Abstract
Cas12a (Cpf1) is a CRISPR-associated nuclease with broad utility for synthetic genome engineering, agricultural genomics, and biomedical applications. While bacteria harboring CRISPR-Cas9 or CRISPRCas3 adaptive immune systems sometimes acquire mobile genetic elements encoding anti-CRISPR proteins that inhibit Cas9, Cas3, or the DNA-binding Cascade complex, no such inhibitors have been found for CRISPR-Cas12a. Here we employ a comprehensive bioinformatic and experimental screening approach to identify three different inhibitors that block or diminish CRISPR-Cas12a-mediated genome editing in human cells. We also find a widespread connection between CRISPR self-targeting and inhibitor prevalence in prokaryotic genomes, suggesting a straightforward path to the discovery of many more anti-CRISPRs from the microbial world.
CRISPR-Cas systems represent the only known adaptive mechanism by which prokaryotes protect themselves from biological attackers (1). Although diverse in composition, all CRISPR-Cas pathways employ RNA-guided enzymes that recognize and destroy foreign nucleic acids, commonly double-stranded DNA (2). The ease of changing the RNA guide molecule, and hence the DNA targeting specificity, has enabled use of CRISPR-Cas9 and Cas12 for programmable genome editing in a wide range of cells and organisms (3, 4). To control Cas9, bacterial inhibitors referred to as anti-CRISPRs (Acrs) have been found to limit or block Cas9 functions (5–9). However, these inhibitors have been found only sporadically, and no such inhibitors have been reported for Cas12a (Cpf1).
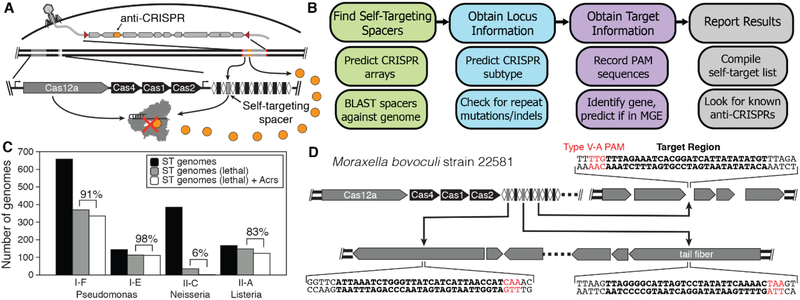
The known CRISPR-Cas inhibitors have been identified either through isolation of CRISPR-resistant phages (9–13) or by proximity to anti-CRISPR associated (aca) genes (14, 15). As an alternative inhibitor discovery strategy, the presence of stable self-targeting CRISPR sequences has been proposed as a potential indicator of genomes or mobile genetic elements (MGEs) harboring CRISPR inhibitors (Fig. 1A) (16, 17). Self-targeting CRISPR sequences in CRISPR arrays are expected to be lethal to the host cell by directing cleavage and subsequent degradation of the microbial genome (18). However, such self-targeting CRISPR sequences could exist in cells harboring CRISPR inhibitors (16). To test whether CRISPR inhibitors can be discovered systematically by flagging CRISPR self-targeting genomes, we built a bioinformatic pipeline to search across the NCBI prokaryotic sequence database to locate self-targeting examples within predicted CRISPR arrays (Fig. 1B). The Self-Targeting Spacer Searcher (STSS) first predicts all possible CRISPR arrays using the CRISPR Recognition Tool (CRT) (19) and BLASTs each spacer against the host genome and any associated plasmids. Additionally, STSS collects information to gauge the likelihood that the self-targeting sequence would be lethal to the organism and if the target sequence occurs in a MGE (fig. S1) (20, 21).
Fig. 1. Bioinformatic approach for discovering Acr genes.
(A) Anti-CRISPRs allow survival of cells containing self-targeting CRISPR arrays. (B) STSS finds self-targeting CRISPR spacers in genomic DNA, predicts the type of CRISPR system involved, and obtains information about the targeted sequence. (C) A large percentage of genomes containing self-targeting (ST) spacers predicted to be lethal contain previously-identified Acr genes. (D) Moraxella bovoculi strain 22581 contains three self-targeting spacers in two different prophages in the genome. All of the protospacers contain a TTV protospacer-adjacent motif (PAM) (22).
Using STSS, we collected self-targeting data for 150,291 genomes, observing 22,125 cases of predicted self-targets, representing 8,917 unique sequences across 9,155 genomes (fig. S2 and data S1). Focusing initially on three species in which multiple Acrs have been previously identified (Pseudomonas aeruginosa, Listeria monocytogenes, and Neisseria meningitidis), we determined the number of genomes that contained at least one lethal self-targeting CRISPR spacer, and the number of those genomes that also contained an Acr using a blastp search (Fig. 1C). In N. meningitidis only 6% of the genomes were observed to contain a potential anti-CRISPR, while in P. aeruginosa and L. monocytogenes the number exceeded 80% or 90%. The self-targeting genomes devoid of known Acrs may also contain inhibitors that have yet to be discovered, especially in N. meningitidis where the number of self-targeting genomes is high but the number containing known Acrs is low.
Based on our observation that self-targeting genomes frequently contain CRISPR inhibitors, we sought to determine whether screening genes in genomes containing self-targeting spacers could uncover new inhibitors. We focused our efforts on the CRISPR-Cas12a system (22–24), which has so far eluded discovery of inhibitory proteins despite the increasing use of Cas12a in gene editing and diagnostic applications (22, 25, 26). From the STSS results, we identified four strains of Moraxella bovoculi that contain self-targeting CRISPRCas12a systems as top candidates for containing anti-CRISPRs (see Methods). These strains each contained at least one perfectly matched self-targeting sequence in or near a predicted MGE with a correct TTV PAM sequence and intact Cas12 open reading frame, which should render the self-targeting spacers lethal in the absence of anti-CRISPRs (Fig. 1D and fig. S3) (27).
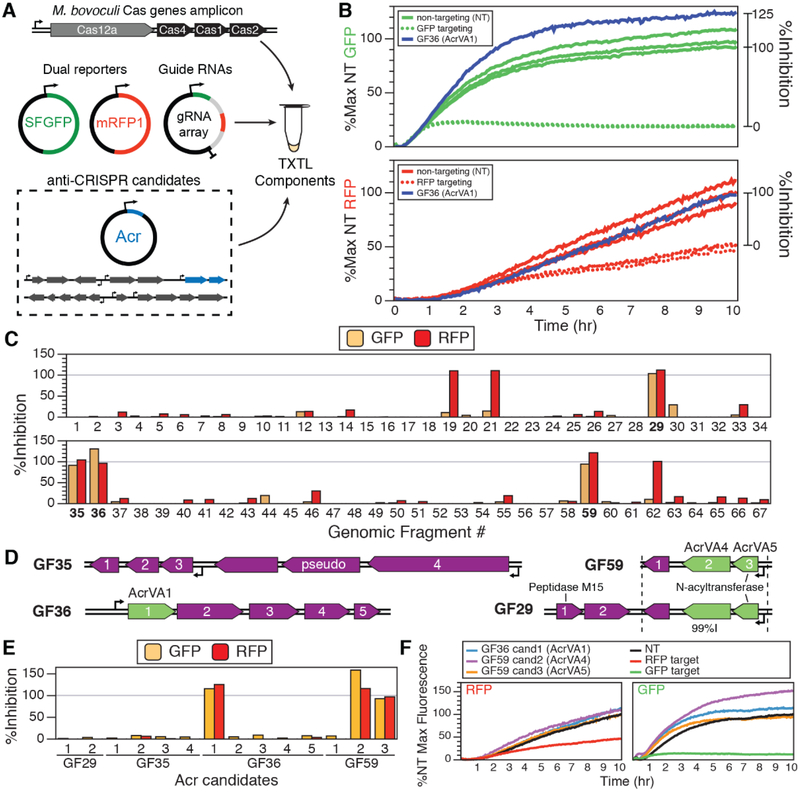
To test whether the Moraxella genomes encode type V-A anti-CRISPRs (AcrVA), we employed a cell-free transcription-translation (TXTL) system (28, 29) to express gene products from Moraxella genomic fragments. As an initial test of M. bovoculi Cas12a (MbCas12a) protein activity, we PCR-amplified a genomic fragment containing the promoter region and all of the Cas proteins (Cas12a, Cas1, Cas2, and Cas4) from M. bovoculi strain 22581. This amplicon was added to TXTL reactions with two reporter plasmids encoding green or red fluorescent protein (GFP or RFP) (Fig. 2A). When supplied with CRISPR RNAs (crRNAs) with base pairing complementary to the GFP and RFP genes, the presence of the MbCas12a-containing genomic fragment greatly reduced expression of both reporters (Fig. 2B and fig. S4). This result suggests that MbCas12 is active in M. bovoculi, suggesting the existence of CRISPR inhibitor(s) to prevent the self-targeting spacers from killing the cell.
Fig. 2. TXTL screening for Acr gene candidates.
(A) Overview of the transcription-translation (TXTL) reaction. DNA expressing Cas12a, two fluorescent reporters, and two gRNAs are mixed with or without DNA potentially containing Acr genes. (B) Cleavage of the reporter plasmid results in a reduced fluorescent output that is rescued by Acr genes (Acr-absent data in triplicate). (C) Amount of relative inhibition observed for 67 genomic fragments (GFs) across three self-targeting M. bovoculi strains. Four GFs (bold) exhibited inhibition in both fluorescence channels. (D) Genomic fragments GF29, GF35, GF36, and GF59 (99% nucleotide identity to GF29) exhibited high levels of expression for both reporters. (E) Testing the individual genes from the fragments in (D) (table S2) resulted in the identification AcrVA1 (GF36 candidate 1), AcrVA4 (GF59 candidate 2), and AcrVA5 (GF59 candidate 3). (F) Kinetic TXTL data for the AcrVA genes measured over the course of 10 hours of gene expression.
To identify potential AcrVA-encoding genes, we used a directed screening approach to search the predicted MGEs within three of the M. bovoculi strains (strain 283689 was unavailable) containing self-targeting sequences from a type V-A CRISPR array. Interestingly, we also observed 13 self-targeting CRISPR type I-C spacers in strain 58069 that strongly suggest the presence of I-C anti-CRISPRs in that strain (fig. S5). For each of the M. bovoculi genomes, pairs of PCR primers were used to make overlapping ~2–10 kb amplicons spanning all of the predicted MGEs in the three strains (generally excluding highly similar sequences) (table S1). These genomic fragments (GFs) were then added to the TXTL cleavage reactions described above.
From a total of 67 GFs that we tested for type V-A CRISPR inhibition activity, four correlated with increased levels of gene expression for both reporters (Fig. 2, C and D, and figs. S6 to S8). We then cloned the individual open reading frames within these fragments (Fig. 2D and table S3) downstream of the Ptet promoter to separately induce transcription and translation of each gene and assessed them for CRISPR inhibition activity using the TXTL Cas12a cleavage assay. From the pool of candidates, three proteins supported high levels of dual reporter gene expression (Fig. 2, E and F, and fig. S9): GF36 candidate 1, GF59 candidate 2, and GF59 candidate 3, hereafter referred to as AcrVA1, AcrVA4, and AcrVA5, respectively, to complement the other AcrVA genes discovered concurrently with this work (30).
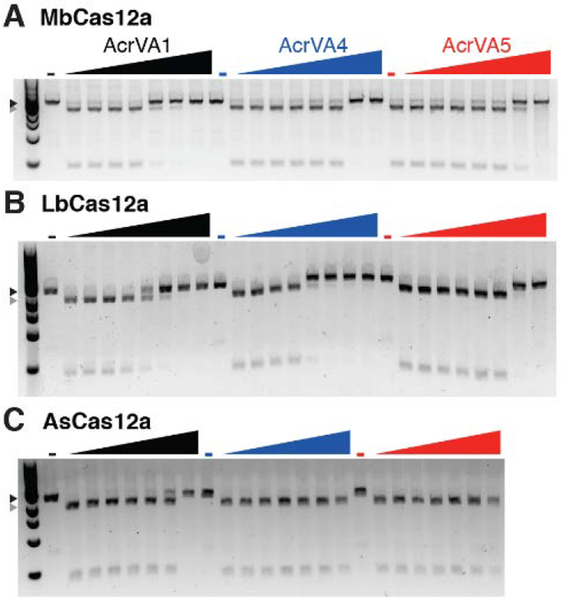
To confirm the CRISPR inhibition activity of AcrVA1, AcrVA4, and AcrVA5 in vitro, we first overexpressed and purified each putative Cas12a inhibitor, MbCas12a, and two additional Cas12a enzymes (AsCas12a and LbCas12a) commonly used in genome editing or DNA detection applications (fig. S10) (25, 26). We then generated crRNA-protein (RNP) complexes for each of the purified Cas12a enzymes and added the RNPs to a linearized target plasmid that was preincubated with increasing concentrations of each candidate AcrVA protein. We observed that AcrVA1 inhibited DNA cleavage by all three Cas12a enzymes, with the strongest inhibition observed for MbCas12a and weakest observed for As-Cas12a (Fig. 3). AcrVA4 and AcrVA5 inhibited dsDNA cleavage for both MbCas12a and LbCas12a, but did not inhibit AsCas12a. Interestingly, we also observed that AcrVA4 more strongly inhibited the MbCas12a from strain 58069 (fig. S11) than the MbCas12a from strain 22581 (Fig. 3A), and that AcrVA5 was unable to inhibit the MbCas12a from strain 58069 (fig. S11). None of the AcrVA proteins inhibited S. pyogenes Cas9 (SpyCas9) cleavage (fig. S12).
Fig. 3. Biochemical validation of AcrVA inhibitors.
(A) Moraxella bovoculi Cas12a (MbCas12a) in vitro dsDNA cleavage is inhibited by increasing concentrations of AcrVA1, AcrVA4, and AcrVA5 (0 – 1.25 μM; see Methods). (B) LbCas12a, a Cas12a commonly used for gene editing and diagnostics (4, 22, 26), is also inhibited by all three AcrVA proteins. (C) High concentrations of AcrVA1 inhibit AsCas12a-mediated dsDNA cleavage, but AcrVA4 and AcrVA5 have no effect. Triangles indicate uncleaved (black) or cleaved (gray) DNA.
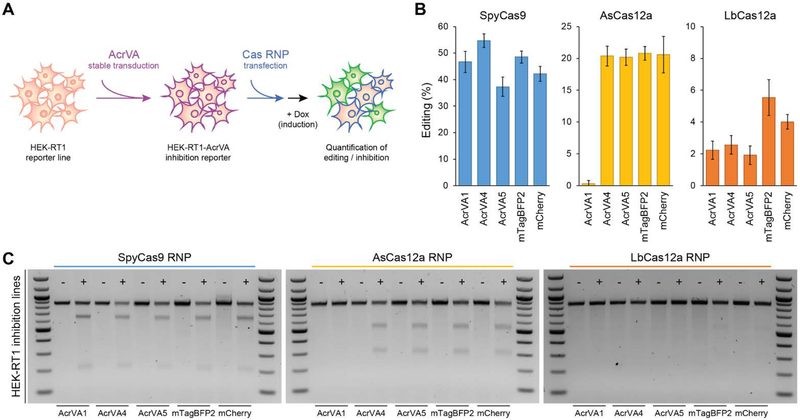
Having confirmed robust DNA cleavage inhibition by AcrVA1, AcrVA4, and AcrVA5 using purified protein samples, we next tested whether these Cas12a inhibitors could block or reduce Cas12a-mediated genome editing in human cells. We cloned each AcrVA candidate, AcrIIA4 (a SpyCas9 inhibitor), or negative controls, into a lentiviral expression vector and stably transduced HEK293T-derived genome editing reporter cells containing a doxycycline-inducible GFP marker. Purified AsCas12a, LbCas12a, MbCas12a, or SpyCas9 protein was assembled with a GFP-targeting guide RNA and transfected into the AcrVA-expressing reporter cell lines (Fig. 4A). At 24 hours post RNP delivery, cells were induced by doxycycline for another 24 hours before quantifying editing efficiency by flow cytometry (Fig. 4B and fig. S13A) and a T7 endonuclease 1 assay (Fig. 4C and fig. S13B). We observed high levels of genome editing induced by SpyCas9 with no inhibition by any of the AcrVA proteins or negative controls, but virtually complete inhibition by AcrIIA4. We also observed strong Cas12a RNP editing inhibition that generally matched the in vitro cleavage results. Mirroring their biochemical behavior, AcrVA1 provided the broadest inhibition of Cas12a and fully blocked AsCas12a with efficiencies comparable to AcrIIA4’s inhibition of SpyCas9. AcrVA4 and AcrVA5 only inhibited LbCas12a. RNP-based delivery of MbCas12a did not edit efficiently enough to determine the effectiveness of the AcrVA genes on its activity (fig. S13), consistent with previous findings (22).
Fig. 4. AcrVAs robustly inhibit genome editing by specific CRISPR-Cas12a nucleases in mammalian cells.
(A) Overview of the editing reporter assay in human cells. (B) Quantification of genome editing in reporter cell lines stably expressing the indicated CRISPR-Cas12a inhibitors (AcrVAs) or a control (mTagBFP2, mCherry). The scale of each plot is adjusted to compensate for differences in editing efficiency. Error bars indicate standard deviations of triplicates. (C) Biochemical analysis of AcrVA-mediated inhibition in representative samples shown in (B). Editing was assessed by the T7 endonuclease 1 (T7E1) assay.
Together, these results establish a new approach for systematic discovery and validation of CRISPR-Cas inhibitors hidden within self-targeting genomes. Importantly, the Cas12a inhibitors revealed by this approach are only found within a few genomes within the NCBI database, with AcrVA4 and AcrVA5 being particularly rare genes, only cooccurring with each other and thus intractable to an aca-based search approach (fig. S14). While we expect the extensive set of yet-to-be-analyzed CRISPR self-targeting genomes (data S1) will lead to the discovery of many more Acrs across all CRISPR subtypes, the AcrVAs discovered and validated here provide a toolkit for selective Cas12a regulation both in vitro and in mammalian systems, with the potential to advance synthetic biology, CRISPR diagnostics, and therapeutic genome editing.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rachel Lew for technical support, David Burstein for Cas protein HMMs, Mary West and the CIRM/QB3 Shared Stem Cell Facility/High-Throughput Screening Facility for flow cytometry equipment support, Adair Borges for discussions on self-targeting in Pseudomonas, and Doudna lab members for thoughtful discussion.
Funding: The authors acknowledge financial support from the Defense Advanced Research Projects Agency (DARPA) (award HR0011-17-2-0043 to J.A.D.), the Paul G. Allen Frontiers Group and the National Science Foundation (MCB-1244557 to J.A.D.). C.F. is supported by a US National Institutes of Health K99/R00 Pathway to Independence Award (K99GM118909, R00GM118909) from the National Institute of General Medical Sciences (NIGMS). J.A.D. is an investigator of the Howard Hughes Medical Institute (HHMI), and this study was supported in part by HHMI; J.A.D is also a Paul Allen Distinguished Investigator.
Footnotes
Competing interests: J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences, a scientific adviser to Caribou, Intellia, Scribe, Synthego, Metagenomi, Inari, and eFFECTOR Therapeutics, and a director of Driver and Johnson & Johnson. The Regents of the University of California have patents pending for CRISPR related technologies on which the authors are inventors.
Data and materials availability: All data are available in the main text or the supplementary materials. Plasmids for expression of AcrVA1, AcrVA4, AcrVA5, and MbCas12 are available on Addgene (IDs 115653–115664, 115669–115670, and 115795–115797) under a Uniform Biological Material Transfer Agreement. Code for STSS is available on GitHub. M. bovoculi strains are available from the University of Nebraska (strain 58069) or the US Meat Animal Research Center (strains 22581 and 33362) under a Material Transfer Agreement.
SUPPLEMENTARY MATERIALS
www.sciencemag.org/cgi/content/full/science.aau5138/DC1
Materials and Methods
Figs. S1 to S14
Tables S1 to S4
Data S1
REFERENCES AND NOTES
- 1.Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJJ, CRISPR-Cas: Adapting to change. Science 356, eaal5056 (2017). doi: 10.1126/science.aal5056 [DOI] [PubMed] [Google Scholar]
- 2.Plagens A, Richter H, Charpentier E, Randau L, DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol. Rev 39, 442–463 (2015). doi: 10.1093/femsre/fuv019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fellmann C, Gowen BG, Lin P-C, Doudna JA, Corn JE, Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov 16, 89–100 (2017). doi: 10.1038/nrd.2016.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright AV, Nuñez JK, Doudna JA, Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell 164, 29–44 (2016). doi: 10.1016/j.cell.2015.12.035 [DOI] [PubMed] [Google Scholar]
- 5.Shin J, Jiang F, Liu J-J, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA, Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv 3, e1701620 (2017). doi: 10.1126/sciadv.1701620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H, Patel DJ, Inhibition mechanism of an anti-CRISPR suppressor AcrIIA4 targeting SpyCas9. Mol. Cell 67, 117–127.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong D, Guo M, Wang S, Zhu Y, Wang S, Xiong Z, Yang J, Xu Z, Huang Z, Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein. Nature 546, 436–439 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Basgall EM, Goetting SC, Goeckel ME, Giersch RM, Roggenkamp E, Schrock MN, Halloran M, Finnigan GC, Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology 164, 464–474 (2018). doi: 10.1099/mic.0.000635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hynes AP, Rousseau GM, Agudelo D, Goulet A, Amigues B, Loehr J, Romero DA, Fremaux C, Horvath P, Doyon Y, Cambillau C, Moineau S, Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun 9, 2919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR, Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432 (2013). doi: 10.1038/nature11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pawluk A, Bondy-Denomy J, Cheung VHW, Maxwell KL, Davidson AR, A new group of phage anti-CRISPR genes inhibits the type I-E CRISPR-Cas system of Pseudomonas aeruginosa. mBio 5, e00896 (2014). doi: 10.1128/mBio.00896-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes AP, Rousseau GM, Lemay M-L, Horvath P, Romero DA, Fremaux C, Moineau S, An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcus pyogenes Cas9. Nat. Microbiol 2, 1374–1380 (2017). doi: 10.1038/s41564-017-0004-7 [DOI] [PubMed] [Google Scholar]
- 13.He F, Bhoobalan-Chitty Y, Van LB, Kjeldsen AL, Dedola M, Makarova KS, Koonin EV, Brodersen DE, Peng X, Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat. Microbiol 3, 461–469 (2018). doi: 10.1038/s41564-018-0120-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawluk A, Staals RHJ, Taylor C, Watson BNJ, Saha S, Fineran PC, Maxwell KL, Davidson AR, Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol 1, 16085 (2016). doi: 10.1038/nmicrobiol.2016.85 [DOI] [PubMed] [Google Scholar]
- 15.Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, Davidson AR, Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838.e9 (2016). doi: 10.1016/j.cell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauch BJ, Silvis MR, Hultquist JF, Waters CS, McGregor MJ, Krogan NJ, Bondy-Denomy J, Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 168, 150–158.e10 (2017). doi: 10.1016/j.cell.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bondy-Denomy J, Protein inhibitors of CRISPR-Cas9. ACS Chem. Biol 13, 417–423 (2018). doi: 10.1021/acschembio.7b00831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heussler GE, O’Toole GA, Friendly fire: Biological functions and consequences of chromosomal targeting by CRISPR-Cas systems. J. Bacteriol 198, 1481–1486 (2016). doi: 10.1128/JB.00086-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, Hugenholtz P, CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8, 209 (2007). doi: 10.1186/1471-2105-8-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS, PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res 44, W16–W21 (2016). doi: 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson CM, Lau BY, Williams KP, Islander: A database of precisely mapped genomic islands in tRNA and tmRNA genes. Nucleic Acids Res 43, D48–D53 (2015). doi: 10.1093/nar/gku1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F, Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771 (2015). doi: 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV, Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397 (2015). doi: 10.1016/j.molcel.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF, New CRISPR-Cas systems from uncultivated microbes. Nature 542, 237–241 (2017). doi: 10.1038/nature21059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F, Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018). doi: 10.1126/science.aaq0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA, CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018). doi: 10.1126/science.aar6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickey AM, Loy JD, Bono JL, Smith TPL, Apley MD, Lubbers BV, DeDonder KD, Capik SF, Larson RL, White BJ, Blom J, Chitko-McKown CG, Clawson ML, Large genomic differences between Moraxella bovoculi isolates acquired from the eyes of cattle with infectious bovine keratoconjunctivitis versus the deep nasopharynx of asymptomatic cattle. Vet. Res 47, 31 (2016). doi: 10.1186/s13567-016-0316-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall R, Maxwell CS, Collins SP, Jacobsen T, Luo ML, Begemann MB, Gray BN, January E, Singer A, He Y, Beisel CL, Noireaux V, Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell 69, 146–157.e3 (2018). doi: 10.1016/j.molcel.2017.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun ZZ, Hayes CA, Shin J, Caschera F, Murray RM, Noireaux V, Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J. Vis. Exp 2013, e50762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marino ND, Zhang JY, Borges AL, Sousa AA, Leon LM, Rauch BJ, Walton RT, Berry JD, Joung JK, Kleinstiver BP, Bondy-Denomy J, Discovery of widespread type I and type V CRISPR-Cas inhibitors. Science 10.1126/science.aau5174 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy SR, Accelerated profile HMM searches. PLOS Comput. Biol 7, e1002195 (2011). doi: 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV, An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol 13, 722–736 (2015). doi: 10.1038/nrmicro3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV, Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol 15, 169–182 (2017). doi: 10.1038/nrmicro.2016.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539–539 (2011). doi: 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Ye Y, Not all predicted CRISPR-Cas systems are equal: Isolated cas genes and classes of CRISPR like elements. BMC Bioinformatics 18, 92 (2017). doi: 10.1186/s12859-017-1512-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lange SJ, Alkhnbashi OS, Rose D, Will S, Backofen R, CRISPRmap: An automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res 41, 8034–8044 (2013). doi: 10.1093/nar/gkt606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alkhnbashi OS, Costa F, Shah SA, Garrett RA, Saunders SJ, Backofen R, CRISPRstrand: Predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics 30, i489–i496 (2014). doi: 10.1093/bioinformatics/btu459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F, C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016). doi: 10.1126/science.aaf5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA, Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339.e5 (2018). doi: 10.1016/j.molcel.2018.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH, CDD: NCBI’s conserved domain database. Nucleic Acids Res 43, D222–D226 (2015). doi: 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Heidrich N, Ampattu BJ, Gunderson CW, Seifert HS, Schoen C, Vogel J, Sontheimer EJ, Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol. Cell 50, 488–503 (2013). doi: 10.1016/j.molcel.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall R, Maxwell CS, Collins SP, Beisel CL, Noireaux V, Short DNA containing χ sites enhances DNA stability and gene expression in E. coli cell-free transcription-translation systems. Biotechnol. Bioeng 114, 2137–2141 (2017). doi: 10.1002/bit.26333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IAM, Kwon JS, Guan Y, Sinha N, Zuber J, An optimized microRNA backbone for effective single-copy RNAi. Cell Reports 5, 1704–1713 (2013). doi: 10.1016/j.celrep.2013.11.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.