Abstract
Nonsyndromic cleft lip with or without cleft palate (NSCL/P) is the most common craniofacial birth defect in humans and is notable for its apparent sexual dimorphism where approximately twice as many males are affected as females. The sources of this disparity are largely unknown, but interactions between genetic and sex effects are likely contributors. We examined gene-by-sex (G x S) interactions in a worldwide sample of 2,142 NSCL/P cases and 1,700 controls recruited from 13 countries. First, we performed genome-wide joint tests of the genetic (G) and G x S effects genome-wide using logistic regression assuming an additive genetic model and adjusting for 18 principal components of ancestry. We further interrogated loci with suggestive results from the joint test (p < 1.00 × 10−5) by examining the G x S effects from the same model. Out of the 133 loci with suggestive results (p < 1.00 × 10−5) for the joint test, we observed one genome-wide significant G x S effect in the 10q21 locus (rs72804706; p = 6.69 × 10−9; OR = 2.62 [1.89, 3.62]) and 16 suggestive G x S effects. At the intergenic 10q21 locus, the risk of NSCL/P is estimated to increase with additional copies of the minor allele for females, but the opposite effect for males. Our observation that the impact of genetic variants on NSCL/P risk differs for males and females may further our understanding of the genetic architecture of NSCL/P and the sex differences underlying clefts and other birth defects.
Keywords: Oral Facial Cleft, cleft lip, cleft palate, genetic risk
Introduction
Orofacial clefts (OFCs) are one of the most commonly occurring congenital defects worldwide, affecting 1 in 700 births (Leslie & Marazita, 2013). Incidence rates are vary across populations; to the best of our knowledge, populations with African ancestry have the lowest rates among live births (~1/2500), European populations have intermediate rates (~1/1000), and Asian populations have the highest rates (~1/500) (Mossey, 2007). OFCs are classified as syndromic or nonsyndromic based on the presence or absence, respectively, of additional structural or cognitive abnormalities (Dixon et al., 2011). OFCs are highly heritable (50–80%) based on concordance rates in monozygotic twins (Grosen et al., 2011). This suggests a strong genetic component to the etiology of OFCs.
Approximately 70% of OFCs are considered nonsyndromic and have a multifactorial etiology where genetics, environmental exposures, and their interactions have been shown to modify risk (Dixon et al., 2011). There have been nine independent genome wide association studies (GWAS) and two meta-analyses that have identified at least 25 genetic loci that contribute to risk for OFCs [(Beaty et al., 2010; Beaty et al., 2011; Beaty et al., 2013; Birnbaum et al., 2009; Grant et al., 2009; Leslie et al., 2017; Leslie, Carlson, et al., 2016; Leslie, Liu, et al., 2016; Ludwig et al., 2012; Sun et al., 2015; Wolf et al., 2015; Yu et al., 2017)]. GWAS has been a useful approach for identifying genetic risk factors for OFCs, however, these studies cumulatively explain only 25–30% of the heritable risk of OFCs in any population (Leslie, Carlson, et al., 2016; Yu et al., 2017).
A likely explanation for the missing heritability of OFCs are non-additive effects, which include interactions with environmental exposures or other genes. Environmental risk factors include maternal tobacco use (M. Shi et al., 2008), low maternal folic acid (Yazdy et al., 2007), and maternal alcohol use. Incorporation of these environmental exposures into genetic studies revealed several gene-environment interactions. These include a 3- to 6-fold increased risk for OFCs for fetuses with null alleles of glutathione s-transferases (GSTT1 and GSTM1) and whose mothers smoked during pregnancy (Lammer et al., 2005; Min Shi et al.). In genome-wide studies, several gene-environment (G × E) interactions have been identified (Wu et al., 2010; Wu et al., 2014). Until recently, G × E interactions were the only genome-wide significant association signals for cleft palate (Beaty et al., 2011; Leslie, Liu, et al., 2016).
An understudied factor has been the contribution of sex to OFC risk. Males are twice as likely to have CL or CLP than females (Marazita, 2012) and females are twice as likely to have CP than males, which implies that cleft type and incidence are sexually dimorphic. Explanations for these incidence rates have been elusive, but several genetic models could explain the reduced risk for CL/P in females. First, there may be specific susceptibility factors on the X or Y chromosomes that affect male-CL/P risk, but not female-CL/P risk, due to the compensatory second copy of the X chromosome or lack of a Y chromosome in females. There are several notable candidate gene association signals on the X-chromosome, including TBX22, associated with CP (Marcano et al., 2004; Suphapeetiporn et al., 2007), and EFNB1 and DMD (Patel et al., 2013). Several other loci have been identified using a variety of methods and genetic models (Jugessur et al., 2012; Patel et al., 2013; Skare et al., 2017). X-chromosome inactivation has been investigated in female discordant CL/P twin and sibling pairs, suggesting a role for this mechanism in CL/P (Kimani et al., 2007). Second, there may be different liability thresholds in which the same risk alleles affect males and females equally, but females have a higher threshold that requires a higher load of risk alleles or highly penetrant mutations. Finally, risk factors could be located on the autosomes with differential effects in males and females, perhaps due to hormonally-mediated dimorphism. The third model is the focus of the present study as such gene-by-sex (G × S) interactions have not yet been directly tested.
It is well established that a standard case-control analysis has low power to detect interactions because of small numbers of cases or controls when stratified by the exposure. Alternative approaches have been developed for testing interactions in the context of genome-wide association analyses. We elected to perform a two-step procedure to gain power by enriching for possible G × S interactions by first jointly testing for genetic main effect and G × S interaction (Dai et al., 2012; Kraft et al., 2007). We therefore sought to determine how sex contributes to OFC sex-specific risk with a genome-wide analysis of G × S interactions, and potentially to identify new OFC risk loci, using our large, multi-ethnic OFC population.
Methods
Study Population:
The subjects for this study were drawn from our large, multi-ethnic OFC cohort we described previously (Leslie, Carlson, et al., 2016). We selected 5,984 subjects from 18 recruitment sites, located in Africa, Asia, Latin America and North America (Table 1). The recruitment sites were a part of genetic studies being conducted by the University of Iowa and University of Pittsburgh Center for Craniofacial and Dental Genetics. All sites had local Institutional Review Board approval and subjects were recruited using informed consent procedures. All subjects consented to the use of their DNA in genomic studies. Cases were defined as unrelated individuals with nonsyndromic cleft lip with or without cleft palate (CL/P). Controls were defined as unrelated subjects with no personal or family history of any structural or developmental anomaly. Sex was confirmed genetically for all subjects as part of quality control procedures.
Table 1:
Study Population for the Genome-Wide Interaction Analyses
| Cleft Type | Males | Females | Total |
|---|---|---|---|
| CL/P | 1293 | 849 | 2142 |
| CL | 251 | 199 | 450 |
| CLP | 1042 | 650 | 1692 |
| Controls | 713 | 987 | 1700 |
Genotyping and quality control:
The methods for genotyping, quality control, imputation, and deriving principal components (PCs) of ancestry have been previously described (Leslie, Carlson, et al., 2016). Briefly, samples were genotyped for ~580K SNPs at the Center for Inherited Research at John Hopkins University using the Illumina HumanCore+Exome chip. Approximately 97% of the genotyped SNP’s passed quality control leaving a total of 539,473 genotyped SNPs. Low minor allele frequency (MAF <5%) SNPs were excluded from analysis. IMPUTE2 software was utilized for imputation of 34,985,077 unobserved polymorphisms using the multiethnic 1000 Genomes Project Phase 3 reference panel. Imputed genotype probabilities were converted to most-likely genotypes using GTOOL; only most-likely genotypes with probabilities > 0.9 were retained for statistical analysis. Imputed SNPs with INFO scores < 0.5 or those deviating from Hardy-Weinberg equilibrium (p < 1.00 × 10−4 in European controls) were also excluded from the analysis. Population structure was investigated by using a principal components analysis using the R package SNPRelate following the approach introduced by Patterson et al. (2006). The University of Washington Genetics Coordinating Center released a full online report on the data cleaning, quality assurance, ancestry analyses and imputation for this study (http://www.ccdg.pitt.edu/docs/Marazita_ofc_QC_report_feb2015.pdf, last accessed April 25, 2016).
Statistical Analysis:
We examined potential gene-by-sex interactions following a case-control approach first introduced by Kraft et. al (Kraft et al., 2007). In the first step, logistic regression examining main effects of genotype (using the additive genetic model) and sex, as well as the interaction between them, was performed using PLINK (v1.9) (Purcell et al., 2007). Eighteen principal components of ancestry were identified and used as covariates in the analysis. A two degree of freedom likelihood ratio test was used to assess the joint association of the genetic and gene-by-sex components. SNPs demonstrating at least moderate evidence of association (i.e. p-value < 1.00 × 10−5) in the two degree of freedom test were then individually examined in the second step. Because there are very strong main effects of genotype for CL/P, we then interrogated the G × S term (from the same model) to identify significant interactions, using a genome-wide significance threshold of 5.00 × 10−8 and a suggestive threshold of 1.00 × 10-5. Overall, this approach examines potential genetic risk factors for OFC which differ between males and females.
Results
We previously performed conventional tests of association to identify marginal gene (G) effects increasing risk for NSCL/P (Leslie, Carlson, et al., 2016). Here we carried out a genome-wide screen for G × S interaction where we first examined the 2df test for gene (G) and gene-sex (G × S) interaction. Our analysis identified four loci that achieved genome-wide significance in the 2df test. Three of these loci (IRF6 on 1q32, 8q24, and NTN1 on 17p13) showed very strong G effects, consistent with our previous analysis of this dataset (Leslie, Carlson, et al., 2016). The fourth locus, on 10q21, represented a new signal (lead SNP rs72804706, p2df = 3.67 × 10−8), driven by a strong G × S interaction.
We then examined the test for G × S interaction alone for all SNPs demonstrating at least suggestive evidence of association in the 2df test. Because the goal of this study was to identify interactions, we focused on signals showing evidence of G × S interaction (Figure 1). 128 loci containing 1,649 SNPs surpassed a suggestive p-value threshold in the 2df test. Of these, 252 SNPs from 17 loci showed evidence of a suggestive statistical G × S interaction (pG×S < 1.00 × 10−5), with one locus demonstrating a genome-wide significant interaction effect (10q21, lead SNP rs72804706, pG×S = 6.69 × 10−9; Table 2). Interestingly, we did not find evidence of sex-specific effects for any previously identified NSCL/P risk loci.
Figure 1.
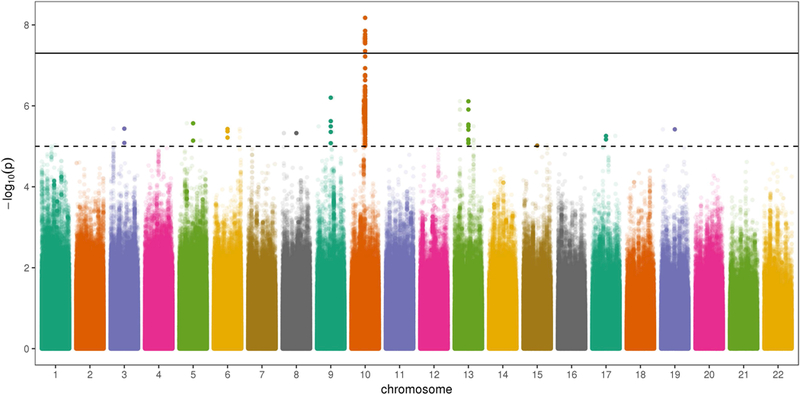
Manhattan plot displaying the p-values from the gene-by-sex effect GWAS. Bolded points indicate a variant for which p-value for the two degree of freedom joint test of gene and gene-by-sex effect was less than 1.00 × 10−5.
Table 2. 17 loci with evidence for G × S interactions.
| Locus | SNP | Chrom. | Nuc. Position (hg19) |
Closest Gene |
A1 | A2 | MAF | P.2df | P.G×S |
|---|---|---|---|---|---|---|---|---|---|
| 3p24.3 | rs12490991 | 3 | 16457095 | RFTN1 | A | G | 0.308 | 1.85E-05 | 3.66E-06 |
| 5p13.1 | rs72753006 | 5 | 42310935 | GHR | C | G | 0.080 | 1.53E-05 | 2.70E-06 |
| 5q31.3 | rs34993514 | 5 | 139651948 | PFDN1 | A | AT | 0.425 | 3.46E-05 | 7.25E-06 |
| 6q26 | rs6932653 | 6 | 163666803 | PACRG | C | C | 0.425 | 1.26E-05 | 3.74E-06 |
| 8p23.2 | rs2623571 | 8 | 3572809 | CSMD1 | T | T | 0.109 | 2.75E-05 | 4.72E-06 |
| 9p24.1 | rs148091490 | 9 | 4950144 | JAK2 | C | T | 0.079 | 1.89E-05 | 3.22E-06 |
| 9q21.32 | rs1246259 | 9 | 86766132 | SLC28A3 | A | G | 0.328 | 1.20E-05 | 2.39E-06 |
| 9q22.1 | rs11142081* | 9 | 90689210 | SPATA31C2 | T | G | 0.137 | 2.09E-06 | 6.29E-07 |
| 10q21.1a | rs370068281 | 10 | 55079463 | PCHD15 | ATG | A | 0.165 | 1.28E-05 | 2.06E-06 |
| 10q21.1b | rs72804706 | 10 | 59564480 | IPMK | C | T | 0.102 | 3.67E-08 | 6.69E-09 |
| 13q13.3 | rs77590619 | 13 | 36148527 | NBEA | G | A | 0.256 | 4.78E-06 | 7.72E-07 |
| 13q21.33 | rs17087328 | 13 | 71141833 | LINC00438 | T | C | 0.089 | 4.39E-05 | 8.23E-06 |
| 13q31.3 | rs28793707 | 13 | 86486416 | SLITRK6 | T | C | 0.163 | 1.64E-05 | 3.20E-06 |
| 15q26.1 | rs11855106 | 15 | 93599565 | RGMA | G | A | 0.376 | 4.24E-05 | 9.59E-06 |
| 17q21.31 | -- | 17 | 42535623 | GPATCH8 | GA | G | 0.106 | 2.57E-05 | 6.76E-06 |
| 17q25.1 | rs4479318 | 17 | 70906038 | SLC39A11 | T | T | 0.481 | 2.74E-05 | 5.54E-06 |
| 19p13.3 | rs7247005 | 19 | 3416299 | NFIC | G | A | 0.221 | 1.41E-05 | 3.81E-06 |
indicates genotyped SNPs, others are imputed
The 10q21 locus included 220 SNPs with evidence of G × S interaction (Figure 2B). For lead SNP rs72804706, we estimated the odds of CL/P to be 28% lower in males for each additional copy of the T allele. By contrast, the odds of CL/P in females were estimated to be 81% higher with each additional copy. However, it is worth noting that males had much higher estimated risk of CL/P than females for individuals with the homozygous reference genotype; the interaction effect is realized in the increasing risk for females with copies of the risk allele, T (Figure 2E). Post-hoc power was estimated to be 75% for the effects observed with rs72804706 (Kooperberg, 2015).
Figure 2.
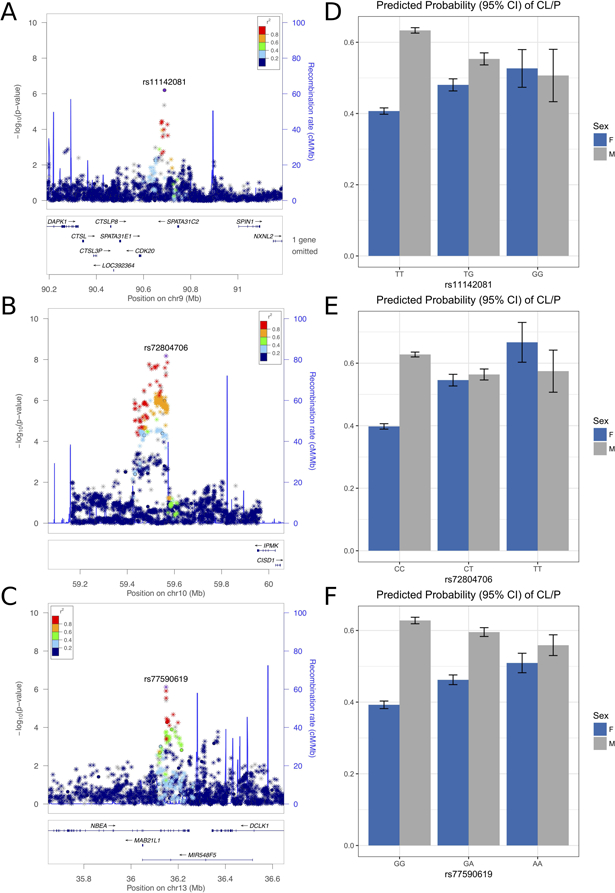
Results for the 9q22.1 (A,D), 10q21.1 (B,E), and 13q13.3 (C,F) loci. Regional association plot show results for the gene-by-sex effect model. Points are color-coded based on linkage disequilbrium (r2) in Europeans. Plots were generated with LocusZoom (Pruim et al., 2010). Bar charts of the predicted probability of CL/P are shown, stratified by sex and adjusting for principal components of ancestry. Plots were generated with the ggplot2 package in R (R Core Team, 2017; Wickham, 2009).
Sixteen loci had suggestive evidence of G × S interactions, including loci on 9q22.1 and 13q13.3 with G × S interactions approaching genome-wide significance (p < 1.00 × 10−7; Figure 2A,C). At 9q22.1, for lead SNP rs11142081, we estimated the odds of CL/P to be 28% higher in females and 31% lower in males for each additional copy of the G allele (Figure 2D). At 13q13.3, for lead SNP rs77590619, the odds of CL/P were estimated to be 28% higher in females and 21% lower in males for each copy of the A allele (Figure 2F). The findings for these loci are similar to that for 10q21, in that males had a much higher risk of CL/P for individuals with the reference homozygous genotype.
Because the incidence of CL/P among males and females is opposite the incidence of CP (CP being more common among females), we next wanted to determine if the direction of the effect for our top SNPs was the same among CP cases. Because most genetic studies have focused on CL/P, the samples sizes are still quite small for CP (Leslie, Liu, et al., 2016). As interaction tests require much larger samples sizes to maintain statistical power than tests for main effects alone, we performed a qualitative analysis of the unadjusted minor allele frequencies of these SNPs in CP cases and controls (Table 3). The frequencies of the lead SNP at 10q21 did not suggest an interaction for CP, but the frequencies of 13q13.3 and 9q22.1 SNPs were consistent with the trends we observed in the CL/P analysis (higher frequencies among female cases and male controls).
Table 3. Minor allele frequencies for top CL/P loci in CP cases.
| rs72804706 | rs11142081 | rs77590619 | ||||
|---|---|---|---|---|---|---|
| N | MAF | N | MAF | N | MAF | |
| Male CP | 97 | 0.082 | 98 | 0.097 | 97 | 0.242 |
| Female CP | 145 | 0.066 | 145 | 0.155 | 138 | 0.290 |
| Male Controls | 697 | 0.123 | 713 | 0.158 | 705 | 0.271 |
|
Female Controls |
973 | 0.079 | 987 | 0.126 | 967 | 0.231 |
Discussion
Our results support interaction between SNPs in autosomal genes and sex leading to higher risk of CL/P. This finding is not surprising as incidence rates of CL/P differ by sex, however, this is the first study to use a genome-wide approach to identify the genetic contribution to this phenomenon. Outside of the OFC literature, there is substantial evidence that genetic variation has sex-dependent effects; a 2014 study catalogued 33 sex-dependent loci in 26 medically-relevant traits (Gilks et al., 2014). The well-documented advantages of the unbiased GWAS are useful in studies such as ours, where the mechanisms of the CL/P male bias are not understood. Consequently, this study should be viewed as hypothesis generating because gene-by-sex interactions identify modifiers whose biological impact could occur far upstream of craniofacial development. Model systems provide evidence for some of these mechanisms, such as sex-specific trans-eQTLs (Derome et al., 2008) and sex-specific epistasis (Bhasin et al., 2008), but the biochemical causes of these effects likely involve sex hormones, sex-biased methylation (McCarthy et al., 2014), and/or interactions with sex chromosomes. Given the diversity of possible mechanisms, it is impossible to adequately explore or discuss each locus, so we will only briefly discuss what is known about the genes located closest to the top three association signals.
The primary finding of this study was a novel locus at 10q21.1 with a genome-wide significant G × S interaction for multiple SNPs. The lead SNP, rs72804706, is located in a large intergenic region 400kb upstream of inositol polyphosphate multikinase (IPMK). Within 500kb either side of the lead SNP are formally significant and suggestive association signals from genome-wide association studies covering a wide range of traits and disease including diabetic retinopathy, aggressiveness in attention deficit hyperactivity disorder, alcohol dependence, cardiovascular traits, and inflammatory bowel disease (MacArthur et al., 2017). Further, a truncation mutation in IPMK was identified by linkage and exome sequencing in a large family with small intestinal carcinoids (Sei et al., 2015). Although it is not known if any of the associated nearby SNPs regulate IPMK, the variety of disease process is consistent with the diverse roles of IPMK. IPMK is considered to be a pleiotropic protein with both kinase and non-catalytic functions in multiple cellular processes, including transcriptional coactivation, corepression, and cell signaling (Kim et al., 2017; Xu et al., 2013). IPMK-null mice die early in embryonic development and have multiple morphological defects, including neural tube misfolding (Frederick et al., 2005). Tissue-specific knockouts will be needed to dissect the specific roles of IPMK in OFCs and other disease. Among our other findings are loci at 13q13.3, and 9q22.1. The 13q13.3 lead SNP is located within an intron of neurobeachin (NBEA), a synaptic scaffolding protein associated with autism (Castermans et al., 2003) and migraine in bipolar disorder (Jacobsen et al., 2015). Little is known about spermatogenesis-associated protein 31C2 (SPATA31C2), the proximal gene at the 9q22.1 locus. The implicated variants from our analyses in these three regions are associated with higher risk of CL/P for females carrying the minor risk allele; this trend is not present in males, who have higher incidence of CL/P than females.
We did not perform a formal interaction analysis for CP, but found a similar trend in unadjusted minor allele frequencies for our associated SNPs for the 9q22.1 and 13q13.3 loci. This would suggest that these loci we identified do not completely explain the bias in CL/P and CP incidence in males and females because if they did, we would expect no effect or the opposite effect in CP. Thus, the alleles at these loci may increase risk for any kind of OFC in sex-specific manner. Although the female palate is known to close approximately one week later than the male palate, increasing risk for CP and possibly explaining the female preponderance of CP (Burdi & Silvey, 1969), genetic variants may contribute to this delay so formal interaction studies should be performed in this OFC subtype. Altogether our current findings demonstrate the capability of genome-wide interaction studies to identify sex-specific effects, but more research is warranted to investigate the underlying sex discrepancy in the incidence of both CL/P and CP.
Acknowledgments
We gratefully acknowledge the participation of the families, field staff, and collaborators around the world who made this project possible. This work was supported by grants from the National Institutes of Health (NIH): R00-DE025060 [EJL], X01-HG007485 [MLM, EF], R01-DE016148 [MLM, SMW], U01-DE024425 [MLM], R37-DE008559 [JCM, MLM], R01-DE009886 [MLM], R21-DE016930 [MLM], R01-DE014667 [LM], R01-DE012472 [MLM], R01-DE011931 [JTH], R01-DE011948 [KC], U01-DD000295 [GLW], K99-DE024571 [CJB]; and PICT-2016–3869 [FAP].], S21-MD001830 [CJB], U54-MD007587 [CJB]). Additional support was provided by the Philippine Band of Mercy, the Hope Foundation Inc., Operation Smile, and the Noordhoff Craniofacial Foundation Philippines Inc. Genotyping and data cleaning were provided via an NIH contract to the Johns Hopkins Center for Inherited Disease Research: HHSN268201200008I.
Grant Numbers:
R00-DE025060, X01-HG007485, R01-DE016148, U01-DE024425, R37-DE008559, R01-DE009886, R21-DE016930, R01-DE014667, R01-DE012472, R01-DE011931, R01-DE011948, U01-DD000295, K99-DE024571; PICT-2016–3869, S21-MD001830, U54-MD007587
References
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, Redett RA, Raymond G, Schwender H, Jin SC, Cooper ME, Dunnwald M, Mansilla MA, Leslie E, Bullard S, Lidral AC, Moreno LM, Menezes R, Vieira AR, Petrin A, Wilcox AJ, Lie RT, Jabs EW, Wu-Chou YH, Chen PK, Wang H, Ye X, Huang S, Yeow V, Chong SS, Jee SH, Shi B, Christensen K, Melbye M, Doheny KF, Pugh EW, Ling H, Castilla EE, Czeizel AE, Ma L, Field LL, Brody L, Pangilinan F, Mills JL, Molloy AM, Kirke PN, Scott JM, Arcos-Burgos M, & Scott AF (2010). A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet, 42(6), 525–529. 10.1038/ng.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, Murray T, Redett RJ, Fallin MD, Liang KY, Wu T, Patel PJ, Jin SC, Zhang TX, Schwender H, Wu-Chou YH, Chen PK, Chong SS, Cheah F, Yeow V, Ye X, Wang H, Huang S, Jabs EW, Shi B, Wilcox AJ, Lie RT, Jee SH, Christensen K, Doheny KF, Pugh EW, Ling H, & Scott AF (2011). Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genetic Epidemiology, 35(6), 469–478. 10.1002/gepi.20595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Taub MA, Scott AF, Murray JC, Marazita ML, Schwender H, Parker MM, Hetmanski JB, Balakrishnan P, Mansilla MA, Mangold E, Ludwig KU, Noethen MM, Rubini M, Elcioglu N, & Ruczinski I (2013). Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum Genet, 132(7), 771–781. 10.1007/s00439-013-1283-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasin JM, Chakrabarti E, Peng DQ, Kulkarni A, Chen X, & Smith JD (2008). Sex specific gene regulation and expression QTLs in mouse macrophages from a strain intercross. PLoS One, 3(1), e1435 10.1371/journal.pone.0001435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de Assis N, Alblas MA, Barth S, Freudenberg J, Lauster C, Schmidt G, Scheer M, Braumann B, Berge SJ, Reich RH, Schiefke F, Hemprich A, Potzsch S, Steegers-Theunissen RP, Potzsch B, Moebus S, Horsthemke B, Kramer FJ, Wienker TF, Mossey PA, Propping P, Cichon S, Hoffmann P, Knapp M, Nothen MM, & Mangold E (2009). Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet, 41(4), 473–477. 10.1038/ng.333 [DOI] [PubMed] [Google Scholar]
- Burdi AR, & Silvey RG (1969). Sexual differences in closure of the human palatal shelves. Cleft Palate J, 6, 1–7. [PubMed] [Google Scholar]
- Castermans D, Wilquet V, Parthoens E, Huysmans C, Steyaert J, Swinnen L, Fryns JP, Van de Ven W, & Devriendt K (2003). The neurobeachin gene is disrupted by a translocation in a patient with idiopathic autism. J Med Genet, 40(5), 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai JY, Kooperberg C, Leblanc M, & Prentice RL (2012). Two-stage testing procedures with independent filtering for genome-wide gene-environment interaction. Biometrika, 99(4), 929–944. 10.1093/biomet/ass044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derome N, Bougas B, Rogers SM, Whiteley AR, Labbe A, Laroche J, & Bernatchez L (2008). Pervasive sex-linked effects on transcription regulation as revealed by expression quantitative trait loci mapping in lake whitefish species pairs (Coregonus sp., Salmonidae). Genetics, 179(4), 1903–1917. 10.1534/genetics.107.086306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, & Murray JC (2011). Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet, 12(3), 167–178. 10.1038/nrg2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick JP, Mattiske D, Wofford JA, Megosh LC, Drake LY, Chiou ST, Hogan BL, & York JD (2005). An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc Natl Acad Sci U S A, 102(24), 8454–8459. 10.1073/pnas.0503706102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks WP, Abbott JK, & Morrow EH (2014). Sex differences in disease genetics: evidence, evolution, and detection. Trends Genet, 30(10), 453–463. 10.1016/j.tig.2014.08.006 [DOI] [PubMed] [Google Scholar]
- Grant SF, Wang K, Zhang H, Glaberson W, Annaiah K, Kim CE, Bradfield JP, Glessner JT, Thomas KA, Garris M, Frackelton EC, Otieno FG, Chiavacci RM, Nah HD, Kirschner RE, & Hakonarson H (2009). A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J Pediatr, 155(6), 909–913. 10.1016/j.jpeds.2009.06.020 [DOI] [PubMed] [Google Scholar]
- Grosen D, Bille C, Petersen I, Skytthe A, Hjelmborg J, Pedersen JK, Murray JC, & Christensen K (2011). Risk of oral clefts in twins. Epidemiology, 22(3), 313–319. 10.1097/EDE.0b013e3182125f9c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen KK, Nievergelt CM, Zayats T, Greenwood TA, Anttila V, Akiskal HS, Bi GSC, Consortium IHG, Haavik J, Bernt Fasmer O, Kelsoe JR, Johansson S, & Oedegaard KJ (2015). Genome wide association study identifies variants in NBEA associated with migraine in bipolar disorder. J Affect Disord, 172, 453–461. 10.1016/j.jad.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Skare O, Lie RT, Wilcox AJ, Christensen K, Christiansen L, Nguyen TT, Murray JC, & Gjessing HK (2012). X-linked genes and risk of orofacial clefts: evidence from two population-based studies in Scandinavia. PLoS One, 7(6), e39240 10.1371/journal.pone.0039240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Ahn H, Kim MG, Lee H, & Kim S (2017). The Expanding Significance of Inositol Polyphosphate Multikinase as a Signaling Hub. Mol Cells, 40(5), 315–321. 10.14348/molcells.2017.0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimani JW, Shi M, Daack-Hirsch S, Christensen K, Moretti-Ferreira D, Marazita ML, Field LL, Canady JW, & Murray JC (2007). X-chromosome inactivation patterns in monozygotic twins and sib pairs discordant for nonsyndromic cleft lip and/or palate. Am J Med Genet A, 143A(24), 3267–3272. 10.1002/ajmg.a.32098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooperberg CH, L. (2015). powerGWASinteraction: Power Calculations for GxE and GxG Interactions for GWAS [Google Scholar]
- Kraft P, Yen YC, Stram DO, Morrison J, & Gauderman WJ (2007). Exploiting gene-environment interaction to detect genetic associations. Hum Hered, 63(2), 111–119. 10.1159/000099183 [DOI] [PubMed] [Google Scholar]
- Lammer EJ, Shaw GM, Iovannisci DM, & Finnell RH (2005). Maternal smoking, genetic variation of glutathione s-transferases, and risk for orofacial clefts. Epidemiology, 16(5), 698–701. [DOI] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Butali A, Buxo CJ, Castilla EE, Christensen K, Deleyiannis FW, Leigh Field L, Hecht JT, Moreno L, Orioli IM, Padilla C, Vieira AR, Wehby GL, Feingold E, Weinberg SM, Murray JC, Beaty TH, & Marazita ML (2017). Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum Genet, 136(3), 275–286. 10.1007/s00439-016-1754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, McHenry T, Resick J, Sanchez C, Jacobs J, Emanuele B, Vieira AR, Neiswanger K, Lidral AC, Valencia-Ramirez LC, Lopez-Palacio AM, Valencia DR, Arcos-Burgos M, Czeizel AE, Field LL, Padilla CD, Cutiongco-de la Paz EM, Deleyiannis F, Christensen K, Munger RG, Lie RT, Wilcox A, Romitti PA, Castilla EE, Mereb JC, Poletta FA, Orioli IM, Carvalho FM, Hecht JT, Blanton SH, Buxo CJ, Butali A, Mossey PA, Adeyemo WL, James O, Braimah RO, Aregbesola BS, Eshete MA, Abate F, Koruyucu M, Seymen F, Ma L, de Salamanca JE, Weinberg SM, Moreno L, Murray JC, & Marazita ML (2016). A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum Mol Genet, 25(13), 2862–2872. 10.1093/hmg/ddw104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Liu H, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, Jain D, Laurie CC, Doheny KF, McHenry T, Resick J, Sanchez C, Jacobs J, Emanuele B, Vieira AR, Neiswanger K, Standley J, Czeizel AE, Deleyiannis F, Christensen K, Munger RG, Lie RT, Wilcox A, Romitti PA, Field LL, Padilla CD, Cutiongco-de la Paz EM, Lidral AC, Valencia-Ramirez LC, Lopez-Palacio AM, Valencia DR, Arcos-Burgos M, Castilla EE, Mereb JC, Poletta FA, Orioli IM, Carvalho FM, Hecht JT, Blanton SH, Buxo CJ, Butali A, Mossey PA, Adeyemo WL, James O, Braimah RO, Aregbesola BS, Eshete MA, Deribew M, Koruyucu M, Seymen F, Ma L, de Salamanca JE, Weinberg SM, Moreno L, Cornell RA, Murray JC, & Marazita ML (2016). A Genome-wide Association Study of Nonsyndromic Cleft Palate Identifies an Etiologic Missense Variant in GRHL3. Am J Hum Genet, 98(4), 744–754. 10.1016/j.ajhg.2016.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, & Marazita ML (2013). Genetics of cleft lip and cleft palate. Am J Med Genet C Semin Med Genet, 163C(4), 246–258. 10.1002/ajmg.c.31381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, Becker J, Herberz R, AlChawa T, Nasser E, Bohmer AC, Mattheisen M, Alblas MA, Barth S, Kluck N, Lauster C, Braumann B, Reich RH, Hemprich A, Potzsch S, Blaumeiser B, Daratsianos N, Kreusch T, Murray JC, Marazita ML, Ruczinski I, Scott AF, Beaty TH, Kramer FJ, Wienker TF, Steegers-Theunissen RP, Rubini M, Mossey PA, Hoffmann P, Lange C, Cichon S, Propping P, Knapp M, & Nothen MM (2012). Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet, 44(9), 968–971. 10.1038/ng.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, Pendlington ZM, Welter D, Burdett T, Hindorff L, Flicek P, Cunningham F, & Parkinson H (2017). The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res, 45(D1), D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML (2012). The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet, 13, 263–283. 10.1146/annurev-genom-090711-163729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcano AC, Doudney K, Braybrook C, Squires R, Patton MA, Lees MM, Richieri-Costa A, Lidral AC, Murray JC, Moore GE, & Stanier P (2004). TBX22 mutations are a frequent cause of cleft palate. J Med Genet, 41(1), 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy NS, Melton PE, Cadby G, Yazar S, Franchina M, Moses EK, Mackey DA, & Hewitt AW (2014). Meta-analysis of human methylation data for evidence of sex-specific autosomal patterns. BMC Genomics, 15, 981 10.1186/1471-2164-15-981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey P (2007). Epidemiology underpinning research in the aetiology of orofacial clefts. Orthod Craniofac Res, 10(3), 114–120. 10.1111/j.1601-6343.2007.00398.x [DOI] [PubMed] [Google Scholar]
- Patel PJ, Beaty TH, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, Wu T, Murray T, Rose M, Redett RJ, Jin SC, Lie RT, Wu-Chou YH, Wang H, Ye X, Yeow V, Chong S, Jee SH, Shi B, & Scott AF (2013). X-linked markers in the Duchenne muscular dystrophy gene associated with oral clefts. Eur J Oral Sci, 121(2), 63–68. 10.1111/eos.12025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, & Reich D (2006). Population structure and eigenanalysis. PLoS Genetics, 2(12), e190 10.1371/journal.pgen.0020190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, & Willer CJ (2010). LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26(18), 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, & Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet, 81(3), 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2017). R: A language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Sei Y, Zhao X, Forbes J, Szymczak S, Li Q, Trivedi A, Voellinger M, Joy G, Feng J, Whatley M, Jones MS, Harper UL, Marx SJ, Venkatesan AM, Chandrasekharappa SC, Raffeld M, Quezado MM, Louie A, Chen CC, Lim RM, Agarwala R, Schaffer AA, Hughes MS, Bailey-Wilson JE, & Wank SA (2015). A Hereditary Form of Small Intestinal Carcinoid Associated With a Germline Mutation in Inositol Polyphosphate Multikinase. Gastroenterology, 149(1), 67–78. 10.1053/j.gastro.2015.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Christensen K, Weinberg CR, Romitti P, Bathum L, Lozada A, Morris RW, Lovett M, & Murray JC Orofacial Cleft Risk Is Increased with Maternal Smoking and Specific Detoxification-Gene Variants. The American Journal of Human Genetics, 80(1), 76–90. 10.1086/510518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Wehby GL, & Murray JC (2008). Review on genetic variants and maternal smoking in the etiology of oral clefts and other birth defects. Birth Defects Res C Embryo Today, 84(1), 16–29. 10.1002/bdrc.20117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skare O, Gjessing HK, Gjerdevik M, Haaland OA, Romanowska J, Lie RT, & Jugessur A (2017). A new approach to chromosome-wide analysis of X-linked markers identifies new associations in Asian and European case-parent triads of orofacial clefts. PLoS One, 12(9), e0183772 10.1371/journal.pone.0183772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, Du Y, Wang M, Lan F, Hu Z, Wang G, Jiang M, Ma J, Zhang X, Ma H, Ma J, Zhang W, Huang Q, Zhou Z, Ma L, Li Y, Jiang H, Xie L, Jiang Y, Shi B, Cheng J, Shen H, Wang L, & Yang Y (2015). Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun, 6, 6414 10.1038/ncomms7414 [DOI] [PubMed] [Google Scholar]
- Suphapeetiporn K, Tongkobpetch S, Siriwan P, & Shotelersuk V (2007). TBX22 mutations are a frequent cause of non-syndromic cleft palate in the Thai population. Clin Genet, 72(5), 478–483. 10.1111/j.1399-0004.2007.00891.x [DOI] [PubMed] [Google Scholar]
- Wickham H (2009). ggplot2: Elegant Graphics for Data Analysis: Springer-Verlag; New York. [Google Scholar]
- Wolf ZT, Brand HA, Shaffer JR, Leslie EJ, Arzi B, Willet CE, Cox TC, McHenry T, Narayan N, Feingold E, Wang X, Sliskovic S, Karmi N, Safra N, Sanchez C, Deleyiannis FW, Murray JC, Wade CM, Marazita ML, & Bannasch DL (2015). Genome-wide association studies in dogs and humans identify ADAMTS20 as a risk variant for cleft lip and palate. PLoS Genetics, 11(3), e1005059 10.1371/journal.pgen.1005059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Liang KY, Hetmanski JB, Ruczinski I, Fallin MD, Ingersoll RG, Wang H, Huang S, Ye X, Wu-Chou YH, Chen PK, Jabs EW, Shi B, Redett R, Scott AF, & Beaty TH (2010). Evidence of gene-environment interaction for the IRF6 gene and maternal multivitamin supplementation in controlling the risk of cleft lip with/without cleft palate. Hum Genet, 128(4), 401–410. 10.1007/s00439-010-0863-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Schwender H, Ruczinski I, Murray JC, Marazita ML, Munger RG, Hetmanski JB, Parker MM, Wang P, Murray T, Taub M, Li S, Redett RJ, Fallin MD, Liang KY, Wu-Chou YH, Chong SS, Yeow V, Ye X, Wang H, Huang S, Jabs EW, Shi B, Wilcox AJ, Jee SH, Scott AF, & Beaty TH (2014). Evidence of gene-environment interaction for two genes on chromosome 4 and environmental tobacco smoke in controlling the risk of nonsyndromic cleft palate. PLoS One, 9(2), e88088 10.1371/journal.pone.0088088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Paul BD, Smith DR, Tyagi R, Rao F, Khan AB, Blech DJ, Vandiver MS, Harraz MM, Guha P, Ahmed I, Sen N, Gallagher M, & Snyder SH (2013). Inositol polyphosphate multikinase is a transcriptional coactivator required for immediate early gene induction. Proc Natl Acad Sci U S A, 110(40), 16181–16186. 10.1073/pnas.1315551110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdy MM, Honein MA, & Xing J (2007). Reduction in orofacial clefts following folic acid fortification of the U.S. grain supply. Birth Defects Res A Clin Mol Teratol, 79(1), 16–23. 10.1002/bdra.20319 [DOI] [PubMed] [Google Scholar]
- Yu Y, Zuo X, He M, Gao J, Fu Y, Qin C, Meng L, Wang W, Song Y, Cheng Y, Zhou F, Chen G, Zheng X, Wang X, Liang B, Zhu Z, Fu X, Sheng Y, Hao J, Liu Z, Yan H, Mangold E, Ruczinski I, Liu J, Marazita ML, Ludwig KU, Beaty TH, Zhang X, Sun L, & Bian Z (2017). Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat Commun, 8, 14364 10.1038/ncomms14364 [DOI] [PMC free article] [PubMed] [Google Scholar]


