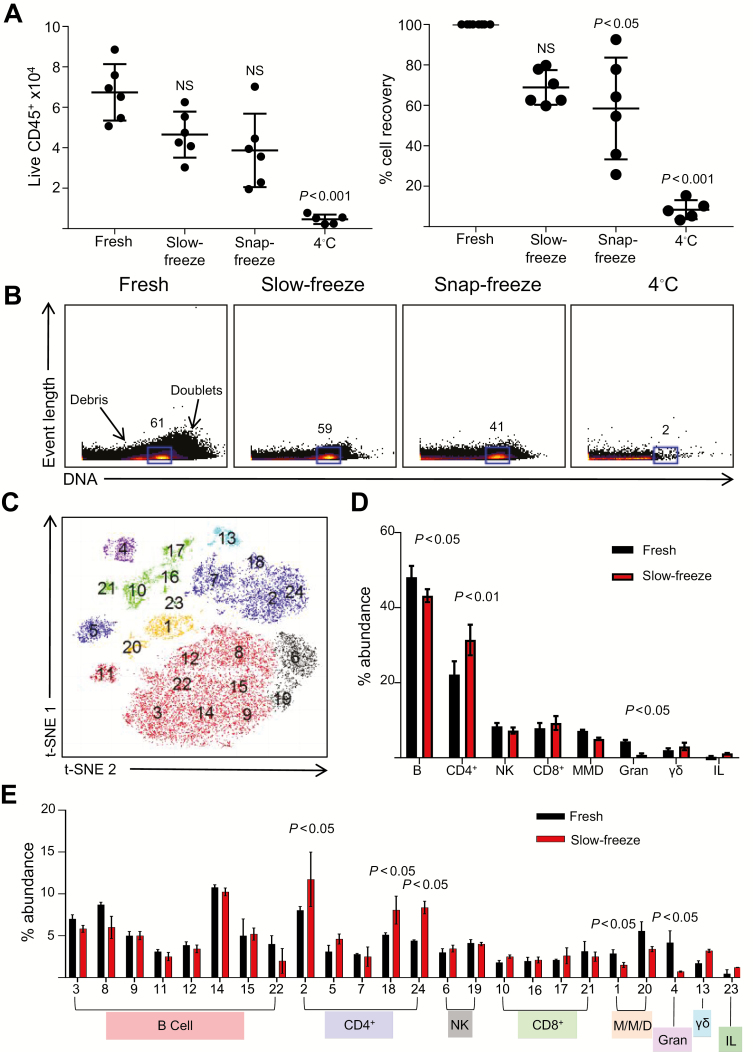
The use of intestinal biopsies for mechanism of action studies and translational research has been limited by factors such as processing time, variability of local technical expertise, and infrastructure requirements, particularly when compared with the relative ease of cell isolation from peripheral blood. These logistical challenges are most significant for study sites without immediate access to wet laboratory facilities. Inclusion of such sites demands easy processing and transport of samples to wet laboratories with the necessary infrastructure. Thus, we assessed the effects of different tissue preservation protocols on cell viability and cellular phenotypes of biopsy-derived cells. Data were obtained from intestinal biopsies processed promptly after harvest, or following slow-freezing, snap-freezing, or overnight storage of intact biopsies at 4°C. Total cell recovery, and live CD45positive events acquired, decreased by 30%–90% with the different preservation methods (Fig. 3A). Slow-freezing of intact biopsies provided the most effective cryopreservation, allowing for approximately 70% live cell recovery, compared with freshly processed biopsies. In contrast, snap-freezing led to ~50% recovery and large variations. Overnight storage at 4°C resulted in loss of >90% of cells. Identification of nucleated cells based on event length and DNA intercalator labeling revealed similar percentages between freshly processed and slow-frozen biopsies, whereas snap-frozen biopsies showed further cell decrease, combined with increased cellular debris and nearly total cell loss after 4°C storage (Fig. 3B). Similar data were also obtained using terminal ileal biopsies (Supplementary Fig. 2).
FIGURE 3.
Slow-freezing allows for long-term storage of intestinal biopsies and provides robust recovery of viable immune cells. Six colonic biopsies were either processed immediately after harvest or after storage by slow-freezing, snap-freezing, or overnight storage at 4°C. A, Total CD45positive events acquired and viable cell recovery after storage under indicated conditions (mean ± SD from 5–6 donors, analyzed using the Kruskal-Wallis test followed by Dunn’s multiple comparisons test). Percentage cell recovery was calculated as yield derived from frozen biopsies as a proportion of cells derived from fresh biopsies, in paired samples. B, Representative event length vs DNA stains of cells under the indicated conditions. Gates display percentages of nucleated cells. Labels indicate doublets (high DNA staining) or cellular fragments/debris (low DNA staining). C, t-SNE map of live CD45positive events derived from fresh and slow-frozen biopsies. Numbers display individual subsets identified by ClusterX analysis. Colors of clusters represent lineages, as displayed in (D). D, Percentage abundance of the 8 major lineages identified previously, derived from the phenotype of subsets identified in (C). E, Percentage abundance of the individual subsets identified in (C)/(D), derived from either fresh or slow-frozen biopsies.
Next, we examined the effects of slow-freezing on the recovery of LPMC subsets, compared with freshly processed biopsies, and whether any populations were lost due to the storage protocol. The ClusterX automated cluster identification tool was utilized for this purpose.27 This analytical technique produces a t-SNE map of the data and automatically identifies subpopulations within the data set. Twenty-four individual subsets were identified within the CD45positive cells from fresh and slow-frozen biopsy samples (Fig. 3C), using markers shown in Supplementary Table 1. For simplicity, the phenotypes of the 24 subsets were identified and grouped into the major immune cell lineages (Fig. 3D). The subsets whose abundance was negatively affected by slow-freezing included primarily CD66b+ granulocytes, followed by monocytes/macrophages/dendritic cells (DCs; CD11b+/CD11c+/CD14+), CD19+/CD38+ B cells, and CD56+ natural killer (NK) cells. Subsequently, as a proportion of total events, CD4+ T cells, CD8+ T cells, and innate lymphocytes (Lin-CD7+) were overrepresented after slow-freezing, compared with fresh samples.
Despite significant changes in the abundance of a several lineages, all 24 subsets identified by ClusterX were present in both freshly isolated samples and slow-frozen colonic samples (Fig. 3E). For a large proportion of the 24 subsets identified, fresh and frozen biopsies displayed a similar representation of subsets in terms of percentage abundance. However, several specific subsets showed significant increases (2, 18, and 24) or decreases (1 and 4), as a proportion of the total cells isolated. The increased abundance of these CD4+ T-cell subsets occurred alongside decreases in monocytic and granulocyte subsets, as previously described. Slow-freezing of PBMCs is widely utilized for preservation of samples33; thus a similar analysis was conducted on PBMCs, acquired fresh after isolation or following slow-freezing (Supplementary Fig. 3). It was encouraging that the effects observed in PBMCs were similar to those of colonic samples, with changes in abundance of all subsets after freezing, as compared with matched fresh samples. In addition, several alternative protocols, including different freezing buffers, and alternative thawing protocols did not appear to affect cell yields or viability of slow-frozen biopsies (Supplementary Fig. 4), indicating that the protocol presented here allowed for robust cell recovery. In addition, the mechanical dissociation of biopsies before freezing had a small but significant negative effect on cell yield, in contrast to the beneficial effect of biopsy mincing immediately before digestion. Overall, in terms of viable cellular recovery and composition, slow-freezing of intact biopsies represents an adequate method of preserving biopsy samples for centralized cell isolation, staining, and mass cytometry.
Cryopreservation of Stained Samples for Delayed Mass Cytometry Acquisition
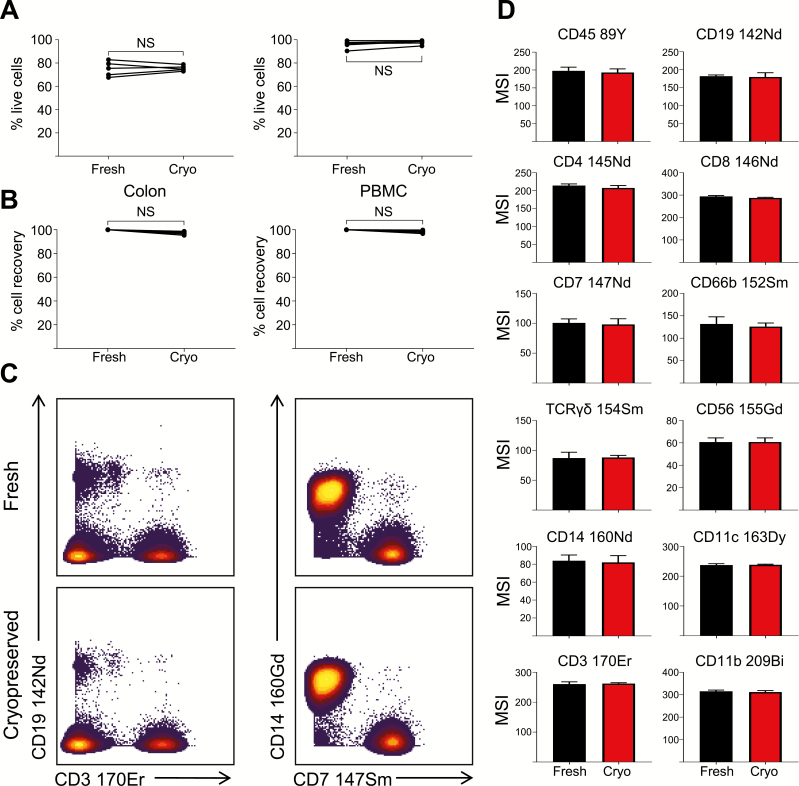
Collection of tissue samples during clinical trials can often be unpredictable and irregular. The ability to store fully prepared samples for later acquisition could be beneficial for data reproducibility and would provide additional flexibility for centralized sample analysis. Recently, a protocol for the preservation of labeled PBMCs for later acquisition by mass cytometry was described.25 Given the flexibility that this protocol would provide, we assessed the effects of storing colonic cells and PBMCs after labeling with metal-conjugated antibodies, compared with samples analyzed immediately after staining. Total cell recovery (Fig. 4A) and viability (Fig. 4B) were unaffected by cryopreservation of both colonic cell samples and PBMCs. In addition, the signal intensity of several major immune cell markers remained unaffected by cryopreservation at –80°C for up to 2 months (Fig. 4C/D). Given the consistency of the data obtained, cryopreservation of labeled cells would allow for the collection and storage of an entire patient cohort for simultaneous analysis.
FIGURE 4.
Cryopreservation maintains the integrity of stained samples for delayed acquisition. Percentage live cells (A) and total CD45positive cells (B) acquired after isolation from either colonic biopsies or peripheral blood, and acquired immediately after staining (Fresh) or after cryopreservation (Cryo) at –80°C for up to 2 months. Data points represent individual donors. Percentage cell recovery was calculated as yield derived from cryopreserved cells as a proportion of yield in fresh samples, in paired sample sets. C, Representative biaxial plots of expression of indicated lineage markers, from fresh or cryopreserved samples. F, Median signal intensity (MSI) of major lineage markers in fresh or cryopreserved samples after staining. Mean ± SD of 5 individual donors, analyzed using the Wilcoxon test.
DISCUSSION
Mass cytometry is a powerful new tool for deep immunophenotyping intestinal immune cell subsets with unprecedented resolution, while efficiently using minimal amounts of patient-derived tissue. This study was designed to evaluate the implementation of mass cytometry as an analytical tool during mechanism of action studies and clinical trials, to potentially complement clinical end points and provide an opportunity to assess drug efficacy within the target tissue. Presented here are solutions to several logistical challenges involved in the analysis of intestinal cellular subsets by mass cytometry. We found that adequate cell numbers can be isolated from intestinal pinch biopsies in a short time frame, and as few as 2 colon or 4 ileal biopsies are sufficient for robust identification of most major immune cell subsets. Biopsy preservation by slow-freezing represents a solution for sample acquisition and tissue transport to a wet laboratory facility, as well as allowing for centralized processing of samples at a convenient time, whether soon after collection or once the entire patient cohort has been gathered. To supplement this, labeled cells may also be cryopreserved before mass cytometry analysis, providing even greater flexibility to the overall workflow.
During an endoscopy, collection of additional biopsies for research purposes adds to procedure time. Therefore, maximizing the cell yields obtained from the least amount of biopsy material is necessary. We were able to obtain viable cells from as few as 2 colonic biopsies or 4 ileal biopsies, which were sufficient to identify most major immune cell lineages in an unbiased manner. A prior study found that as few as 10,000 starting events were sufficient for analysis of multiple immune cell lineages,18 and similarly, the number of events obtained from 2–3 duodenal or rectal biopsies was sufficient for in-depth analysis of cellular subsets.24 The protocols presented here require minimal patient-derived material and can be completed in less than 2 hours, minimizing potential phenotypic changes that may occur during prolonged manipulation and providing insight into intestinal immune cell composition, in as close as possible to in vivo conditions.
One major logistical challenge when harvesting patient-derived material during clinical trials is tissue transport, as trial sites may not have wet laboratory facilities for tissue processing. Ideally, transport must be accomplished as quickly as possible to ensure minimal sample degradation. In addition, depending on the time of acquisition, the material may not be processed immediately after delivery. Although snap-freezing also allows for recovery of live cells, slow-freezing provides a more uniform result, without much more added effort. It is important to note that the effect of slow-freezing on cells of the myelo-monocytic lineage is not negligible. Adequate isolation of these cells should ideally be performed without delay. The ability to freeze biopsies by adapting the slow-freezing procedure commonly utilized for PBMCs provides the flexibility that allows for sample collection at multiple centers and centralized tissue processing, a prerequisite for the incorporation of mass cytometry analyses into multicenter trials. However, further improvements on this protocol have not been examined, and as such, there may be additional ways to optimize cell recovery and phenotype preservation. In addition, only surface phenotype was assessed during this study, and the ability to stimulate cells for intracellular cytokine or transcription factor analysis after slow-freezing was not examined. Given the alterations in abundance and phenotype of some subsets following slow-freezing, every marker of interest should be examined after preservation to ensure accurate representation of antigens. An added advantage of CyTOF is that cryopreservation of stained samples allows for indefinite delay of sample analysis, beyond what is possible with flow cytometry given the absence of metal decay.34 This permits later sample analysis upon instrument malfunction26 and, more importantly, enables longitudinal sample collection for coordinated analysis and avoidance of known batch effects.26, 35
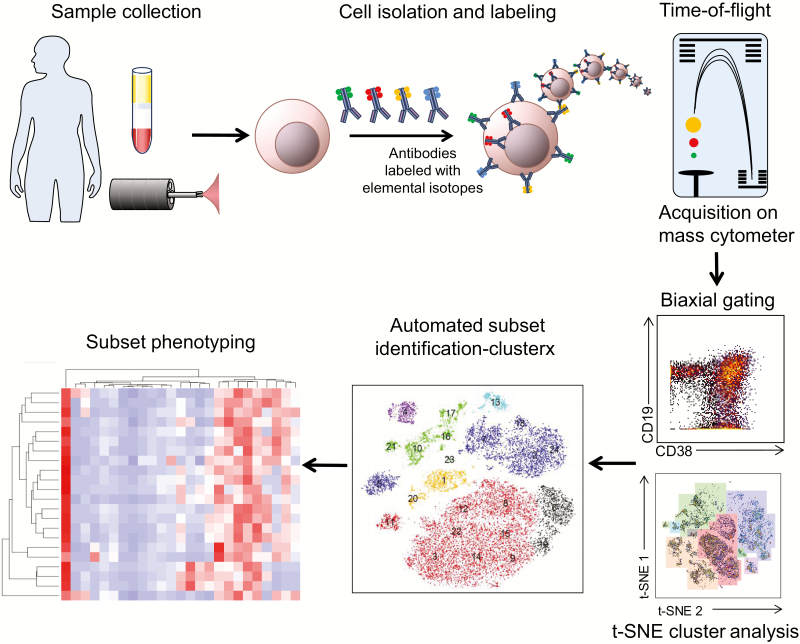
A proposed overall workflow for the implementation of mass cytometry in IBD research, from sample collection to data analysis, is shown in Figure 5. The scientific benefit of utilizing CyTOF for analysis of human samples has already been shown,20–22, 24 and the logistics are hereby simplified.
FIGURE 5.
Overview of mass cytometry sample processing and analyses for IBD studies. Workflow of a CyTOF study, from sample acquisition to analysis. Peripheral blood and intestinal pinch biopsies are processed for cell isolation. Single-cell suspensions are stained with metal-conjugated antibodies and acquired on a mass cytometer. Data analysis techniques include traditional biaxial gating, followed by complex analysis tools including t-SNE and ClusterX.
Overall, we envision mass cytometry as a novel technology with the potential of revolutionizing our understanding of IBD pathogenesis. As with any new technology, there are associated limitations, such as slow acquisition time compared with flow cytometry, inability to sort cells, and upfront cost of instrumentation, which are described in the literature.35 However, these limitations are outweighed by the comprehensive nature of the data that can be obtained from finite human material, as described here and elsewhere.18, 24 For the IBD field in the immediate term, this technique may allow identification of critical pathogenic cellular effectors that infiltrate the intestinal lamina propria during a flare and disappear after mucosal healing. It may also allow identification of critical cellular determinants of disease activity detectable in peripheral blood that may serve as cellular biomarkers and assist clinicians with treatment decisions. Mass cytometry may be further incorporated into objective biologic mechanism of action readouts, based on a drug’s proposed mechanism of action within tissues. Finally, we envision that the application of this novel technology to gastrointestinal research should not only impact IBD but also other gastrointestinal inflammatory conditions where inflammatory cells are critical for pathogenesis.
SUPPLEMENTARY DATA
Supplementary data is available at Inflammatory Bowel Diseases online.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ms. Gelareh Gangi for assistance with maintenance of the institutional review board protocol and Ms. Helen Le and Ms. Jennifer Neill for assistance with informed consents.
Conflicts of interest: The authors disclose no relevant conflicts.
Supported by: This work is funded by grants from the National Institutes of Health (DK108670) and VA merit I01 BX003436 to J.R.N.; by Shared Instrumentation Grant (SIG) Program (S10): CyTOF Mass Cytometer S10 OD018499-01 to L.J.I.; and by support from the Chiba University–UC San Diego Program in Mucosal Immunology, Allergy and Vaccines.
Author contributions: C.J.T., J.R.N.: study concept and design; P.D., B.S.B., S.B.H., W.J.S., D.P., J.R.N.: sample acquisition; C.J.T., T.P.J., E.E., T.K., J.D.B.: acquisition of data; C.J.T., T.P.J., E.E., B.C., T.K., J.D.B., J.L.: analysis and interpretation of data; C.J.T., T.P.J., B.C., J.R.N.: drafting of the manuscript; P.D., B.S.B., J.L., L.E., P.B.E., J.R.N.: critical revision of the manuscript; C.J.T., E.E., B.C., J.L.: statistical analysis.
REFERENCES
- 1. M’Koma AE. Inflammatory bowel disease: an expanding global health problem. Clin Med Insights Gastroenterol. 2013;6:33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Danese S, Fiocchi C, Panés J. Drug development in IBD: from novel target identification to early clinical trials. Gut. 2016;65:1233–1239. [DOI] [PubMed] [Google Scholar]
- 3. Currò D, Pugliese D, Armuzzi A. Frontiers in drug research and development for inflammatory bowel disease. Front Pharmacol. 2017;8:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sandborn WJ, Feagan BG, Hanauer SB, et al. A review of activity indices and efficacy endpoints for clinical trials of medical therapy in adults with Crohn’s disease. Gastroenterology. 2002;122:512–530. [DOI] [PubMed] [Google Scholar]
- 5. D’Haens G, Sandborn WJ, Feagan BG, et al. A review of activity indices and efficacy end points for clinical trials of medical therapy in adults with ulcerative colitis. Gastroenterology. 2007;132:763–786. [DOI] [PubMed] [Google Scholar]
- 6. Levesque BG, Sandborn WJ, Ruel J, et al. Converging goals of treatment of inflammatory bowel disease from clinical trials and practice. Gastroenterology. 2015;148:37–51.e1. [DOI] [PubMed] [Google Scholar]
- 7. Sandborn WJ, Ghosh S, Panes J, et al. Study A3921043 Investigators A phase 2 study of tofacitinib, an oral Janus kinase inhibitor, in patients with Crohn’s disease. Clin Gastroenterol Hepatol. 2014;12:1485–1493.e2. [DOI] [PubMed] [Google Scholar]
- 8. Panés J, Sandborn WJ, Schreiber S, et al. Tofacitinib for induction and maintenance therapy of Crohn’s disease: results of two phase IIb randomised placebo-controlled trials. Gut. 2017;66:1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sandborn WJ, Lee SD, Tarabar D, et al. Anti-MAdCAM-1 antibody (PF-00547659) for active refractory Crohn’s disease: results of the OPERA study. Gastroenterology. 2015;148:S–S162. [Google Scholar]
- 10. Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Irish JM. Beyond the age of cellular discovery. Nat Immunol. 2014;15:1095–1097. [DOI] [PubMed] [Google Scholar]
- 12. Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qiu P, Simonds EF, Bendall SC, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Maaten L, Hinton GE. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 15. van der Maaten L. Accelerating t-SNE using tree-based algorithms. J Mach Learn Res. 2014;15:3221–3245. [Google Scholar]
- 16. Amir ED, Davis KL, Tadmor MD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature Biotechnol. 2014;31:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bruggner RV, Bodenmiller B, Dill DL, et al. Automated identification of stratifying signatures in cellular subpopulations. Proc Natl Acad Sci U S A. 2014;111:E2770–E2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yao Y, Liu R, Shin MS, et al. CyTOF supports efficient detection of immune cell subsets from small samples. J Immunol Methods. 2014;415:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGowan I, Anton PA, Elliott J, et al. Exploring the feasibility of multi-site flow cytometric processing of gut associated lymphoid tissue with centralized data analysis for multi-site clinical trials. PLoS One. 2015;10:e0126454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guilliams M, Dutertre CA, Scott CL, et al. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity. 2016;45:669–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong MT, Ong DE, Lim FS, et al. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity. 2016;45:442–456. [DOI] [PubMed] [Google Scholar]
- 22. Chevrier S, Levine JH, Zanotelli VRT, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169:736–749.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Newell EW, Sigal N, Nair N, et al. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31:623–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Unen V, Li N, Molendijk I, et al. Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity. 2016;44:1227–1239. [DOI] [PubMed] [Google Scholar]
- 25. Sumatoh HR, Teng KWW, Cheng Y, et al. Optimization of mass cytometry sample cryopreservation after staining. Cytometry Part A. 2016;12:1–14. [DOI] [PubMed] [Google Scholar]
- 26. Finck R, Simonds EF, Jager A, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen H, Lau MC, Wong MT, et al. Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput Biol. 2016;12:e1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Babicki S, Arndt D, Marcu A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fiocchi C, Youngman KR.. Isolation of Human Intestinal Mucosal Mononuclear Cells. New York: John Wiley & Sons; 1996. Current Protocols in Immunology; vol 1 (suppl 19). [DOI] [PubMed] [Google Scholar]
- 30. Preza GC, Yang OO, Elliott J, et al. T lymphocyte density and distribution in human colorectal mucosa, and inefficiency of current cell isolation protocols. PLoS One. 2015;10:e0122723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bowcutt R, Malter LB, Chen LA, et al. Isolation and cytokine analysis of lamina propria lymphocytes from mucosal biopsies of the human colon. J Immunol Methods. 2015;421:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rajamohan D, Kalra S, Duc Hoang M, et al. Automated electrophysiological and pharmacological evaluation of human pluripotent stem cell-derived cardiomyocytes. Stem Cells Dev. 2016;25:439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weinberg A, Song LY, Wilkening C, et al. Pediatric ACTG Cryopreservation Working Group Optimization and limitations of use of cryopreserved peripheral blood mononuclear cells for functional and phenotypic T-cell characterization. Clin Vaccine Immunol. 2009;16:1176–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pinto LA, Trivett MT, Wallace D, et al. Fixation and cryopreservation of whole blood and isolated mononuclear cells: influence of different procedures on lymphocyte subset analysis by flow cytometry. Cytometry B Clin Cytom. 2005;63:47–55. [DOI] [PubMed] [Google Scholar]
- 35. Newell EW, Cheng Y. Mass cytometry: blessed with the curse of dimensionality. Nat Immunol. 2016;17:890–895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.