Abstract
Biomarkers are nucleic acids, proteins, single-cells, or small molecules in human tissues or biological fluids whose reliable detection can be used to confirm or predict disease and disease states. Sensitive detection of biomarkers is therefore critical in a variety of applications including disease diagnostics, therapeutics, and drug screening. Unfortunately for many diseases, low abundance of biomarkers in human samples and low sample volumes render standard benchtop platforms like 96-well plates ineffective for reliable detection and screening. Discretization of bulk samples into a large number of small volumes (fL-nL) via droplet microfluidic technology offers a promising solution for high-sensitivity and high-throughput detection and screening of biomarkers. Several microfluidic strategies exist for high-throughput biomarker digitization into droplets, and these strategies have been utilized by numerous droplet platforms for nucleic-acid, protein, and single-cell detection and screening. While the potential of droplet based platforms has led to burgeoning interest in droplets, seamless integration of sample preparation technologies and automation of platforms from biological sample to answer remain critical components that can render these platforms useful in the clinical setting in the near future.
Graphical abstract
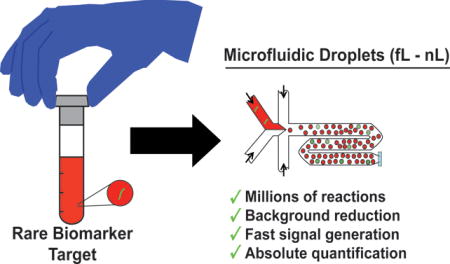
High-sensitivity and high-throughput microfluidic droplet platforms promise rapid and quantitative detection and screening of disease biomarkers from clinical samples
1. Introduction
Biomarkers are disease related cellular and molecular changes in tissues or bodily fluids, and their reliable detection and quantification is of utmost importance for clinical diagnostic and therapeutic applications (Strimbu & Tavel, 2011). Nucleic acids, proteins, single-cells, and small molecules within a tissue, cells, or biological fluid all comprise types of biomarkers found in the human body. Detection of changes, additions, or deletions of these biomarkers may serve as surrogate clinical endpoints that can be used to confirm and even predict disease and disease states (Aronson, 2005). As a result, biomarkers are instrumental in a variety of applications including point-of-care diagnostics, drug screening, and medication therapy management. Unfortunately for many diseases, biomarkers may exist at very low quantities in human samples and are often undetectable by standard benchtop techniques. Moreover, most standard protocols are largely incapable of detecting biomarkers directly from patient samples, and therefore require complex multi-step sample preparation protocols that are time consuming and can delay patient care. Indeed, the development of highly sensitive methods for detection and screening of biomarkers can not only improve clinical outcomes, but also plays a pivotal role in the increasing prevalence of preventative and personalized medicine (Rinaldi, 2011).
Since its inception in the late 1990s, microfluidics has been a popular approach for the detection of disease biomarkers due to its ability of precisely manipulating sub-microliters of samples and accelerating biochemical reactions. Advances in using various formats of microfluidic technologies for molecular and cellular biomarker detection have been extensively reviewed in literature (Choi, Ng, Fobel, & Wheeler, 2012; Chou, Lee, Yang, Huang, & Lin, 2015; Hung, Wu, Hsieh, & Lee, 2014; Nahavandi et al., 2014; Niu & deMello, 2012; Valérie Taly, Pekin, Abed, & Laurent-Puig, 2012a; Zec, Shin, & Wang, 2014; Y. Zhang & Nguyen, 2017). Notably, researchers have developed surface droplet platforms (Y. Zhang & Nguyen, 2017) whereby individual droplets containing the biomarker of interest may be manipulated (cut, moved, mixed, merged, etc. (Cho, Moon, & Kim, 2003)) in an open planar array by pneumatic (C. J. Huang, Fang, Ke, Chou, & Yang, 2014), magnetic (Khaw et al., 2016; Pipper, Zhang, Neuzil, & Hsieh, 2008; Y. Zhang et al., 2011; Y. Zhang & Wang, 2013), electric (Cho et al., 2003; Miller & Wheeler, 2008; Nelson & Kim, 2012), optoelectronic (Park, Teitell, & Chiou, 2010), or acoustic (Guttenberg et al., 2005) forces. Researchers have also developed static microchamber devices (Zec et al., 2014) for passively partitioning bulk samples into finite nanoliter (Matsubara et al., 2005), picoliter, and even femtoliter volume (Rondelez et al., 2005) wells for further analysis. Both of these approaches, however, have been limited either by sensitivity in the case of surface droplet platforms or volumetric throughput in the case of microchambers.
More recently, a special class of microfluidics known as droplet microfluidics has emerged as a popular approach for the detection and screening of biomarkers. Instead of shrinking the volume of a single bulk sample, as is the case for other microfluidic approaches, droplet microfluidic devices can discretize the bulk sample into thousands to millions of microdroplets – each of which serves as an isolated reaction chamber – and thus has the potential for achieving high-sensitivity and high-throughput detection. The significant reduction in volume facilitates an equivalent reduction in background and a drastic increase in the local concentration of the biomarker of interest. This in turn increases the signal to background ratio from each isolated reaction and consequently increases the overall sensitivity of the assay. Furthermore, these microfluidic devices can potentially facilitate rapid digitization of biomarkers in entire samples into thousands to millions of microdroplets. Such high-throughput operation can be beneficial not only for detecting exceedingly rare biomarkers in a sample, but it can also enhance screening assays by increasing the number of potential conditions screened per sample. Ultimately, the high sensitivity and high throughput afforded by droplets offer the promise of clinical sample-to-answer analysis with potentially improved dynamic range compared to bulk assays.
In this article, we focus our discussion on promising microfluidic droplet platforms and review their recent developments and state-of-the-art for biomarker detection and screening, with particular emphasis on their potential clinical applicability. We first discuss the importance and prevalence of biomarker detection and screening for clinical diagnostic and therapeutic applications. Next, we review microfluidic strategies for sample digitization, high-throughput droplet generation, and workflow integration. We then present existing high-throughput droplet platforms for nucleic-acid, protein, and single-cell detection and screening, and discuss the advantages, disadvantages, and the clinical potential of these platforms. Finally, we discuss strategies for platform automation, integrated sample preparation, and throughput maximization and the potential of these droplet platforms to be adopted in a clinical setting.
2. BIOMARKERS FOR DIAGNOSTIC AND THERAPEUTIC APPLICATIONS
Biomarkers have been used for the diagnosis of diseases in as early as the 19th century when scientists began isolating and culturing pathogenic bacteria from infected patients to confirm or rule out an infection. The formulation of Koch’s postulates in 1884 was pivotal in establishing a causative relationship between an isolated microorganism and the host’s disease (Micheel & Ball, 2010) and demonstrating that the isolated pathogen can serve as a biomarker for infection. With the research and discoveries in the human genome, proteome, and metabolome, the 20th century witnessed the advent of molecular diagnostics. As a result, the prognostic and diagnostic utility of nucleic acids, proteins, and small-molecules, became apparent. For example, the detection of the rpoB gene in human sputum samples offers a much faster alternative for the detection of pathogenic Mycobacterium tuberculosis than traditional culture based methods, which can take as long as several weeks (Boehme et al., 2010). Another example is the prominent protein biomarker, prostate-specific antigen (PSA), which has been commonly used for early detection and screening for prostate cancer since its approval by the FDA in 1984 (De Angelis, Rittenhouse, Mikolajczyk, Blair Shamel, & Semjonow, 2007).
In addition to providing diagnostic information, biomarkers serve as clinical endpoint surrogates for determining therapeutic efficacy of drugs and treatments. For patients infected with HIV, routine monitoring of viral load is necessary for guiding antiretroviral therapy. The viral load is typically determined by quantifying the prevalence of HIV nucleic acid target biomarkers (eg: gag and pol (Luft, Gill, & Church, 2011)) in patient samples. For patients with bacterial infections, antimicrobial susceptibility tests (AST) are commonly performed to acquire drug sensitivity profiles for pathogens. Here, the pathogen cells of interest (in this case, the biomarker of infection) are isolated from the patient sample and grown directly in the presence of various antibiotics to determine their sensitivity/resistance (Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement, 2014). The resulting information is then used by physicians to determine an appropriate therapeutic regimen for the infected patient. The benefit of biomarkers as clinical endpoint surrogates can also be leveraged in drug efficacy and/or toxicity screening. In the case of cancer, known genetic biomarkers (HER-2, EGFR, KRAS, etc. (Garnett et al., 2012)) as well as small-molecule metabolites (Sawyers, 2008) are commonly monitored in drug screening assays, wherein cancer cells are subject to various types and concentrations of drugs.
In order to be of practical utility to the clinician, methods for detecting and screening biomarkers must be highly sensitive and highly quantitative (Strimbu & Tavel, 2011). For many diseases, biomarkers exist in such small quantities that accurate quantification is unachievable by conventional laboratory methods. For example, in the case of sepsis, the bacterial concentration in blood may be as low as 1 CFU/mL. The low concentration here requires a lengthy culturing step to detect the pathogenic bacteria, thus delaying definitive diagnosis and targeted treatment and causing poor clinical outcomes and undesired morbidity (Mancini et al., 2010). Likewise, most HIV viral load tests have a limit of detection of approximately 50 copies/mL. Accurate quantification of low levels of viremia (<50 copies/mL – 200 copies/mL) is critical to guiding antiretroviral therapy and preventing virological failure in patients with AIDS (DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents, 2016; Doyle et al., 2014).
Ideally, clinically relevant biomarker detection and screening platforms must also be able to seamlessly process patient samples in a simple sample-to-answer format. Patient samples may include blood, plasma, urine, cerebrospinal fluid, feces, sputum, peritoneal fluid or any other source containing the biomarker of interest (Nahavandi et al., 2014). However, when working with such clinical samples, two important challenges remain to be overcome – sensitivity and volume. It is known that background cells, nucleic acids, and proteins in clinical samples can interfere or inhibit the detection and quantification of the biomarker of interest (Davenport et al., 2017; Mach, Wong, & Liao, 2011). To address this challenge, protocols for the extraction and purification of biomarkers have been developed. Although such sample preparation steps may enhance assay reliability and ensure detectable signal over the background, they render the assay more complex and lengthen the assay turnaround time, which is undesirable in clinical settings. The second issue when working with clinical samples is that often, these samples may be precious and in limited quantity. Most screening workflows necessitate a single sample to be subject to multiple conditions, and volume limitations can ultimately limit the resolution of biomarker screening. Ultimately, a methodology that minimizes the effect of sample background, improves the sensitivity of biomarker detection, and improves the resolution of biomarker screening is desired.
3. MICROFLUIDIC PLATFORMS FOR SAMPLE DISCRETIZATION
Discretization of samples into thousands to millions of isolated reactions offers a potential solution to increasing sensitivity as well as throughput when detecting biomarkers (Valérie Taly, Pekin, Abed, & Laurent-Puig, 2012b; Zec et al., 2014) (Figure 1). Traditional bulk platforms for detection and screening of biomarkers, like 96-well plates, require relatively large volumes (μL-mL) and can therefore achieve at most tens of parallel reactions from a single sample. Bulk analyses become more problematic when measuring rare biomarkers accompanied by a high background in the clinical sample matrix. The relatively low biomarker concentration results in slower generation of detectable signal over the high local background, and the uncertainty in biomarker concentration necessitates cumbersome calibration curves for enabling relative quantification. In contrast, discretization of the entire sample into millions of small-volume (femtoliter to nanoliter) reactions suspended in an oil phase enables higher sensitivity and higher throughput for biomarker detection and screening. Discretization of samples into small volumes can facilitate digitization of biomarkers into isolated reaction compartments. The small volume drastically reduces background and increases the local concentration of the biomarker of interest. This in turn allows faster signal turnaround than bulk methods. Detection of these single-biomarker compartments can result in absolute quantification of the biomarker with single-copy sensitivity, obviating the need for calibration curves. Finally, running potentially millions of single-biomarker reactions from an entire sample in parallel can not only help detect rare targets but it can also enhance sample screening assays to accommodate more conditions per sample.
Figure 1. Discretization facilitates high-throughput, high-sensitivity, rapid, and quantitative analysis for rare biomarkers in a sample.
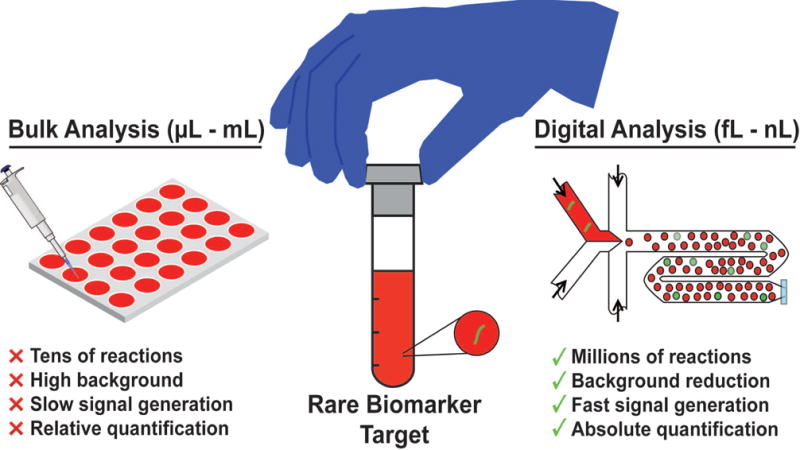
For effective detection of a biomarker of interest, conventional bulk analysis (left) is restricted to a few replicate reactions between microliters and milliliters each, wherein signal can be drowned out by high concentration of sample background, limiting overall sensitivity and speed. In contrast, digitization of sample (right) into femtoliter to nanoliter volume droplets facilitates background reduction and subsequently greater sensitivity and speed. Furthermore, encapsulation of single targets into these droplets allow for absolute quantification of rare targets.
The benefit of water-in-oil droplets for digitizing biomarkers was expounded as early as 2003 by Vogelstein and Kinzler in their seminal work describing the BEAMing protocol (Dressman, Yan, Traverso, Kinzler, & Vogelstein, 2003). In BEAMing, PCR reagents, primer functionalized magnetic beads and nucleic acid templates are manually stirred in an oil/detergent mixture to create microemulsion of droplets (M. Li, Diehl, Dressman, Vogelstein, & Kinzler, 2006). Critically, the input concentrations are selected such that each droplet within the microemulsion contains either a bead, or a single copy of nucleic acid template, or a bead along with a single copy of nucleic acid template. Following PCR amplification, beads that are co-encapsulated with a DNA template contain amplified copies of the template. These beads are then magnetically purified and tagged specifically with uniquely colored fluorescent antibodies based on their template sequence. Finally, the fluorescent beads can be counted using flow cytometry to reveal minor variants in a DNA population.
Microfluidic droplet platforms have subsequently presented an improved method for sample discretization and analysis. Specifically, BEAMing employed cumbersome and uncontrollable bulk methods for generation of emulsion, leading to polydisperse droplets and subsequently non-uniform reaction conditions therein. In contrast, microfluidic technologies enable the manipulation of very small volumes of fluids using channels and chambers with dimensions on the order of tens to hundreds of micrometers (Whitesides, 2006). The ability to manipulate small fluidic volumes is critical to discretizing bulk samples in a monodisperse manner. Therefore, microfluidics presents an effective framework for high-sensitivity single-biomarker analysis. Furthermore, by significantly reducing reaction volumes in microfluidic devices, reagent costs may be significantly lowered. Such platforms may be further amenable to process automation, streamlining the process from sample to answer, and obviating the need for human interference and unnecessary labor costs. In order to achieve high-throughput sample discretization, researchers have developed several high-throughput methods for microfluidic droplet generation. In the following section, we present notable examples of droplet generation platforms and evaluate their relative throughput and control of droplet size, content, and motion.
3.1 Active and passive methods for high-throughput generation of microfluidic droplets
Microfluidic droplets can be generated via active or passive methods (Figure 2). Active platforms enable on-demand generation of droplets with programmable control of droplet size, content, and droplet motion (“Control” in Figure 2). However, greater control generally comes at the expense of droplet throughput (“Throughput” in Figure 2), as most active designs employ moving parts that require timed and controlled actuation. In contrast passive droplet generation platforms employ non-moving structures that can disturb the interfacial tensions between oil and aqueous phases to achieve high-throughput droplet generation, but at the expense of droplet control. Herein, we highlight examples of active and passive platforms that can be used for single-biomarker discretization.
Figure 2. Methods for high-throughput droplet generation.
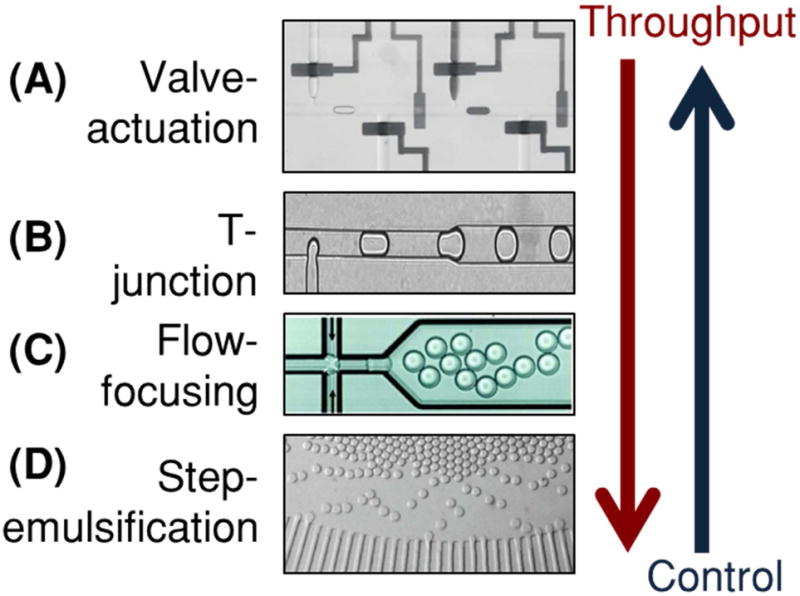
(A) Valve-actuation involves pneumatically pulsing PDMS “Quake” valves to generate droplets. They offer greater control of droplet size, content, and motion, but are limited in generation speed and throughput (Reprinted from (Guo et al., 2010) with the permission of AIP Publishing). (B) Cross-flow devices feature a “T-junction” where an aqueous stream meets a flowing continuous phase to generate droplets. (Reprinted with permission from (Zagnoni, Anderson, & Cooper, 2010). Copyright 2010 American Chemical Society) (C) Flow-focusing devices are most commonly used for droplet generation and feature a junction where a flowing aqueous stream is sheared by two perpendicularly intersecting streams of the continuous phase (Hindson et al., 2011). (D) Step-emulsification devices feature a 3-D step where an aqueous stream enters into a much larger oil reservoir, creating a droplet. (Reprinted from (R. Dangla et al., 2013). Copyright 2012 National Academy of Sciences). Step-emulsifiers may be parallelized to generate droplets at very high speeds, but are more difficult to control in content and movement.
Active designs enable on-demand generation of droplets with good control of droplet size, content and motion. These platforms typically require pre-programmed instrumentation for controlling moving parts within a microfluidic device that enable droplet control. Pneumatic “Quake” valves are one means for mechanically assembling and generating droplets (Unger, Chou, Thorsen, Scherer, & Quake, 2000) (Figure 2A). These PDMS microvalves sit above a microfluidic channel, straddling the width of the channel. When pressurized, the valves deform into the channel, constricting the flow of liquid. By selectively pressurizing and de-pressurizing a valve, droplets of water in oil can be generated with a high degree of control on droplet size (Guo et al., 2010). Furthermore, by using multiple inlets of valve controlled channels in parallel, droplets can be injected with precise volumes of reagents, and therefore droplet content can be precisely controlled for each assembled droplet (Zec, Rane, & Wang, 2012; Zeng, Li, Su, Qin, & Lin, 2009). In addition, if each droplet generated using valves spans the width of the channel it resides in, it will keep its position in a train of generated droplets. This enables one to spatially barcode an individual droplet, where the droplet’s position in a droplet train uniquely identifies the droplet and its content (Tushar D. Rane, Zec, & Wang, 2015). While pneumatic valve-based platforms confer additional control of droplets, they are traditionally limited in throughput by the actuation time of each microvalve and the minimal spacing between subsequent droplets to avoid coalescence. Other methods for active generation of droplets include the use of magnetic (Tan, Nguyen, Yobas, & Kang, 2010), optothermal (Park, Wu, Chen, Teitell, & Chiou, 2011), piezoelectric (J. Xu & Attinger, 2008), and surface acoustic forces (Schmid & Franke, 2013, 2014). While many of these methods facilitate generation of droplets on-demand, they do not provide the high level control of droplet size and content as do pneumatic valve-based methods.
In contrast to active droplet generation, passive designs can achieve high throughput but with minimal control of droplet size, content, and movement. Passive droplet devices employ micro-structures and micro-constrictions that can disturb the interfacial tensions between co-flowing streams of oil and aqueous samples in order to generate uniform dispersions of the aqueous phase in the continuous oil phase (P. Zhu & Wang, 2017). Typically, passive droplet devices require no moving parts and minimal instrumentation (eg: syringe pumps, pressure regulators, etc.), solely for controlling the flow rates or pressures of the input fluids. In 2001, Thorsen et al. reported a microfabricated “T-junction” channel geometry whereby a perpendicularly intersecting stream of oil/surfactant mixture could be used to generate monodisperse water droplets, depending on the relative flow rates of the oil and aqueous stream (Thorsen, Roberts, Arnold, & Quake, 2001) (Figure 2B). Notably, the high shear forces created at the intersection of the water and oil phases helps break the interfacial tension of the aqueous phase (Garstecki, Fuerstman, Stone, & Whitesides, 2006). This then creates a localized break of the aqueous phase that proceeds downstream as a spherical droplet. T-junction devices feature low coefficients of variation in droplet size and throughputs on the order of a few kHz (Beer et al., 2008; J. H. Xu, Li, Tán, Wang, & Luo, 2006; P. Zhu & Wang, 2017). Following the same principle, the “flow-focusing” channel geometry was designed (Anna, Bontoux, & Stone, 2003; Dreyfus, Tabeling, & Willaime, 2003; Gupta, Matharoo, Makkar, & Kumar, 2014) (Figure 2C). In these devices, the continuous oil phase is used to squeeze the dispersed aqueous phase laterally into a neck, where droplet formation occurs. Critically, the size of droplets generated from flow-focusing devices is related to the dimensions of the flow-focusing junction as well as the relative flow-rates between the continuous and dispersed phases (Stan, Tang, & Whitesides, 2009). Droplet generation rates as high as tens of kHz have been reported using flow-focusing devices (Pekin et al., 2011). For simpler device operation and potentially higher throughput droplet generation, the step-emulsification droplet generator was designed (R. Dangla, Kayi, & Baroud, 2013; Rémi Dangla, Fradet, Lopez, & Baroud, 2013; Z. Li, Leshansky, Pismen, & Tabeling, 2015) (Figure 2D). In these devices, a physical step or terrace separates the aqueous channel from a relatively large reservoir containing the oil phase. When the aqueous phase is pushed across the step at high flow rates, the drastic change in interfacial tension between the aqueous channel and the reservoir triggers droplet pinch off. Step-emulsification devices typically do not require co-flow of two phases, as only the aqueous phase is driven to produce droplets (Dutka, Opalski, & Garstecki, 2016). These devices are less resistant to variations in the aqueous flow rate, and the single-input nature of these devices make them amenable to parallelization for increased throughputs, without the need for channel distribution layers or multiple inlets (Ofner et al., 2016; Postek, Kaminski, & Garstecki, 2017). However, the lack of control on the continuous phase can result in lack of control on droplet movement following droplet generation. Other methods for passive droplet generation include the V-junction (Ding, Casadevall i Solvas, & deMello, 2015), microchannel emulsification (Sugiura, Nakajima, & Seki, 2002), cross-interface emulsification (P. Xu, Zheng, Tao, & Du, 2016), rapid emulsification (S. C. Kim et al., 2017), and membrane emulsification (Nakashima, Shimizu, & Kukizaki, 2000). Passive methods generally feature very high throughputs, up to 110 000 droplets/s (J. Lim et al., 2015), and may be parallelized for increased throughput. However, they remain limited in customizability of droplet size, content and control of droplet position during and after droplet generation.
3.2 The 3-step droplet workflow: biomarker encapsulation, incubation, detection
Once a droplet generation platform is chosen, there are three important steps that need to be implemented for successful biomarker detection and screening: (i) single-biomarker encapsulation in droplets, (ii) incubation of droplets to produce detectable signal, and (iii) detection of the signal within droplets. For most droplet platforms, encapsulation of biomarker in droplets occurs during droplet generation. The efficiency of encapsulation of biomarkers into droplets is dependent on the starting concentration of the biomarker and the size of the droplet. Passive encapsulation of biomarkers into droplets can be modeled as a Poisson process (Collins, Neild, deMello, Liu, & Ai, 2015; Sykes et al., 1992). Therefore the probability of encapsulating exactly x biomarker in each droplet can be described by Equation 1.
| (1) |
Here, represents the input concentration of the biomarker of interest, and represents the volume of the droplets being generated. Practically, for highly accurate quantification, it is important to minimize the occurrence of multiple biomarkers in a droplet (i.e., doublets) (Guan, Chen, Rane, & Wang, 2015). As such, it is imperative to operate in the “digital” Poisson regime where the expected number of biomarkers per droplet (or ) should be much less than 1. For example, ensures that the vast majority of generated droplets contains 0 or 1 copy of the biomarker of interest, and only a minute 0.47% of droplets contain 2 or more biomarkers.
Following passive encapsulation of biomarkers in droplets, droplets may be moved off-chip or into an on-chip module for the next step of the workflow, incubation, during which critical biochemical reactions and assays such as nucleic acid hybridization, PCR, substrate catalysis, and bacterial growth take place. In most cases, every step in the droplet workflow is a standalone part of the droplet platform that requires initiation by the user (eg: moving droplets into an incubation device). Each step may or may not require a separate device, but certainly requires user intervention and initiation. In most reported droplet platforms, droplets are typically generated (step 1) (Figure 3Ai) and collected in external tubes, which may be heated off-chip (step 2) in an incubator or thermocycler (Figure 3Aii). Following incubation, the droplets are reinjected into detection devices where the biochemical reaction products formed during incubation and retained within droplets are measured (step 3) with a detector (Figure 3Aiii). Notably, some droplet platforms that perform multi-step assays may require more than 3 user-initiated workflow steps prior to detection (S. C. Kim et al., 2017; Novak et al., 2011). For droplet detection, droplet platforms have predominantly employed fluorescence-based assays and a sequential, in-line flow-based detection approach. These droplet detection technologies typically consist of an excitation source and a photon detector. When fluorophores are excited using a laser source, the detection scheme is commonly referred to as laser-induced fluorescence (LIF) (Y. Zhu & Fang, 2013). LIF detectors are commonly employed in droplet platforms and may include a photodiode (Tanyeri, Perron, & Kennedy, 2007), avalanche photodiode (APD) (Kaushik et al., 2017; Nguyen, Lassemono, & Chollet, 2006), or photomultiplier tube (PMT) (Liu et al., 2016) to convert incident photons into electric current. To facilitate higher sensitivity, single-molecule detection within droplets, Rane and Puleo et al. presented a cylindrical illumination confocal spectroscope (CICS), wherein a sheet-like illumination volume is used to span the entirety of the detection volume and maximize intra-droplet detection efficiency (T. D. Rane et al., 2010). Whereas, LIF and CICS only allow for serial measurements of droplets, a higher-throughput alternative was developed by Kim et al., wherein an LED excitation source was coupled to a CMOS sensor that rested underneath 16 parallel detection channels (M. Kim et al., 2015). Droplets were imaged as they flowed by, and the authors reported detection throughput as high as 250000 droplets/s using this platform. Recently, label-free droplet interrogation methods like Raman spectroscopy (Cristobal et al., 2006; Luther, Will, & Braeuer, 2014), surface-enhance Raman spectroscopy (März, Henkel, Cialla, Schmitt, & Popp, 2011), surface-enhanced resonance Raman spectroscopy (Cecchini et al., 2011; Syme, Martino, Yusvana, Sirimuthu, & Cooper, 2012), and impedance spectroscopy (Axt, Hsieh, Nalayanda, & Wang, 2017; Kemna, Segerink, Wolbers, Vermes, & van den Berg, 2013; Niu, Zhang, Peng, Wen, & Sheng, 2007) have been developed. While these methods have been successfully used to demonstrate sensitive detection of chemical analytes within droplets, they are yet to be widely used for high-throughput detection and screening of disease biomarkers. Several challenges including operational complexity, matrix effect, droplet surface effect, and portability must be overcome to improve widespread adoption of these label-free detection technologies (Chrimes, Khoshmanesh, Stoddart, Mitchell, & Kalantar-zadeh, 2013).
Figure 3. Fragmented and integrated droplet platforms.
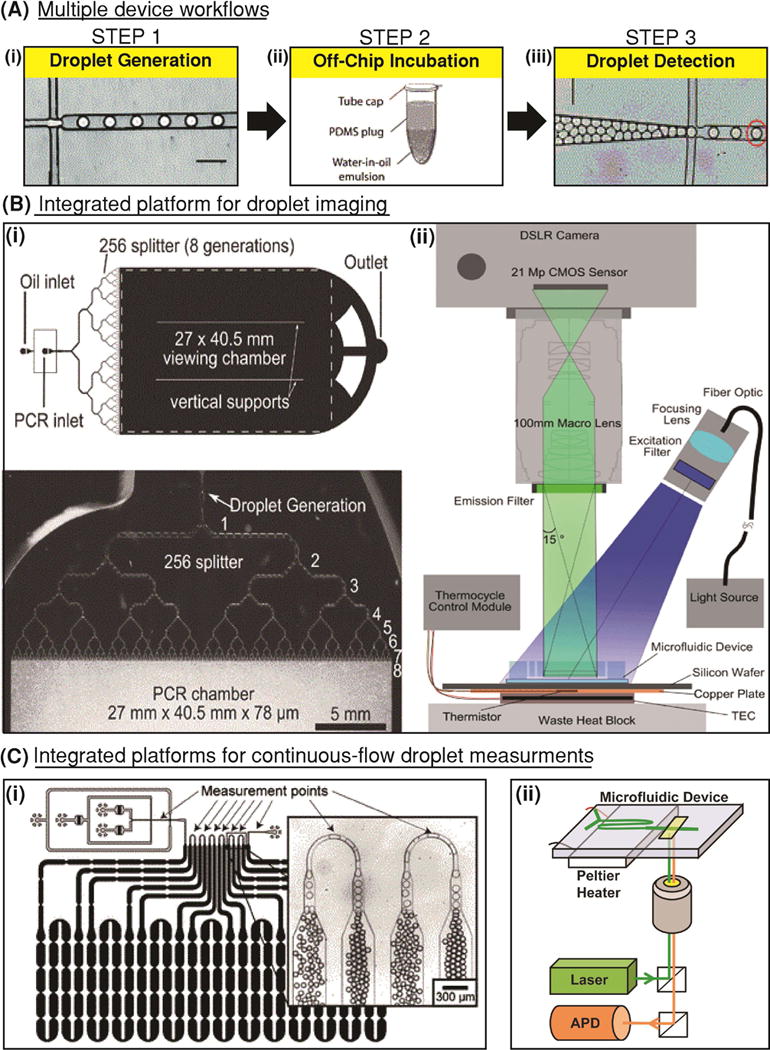
(A) Most droplet platforms utilize a separate device for (i) droplet generation, (ii) a thermocycler or incubator for droplet incubation, and (iii) another device for droplet detection. (Adapted from (Pekin et al., 2011) with permission of The Royal Society of Chemistry) (B) Imaging-based integrated platforms use (i) a single device for droplet generation and incubation. Droplets are incubated in an on-chip droplet reservoir that rests on a heat block, and (ii) real-time imaging or microscopy may be used to observe these droplets. (Reproduced from (Hatch, Fisher, Tovar, et al., 2011) with permission of The Royal Society of Chemistry). (C) Integrated platforms that facilitate continuous-flow detection of droplets typically contain (i) delay lines for on-chip droplet incubation. Often, these delay lines may contain constrictions to reduce variability in droplet speed through the incubation region. (Reproduced from (Frenz et al., 2009) with permission of The Royal Society of Chemistry). These constrictions also serve as points for droplet detection where individual droplets can be sequentially measured. (ii) Integrated continuous flow platforms may feature a Peltier heater to heat the incubation line and a laser excitation source and APD detector for continuous measurements of droplet fluorescence. (Reprinted from Biosensors and Bioelectronics, 97, Kaushik and Hsieh et al., Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform, 260-266, Copyright 2017, with permission from Elsevier.) (Kaushik et al., 2017).
3.3 The integrated workflow
Although the 3-step workflow has remained common for many years, researchers have attempted to improve upon this fragmented and cumbersome approach by integrating the 3 steps in the workflow to facilitate ease of operation and even automation. As a step in this direction, some platforms have employed 2-step workflows that make use of a device for droplet generation and a separate droplet collection chamber for integrated droplet incubation and detection (Kang et al., 2014). However, full integration in a monolithic device can only be achieved by connecting droplet generation and droplet detection by an on-chip chamber/reservoir or channel/delay line where droplets may be incubated for an appropriate duration. To this end, Hatch et al. designed a device that redirects generated droplets into an on-chip reservoir (Hatch, Fisher, Tovar, et al., 2011), where droplets may either form a 2-D planar array or a 3-D packed stack (Hatch, Fisher, Pentoney, Yang, & Lee, 2011). In the reservoir, droplets were collectively heated (Figure 3Bi). An LED excitation source and a wide-field camera were used for real-time fluorescence detection across all droplets (Figure 3Bii). While such a platform integrates the functions of droplet generation, incubation, and detection, its throughput and dynamic range are ultimate limited by the volume of the on-chip reservoir, the field of view of the imaging system, as well as the resolution of the imaging camera. For increased flexibility with sample volume, an integrated design compatible with continuous flow operation is desired. In continuous-flow operation, droplet generation, incubation, and detection can be performed in parallel. In order to facilitate continuous-flow incubation, an on-chip delay line may be employed. A noteworthy example is the delay line designed by Frenz et al. (Frenz, Blank, Brouzes, & Griffiths, 2009), which features narrow constrictions to obviate Taylor dispersion of droplets (Taylor, 1934). Due to parabolic flow profiles within a channel, droplets in the center of the channel travel faster than droplets closer to the edge. This results in high variation in incubation time for droplets over larger durations of incubation. By placing equidistant constrictions throughout length of the incubation channel, droplets are continuously redistributed through the width of the delay lines, therefore reducing the variation in incubation duration (Figure 3Ci). Following on-chip incubation, droplets can be detected sequentially in continuous flow by creating a constriction in the delay line where only a single droplet may pass through at a time. Kaushik and Hsieh et al. utilized an on-chip delay line with a detection constriction for individually interrogating droplets during continuous-flow operation (Kaushik et al., 2017). In this work, the microfluidic device was directly connected to supporting instrumentation, which consisted of a Peltier heating device, an optical excitation source and a fluorescence detector (Figure 3Cii). These works highlight some of the advantages of platforms with an integrated workflow. By parallelizing droplet generation, incubation, and detection in a continuous flow, these devices reduce assay idle time and manual intervention, making them more amenable to process automation and potential adoption in clinical settings.
4. DETECTION AND SCREENING OF NUCLEIC ACID BIOMARKERS
Several microfluidic droplet platforms have been developed for detecting nucleic acid markers specific to cancer and infectious diseases, among other applications. An early implementation of droplet-based PCR from Beer et al. showed the benefit of confining single-molecules in 10 pL droplets in order to reduce background and determine signal over background 20 cycles faster than traditional tube-based PCR reactions (Beer et al., 2007). Since then, platforms, both commercial and custom, have been developed that have demonstrated high speed, high sensitivities and high throughputs for a variety of applications. Herein we present droplet platforms developed for quantifying oncogenic mutations, for rapid detection of low-abundance viral nucleic acids, and for amplification-based and amplification-free detection of bacterial RNA and DNA. Table 1 provides a brief summary of nucleic acid biomarkers, along with their associated diseases of concern, that have been detected using high-throughput droplet platforms. Additionally, we make note of the droplet generation format used by the method, the approximate number of droplets processed per experiment, and the number of distinct workflow steps as metrics to evaluate the droplet device and platform. In order to evaluate sensitivity achieved by these methods, we also include the volume of droplets employed, the assay used, and, the input sample into the system, followed by associated references.
Table 1.
Summary of droplet platforms applied to nucleic acids biomarker detection and screening
| Biomarker | Disease | Droplet Format | Throughput (per run) | Workflow steps | Droplet volume | Assay | Clinical sample | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| BRAF, EGFR, TP53 | Cancer | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | DNA extracted from plasma, FFPE samples | (Chang-Hao Tsao et al., 2015; Héritier et al., 2016; Hindson et al., 2011; Oxnard et al., 2014; Reid, Freeman, Millward, Ziman, & Gray, 2015; Sacher et al., 2016; Sanmamed et al., 2015; Takahama et al., 2016; Thress et al., 2015; van Ginkel, Huibers, van Es, de Bree, & Willems, 2017; Watanabe et al., 2015; G. Zhu et al., 2015; Zonta et al., 2016) |
| erbB2/HER2 | Breast cancer | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | DNA extracted from FFPE samples | (Belgrader et al., 2013; Heredia et al., 2013; Kinugasa et al., 2015; Otsuji et al., 2016) |
| KRAS | Colorectal cancer, lung cancer | Flow-focusing | ~ 106 | 3 | 4 pL, 9 pL | PCR | DNA extracted from plasma, FFPE samples | (Laurent-Puig et al., 2015; Pekin & Taly, 2017; Valerie Taly et al., 2013; Zhong et al., 2011) |
| pAdeasy-1 | Viral infection | Flow-focusing | Cont. flow | 1 | 65 pL | PCR | NA | (Kiss et al., 2008) |
| MS2 RNA | Viral infection | T-junction | ~ 102 | 1 | 70 pL | PCR | MS2 virions | (Beer et al., 2008) |
| HTLV-1 tax | Viral infection | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | DNA extracted from T cells in blood and CSF | (Brunetto et al., 2014) |
| HBV DNA | Hepatitis B | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | DNA extracted from FFPE samples | (J. T. Huang et al., 2015) |
| HCV RNA | Hepatitis C | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | RNA extracted from serum followed by reverse transcription in bulk | (Mukaide et al., 2014) |
| HCMV DNA | Viral infection | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | whole virus in cell lysate and Tris-HCl buffer | (Pavšič, Žel, & Milavec, 2016) |
| CCL3L1 | AIDS | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | NA | (Hindson et al., 2011) |
| HIV-1 RNA | AIDS | Flow-focusing | ~ 104 | 3 | ~ 1 nL | RT-PCR | RNA extracted from patient PBMC samples | (Kiselinova et al., 2014) |
| HIV-1 DNA (pol, 2-LTR) | AIDS | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | DNA extracted from patient blood | (Strain et al., 2013) |
| HPV DNA | Ano-genital carcinoma | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | DNA isolated from patient serum | (Jeannot et al., 2016) |
| H5N1 RNA | Influenza | Cross interface emulsification | ~ 103 | 3 | 200 pL - 4 nL | LAMP | RNA extracted from virus followed by reverse transcription | (Hu, Xu, Luo, He, & Du, 2017) |
| S. aureus nuc and mecA genes | Bacterial Infection | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | gDNA extracted from nasal swabs, S. aureus spiked into nasal swab, PBS | (Kelley, Cosman, Belgrader, Chapman, & Sullivan, 2013; Luo, Li, Yang, Yu, & Wei, 2017) |
| M. tuberculosis rpoB gene | Tuberculosis | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | gDNA extracted from culture | (Devonshire et al., 2015) |
| L. monocytogenes DNA | Bacterial Infection | Flow-focusing | ~ 104 | 1,3 | 140 pL, 1 nL | PCR | NA | (Bian et al., 2015; Witte, Fister, Mester, Schoder, & Rossmanith, 2016) |
| N. gonorrhea DNA | Bacterial Infection | Flow-focusing | Cont. flow | 1 | 10 pL | LAMP | gDNA extracted from N. gonorrhea | (Rane, TD; Chen, L; Zec, HC; Wang, 2014) |
| L. monocytogenes DNA | Bacterial Infection | Step-emulsification | ~ 103 | 1 | 1.5 nL | RPA | gDNA extracted from L. monocytogenes | (Schuler et al., 2015) |
| C. trachomatis DNA | Bacterial Infection | Flow-focusing | ~ 106 | 1 | 50 pL | PCR | NA | (Hatch, Fisher, Tovar, et al., 2011) |
| N. gonorrhea 16s rRNA | Bacterial Infection | Flow-focusing | Cont. flow | 1 | 10 pL | Enzyme assay | RNA extracted from N. gonorrhea | (Guan et al., 2015) |
| E. coli lacZ gene | Bacterial Infection | Flow-focusing | ~ 106 | 3 | 15 pL | HRCA | NA | (Mazutis et al., 2009) |
| SMN1, SMN2 | Spinal muscular atrophy | Flow-focusing | ~ 104 | 3 | ~ 1 nL | PCR | NA | (Zhong et al., 2011) |
4.1 Quantification of rare oncogenic mutations in droplets
Droplet-based PCR platforms have been used extensively for the analysis of a few well-known genetic markers for cancer. Specifically, commercial digital PCR platforms, developed by companies like BioRad and RainDance, have recently achieved experimental throughputs up to ~104 – 106 droplet reactions per sample. This has resulted in drastically improved limits of detection for droplet platforms, which has enabled its evaluation for use in clinical diagnostics (Baker, 2012), specifically for detection and screening of genetic mutations specific to cancer. For example, BioRad’s ddPCR platform was first used to detect the melanoma-linked BRAF V600E mutation for mutant fractions as low as 0.001%, 1000 times lower than real-time PCR (Hindson et al., 2011). Since its commercialization, the ddPCR platform has been used to detect and quantify mutations in EGFR for lung cancer (Takahama et al., 2016; Thress et al., 2015), quantify expression of erbB2 for breast cancer (Heredia et al., 2013), and quantify mutations in TP53 for head and neck cancer (van Ginkel et al., 2017) among other applications (Beltrame et al., 2015; Guttery et al., 2015). A multiplexed digital PCR platform was presented by RainDance Technologies for simultaneously quantifying 6 distinct somatic mutations relevant to colorectal cancer in the KRAS oncogene (Pekin et al., 2011). By employing picoliter volume droplets, this platform was able to screen millions of reactions from one sample with reduced reagent consumption per sample. Moreover, by optically indexing unique probe droplets with varying concentrations of fluorescent dye and passively merging the probe droplets with sample droplets, the authors were able to screen each sample for six mutations simultaneously (Figure 4A). RainDance Technologies’ commercial RainDrop platform has been applied clinically for detecting mutations in KRAS for colorectal cancer (Valerie Taly et al., 2013) as well as EGFR for lung cancer (G. Zhu et al., 2015), BRAF for Langerhans cell histocytosis (Héritier et al., 2016), and a 5-plex genotyping assay for spinal muscular atrophy (Zhong et al., 2011). Unfortunately, almost all implementations of droplet digital PCR with clinical samples have relied on nucleic acid extraction and purification as a separate benchtop step prior to droplet generation. To further automate and integrate DNA extraction into the microfluidic PCR workflow, several microfluidic technologies have been developed, making use of silica beads (Shin, Zhang, & Wang, 2014; Y. Zhang, Park, Yang, & Wang, 2010), glass pillars (Wu et al., 2011), sol-gels (Breadmore et al., 2003), and isolation chambers (Easley et al., 2006). Yet none of these platforms have been integrated into droplet digital PCR workflows. Therefore, for fully realizing the sample-in-answer-out potential of these droplet platforms, further integration of sample preparation methods is warranted.
Figure 4. Droplet-based detection of nucleic acid biomarkers.
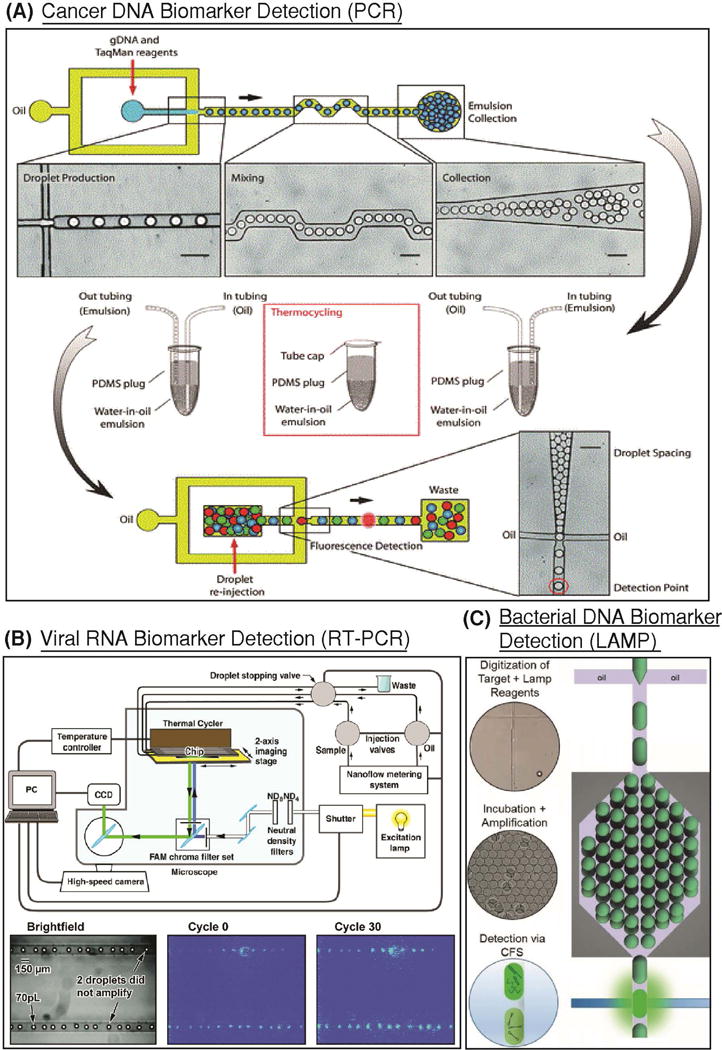
(A) Droplet-based digital PCR was used to screen for mutations in the KRAS oncogene. (Reproduced from (Pekin et al., 2011) with permission of The Royal Society of Chemistry). TaqMan probes specific for the wild-type and mutant genes were encapsulated in droplets that contain at most one haploid genome. The emulsion was then thermocycled off-chip, and reinjected for fluorescence detection. By optically coding droplet groups, parallel analysis of six mutations in KRAS were detected simultaneously using this platform. (B) An integrated platform for viral RNA detection using RT-PCR was developed, where 70 pL droplets containing MS2 virions and RT-PCR reagents were immobilized in microfluidic channels. An integrated thermal cycler provided temperatures necessary for reverse transcription and PCR, and a CCD camera was used for fluorescence detection. (Reprinted with permission from (Beer et al., 2008). Copyright 2007 American Chemical Society.) (C) Loop-mediated isothermal amplification was demonstrated for detection and quantification of N. gonorrhoeae gDNA down to 600 copies per μL. Critically, the assay was conducted in continuous-flow in an integrated platform that facilitated digitization of targets, on-chip incubation, and detection. (Reproduced from (Rane, TD; Chen, L; Zec, HC; Wang, 2014) with permission of The Royal Society of Chemistry).
Droplet based PCR platforms have also been used successfully to analyze liquid biopsies, an emerging, minimally invasive method for cancer diagnosis. Traditionally, nucleic acid detection for cancer necessitated the use of tissue samples extracted from tumorous tissue, which is often invasive and sometimes impossible due to physiologically inaccessible tumor sites. In contrast, liquid biopsies such as patient blood, cerebrospinal fluid, sputum, or other biological fluids, which may contain trace amounts of potential biomarkers such as circulating tumor DNA (cfDNA), circulating tumor cells (CTCs), or tumor-derived exosomes and micro vesicles, present a minimally invasive surrogate for direct tumor extraction and thus an increasingly popular approach for cancer diagnosis (Crowley, Di Nicolantonio, Loupakis, & Bardelli, 2013). To this end, commercial droplet PCR platforms have been used successfully to detect circulating DNA in blood plasma for skin, lung, breast, and gastric cancer, among others (Chang-Hao Tsao et al., 2015; Oxnard et al., 2014; Reid et al., 2015; Sanmamed et al., 2015; Takahama et al., 2016; Thress et al., 2015), and in CSF for monitoring progression of brain tumors (De Mattos-Arruda et al., 2015). Notably, ddPCR has been able to detect oncogenic mutations as rare as a few copies per milliliter (Oxnard et al., 2014), and has been touted as being 200 times more sensitive than bulk TaqMan-based mutation assays like castPCR (Reid et al., 2015). While these methods to quantify circulating DNA show great promise, ultimately, as is the case with tumor tissue-derived DNA, seamless integration of a DNA isolation module is important in making these droplet platforms further amenable to the clinical setting.
4.2 Sensitive and rapid detection of viral nucleic acids
BioRad’s commercial ddPCR platform has been evaluated clinically for nucleic acid detection of viral infections including human immunodeficiency virus (HIV) (Kiselinova et al., 2014; Strain et al., 2013), hepatitis B (HBV) (J. T. Huang et al., 2015), hepatitis C (HCV) (Mukaide et al., 2014), human cytomegalovirus (HCMV) (Pavšič et al., 2016), human T-cell lymphotropic virus (HTLV) (Brunetto et al., 2014), and human papillomavirus (HPV) (Jeannot et al., 2016). While benefitting from highly precise (5 × lower CV for HIV target) and accurate (20 × accuracy for HIV target) quantification compared to traditional qPCR methods (Strain et al., 2013), ddPCR is yet to be successfully demonstrated for detection of low-abundance targets (<50 copies/mL) from clinical samples. Specifically, researchers have reported low numbers of false positive droplets in some viral samples, resulting in over-quantification of low-level viremia (Trypsteen, Kiselinova, Vandekerckhove, & De Spiegelaere, 2016). Researchers have therefore called for an accurate determination of droplet fluorescence threshold when working with the ddPCR platform (Ruelle, Yfantis, Duquenne, & Goubau, 2014; Trypsteen et al., 2015). An additional area of improvement for ddPCR platforms remains process integration, as ddPCR platforms have primarily relied on fragmented workflows, requiring separate steps and instruments for droplet generation, thermal cycling, and detection.
Custom droplet PCR platforms have also been developed to detect viral nucleic acids with a focus on speed, quantification, and integration. Kiss et al. developed an integrated device to digitally amplify and quantify adenoviral DNA in 65 pL droplets. Notably, the combination of the small droplet volume and the integration of all workflow steps in the same device allowed the authors to screen millions of reactions in continuous flow and detect signal from amplification in as little as 35 min. (Kiss et al., 2008). Moreover, multiple droplet interrogation points in the device allowed for generation of real-time PCR fluorescence curves. While this work demonstrated quantification of the pAdeasy-1 adenoviral vector, for more clinically relevant viral load tests, like that of HIV-1 and HCV, a sensitive RNA quantification platform is desired. To this end, Rački et al. developed a rotavirus quantification platform that performed off-chip viral RNA extraction and reverse-transcription followed by droplet digital PCR in order to quantify viral RNA in wastewater effluent (Rački, Morisset, Gutierrez-Aguirre, & Ravnikar, 2014). A more integrated platform compared to the previous work was developed by Beer et al. in order to detect the bacteriophage MS2 (Beer et al., 2008). Seventy pL droplets containing RT-PCR mix and single virions were first generated via T-junction geometry. The droplets were then immobilized in channels on-chip and the chip was mounted on a Peltier thermal cycler. Real-time detection of the droplets was conducted using a CCD camera over the course of ~1 h. While fully integrated and automated, the relatively small device footprint limited its throughput to roughly 102 droplets per experiment (Figure 4B).
4.3 Amplification-based detection of bacterial nucleic acids
A vast array of microfluidic approaches have been applied to bacterial nucleic acid detection and quantification via PCR amplification. Commercial platforms such as BioRad’s ddPCR have been used to detect and quantify DNA from pathogens in low abundance (as low as 103 CFU/mL (Luo et al., 2017)) faster and with higher sensitivity than bulk plating (Devonshire et al., 2015; Kelley et al., 2013; Luo et al., 2017). In pursuit of increased process integration, high-throughput platforms employing integrated fluorescence detection have been presented. A noteworthy demonstration of high-throughput and rapid detection of bacterial DNA via integrated imaging was presented by Hatch et al. Herein, droplets containing PCR reagents and C. trachomatis DNA templates were generated and collected in a large incubation/imaging chamber. Wide-field microscopy of the chamber enabled real-time PCR quantification of bacterial DNA from 106 droplets simultaneously per experiment. Notably, the authors were able to screen 50 μL of sample in ~65 min. and were able to quantify bacterial DNA down to 0.4 copies per μL (Hatch, Fisher, Tovar, et al., 2011).
Isothermal amplification platforms have been recently developed to reduce constraints on platform instrumentation and device design. In this pursuit, Rane et al. developed a high-throughput, continuous-flow droplet platform for detection of bacterial DNA (Rane, TD; Chen, L; Zec, HC; Wang, 2014). Here, loop-mediated isothermal amplification (LAMP) was used for amplifying and detecting single copies of target genes purified from samples containing Neisseria gonorrhea (Figure 4C). Critically, using this approach, the authors were able to screen 10 μL of sample in less than 110 min. and were able to quantify bacterial DNA down to 600 copies per μL. The authors noted that further improvement in the limit of detection is possible by reducing nonspecific amplification by optimizing the LAMP assay in the droplet system. In this work the choice of a single-step isothermal amplification protocol simplified the instrumentation needs of the platform and eased concerns of droplet stability at higher temperatures. Similar isothermal droplet platforms have been utilized for detection and analysis of bacterial DNA by employing assays such as hyperbranched rolling circle amplification (Mazutis et al., 2009), recombinase polymerase amplification (Schuler et al., 2015), and most recently, multiple displacement amplification (Rhee, Light, Meagher, & Singh, 2016).
4.4 Amplification-free detection of bacterial nucleic acids
Genetic amplification techniques for detecting nucleic acids (eg: PCR, LAMP), while sensitive and quantitative, are often limited by amplification efficiency, susceptibility to contamination, and thermocycling duration, making them harder to implement in the clinic (Millar, Xu, & Moore, 2007). Volume reduction and background suppression using droplets presents a unique opportunity to detect nucleic acids without the need for target or signal amplification. Amplification-free assays for detecting nucleic acids therefore offer a more clinically amenable alternative to amplification techniques. Guan et al. developed such an amplification-free platform for absolute quantification of bacterial rRNA purified from clinical isolates of N. gonorrhea (Guan et al., 2015). Here, an enzyme-linked oligonucleotide hybridization assay (ELOHA) for absolute quantification of RNA molecules was developed, where RNA molecules were allowed to hybridize onto DNA capture probes that were conjugated on to magnetic beads. Enzyme-labeled detection probes were then hybridized to the captured RNA. The beads were then encapsulated into microfluidic droplets along with a fluorogenic substrate for downstream detection and quantification. ELOHA-based quantification of nucleic acid containing beads achieved high degree of accuracy (CV<10%) over a relatively wide dynamic range across 3 orders of magnitude (Figure 5).
Figure 5. Amplification-free quantification of RNA using droplet digital enzyme-linked oligonucleotide hybridization assay (ELOHA) (Reprinted from (Guan et al., 2015) under the terms of the Creative Commons Attribution 4.0 License).
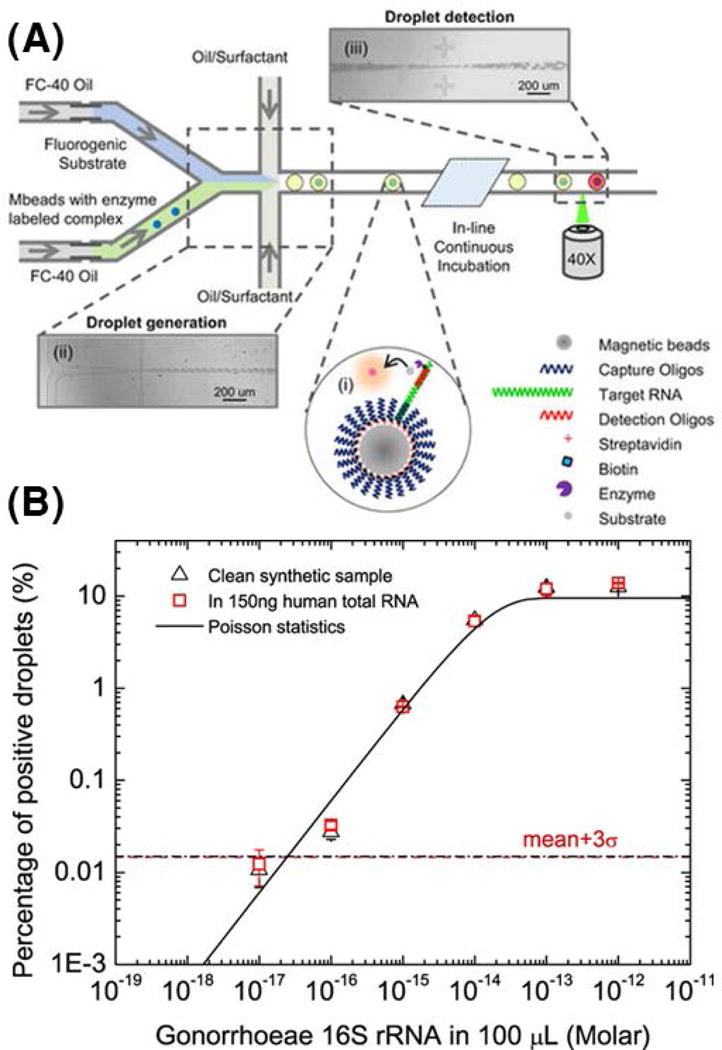
(A) A sandwiched complex consisting of capture oligo-coated magnetic beads hybridized to a single molecule of target RNA which is then hybridized to an enzyme-linked detection oligo is co-flowed into droplets along with corresponding enzyme substrate. Following in-line incubation, droplets containing single RNA exhibit strong fluorescence, whereas droplets without the sandwiched complex exhibit weak fluorescence. (B) By interrogating ~106 droplets, the percentage of highly fluorescent droplets can be used to quantify input concentration of 16S rRNA from N. gonorrhoeae down to 600 molecule copies in 100 μL of sample.
5. DETECTION AND SCREENING OF PROTEIN BIOMARKERS
Herein, we present microfluidic platforms that have been developed for the detection of low-abundance protein biomarkers and for multiplexed screening and analysis of protein biomarkers, applied to diseases such as cancer, bacterial infections, diabetes, and endometriosis, among others. In order to provide the readers with metrics to evaluate each droplet platform, Table 2 provides a brief summary of protein biomarkers analyzed along with the droplet generation format implemented, the approximate experimental throughput, and the number workflow steps. Additionally, in order to evaluate the sensitivity achieved by each platform, we list the associated droplet volume, detection assay, and input sample, along with associated references. Input samples containing direct clinical biological fluid are highlighted in red.
Table 2.
Summary of droplet platforms applied to protein biomarker detection and screening
| Biomarker | Disease | Droplet Format |
Through put (per run) |
Workfl ow steps |
Dropl et volu me |
Assay | Clinical Sample |
Reference(s ) |
|---|---|---|---|---|---|---|---|---|
| PSA | Prostate cancer | Flow-focusing | ~ 105 | 1 | ~ 35 fL | Bead-based ELISA | NA | (Shim et al., 2013) |
| Matrix Metalloproteinase | Cancer | Valve-actuation | Cont. flow | 1 | 180 pL, 10 nL | Enzyme assay | NA | (Jambovane, Kim, Duin, Kim, & Hong, 2011; Tushar D. Rane et al., 2015) |
| Matrix Metalloproteinase | Endometriosis; breast cancer | Flow-focusing | ~ 103 | 1 | 60 pL | Enzyme assay | Peritoneal fluid supernatant from 12Z cell line; supernatant from MDA-MB-231 cell line | (Chen et al., 2011; Ng, Miller, Jing, Lauffenburger, & Chen, 2015) |
| β-galactosidase | Bacterial infection | Flow-focusing | ~ 106 | 1 | ~ 100 pL, ~ 35 fL | Enzyme assay | NA | (J. Lim et al., 2013, 2015; Shim et al., 2013) |
| Cellobiohydrolase | Bacterial infection | Flow-focusing | ~ 104 | 1 | 37 pL | Enzyme assay | NA | (Najah et al., 2013) |
| Glucose oxidase | Diabetes | Flow-focusing | Cont. flow | 1 | 62 nL | Enzyme assay | Glucose spiked into blood serum | (Gu et al., 2014) |
| β-glucosidase, catalase, C-reactive protein | Infection, inflammation, cancer | T-junction, valve-actuation, flow-focusing | ~ 106 | 3,1 | ~ 13 fL, NA, ~ 20 pL | Enzyme assay, antibody conjugation | NA | (Arayanarakool, Shui, Kengen, van den Berg, & Eijkel, 2013; Han, Li, Huang, & Zheng, 2009; Tang & Shum, 2016) |
5.1 Detection of low-abundance protein biomarkers
Enzyme-linked immunosorbent assays (ELISA) have long been the gold-standard for sensitive detection of proteins; however, droplet platforms offer the opportunity for improving their limit of detection. In ELISA, proteins linked with reporter enzymes are conjugated to immobilized antibodies on the surface of a planar substrate (eg: well-plate) or suspended microbead (Tighe, Ryder, Todd, & Fairclough, 2015). Following protein capture, a fluorogenic substrate is introduced into the mix and proteins can be detected by enzyme-substrate cleavage after incubation. Alternatively, a labeled secondary antibody may be used for detection. Commercial immunoassay platforms have focused on optically coding microbeads or customizing planar microarrays (Leng et al., 2008; C. T. Lim & Zhang, 2007) in order to multiplex analyte detection. Most of these platforms are expensive due to the inherent complexity in fabrication of beads or microarrays and require large supporting instrumentation. For this reason, most researchers continue to rely on 96-well plates, with a greater focus on achieving assay sensitivity and specificity than multiplexability (Ellington, Kullo, Bailey, & Klee, 2010). Bulk ELISA assays are typically limited in sensitivity to picomolar concentrations or above. For many diseases like prostate cancer, protein biomarkers may exist in much lower abundance in biological samples (Hanash, Baik, & Kallioniemi, 2011). To address this need, Shim et al. developed a femtoliter droplet platform for single-molecule counting immunoassays (Shim et al., 2013). In this work, antibody-functionalized capture beads were first mixed with the target protein in phosphate-buffered saline such that each bead contained zero or one copy of the target analyte. Next, a detection antibody was conjugated to the captured proteins followed by the streptavidin-conjugated β-galactosidase reporter enzyme. For each sample, 2 × 104 ultra-small droplets (35 fL) were generated that contained bead-analyte-enzyme complex and a fluorogenic substrate. Upon droplet formation, beads containing the analyte-enzyme complex mixed with the fluorogenic substrate and started producing fluorescence. After a short 10-min. on-chip incubation step, signal from single-molecule containing droplets could be detected via imaging using a CCD. The authors used their platform to detect prostate-specific antigen (PSA), an important protein biomarker for prostate cancer, as low as 46 fM in concentration, approximately 2 orders of magnitude lower than conventional bulk ELISA. The authors further point out their ability to simply scale up their incubation region and detection format to allow for an increased dynamic range and potentially lower limit of detection (Figure 6A).
Figure 6. Droplet based detection of protein biomarkers.
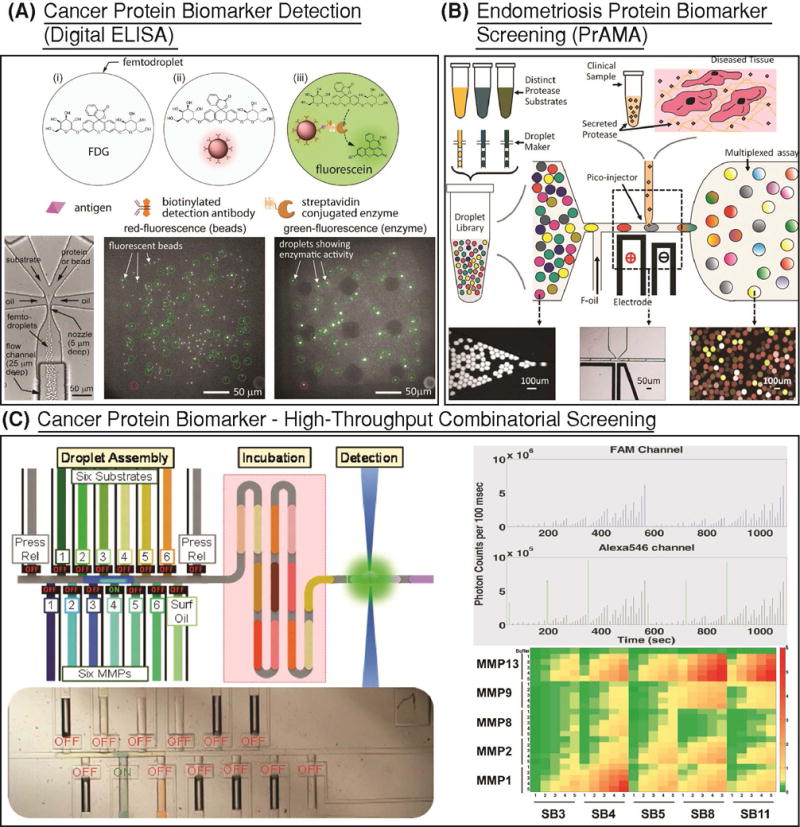
(A) A bead based sandwich assay, similar to ELISA, was developed for quantifying prostate specific antigen, a prostate cancer biomarker, in buffer (Shim et al., 2013). The target protein is tagged with a reporter enzyme and hybridized to the bead. Beads and substrate are co-flown to generate femtoliter droplets. After 10 min. of incubation, this platform is able to detect protein biomarker concentrations as low as 46 fM. Reprinted with permission from (Rane, TD; Chen, L; Zec, HC; Wang, 2014). Copyright 2013 American Chemical Society.) (B) Matrix metalloproteinase (MMP) biomarkers secreted into the peritoneal fluid of clinical endometriosis tissue sample was combined with a droplet library of 4 FRET based protease substrates with and without specific inhibitors and incubated on chip. Following at least 3 h of fluorescence monitoring, proteolytic activity matrix analysis (PrAMA) was conducted to assay protease activity within droplets. (Reprinted with permission from (Chen et al., 2013). Copyright 2013 American Chemical Society.) (C) In order to scale up PrAMA analysis to a larger set of MMPs and substrates, a high-throughput combinatorial valve-based platform was developed (Tushar D. Rane et al., 2015). 650 unique combinations of MMPs and substrates were screened in continuous flow following 12 min. incubation using this approach.
In addition to ELISA, droplet technology has delivered platforms for sensitive enzyme kinetic analysis and quantification. Notably, the improved control of sample reaction time afforded by droplet platforms offers the opportunity for more controlled and more accurate kinetic analysis. Indeed, droplets have been used to determine dilute concentrations of enzymes like β-glucosidase (with single molecule sensitivity) (Arayanarakool et al., 2013), β-galactosidase (J. Lim et al., 2013, 2015; Shim et al., 2013), and catalase (Han et al., 2009). One such enzyme detection droplet platform was used to investigate glucose oxidase reaction kinetics in order to determine the concentration of glucose in a sample (Gu et al., 2014). Notably, the authors could quantify glucose spiked into human blood serum with high precision in the normal human physiological range (4.0 to 6.0 mM) as well as in the elevated range (≥ 8.0 mM) for diabetes. Such demonstrations of sensitive and rapid enzyme detection can be used to further expedite the detection of a vast array of analytes, including proteins by incorporation into droplet based ELISA workflows.
5.2 Multiplexed screening of enzyme kinetics
Passively generated flow-focusing droplet platforms have been developed for performing high-throughput assays for protein screening applications. Due to the significantly reduced volumes and increased automation afforded by microfluidic droplet platforms, reagent costs and operation costs can be significantly reduced for high-throughput screening. Chen et al. showcased the utility of droplets for detection of low levels of secreted proteases (Chen et al., 2011) as well as multiplexed protease activity matrix analysis (PrAMA) for matrix metalloproteinase (MMP) secreted from the MDA-MB-231 breast cancer cell line (Ng et al., 2015) and clinical endometriosis tissue (Chen et al., 2013). In the latter work, multiple groups of droplets containing distinct short FRET peptide substrates were generated and pooled into a common library. Each substrate droplet group was optically barcoded by a unique concentration of fluorescent dye. The fluorogenic substrate droplets were then merged with droplets containing proteases from clinical peritoneal fluid (PF). Using this method, kinetics of proteases within the PF samples were monitored against 9 unique substrates. The resulting reaction rates from each enzyme-substrate reaction in droplets was then used to determine the most active proteases present in the biological sample, following PrAMA analysis (Figure 6B).
For greater control and multiplexing in kinetic enzyme analyses, active pneumatic valve-based platforms have been developed by researchers. Jambovane et al. presented the potential of using a valve-based platform for controlled dilutions of 2 MMPs against 1 substrate (Jambovane et al., 2011). Using this platform, they performed detailed kinetic analysis and evaluated efficiency for MMP-2 and MMP-9 in a sample. Rane et al. developed a higher-throughput controllable valve-based platform for PrAMA analysis with increased multiplexing capability (Tushar D. Rane et al., 2015) (Figure 6C). Herein, by using PDMS “Quake” valves, the authors were able to generate a nanoliter droplet train of unique MMP and substrate combinations. Following droplet assembly, the droplet train proceeded to on-chip incubation for 12 min. followed by continuous flow droplet measurements using an in-line confocal spectroscope. Critically, instead of preloading droplets with optical barcodes, the platform was designed and programmed to maintain a predetermined spatially indexed droplet train that maintained its sequence from droplet generation through droplet detection. This high-throughput combinatorial platform was used to screen 650 unique combinations of MMPs and substrates.
6. DETECTION AND SCREENING OF SINGLE-CELL DERIVED BIOMARKERS
Several droplet microfluidic platforms exist for the detection and screening of single-cell derived biomarkers associated with cancer and infectious diseases. For these diseases, certain cells in the body may contain specific information about the underlying disease, and therefore, the cells themselves can be used as biomarker for disease. In order to detect these cells, researchers may employ the nucleic acids or proteins in the cells as markers for the cells. Furthermore, by studying a population of disease-specific cells, we may obtain additional information regarding the heterogeneity inherent to the population, which may itself be a marker for disease or disease state. To this end, we present droplet platforms developed for analysis of cellular heterogeneity of diseased tissue, for detection and quantification of low-abundance bacterial pathogens, and for detection of low-abundance cell-surface and cell-secreted protein biomarkers. Table 3 summarizes existing droplet platforms used to analyze single-cell derived biomarkers and their associated diseases. As metrics to evaluate the droplet platforms, we also present the droplet generation format, approximate experimental throughput, and the number of workflow steps needed. Finally, as metrics to evaluate sensitivity, we present the volume of droplets employed, the bioassay used for detection, the input sample, and associated references. Input samples containing direct clinical biological fluid are highlighted in red.
Table 3.
Summary of droplet platforms applied to single-cell derived biomarker detection and screening
| Biomarker | Disease | Droplet Format | Throughput (per run) | Workflow steps | Droplet volume | Assay | Clinical Sample | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| t(14;18) | Lymphatic cancer | Flow-focusing | ~ 103 | 4 (additional step to elute beads containing amplicons from droplet) | 3 nL | PCR | RL cell line | (Novak et al., 2011) |
| VCaP gDNA | Prostate cancer | Rapid emulsification | ~ 105 | 4 (additional step to isolate single-cell and lyse off-chip) | ~ 30 pL | MDA | VCaP cell line | (S. C. Kim et al., 2017) |
| anti-TNF-alpha | Cancer | Flow-focusing | ~ 102 | 3 | ~ 30 pL | Antibody conjugation | 9E10 Hybridoma cell line | (Akbari & Pirbodaghi, 2014) |
| PTPRC | Cancer | Flow-focusing | ~ 104 | 4 (additional step for droplet dilution and RT-PCR mix merging) | ~ 100pL | RT-PCR | Raji and PC3 cell lines | (Eastburn, Sciambi, & Abate, 2013) |
| Matrix Metalloproteinase | Cancer | Flow-focusing | ~101 | 1 | ~10 pL | Enzyme assay | PC-9 cell line, K-562 cell line, MDA-MB-231 cell line | (Jing et al., 2015; Ng, Miller, Jing, & Chen, 2016) |
| Matrix Metalloproteinase | Cancer, asthma, rheumatoid arthritis | Flow-focusing | ~ 102 | 1 | ~60 pL | Enzyme assay | Leukocytes with RBC debris | (Jing et al., 2016) |
| Rotavirus RNA | Viral infection | Flow-focusing | ~ 104 | 3 | ~ 1 nL | RT-PCR | RNA spiked into surface water samples, then extracted | (Rački et al., 2014) |
| CCR5 | AIDS | Flow-focusing | ~ 105 | 3 | ~ 30 pL | Enzymatic assay | U937 cell line | (Joensson et al., 2009) |
| E. coli DNAzyme | Bacterial infection | Flow-focusing | ~ 108 | 2 | ~ 8 pL | DNAzyme probe | E. coli spiked into 10% donor blood | (Kang et al., 2014) |
| E. coli | Bacterial infection | Flow-focusing | Cont. flow | 1 | ~ 20 pL | Resazurin | E. coli in broth | (Kaushik et al., 2017) |
| E. coli, E. aerogenes | Bacterial infection | Flow-focusing | ~ 104 | 3 | ~ 1 nL | C-12 Resazurin | Bacteria in broth | (O. Scheler et al., 2017; Ott Scheler, Kaminski, Ruszczak, & Garstecki, 2016) |
| E. coli, S. aureus | Bacterial infection | T-junction | ~ 102 | 3 | ~ 4 nL | alamarBlue | Bacteria spiked into 50% plasma | (Boedicker, Li, Kline, & Ismagilov, 2008) |
6.1 Detection of cellular heterogeneity by single-cell derived nucleic acids
By discretizing single-cells from tissues or biological fluids, droplets enable interrogation of diseases with single-cell sensitivity. This results in information regarding cellular population heterogeneity which may be otherwise lost to averaged signal in bulk methods. Cellular genetic, epigenetic, and proteomic heterogeneity is commonly used as a biomarker for cancer and can contain useful information for further characterizing the disease and disease progression (Navin, 2015). Since mutations leading to cancer occur at the single-cell level, a platform with single-cell resolution is essential to detecting and characterizing this heterogeneity associated with cancer. To this end, Novak et al. developed a platform for co-encapsulation of single cells and primer-functionalized beads into agarose droplets (Novak et al., 2011). Following off-chip lysis, protease digestion, and droplet PCR steps, the now amplicon-rich beads were eluted and analyzed. The authors were able to successfully identify cells containing the translocation t(14;18) in mutant to wild-type cell ratios as low as 10%. For further characterization of cellular heterogeneity at the transcriptional level, Eastburn et al. presented a platform for performing single-cell RT-PCR (Eastburn et al., 2013). This multiple step workflow involved first coencapsulating single-cells and lysis buffer into droplets. The cell-lysate containing droplets were then reinjected into a separate device and merged with a larger water droplet for dilution and then injected with RT-PCR mix. The droplets were then collected, thermocycled off-chip, and then detected using a laser excitation source and a PMT detector. The authors were able to differentiate two cell lines based on their transcriptional signature using this method. A more recent innovation in high-throughput single-cell transcriptomic profiling was presented by Macosko et al. (Macosko et al., 2015). In their “DropSeq” methodology, single cells were encapsulated into nanoliter droplets along with lysis buffer and uniquely barcoded beads. Targeting the polyadenylated tail of mammalian mRNA, these beads were designed to capture all mRNA from single-cells. Once captured on beads and tagged with cell-specific barcodes, the mRNA were subject to reverse transcription, PCR, library preparation, and sequencing. Following data analysis, the authors were able to trace each mRNA sequence to its unique cell using the unique bead barcodes. Using this technology, the authors identified 39 unique subpopulations of retinal cells from 44,808 total cells sequenced. With increasing interest in the DropSeq platform and the falling cost of next-generation sequencing platforms, single-cell droplet platforms will play a pivotal role in studying genetic and transcriptomic heterogeneity in cancer and various other diseases in the coming years (Navin, 2015).
6.2 Detection of low-abundance bacterial pathogens by single-cell derived nucleic acids
For infectious diseases, rapid and sensitive detection of pathogens that exist in low abundance in biological samples is of utmost importance to disease diagnostics and subsequent therapy recommendations. To this end, Rane et al. developed an amplification-free platform for detection of pathogen-specific rRNA from single-bacterial cells (Tushar D. Rane, Zec, Puleo, Lee, & Wang, 2012). The authors encapsulated single cells of E. coli into 10 pL volume droplets and used a 16S rRNA specific peptide nucleic acid (PNA) probe in order to count E. coli spiked into buffer. This platform was able to determine E. coli concentrations as low as 1 cell in every 20 droplets (equivalent to 5 × 106 CFU/mL) and included the ability to operate in continuous flow for increased throughput and dynamic range (Figure 7A). To demonstrate the potential of direct analysis of clinical samples, Kang et al. used a DNAzyme sensor for detection of E. coli spiked into 10% blood (Kang et al., 2014). Herein, single bacterial cells were confined in droplets that were approximately 14 pL (30 μm diameter) in volume. After 45 min. of lysis and DNAzyme reaction, the platform could generate a ‘yes’ or ‘no’ result for the presence of the particular bacteria being tested. After 3.5 h, the platform was able to quantify dilutions of E. coli from 1 CFU/mL to 104 CFU/mL. While rapid bacterial detection has been demonstrated for E. coli by leveraging the increased sensitivity of droplets, more work is required in order to expand the capability to these platforms to test the existence of other pathogenic species in samples. Moreover, while the aforementioned platforms have demonstrated pathogen detection using spiked-in concentrations of pre-cultured E. coli (reference strains or clinical isolates in buffer or diluted blood), ultimately a droplet platform that can directly process culture-positive clinical samples is yet to be demonstrated.
Figure 7. Droplet based detection of single-cell biomarkers.
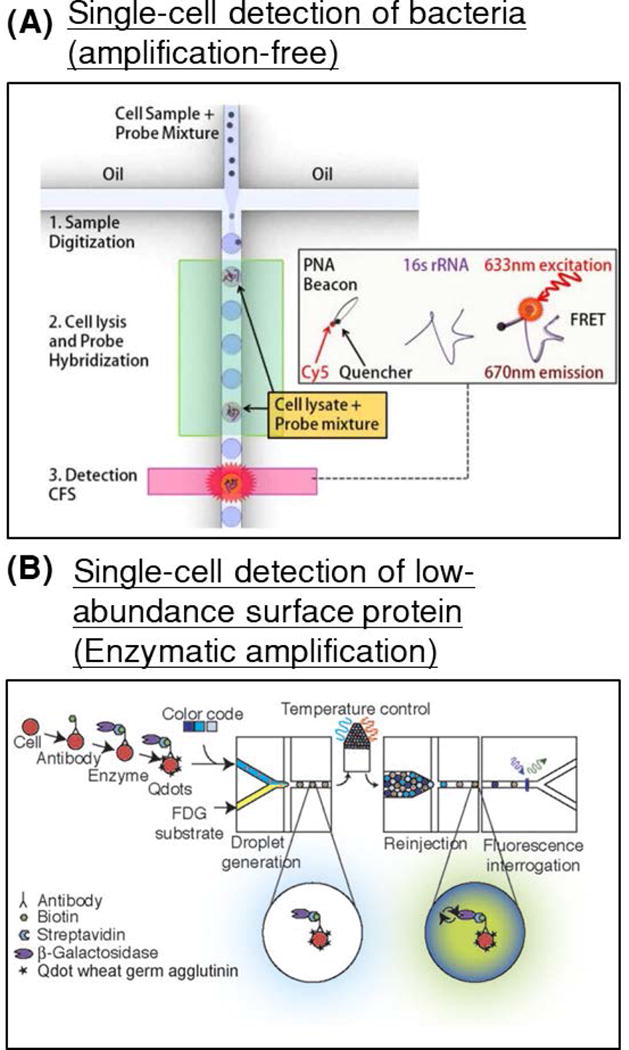
(A) An integrated amplification-free platform for genetic detection of pathogens was developed. Single cells of E. coli were co-encapsulated with peptide nucleic acid beacons, complimentary to a specific region within the 16S rRNA of the pathogen. Following on-chip thermal lysis, rRNA release, and probe hybridization, droplet fluorescence was detected and used to quantify pathogen load within a sample. (Reproduced from (Tushar D. Rane et al., 2012) with permission of The Royal Society of Chemistry). (B) An enzyme-tagged antibody was used to detect the typically low-abundance cell-surface protein biomarker CCR5 (a co-receptor in HIV-1 infection) in U937 cells. (Reprinted from (Joensson et al., 2009) with permission from John Wiley and Sons).
6.3 Detection of low-abundance cell-secreted and cell-surface proteins
By containing diffusible signal from analytes within a confined microenvironment, droplet technology offers a highly sensitive alternative to FACS for detection and sorting single-cell proteins. Proteins secreted from single-cells can be confined within the cellular microenvironment using droplets, and therefore can be used as a target to screen or sort the particular cell of interest (Shembekar, Chaipan, Utharala, & Merten, 2016). This was very simply demonstrated by Mazutis et al. by separating out 9E10 mouse hybridoma cells from K562 human leukemia cells by detecting anti-MYC antibodies secreted by the mouse cells using fluorophore-labeled detection antibodies (Mazutis et al., 2013). Herein, by using droplets of 50 pL volume, the authors were able to generate differentiable signal over background as fast as 15 min. The authors noted that this relatively simple demonstration of separating mouse cells from human cells can be adapted to many applications of human disease to detect secreted molecules from a cell (eg: insulin, cytokines, growth factors, etc.). A notable example of detection of secreted proteins from single leukocytes in order to monitor patient immunity was demonstrated by Jing et al. (Jing et al., 2016). Herein, a suspension of leukocytes in whole blood was subject to red blood cell lysis, centrifugation and resuspension in a buffer solution. The resulting biofluid, containing red blood cell debris among other biochemical contaminants, was entered into a device that facilitated purification of the leukocytes using a deterministic lateral displacement (DLD) pillar array. The purified leukocytes were then encapsulated into droplets along with a protease substrate, and the resulting droplets entered an integrated imaging chamber to monitor protease activity. Low-abundance surface proteins may also be detected using droplets. Joensson et al. presented a method wherein mammalian cells could be screened for the low-abundance surface protein CCR5, a biomarker indicative of HIV-1 infection (Joensson et al., 2009) (Figure 7B). The cells were labeled with enzyme-tagged antibodies, which were mixed with fluorogenic substrates inside 40 μm droplets. Following off-chip incubation, the droplets were reinjected and detected and showed greater resolution for differentiating cells containing CCR5 than traditional FACS-based methods.
6.4 Antimicrobial susceptibility screening by single-cell metabolism
Droplet platforms have shown to be effective for single-cell growth assays. Researchers have cultured cells as diverse as yeast (Boitard et al., 2012), mammalian cell lines (Clausell-Tormos et al., 2008; Köster et al., 2008), bacterial cells [(Boedicker et al., 2008; Kaushik et al., 2017; O. Scheler et al., 2017; Ott Scheler et al., 2016)], as well as multicellular organisms like C. elegans (Clausell-Tormos et al., 2008) and zebrafish (Baret, Beck, Billas-Massobrio, Moras, & Griffiths, 2010) in droplets. Assaying cellular growth in droplets allows one to assess the effects of external variables like drugs, chemical composition, and cellular microenvironment on single-cell growth rapidly and at a high throughput. A notable demonstration of single-cell growth assays has been the implementation of droplet-based antimicrobial susceptibility tests. The current standard phenotypic antimicrobial susceptibility tests (AST) in clinical laboratories include broth dilution, disk diffusion, and gradient diffusion (Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement, 2014), wherein isolated bacteria are directly grown in the presence of various antibiotics to determine their sensitivity or resistance based on bacterial growth after a 16 to 20 h incubation period. Although many strains of bacteria grow rapidly and some (e.g., E. coli) can even replicate in as little as every 20 minutes, conventional AST still requires a lengthy incubation period. This is in part caused by the large volumes in which bacteria are grown and measured (100 μL to 20 mL), which require growing a high number of bacteria to ensure reliable interpretation of bacterial growth. It can be therefore inferred that a reduction of analysis volume can reduce the incubation time necessary for assessing antibiotic resistance. Moreover, this incubation time may be reduced to the timescale of individual bacterial replication if such an event can be reliably observed at the single-cell level. To this end, droplet platforms provide a technology capable of handling small sample volumes and detecting the replication of individual bacteria, which can drastically accelerate phenotypic assessment of antimicrobial susceptibility.
Several droplet-based strategies have emerged in recent years for rapid and sensitive quantification of bacterial growth and antimicrobial susceptibility testing. Scheler et al. presented a platform that utilizes 1 nL volume droplets in order to confine single-bacterial cells (that have been pre-exposed to antibiotics) along with the viability dye C12-resazurin (O. Scheler et al., 2017) (Figure 8Ai). Following 5 h off-chip incubation (Figure 8Aii), the droplets were reinjected into a detection device in order to count the number of fluorescent droplets containing growing bacteria (Figure 8Aiii). A similar approach was implemented by Boedicker et al. to perform antimicrobial susceptibility testing for MSSA and MRSA spiked into 50% blood plasma (Boedicker et al., 2008) (Figure 8B). In that work, a slightly larger droplet volume (4 nL) required 7.5 h off-chip incubation in order to determine the susceptibility and resistance to methicillin. A rapid and integrated approach was presented by Kaushik and Hsieh et al (Kaushik et al., 2017). Here, the authors encapsulated single bacteria in significantly smaller droplets that are 20 pL in volume. The reduced background in the smaller droplets allowed for faster determination of the fluorescence signal generated from bacterial growth. The authors detected the effect of gentamicin on the growth of a susceptible and multi-drug resistant strain of E. coli after only ~1 h, equivalent to only 2-3 bacterial doubling events. For all of the aforementioned droplet-based AST platforms, the authors spiked in fixed concentrations (2 × 106 CFU/mL (O. Scheler et al., 2017), 4 × 105 CFU/mL (Boedicker et al., 2008), 5 × 107 CFU/mL (Kaushik et al., 2017)) of pre-cultured reference bacterial strains into their droplet platforms. While these concentrations were kept relatively high for the purposes of demonstration, Scheler et al. note that they may pre-enrich their sample via methods such as centrifugation, and Boedicker et al. note that they may increase the number of droplets to process lower input concentrations of bacteria. The continuous-flow platform developed by Kaushik et al. has the ability to process potentially unrestricted volume of samples, and therefore can also be used in its current state to process lower concentrations of bacteria. As an additional note, as droplet-based AST platforms become faster and more integrated, more work needs to be done to ensure that these platforms are capable of handling multiple combinations of pathogens and antibiotics.
Figure 8. Droplet based bacterial growth quantification and antibiotic susceptibility testing (AST).
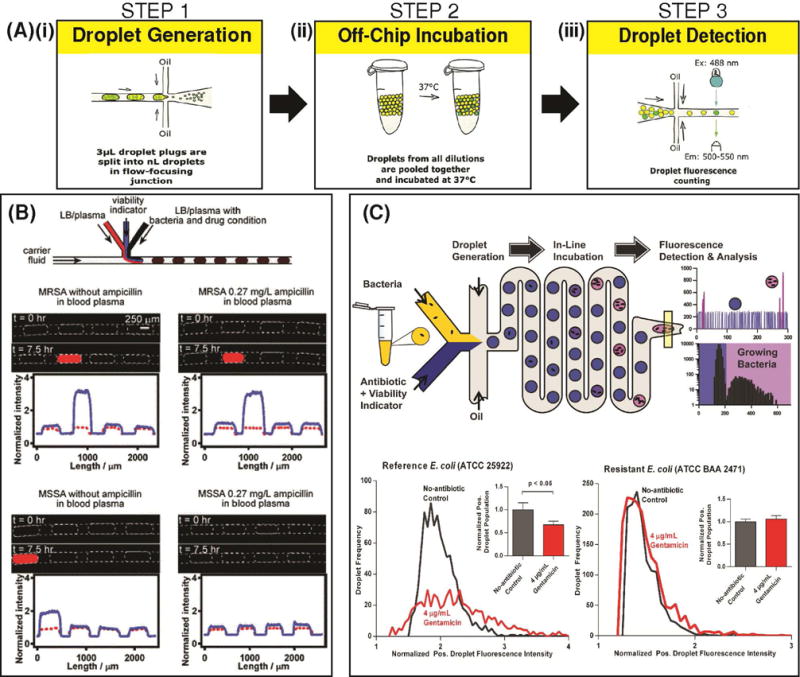
(A) A ddCFU platform was developed to rapidly count viable bacteria within a sample. Critically, this platform followed a fragmented workflow that necessitated separate modules for (i) droplet generation and encapsulation of single bacteria, (ii) off-chip bacterial incubation, and (iii) droplet detection. (Adapted from (O. Scheler et al., 2017) with permission of The Royal Society of Chemistry.). (B) A similar workflow was used to develop a droplet based AST assay. After co-encapsulation of single cells of S. aureus with blood plasma and ampicillin into 4 nL droplets, the pathogen’s susceptibility/resistance to the drug was determined in 7.5 h. (Reproduced from (Boedicker et al., 2008) with permission of The Royal Society of Chemistry.). (C) An integrated one-step platform was developed for bacterial growth analysis and antibiotic susceptibility testing (Kaushik et al., 2017). Critically, the use of much smaller 20 pL droplets in this platform enabled detection of single-cell E. coli growth and its susceptibility/resistance to gentamicin in as little as one hour, equivalent to 2-3 replications of the bacterial cell. (Reprinted from Biosensors and Bioelectronics, 97, Kaushik and Hsieh et al., Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform, 260-266, Copyright 2017, with permission from Elsevier).
7. TOWARD CLINICAL APPLICATIONS
The ultimate goal of developing droplet microfluidic devices is utilizing them for the detection and screening of disease biomarkers, thereby improving the diagnosis and treatment of diseases. Although recent years have seen significant advances in the field of droplet microfluidics, devices that can be implemented in clinical settings to detect and screen disease biomarkers have yet to be reported. Toward achieving this goal, we see several general areas that must be improved to accelerate this process, though we point out that the design of a particular droplet microfluidics platform would be ultimately dictated by its specific application, especially the biomarker(s) of interest and the type of biological sample. Specifically, increasing detection throughput and sensitivity, seamlessly integrating sample preparation methods, increasing multiplexing and screening capacity, and device automation remain crucial objectives which could eventually help in bringing droplet microfluidic platforms to the clinic.
7.1 Increasing detection throughput and sensitivity
Enhancement in the limit of detection (LoD) achievable by droplet microfluidic platforms presents one opportunity for further improvement. Although droplet microfluidic platforms have the capacity to count the number of droplets that encapsulate a single copy of DNA or a single cell, which intuitively suggests single-copy or single-cell sensitivity, the actual analytical sensitivities achieved by these platforms are rarely one copy or one cell in a few to ten mL of biological sample. In other words, for many applications, the single-copy or single-cell sensitivity may not translate directly into copies or cells per mL. Furthermore, droplet microfluidic devices that employ extremely small volume droplets may suffer from low volumetric throughput. The low volumetric throughput consequently limits the capacity to detect exceedingly rare biomarkers, either because only a fraction of the sample would be processed and analysed or because the assay time would be lengthy. For example, to detect 10 pathogenic bacterial cells in 1 mL of blood for the diagnosis of sepsis using 10-pL droplets, 108 droplets will be formed. A typical droplet generation device operating at 103 Hz would take 105 s (or nearly 3 h) to generate such high number of droplets. Perhaps more significant is the issue of background molecules or cells within biological samples, which can still yield false positive counts (Kiselinova et al., 2014; Trypsteen et al., 2016). Such false detection severely hampers the LoD that a platform can achieve. For example, 1 false positive signal in 105 10-pL droplets essentially means the LoD cannot surpass 103 per mL.
There are several routes toward addressing the concerns in detection sensitivity and throughput. First, droplet microfluidic platforms can be selectively applied toward applications with biological samples that have high target concentrations, such as the detection of pathogenic bacteria in urinary tract infections (UTI). For UTI, the bacterial load typically lies between 104 to 108 CFU per mL of urine for infected patients, which can be rapidly detected by droplet microfluidic platforms (Davenport et al., 2017; Mach et al., 2011). Second, increasing the throughput for both droplet generation and droplet detection can improve the assay time and the limit of detection. Methods for increasing the throughput of droplet generation include parallelization of multiple droplet generation modules (R. Dangla et al., 2013; J. Lim et al., 2015; Nisisako & Torii, 2008) and implementation of droplet splitting structures (Hatch, Fisher, Tovar, et al., 2011; Hsieh, Zec, Ma, Rane, & Wang, 2015). For increasing the throughput of droplet detection, leveraging detection modalities that can simultaneously detect many droplets, such as CMOS-based detectors (Hatch, Fisher, Tovar, et al., 2011; M. Kim et al., 2015), presents a viable approach. Optimization between droplet volume and sample volume for each assay may also be important. This is because although smaller droplet volumes can accelerate time-to-detection (Boedicker et al., 2008; Kaushik et al., 2017), they also reduce the volumetric throughput for performing the assay and can consequently lengthen the overall turn-around time if the target biomarker is rare (Rosenfeld, Lin, Derda, & Tang, 2014).
7.2 Integrating sample preparation
Enhancing platform sensitivity requires mechanisms for rejecting assay inhibitors and sources of false-positive signals. To this end, samples may be prepared prior to encapsulation in droplets, such that the background molecules responsible for assay inhibition or false-positive signals are minimized or completely removed. Most current droplet platforms have achieved high sensitivity by relying on some degree of off-chip sample preparation and/or biomarker purification. For example, every droplet platform reviewed here for nucleic acid biomarker detection (Table 1) has relied on off-chip DNA or RNA extraction steps. Likewise, most droplet platforms used for protein detection (Table 2) relied on pre-purified proteins in controlled buffers, and most droplet platforms used for single-cell derived biomarker detection (Table 3) relied on immortalized cell lines in controlled buffers or reference bacterial strains in controlled buffers or broth. For the few droplet platforms that did process clinical biofluids, each required varying degrees of off-chip or on-chip sample preparation. For example, in order to detect secreted proteases from leukocytes, Jing et al. required off-chip red blood cell lysis, centrifugation, and washing steps as well as on-chip separation of cells from background contaminants prior to droplet generation (Jing et al., 2016). In order to demonstrate detection of bacteria in blood, Boedicker et al. input bacteria spiked into 50% plasma that was separated from whole blood off-chip (Boedicker et al., 2008), and Kang et al. spiked bacteria into 10% whole blood that was diluted off-chip (Kang et al., 2014). In both cases, dilution served as a means to reduce the concentration of assay inhibitors and sources of false-positive signals, and for Kang et al., it may have also reduced the likelihood of clogging the droplet device with too many red blood cells.
Although encapsulating biomarkers directly from biological samples appears to be the most straightforward approach to biomarker detection without adding to the assay turnaround time, sample purification and target biomarker concentration may prove useful, especially for samples with exceedingly rare biomarkers. Ideally, choosing (or developing) an assay for biomarker detection that could tolerate the presence of background from clinical biofluids could minimize sample pre-processing. However, practically, some degree of sample preparation may have to be integrated into the droplet platform in order to purify the sample or concentrate the target biomarker prior to droplet generation. For example, integrated sample filtration modules or modules for cell separation like flow-based pillar arrays (Jing et al., 2016) may be used to physically separate the target from its background. Alternatively, nucleic acid- or antibody-coated magnetic beads can be used to separate nucleic acids or protein biomarkers from biological samples and be subsequently encapsulated in droplets for detection (Guan et al., 2015; Shim et al., 2013). Finally, methods for biomarker extraction like glass pillars (Wu et al., 2011), sol-gels (Breadmore et al., 2003), and isolation chambers (Easley et al., 2006) may be integrated as an additional module on-chip. The downsides to these approaches are added assay complexity and difficulty in platform integration. As such, both the advantages and the disadvantages will have to be carefully evaluated for optimal platform performance.
7.3 Increasing multiplexing and screening capacity
Definitive diagnosis of many diseases requires the detection of multiple biomarkers instead of a single biomarker. Consequently, droplet microfluidic platforms can benefit from the ability to perform multiple assays in the same device. To this end, several research groups have attempted to create microfluidic strategies for generating multiple groups of droplets that could contain various conditions (e.g., reagents, substrate, and concentrations) within these devices. For example, a platform presented by Churski et al. constructed libraries of droplets with controllable input concentrations of sample/reagents and controllable volumes at 30 Hz (Churski, Korczyk, & Garstecki, 2010) (Figure 9A). More recently, a device was developed to facilitate the transition of nanoliter plugs that were constructed with controlled combinations of sample/reagents, into picoliter droplets without compromising droplet stability and uniformity (P. Zhang, Kaushik, Hsieh, & Wang, 2017) (Figure 9B). To further increase the number of conditions that can be introduced into the device, an external interface may be integrated with the device. For example, a serial sample loading (SSL) system was developed to create such an interface with conventional 96-well plates and facilitate sample and reagent loading (T. D. Rane, Zec, & Wang, 2012) (Figure 9C). Despite their clinical potential, these technological advances have not yet been integrated into a single platform for performing multiple assays. Therefore, more work to clinically showcase the multiplexability of these platforms is warranted.
Figure 9. Increasing multiplexing and screening capacity of droplet platforms.
(A) A droplet-on-demand system was developed to interface droplet chips with off-chip reservoirs in order to construct libraries of droplets with controllable input concentrations of sample/reagents and controllable volumes at 30 Hz. (Reproduced from (Churski et al., 2010) with permission of The Royal Society of Chemistry.) (B) A device was developed to facilitate the transition of nanoliter plugs that were constructed with controlled combinations of sample/reagents, into picoliter droplets without compromising droplet stability and uniformity. (Copyright 2017 IEEE. Reprinted, with permission, from (P. Zhang et al., 2017)) (C) A serial sample loading (SSL) system was developed to interface droplet platforms with conventional 96-well plates to facilitate automated sample and reagent loading. (Reproduced from (T. D. Rane et al., 2012) with permission of Sage Publishing.)
In biomarker screening assays, it is imperative to challenge samples containing the biomarkers with multiple reagents or substrates. To this end, we previously discussed work by Chen et al. wherein samples of peritoneal fluid were picoinjected into individual droplets of a pooled droplet library containing premade droplets of 9 distinct substrate/inhibitor mixtures for protease activity analysis (Chen et al., 2011). While devices like the one used by Chen et al. could be used to create a large number of monodisperse substrate/inhibitor droplets, since all droplets generated from a single device would contain a homogeneous chemical environment, more substrate/inhibitor droplet devices would be required to increase the multiplexing capacity of this platform. Therefore, one more degree of multiplexing would require one more droplet generation device which could add additional time and complexity to the workflow. Furthermore, since substrate/inhibitor droplets are all pooled together and later merged with the sample individually at random, each substrate droplet group must require a separate barcode in order to effectively demux the resulting data. Thus, as multiplexing capacity increases in such a platform, so do the challenges in speedily deconvoluting data. Rane et al., presented a more flexible and automatable valve-based platform for on-demand generation of droplets that could be assembled in situ with any desired combination and concentration of protease and substrate (Tushar D. Rane et al., 2015). The utility of this platform was demonstrated by screening 650 unique combinations and concentrations of MMPs and substrates. The use of spatial indexing of droplets here allowed for relatively simple deconvolution of the multiplexed assay based on the relative position of each droplet. Unfortunately, this high-throughput platform fell short of detecting clinical samples. Moreover, the significantly larger droplet volumes in this work (10 nL) compared to those used by Chen et al. (60 pL) may not have offered sufficient sensitivity for direct analysis of clinical samples. Therefore, while there has been some advancement in technology, there remains a definite need to develop an optimal platform that can perform high-throughput screening of clinical samples with automated operation.
7.4 Increasing platform automation
Ultimately, in clinical settings, diagnostic assays must be easy to perform with minimal hands-on time. Such requirements necessitate that microfluidic droplet devices and peripheral instrumentation operate collectively as an automated platform. Unfortunately, within the realm of academic research, current platforms almost universally still require significant manual operation (e.g., reagent loading) from experienced researchers. In the commercial sector, although the BioRad platform for ddPCR has shown to be effective for detecting nucleic acid biomarkers, this platform still needs to become more versatile to support the detection and screening of different types of biomarkers. As such, several advances still must be made for increasing platform automation.
Several considerations in the device, the instrumentation, and the interface between devices and instruments should be addressed for developing automated droplet microfluidic platforms. Envisioning the ideal droplet platform, the biological sample is manually loaded into a device (the only manual step of the assay) that is a single-use cartridge, which is then placed into a dedicated external instrument, which controls the fluid flow to perform droplet generation, and subsequent biomarker encapsulation, incubation, and detection in an integrated workflow. Such an integrated workflow is preferred because manual intervention and associated hands-on time can be minimized. An integrated workflow invariably necessitates the capacity to incubate droplets in situ. Incubating droplets within microchannels (delay lines) or microchambers of the device would ensure minimal disturbance to the relatively delicate droplets between the various steps in the assay but also increase the footprint of the device. This constraint may limit the number of conditions that can be tested or the incubation time that can performed in a single device. A potential solution may be to emulate the BioRad platform and use a tube or a segment of tubing that connects between a droplet generation device and a droplet detection device as the incubation reservoir, though such an incubation approach would require additional flow control and introduces challenges in uniform heating for the incubation region. The interface between the device and the instrument must be plug-and-play and allow for robust and efficient injection and switching of reagents in and out of the device. To this end, the needle and tubing based interface that is often used in research settings needs to be improved. Finally, easy-to-use and robust software must also be developed to execute the assay once the device/cartridge is inserted into the instrument and analyse the collected data with statistically significant thresholding. As improvements in sensitivity, throughput, multiplexing capacity, and automation come together, we envision increasing use of droplet based microfluidic platforms for robustly processing patient samples in the clinical setting.
8. Conclusion
In this article, we explore the development of droplet microfluidic platforms toward the detection and screening of disease biomarkers ranging from nucleic acids to proteins to single-cells. Sensitive detection and high-throughput screening of rare biomarkers from biological samples is critical to a variety of applications including disease diagnostics, therapeutics, and drug screening. To this end, droplet microfluidic technology leverages the advantages of sample discretization and as a result has emerged as an effective platform for high-sensitivity and high-throughput detection and screening of biomarkers. Several microfluidic strategies exist for biomarker digitization into droplets, where droplet generation throughput and control of droplet size, content, and motion are important considerations in evaluating each method. These strategies have been used widely to develop platforms for biomarker encapsulation, incubation, and detection. Notably, these platforms have widely relied on fragmented 3-step workflows; however, in pushing for further automation, researchers have presented attempts to integrate these steps and develop continuous-flow platforms. Many of these platforms have been successfully used for nucleic-acid, protein, and single-cell detection and screening, applied to diseases ranging from cancer to infectious diseases. Commercial platforms like BioRad’s ddPCR have now become a turnkey tool for quantitatively detecting nucleic acid biomarkers. Although the potential of droplet microfluidic platforms has led to burgeoning interest and research activities, they have yet to reach the ultimate goal in detecting and screening disease biomarkers in clinical settings. Toward achieving eventual clinical utility, seamless integration of sample preparation technologies and automation of platforms from biological sample to answer are critical components that can render these platforms useful in the clinical setting. With these improvements in place, we envision increasing utility of droplet platforms in clinical diagnostic and therapeutic workflows.
Acknowledgments
This work has been supported by the National Institutes of Health (R01AI117032).
Footnotes
Aniruddha M. Kaushik, 0000-0002-7329-624X
Kuangwen Hsieh, 0000-0003-3730-4406
Tza-Huei Wang, 0000-0002-3540-9354
Contributor Information
Aniruddha M. Kaushik, Department of Mechanical Engineering, Johns Hopkins University
Kuangwen Hsieh, Department of Mechanical Engineering, Johns Hopkins University.
Tza-Huei Wang, Department of Mechanical Engineering, Department of Biomedical Engineering, Johns Hopkins University.
References
- Akbari S, Pirbodaghi T. A droplet-based heterogeneous immunoassay for screening single cells secreting antigen-specific antibodies. Lab on a Chip. 2014;14(17):3275. doi: 10.1039/C4LC00082J. [DOI] [PubMed] [Google Scholar]
- Anna SL, Bontoux N, Stone HA. Formation of dispersions using “flow focusing” in microchannels. Applied Physics Letters. 2003;82(3):364–366. doi: 10.1063/1.1537519. [DOI] [Google Scholar]
- Arayanarakool R, Shui L, Kengen SWM, van den Berg A, Eijkel JCT. Single-enzyme analysis in a droplet-based micro- and nanofluidic system. Lab on a Chip. 2013;13(10):1955. doi: 10.1039/c3lc41100a. [DOI] [PubMed] [Google Scholar]
- Aronson JK. Biomarkers and surrogate endpoints. British Journal of Clinical Pharmacology. 2005;59(5):491–494. doi: 10.1111/j.1365-2125.2005.02435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axt B, Hsieh Y, Nalayanda D, Wang T. Impedance feedback control of microfluidic valves for reliable post processing combinatorial droplet injection. 2017:1–9. doi: 10.1007/s10544-017-0203-2. [DOI] [PubMed]
- Baker M. Digital PCR hits its stride. Nature Methods. 2012;9(6):541–544. doi: 10.1038/nmeth.2027. [DOI] [Google Scholar]
- Baret JC, Beck Y, Billas-Massobrio I, Moras D, Griffiths AD. Quantitative cell-based reporter gene assays using droplet-based microfluidics. Chemistry and Biology. 2010;17(5):528–536. doi: 10.1016/j.chembiol.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Beer NR, Hindson BJ, Wheeler EK, Hall SB, Rose KA, Kennedy IM, Colston BW. On-chip, real-time, single-copy polymerase chain reaction in picoliter droplets. Analytical Chemistry. 2007;79(22):8471–8475. doi: 10.1021/ac701809w. [DOI] [PubMed] [Google Scholar]
- Beer NR, Wheeler EK, Lee-Houghton L, Watkins N, Nasarabadi S, Hebert N, Colston BW. On-chip single-copy real-time reverse-transcription PCR in isolated picoliter droplets. Analytical Chemistry. 2008;80(6):1854–1858. doi: 10.1021/ac800048k. [DOI] [PubMed] [Google Scholar]
- Belgrader P, Tanner SC, Regan JF, Koehler R, Hindson BJ, Brown AS. Droplet digital PCR measurement of HER2 copy number alteration in formalin-fixed paraffin-embedded breast carcinoma tissue. Clinical Chemistry. 2013;59(6):991–994. doi: 10.1373/clinchem.2012.197855. [DOI] [PubMed] [Google Scholar]
- Beltrame L, Di Marino M, Fruscio R, Calura E, Chapman B, Clivio L, Marchini S. Profiling cancer gene mutations in longitudinal epithelial ovarian cancer biopsies by targeted next-generation sequencing: A retrospective study. Annals of Oncology. 2015;26(7):1363–1371. doi: 10.1093/annonc/mdv164. [DOI] [PubMed] [Google Scholar]
- Bian X, Jing F, Li G, Fan X, Jia C, Zhou H, Zhao J. A microfluidic droplet digital PCR for simultaneous detection of pathogenic Escherichia coli O157 and Listeria monocytogenes. Biosensors and Bioelectronics. 2015;74:770–777. doi: 10.1016/j.bios.2015.07.016. Journal Article. [DOI] [PubMed] [Google Scholar]
- Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab on a Chip. 2008;8(8):1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Perkins MD. Rapid molecular detection of tuberculosis and rifampin resistance. The New England Journal of Medicine. 2010;363(11):1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boitard L, Cottinet D, Kleinschmitt C, Bremond N, Baudry J, Yvert G, Bibette J. Monitoring single-cell bioenergetics via the coarsening of emulsion droplets. Proceedings of the National Academy of Sciences. 2012;109(19):7181–7186. doi: 10.1073/pnas.1200894109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breadmore MC, Wolfe KA, Arcibal IG, Leung WK, Dickson D, Giordano BC, Landers JP. Microchip-based purification of DNA from biological samples. Analytical Chemistry. 2003;75(8):1880–1886. doi: 10.1021/ac0204855. [DOI] [PubMed] [Google Scholar]
- Brunetto GS, Massoud R, Leibovitch EC, Caruso B, Johnson K, Ohayon J, Jacobson S. Digital droplet PCR (ddPCR) for the precise quantification of human T-lymphotropic virus 1 proviral loads in peripheral blood and cerebrospinal fluid of HAM/TSP patients and identification of viral mutations. Journal of NeuroVirology. 2014;20(4):341–351. doi: 10.1007/s13365-014-0249-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini MP, Hong J, Lim C, Choo J, Albrecht T, DeMello AJ, Edel JB. Ultrafast surface enhanced resonance raman scattering detection in droplet-based microfluidic systems. Analytical Chemistry. 2011;83(8):3076–3081. doi: 10.1021/ac103329b. [DOI] [PubMed] [Google Scholar]
- Chang-Hao Tsao S, Weiss J, Hudson C, Christophi C, Cebon J, Behren A, Dobrovic A. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Scientific Reports. 2015;5(1):11198. doi: 10.1038/srep11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Miller MA, Sarkar A, Beste MT, Isaacson KB, Lauffenburger DA, Han J. Multiplexed protease activity assay for low-volume clinical samples using droplet-based microfluidics and its application to endometriosis. Journal of the American Chemical Society. 2013;135(5):1645–1648. doi: 10.1021/ja307866z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Sarkar A, Song YA, Miller MA, Kim SJ, Griffith LG, Han J. Enhancing protease activity assay in droplet-based microfluidics using a biomolecule concentrator. Journal of the American Chemical Society. 2011;133(27):10368–10371. doi: 10.1021/ja2036628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Moon H, Kim CJ. Creating, transporting, cutting, and merging liquid droplets by electrowetting-based actuation for digital microfluidic circuits. Journal of Microelectromechanical Systems. 2003;12(1):70–80. doi: 10.1109/JMEMS.2002.807467. [DOI] [Google Scholar]
- Choi K, Ng AHC, Fobel R, Wheeler AR. Digital Microfluidics. Annual Review of Analytical Chemistry. 2012;5(1):413–440. doi: 10.1146/annurev-anchem-062011-143028. [DOI] [PubMed] [Google Scholar]
- Chou WL, Lee PY, Yang CL, Huang WY, Lin YS. Recent advances in applications of droplet microfluidics. Micromachines. 2015 doi: 10.3390/mi6091249. [DOI]
- Chrimes AF, Khoshmanesh K, Stoddart PR, Mitchell A, Kalantar-zadeh K. Microfluidics and Raman microscopy: current applications and future challenges. Chemical Society Reviews. 2013;42(13):5880. doi: 10.1039/c3cs35515b. [DOI] [PubMed] [Google Scholar]
- Churski K, Korczyk P, Garstecki P. High-throughput automated droplet microfluidic system for screening of reaction conditions. Lab Chip. 2010;10(7):816–818. doi: 10.1039/b925500a. [DOI] [PubMed] [Google Scholar]
- Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, Miller OJ, Frenz L, Merten CA. Droplet-Based Microfluidic Platforms for the Encapsulation and Screening of Mammalian Cells and Multicellular Organisms. Chemistry and Biology. 2008;15(5):427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Collins DJ, Neild A, deMello A, Liu AQ, Ai Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip. 2015:3439–3459. doi: 10.1039/C5LC00614G. [DOI] [PubMed]
- Cristobal G, Arbouet L, Sarrazin F, Talaga D, Bruneel JL, Joanicot M, Servant L. On-line laser Raman spectroscopic probing of droplets engineered in microfluidic devices. Lab Chip. 2006;6(9):1140–1146. doi: 10.1039/b602702d. [DOI] [PubMed] [Google Scholar]
- Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature Reviews. Clinical Oncology. 2013;10(8):472–84. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- Dangla R, Fradet E, Lopez Y, Baroud CN. The physical mechanisms of step emulsification. Journal of Physics D: Applied Physics. 2013;46(11):114003. doi: 10.1088/0022-3727/46/11/114003. [DOI] [Google Scholar]
- Dangla R, Kayi SC, Baroud CN. Droplet microfluidics driven by gradients of confinement. Proceedings of the National Academy of Sciences. 2013;110(3):853–858. doi: 10.1073/pnas.1209186110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport M, Mach KE, Shortliffe LMD, Banaei N, Wang TH, Liao JC. New and developing diagnostic technologies for urinary tract infections. Nature Reviews Urology. 2017 doi: 10.1038/nrurol.2017.20. [DOI] [PMC free article] [PubMed]
- De Angelis G, Rittenhouse HG, Mikolajczyk SD, Blair Shamel L, Semjonow A. Twenty Years of PSA: From Prostate Antigen to Tumor Marker. Reviews in Urology. 2007;9(3):113–23. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/17934568\nhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC2002501. [PMC free article] [PubMed] [Google Scholar]
- De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D, Seoane J. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nature Communications. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devonshire AS, Honeyborne I, Gutteridge A, Whale AS, Nixon G, Wilson P, Huggett JF. Highly Reproducible Absolute Quantification of Mycobacterium tuberculosis Complex by Digital PCR. Analytical Chemistry. 2015;87(7):3706–3713. doi: 10.1021/ac5041617. [DOI] [PubMed] [Google Scholar]
- DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. 2016 Retrieved from http://aidsinfo.nih.gov/guidelines.
- Ding Y, Casadevall i Solvas X, deMello A. “V-junction”: a novel structure for high-speed generation of bespoke droplet flows. The Analyst. 2015;140(2):414–421. doi: 10.1039/C4AN01730G. [DOI] [PubMed] [Google Scholar]
- Doyle T, Smith C, Vitiello P, Cambiano V, Johnson M, Owen A, Geretti AM. The importance of HIV RNA detection below 50 copies per mL in HIV-positive patients on antiretroviral therapy: an observational study. The Lancet. 2014;383:S44. doi: 10.1016/S0140-6736(14)60307-X. [DOI] [Google Scholar]
- Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(15):8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus R, Tabeling P, Willaime H. Ordered and Disordered Patterns in Two-Phase Flows in Microchannels. Physical Review Letters. 2003;90(14):144505. doi: 10.1103/PhysRevLett.90.144505. [DOI] [PubMed] [Google Scholar]
- Dutka F, Opalski AS, Garstecki P. Nano-liter droplet libraries from a pipette: step-emulsificator that stabilizes droplet volume against variation in flow rate. Lab Chip. 2016;16(11):2044–2049. doi: 10.1039/C6LC00265J. [DOI] [PubMed] [Google Scholar]
- Easley CJ, Karlinsey JM, Bienvenue JM, Legendre La, Roper MG, Feldman SH, Landers JP. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(51):19272–7. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastburn DJ, Sciambi A, Abate AR. Ultrahigh-Throughput Mammalian Single-Cell Reverse-Transcriptase Polymerase Chain Reaction in Micro fl uidic Drops. 2013 doi: 10.1021/ac402057q. [DOI] [PubMed] [Google Scholar]
- Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: Technical and operational challenges. Clinical Chemistry. 2010 doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed]
- Frenz L, Blank K, Brouzes E, Griffiths AD. Reliable microfluidic on-chip incubation of droplets in delay-lines. Lab Chip. 2009;9(10):1344–1348. doi: 10.1039/B816049J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Benes CH. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garstecki P, Fuerstman MJ, Stone A, Whitesides GM. Formation of droplets and bubbles in a microfluidic T-junction — scaling and mechanism of break-up. 2006:437–446. doi: 10.1039/b510841a. [DOI] [PubMed]
- Gu S, Lu Y, Ding Y, Li L, Song H, Wang J, Wu Q. A droplet-based microfluidic electrochemical sensor using platinum-black microelectrode and its application in high sensitive glucose sensing. Biosensors and Bioelectronics. 2014;55:106–112. doi: 10.1016/j.bios.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Guan W, Chen L, Rane TD, Wang TH. Droplet Digital Enzyme-Linked Oligonucleotide Hybridization Assay for Absolute RNA Quantification. Scientific Reports. 2015;5:13795. doi: 10.1038/srep13795. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Liu K, Ji XH, Ding HJ, Zhang M, Zeng Q, Zhao XZ. Valve-based microfluidic device for droplet on-demand operation and static assay. Applied Physics Letters. 2010;97(23) doi: 10.1063/1.3521283. [DOI] [Google Scholar]
- Gupta A, Matharoo HS, Makkar D, Kumar R. Droplet formation via squeezing mechanism in a microfluidic flow-focusing device. Computers and Fluids. 2014;100:218–226. doi: 10.1016/j.compfluid.2014.05.023. [DOI] [Google Scholar]
- Guttenberg Z, Müller H, Habermüller H, Geisbauer A, Pipper J, Felbel J, Wixforth A. Planar chip device for PCR and hybridization with surface acoustic wave pump. Lab Chip. 2005;5(3):308–317. doi: 10.1039/B412712A. [DOI] [PubMed] [Google Scholar]
- Guttery DS, Page K, Hills A, Woodley L, Marchese SD, Rghebi B, Shaw JA. Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor-positive metastatic breast cancer. Clinical Chemistry. 2015;61(7):974–982. doi: 10.1373/clinchem.2015.238717. [DOI] [PubMed] [Google Scholar]
- Han Z, Li W, Huang Y, Zheng B. Measuring rapid enzymatic kinetics by electrochemical method in droplet-based microfluidic devices with pneumatic valves. Analytical Chemistry. 2009;81(14):5840–5845. doi: 10.1021/ac900811y. [DOI] [PubMed] [Google Scholar]
- Hanash SM, Baik CS, Kallioniemi O. Emerging molecular biomarkers-blood-based strategies to detect and monitor cancer. Nature Reviews Clinical Oncology. 2011 doi: 10.1038/nrclinonc.2010.220. [DOI] [PubMed]
- Hatch AC, Fisher JS, Pentoney SL, Yang DL, Lee AP. Tunable 3D droplet self-assembly for ultra-high-density digital micro-reactor arrays. Lab on a Chip. 2011;11(15):2509–2517. doi: 10.1039/c0lc00553c. Journal Article. [DOI] [PubMed] [Google Scholar]
- Hatch AC, Fisher JS, Tovar AR, Hsieh AT, Lin R, Pentoney SL, Lee AP. 1-Million droplet array with wide-field fluorescence imaging for digital PCR. Lab on a Chip. 2011;11(22):3838. doi: 10.1039/c1lc20561g. Journal Article. [DOI] [PubMed] [Google Scholar]
- Heredia NJ, Belgrader P, Wang S, Koehler R, Regan J, Cosman AM, Karlin-Neumann G. Droplet Digital™ PCR quantitation of HER2 expression in FFPE breast cancer samples. Methods. 2013;59(1):183–186. doi: 10.1016/j.ymeth.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Héritier S, Emile JF, Barkaoui MA, Thomas C, Fraitag S, Boudjemaa S, Donadieu J. BRAF Mutation Correlates With High-Risk Langerhans Cell Histiocytosis and Increased Resistance to First-Line Therapy. Journal of Clinical Oncology. 2016;34(25):3023–3030. doi: 10.1200/JCO.2015.65.9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Colston BW. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical Chemistry. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. Journal Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K, Zec HC, Ma PC, Rane TD, Wang TH. Enhancing throughput of combinatorial droplet devices via droplet bifurcation, parallelized droplet fusion, and parallelized detection. Micromachines. 2015;6(10):1490–1504. doi: 10.3390/mi6101434. [DOI] [Google Scholar]
- Hu Y, Xu P, Luo J, He H, Du W. Absolute Quantification of H5-Subtype Avian Influenza Viruses Using Droplet Digital Loop-Mediated Isothermal Amplification. Analytical Chemistry. 2017;89(1):745–750. doi: 10.1021/acs.analchem.6b03328. [DOI] [PubMed] [Google Scholar]
- Huang CJ, Fang WF, Ke MS, Chou HYE, Yang JT. A biocompatible open-surface droplet manipulation platform for detection of multi-nucleotide polymorphism. Lab on a Chip. 2014;14(12):2057–62. doi: 10.1039/c4lc00089g. [DOI] [PubMed] [Google Scholar]
- Huang JT, Liu YJ, Wang J, Xu ZG, Yang Y, Shen F, Liu SM. Next generation digital PCR measurement of hepatitis B virus copy number in formalin-fixed paraffin-embedded hepatocellular carcinoma tissue. Clinical Chemistry. 2015;61(1):290–296. doi: 10.1373/clinchem.2014.230227. [DOI] [PubMed] [Google Scholar]
- Hung LY, Wu HW, Hsieh K, Lee G Bin. Microfluidic platforms for discovery and detection of molecular biomarkers. Microfluidics and Nanofluidics. 2014;16(5):941–963. doi: 10.1007/s10404-014-1354-6. [DOI] [Google Scholar]
- Jambovane S, Kim DJ, Duin EC, Kim SK, Hong JW. Creation of stepwise concentration gradient in picoliter droplets for parallel reactions of matrix metalloproteinase II and IX. Analytical Chemistry. 2011;83(9):3358–3364. doi: 10.1021/ac103217p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeannot E, Becette V, Campitelli M, Calmejane MA, Lappartient E, Ruff E, Sastre-Garau X. Circulating human papillomavirus DNA detected using droplet digital PCR in the serum of patients diagnosed with early stage human papillomavirus-associated invasive carcinoma. Journal of Pathology: Clinical Research. 2016;2(4):201–209. doi: 10.1002/cjp2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing T, Lai Z, Wu L, Han J, Lim CT, Chen CH. Single Cell Analysis of Leukocyte Protease Activity Using Integrated Continuous-Flow Microfluidics. Analytical Chemistry. 2016;88(23):11750–11757. doi: 10.1021/acs.analchem.6b03370. [DOI] [PubMed] [Google Scholar]
- Jing T, Ramji R, Warkiani ME, Han J, Lim CT, Chen CH. Jetting microfluidics with size-sorting capability for single-cell protease detection. Biosensors and Bioelectronics. 2015;66:19–23. doi: 10.1016/j.bios.2014.11.001. [DOI] [PubMed] [Google Scholar]
- Joensson HN, Samuels ML, Brouzes ER, Medkova M, Uhløn M, Link DR, Andersson-Svahn H. Detection and analysis of low-abundance cell-surface biomarkers using enzymatic amplification in microfluidic droplets. Angewandte Chemie - International Edition. 2009;48(14):2518–2521. doi: 10.1002/anie.200804326. [DOI] [PubMed] [Google Scholar]
- Kang DK, Ali MM, Zhang K, Huang SS, Peterson E, Digman MA, Zhao W. Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nature Communications. 2014;5:5427. doi: 10.1038/ncomms6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik AM, Hsieh K, Chen L, Shin DJ, Liao JC, Wang TH. Accelerating bacterial growth detection and antimicrobial susceptibility assessment in integrated picoliter droplet platform. Biosensors and Bioelectronics. 2017;97:260–266. doi: 10.1016/j.bios.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley K, Cosman A, Belgrader P, Chapman B, Sullivan DC. Detection of methicillin-resistant Staphylococcus aureus by a duplex droplet digital PCR assay. Journal of Clinical Microbiology. 2013;51(7):2033–2039. doi: 10.1128/JCM.00196-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemna EWM, Segerink LI, Wolbers F, Vermes I, van den Berg A. Label-free, high-throughput, electrical detection of cells in droplets. The Analyst. 2013;138(16):4585. doi: 10.1039/c3an00569k. [DOI] [PubMed] [Google Scholar]
- Khaw MK, Ooi CH, Mohd-Yasin F, Vadivelu R, John JS, Nguyen NT. Digital microfluidics with a magnetically actuated floating liquid marble. Lab Chip. 2016;16(12):2211–2218. doi: 10.1039/C6LC00378H. [DOI] [PubMed] [Google Scholar]
- Kim M, Pan M, Gai Y, Pang S, Han C, Yang C, Tang SKY. Optofluidic ultrahigh-throughput detection of fluorescent drops. Lab on a Chip. 2015;15(6):1417–1423. doi: 10.1039/C4LC01465K. [DOI] [PubMed] [Google Scholar]
- Kim SC, Premasekharan G, Clark IC, Gemeda HB, Paris PL, Abate AR. Measurement of copy number variation in single cancer cells using rapid-emulsification digital droplet MDA. Microsystems & Nanoengineering. 2017;3:17018. doi: 10.1038/micronano.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinugasa H, Nouso K, Tanaka T, Miyahara K, Morimoto Y, Dohi C, Yamamoto K. Droplet digital PCR measurement of HER2 in patients with gastric cancer. British Journal of Cancer. 2015;112(10):1652–1655. doi: 10.1038/bjc.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss MM, Ortoleva-Donnelly L, Reginald Beer N, Warner J, Bailey CG, Colston BW, Leamon JH. High-throughput quantitative polymerase chain reaction in picoliter droplets. Analytical Chemistry. 2008;80(23):8975–8981. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster S, Angilè FE, Duan H, Agresti JJ, Wintner A, Schmitz C, Weitz Da. Drop-based microfluidic devices for encapsulation of single cells. Lab on a Chip. 2008;8(7):1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- Laurent-Puig P, Pekin D, Normand C, Kotsopoulos SK, Nizard P, Perez-Toralla K, Taly V. Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with Anti-EGFR therapy. Clinical Cancer Research. 2015;21(5):1087–1097. doi: 10.1158/1078-0432.CCR-14-0983. [DOI] [PubMed] [Google Scholar]
- Leng SX, McElhaney JE, Walston JD, Xie D, Fedarko NS, Kuchel GA. ELISA and Multiplex Technologies for Cytokine Measurement in Inflammation and Aging Research. The Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2008;63(8):879–884. doi: 10.1093/gerona/63.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. BEAMing up for detection and quantification of rare sequence variants. Nature Methods. 2006;3(2):95–97. doi: 10.1038/NMETH850. [DOI] [PubMed] [Google Scholar]
- Li Z, Leshansky AM, Pismen LM, Tabeling P. Step-emulsification in a microfluidic device. Lab on a Chip. 2015;15(4):1023–1031. doi: 10.1039/C4LC01289E. [DOI] [PubMed] [Google Scholar]
- Lim CT, Zhang Y. Bead-based microfluidic immunoassays: The next generation. Biosensors and Bioelectronics. 2007 doi: 10.1016/j.bios.2006.06.005. [DOI] [PubMed]
- Lim J, Caen O, Vrignon J, Konrad M, Taly V, Baret JC. Parallelized ultra-high throughput microfluidic emulsifier for multiplex kinetic assays. Biomicrofluidics. 2015;9(3) doi: 10.1063/1.4919415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Vrignon J, Gruner P, Karamitros CS, Konrad M, Baret JC. Ultra-high throughput detection of single cell beta-galactosidase activity in droplets using micro-optical lens array. Applied Physics Letters. 2013;103 doi: 10.1063/1.4830046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Painter RE, Enesa K, Holmes D, Whyte G, Garlisi CG, Smith CA. High-throughput screening of antibiotic-resistant bacteria in picodroplets. Lab on a Chip. 2016:1636–1643. doi: 10.1039/C6LC00180G. [DOI] [PubMed]
- Luft LM, Gill MJ, Church DL. HIV-1 viral diversity and its implications for viral load testing: review of current platforms. International Journal of Infectious Diseases : IJID : Official Publication of the International Society for Infectious Diseases. 2011;15(10):e661–70. doi: 10.1016/j.ijid.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Luo J, Li J, Yang H, Yu J, Wei H. Journal of Clinical Microbiology. JCM; 2017. Jul, Accurate detection of methicillin-resistant Staphylococcus aureus in mixtures utilizing single bacterial duplex droplet digital PCR; pp. 00716–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther SK, Will S, Braeuer a. Phase-specific Raman spectroscopy for fast segmented microfluidic flows. Lab on a Chip. 2014:2–5. doi: 10.1039/c4lc00428k. [DOI] [PubMed]
- Mach KE, Wong PK, Liao JC. Biosensor diagnosis of urinary tract infections: A path to better treatment? Trends in Pharmacological Sciences. 2011 doi: 10.1016/j.tips.2011.03.001. [DOI] [PMC free article] [PubMed]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, McCarroll SA. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini N, Carletti S, Ghidoli N, Cichero P, Burioni R, Clementi M. The era of molecular and other non-culture-based methods in diagnosis of sepsis. Clinical Microbiology Reviews. 2010 doi: 10.1128/CMR.00043-09. [DOI] [PMC free article] [PubMed]
- März A, Henkel T, Cialla D, Schmitt M, Popp J. Droplet formation via flow-through microdevices in Raman and surface enhanced Raman spectroscopy—concepts and applications. Lab on a Chip. 2011;11(21):3584. doi: 10.1039/c1lc20638a. [DOI] [PubMed] [Google Scholar]
- Matsubara Y, Kerman K, Kobayashi M, Yamamura S, Morita Y, Tamiya E. Microchamber array based DNA quantification and specific sequence detection from a single copy via PCR in nanoliter volumes. Biosensors and Bioelectronics. 2005;20:1482–1490. doi: 10.1016/j.bios.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Mazutis L, Araghi AF, Miller OJ, Baret JC, Frenz L, Janoshazi A, Ryckelynck M. Droplet-based microfluidic systems for high-throughput single DNA molecule isothermal amplification and analysis. Analytical Chemistry. 2009;81(12):4813–4821. doi: 10.1021/ac900403z. [DOI] [PubMed] [Google Scholar]
- Mazutis L, Gilbert J, Ung WL, Weitz DA, Griffiths AD, Heyman JA. Single-cell analysis and sorting using droplet-based microfluidics. Nature Protocols. 2013;8(5):870–891. doi: 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheel CM, Ball JR. Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease. 2010 http://doi.org/10.17226/12869. [PubMed]
- Millar BC, Xu J, Moore JE. Molecular diagnostics of medically important bacterial infections. Current Issues in Molecular Biology. 2007 http://doi.org/17263144. [PubMed]
- Miller EM, Wheeler AR. A digital microfluidic approach to homogeneous enzyme assays. Analytical Chemistry. 2008;80(5):1614–1619. doi: 10.1021/ac702269d. [DOI] [PubMed] [Google Scholar]
- Mukaide M, Sugiyama M, Korenaga M, Murata K, Kanto T, Masaki N, Mizokami M. High-throughput and sensitive next-generation droplet digital PCR assay for the quantitation of the hepatitis C virus mutation at core amino acid 70. Journal of Virological Methods. 2014;207:169–177. doi: 10.1016/j.jviromet.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Nahavandi S, Baratchi S, Soffe R, Tang SY, Nahavandi S, Mitchell A, Khoshmanesh K. Microfluidic platforms for biomarker analysis. Lab on a Chip. 2014;14(9):1496. doi: 10.1039/c3lc51124c. [DOI] [PubMed] [Google Scholar]
- Najah M, Mayot E, Mahendra-Wijaya IP, Griffiths AD, Ladame S, Drevelle A. New glycosidase substrates for droplet-based microfluidic screening. Analytical Chemistry. 2013;85(20):9807–9814. doi: 10.1021/ac4022709. [DOI] [PubMed] [Google Scholar]
- Nakashima T, Shimizu M, Kukizaki M. Particle control of emulsion by membrane emulsification and its applications. Advanced Drug Delivery Reviews. 2000;45(1):47–56. doi: 10.1016/S0169-409X(00)00099-5. [DOI] [PubMed] [Google Scholar]
- Navin NE. The first five years of single-cell cancer genomics and beyond. Genome Research. 2015 doi: 10.1101/gr.191098.115. [DOI] [PMC free article] [PubMed]
- Nelson WC, Kim C-J“CJ”. Droplet Actuation by Electrowetting-on-Dielectric (EWOD): A Review. Journal of Adhesion Science and Technology. 2012;26(12–17):1747–1771. doi: 10.1163/156856111X599562. [DOI] [Google Scholar]
- Ng EX, Miller MA, Jing T, Chen CH. Single cell multiplexed assay for proteolytic activity using droplet microfluidics. Biosensors and Bioelectronics. 2016;81:408–414. doi: 10.1016/j.bios.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Ng EX, Miller MA, Jing T, Lauffenburger DA, Chen CH. Low-volume multiplexed proteolytic activity assay and inhibitor analysis through a pico-injector array. Lab Chip. 2015;15(4):1153–1159. doi: 10.1039/C4LC01162G. [DOI] [PubMed] [Google Scholar]
- Nguyen NT, Lassemono S, Chollet FA. Optical detection for droplet size control in microfluidic droplet-based analysis systems. Sensors and Actuators, B: Chemical. 2006;117(2):431–436. doi: 10.1016/j.snb.2005.12.010. [DOI] [Google Scholar]
- Nisisako T, Torii T. Microfluidic large-scale integration on a chip for mass production of monodisperse droplets and particles. Lab on a Chip. 2008;8(2):287–293. doi: 10.1039/B713141K. [DOI] [PubMed] [Google Scholar]
- Niu X, deMello AJ. Building droplet-based microfluidic systems for biological analysis. Biochemical Society Transactions. 2012;40(4):615–623. doi: 10.1042/bst20120005. [DOI] [PubMed] [Google Scholar]
- Niu X, Zhang M, Peng S, Wen W, Sheng P. Real-time detection, control, and sorting of microfluidic droplets. Biomicrofluidics. 2007;1(4) doi: 10.1063/1.2795392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak R, Zeng Y, Shuga J, Venugopalan G, Fletcher DA, Smith MT, Mathies RA. Single-cell multiplex gene detection and sequencing with microfluidically generated agarose emulsions. Angewandte Chemie - International Edition. 2011;50(2):390–395. doi: 10.1002/anie.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofner A, Moore DG, Rühs PA, Schwendimann P, Eggersdorfer M, Amstad E, Studart AR. High-Throughput Step Emulsification for the Production of Functional Materials Using a Glass Microfluidic Device. Macromolecular Chemistry and Physics. 2016;201600472:1600472. doi: 10.1002/macp.201600472. [DOI] [Google Scholar]
- Otsuji K, Sasaki T, Tanaka A, Kunita A, Ikemura M, Matsusaka K, Seto Y. Use of droplet digital PCR for quantitative and automatic analysis of the HER2 status in breast cancer patients. Breast Cancer Research and Treatment. 2016;162(1):11–18. doi: 10.1007/s10549-016-4092-5. [DOI] [PubMed] [Google Scholar]
- Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O’Connell A, Messineo MM, Jänne PA. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. TL - 20. Clinical Cancer Research. 2014;20VN-r(6):1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Teitell MA, Chiou EPY. Single-sided continuous optoelectrowetting (SCOEW) for droplet manipulation with light patterns. Lab on a Chip. 2010;10(13):1655. doi: 10.1039/c001324b. [DOI] [PubMed] [Google Scholar]
- Park SY, Wu TH, Chen Y, Teitell MA, Chiou PY. High-speed droplet generation on demand driven by pulse laser-induced cavitation. Lab on a Chip. 2011;11(6):1010. doi: 10.1039/c0lc00555j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavšič J, Žel J, Milavec M. Digital PCR for direct quantification of viruses without DNA extraction. Analytical and Bioanalytical Chemistry. 2016;408(1):67–75. doi: 10.1007/s00216-015-9109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekin D, Skhiri Y, Baret JC, Le Corre D, Mazutis L, Ben Salem C, Taly V. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab on a Chip. 2011;11(13):2156–2166. doi: 10.1039/C1LC20128J. Journal Article. [DOI] [PubMed] [Google Scholar]
- Pekin D, Taly V. Droplet-based microfluidics digital PCR for the detection of KRAS mutations. Methods in Molecular Biology. 2017;1547:143–164. doi: 10.1007/978-1-4939-6734-6_12. [DOI] [PubMed] [Google Scholar]
- Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. Clinical Laboratory Standards Institute; Wayne, PA, USA: CLSI; 2014. p. 32. [Google Scholar]
- Pipper J, Zhang Y, Neuzil P, Hsieh TM. Clockwork PCR including sample preparation. Angewandte Chemie - International Edition. 2008;47(21):3900–3904. doi: 10.1002/anie.200705016. [DOI] [PubMed] [Google Scholar]
- Postek W, Kaminski T, Garstecki P. A passive microfluidic system based on step emulsification allows to generate libraries of nanoliter-sized droplets from microliter droplets of varying and known concentration of sample. Lab on a Chip. 2017;17:1323–1331. doi: 10.1039/C7LC00014F. [DOI] [PubMed] [Google Scholar]
- Rački N, Morisset D, Gutierrez-Aguirre I, Ravnikar M. One-step RT-droplet digital PCR: A breakthrough in the quantification of waterborne RNA viruses. Analytical and Bioanalytical Chemistry. 2014;406(3):661–667. doi: 10.1007/s00216-013-7476-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane TD, Chen L, Zec HC, Wang T. Microfluidic continuous flow digital loop-mediated isothermal amplification (LAMP) Lab on a Chip. 2014;15(3):776–782. doi: 10.1039/C4LC00901K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane TD, Puleo CM, Liu KJ, Zhang Y, Lee AP, Wang TH. Counting single molecules in sub-nanolitre droplets. Lab on a Chip. 2010;10(2):161–164. doi: 10.1039/B917503B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane TD, Zec HC, Puleo C, Lee AP, Wang TH. Droplet microfluidics for amplification-free genetic detection of single cells. Lab on a Chip. 2012;12(18):3341. doi: 10.1039/c2lc40537g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane TD, Zec HC, Wang TH. A barcode-free combinatorial screening platform for matrix metalloproteinase screening. Analytical Chemistry. 2015;87(3):1950–1956. doi: 10.1021/ac504330x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane TD, Zec HC, Wang TH. A Serial Sample Loading System: Interfacing Multiwell Plates with Microfluidic Devices. Journal of Laboratory Automation. 2012;17(5):370–377. doi: 10.1177/2211068212455169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid AL, Freeman JB, Millward M, Ziman M, Gray ES. Detection of BRAF-V600E and V600K in melanoma circulating tumour cells by droplet digital PCR. Clinical Biochemistry. 2015;48(15):999–1002. doi: 10.1016/j.clinbiochem.2014.12.007. [DOI] [PubMed] [Google Scholar]
- Rhee M, Light YK, Meagher RJ, Singh AK. Digital Droplet Multiple Displacement Amplification (ddMDA) for Whole Genome Sequencing of Limited DNA Samples. PloS One. 2016;11(5):e0153699. doi: 10.1371/journal.pone.0153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi A. Teaming up for biomarker future. EMBO Reports. 2011;12(6):500–504. doi: 10.1038/embor.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondelez Y, Tresset G, Tabata KV, Arata H, Fujita H, Takeuchi S, Noji H. Microfabricated arrays of femtoliter chambers allow single molecule enzymology. Nature Biotechnology. 2005;23(3):361–365. doi: 10.1038/nbt1072. [DOI] [PubMed] [Google Scholar]
- Rosenfeld L, Lin T, Derda R, Tang SKY. Review and analysis of performance metrics of droplet microfluidics systems. Microfluidics and Nanofluidics. 2014;16(5):921–939. doi: 10.1007/s10404-013-1310-x. [DOI] [Google Scholar]
- Ruelle J, Yfantis V, Duquenne A, Goubau P. Validation of an ultrasensitive digital droplet PCR assay for HIV-2 plasma RNA quantification. Journal of the International Aids Society. 2014;17(4 Suppl 3):19675. doi: 10.7448/IAS.17.4.19675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, Oxnard GR. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncology. 2016;2(8):1014–22. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmamed M, Fernández-Landázuri S, Rodríguez C, Zárate R, Lozano M, Zubiri L, González A. Quantitative cell-free circulating BRAFV600E mutation analysis by use of droplet digital PCR in the follow-up of patients with melanoma being treated with BRAF inhibitors. Clinical Chemistry. 2015;61(1):297–304. doi: 10.1373/clinchem.2014.230235. [DOI] [PubMed] [Google Scholar]
- Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- Scheler O, Kaminski TS, Ruszczak A, Garstecki P. Dodecylresorufin (C12R) Outperforms Resorufin in Microdroplet Bacterial Assays. ACS Applied Materials and Interfaces. 2016;8(18):11318–11325. doi: 10.1021/acsami.6b02360. [DOI] [PubMed] [Google Scholar]
- Scheler O, Pacocha N, Debski PR, Ruszczak A, Kaminski TS, Garstecki P. Optimized droplet digital CFU assay (ddCFU) provides precise quantification of bacteria over a dynamic range of 6 logs and beyond. Lab on a Chip. 2017;17(11):1980–1987. doi: 10.1039/C7LC00206H. [DOI] [PubMed] [Google Scholar]
- Schmid L, Franke T. SAW-controlled drop size for flow focusing. Lab on a Chip. 2013;13(9):1691. doi: 10.1039/c3lc41233d. [DOI] [PubMed] [Google Scholar]
- Schmid L, Franke T. Acoustic modulation of droplet size in a T-junction. Applied Physics Letters. 2014;104(13) doi: 10.1063/1.4869536. [DOI] [Google Scholar]
- Schuler F, Schwemmer F, Trotter M, Wadle S, Zengerle R, von Stetten F, Paust N. Centrifugal step emulsification applied for absolute quantification of nucleic acids by digital droplet RPA. Lab Chip. 2015;15(13):2759–2766. doi: 10.1039/C5LC00291E. [DOI] [PubMed] [Google Scholar]
- Shembekar N, Chaipan C, Utharala R, Merten CA. Droplet-based microfluidics in drug discovery, transcriptomics and high-throughput molecular genetics. Lab on a Chip. 2016;16(8):1314–1331. doi: 10.1039/C6LC00249H. [DOI] [PubMed] [Google Scholar]
- Shim JU, Ranasinghe RT, Smith CA, Ibrahim SM, Hollfelder F, Huck WTS, Abell C. Ultrarapid generation of femtoliter microfluidic droplets for single-molecule-counting immunoassays. ACS Nano. 2013;7(7):5955–5964. doi: 10.1021/nn401661d. [DOI] [PubMed] [Google Scholar]
- Shin DJ, Zhang Y, Wang TH. A droplet microfluidic approach to single-stream nucleic acid isolation and mutation detection. Microfluidics and Nanofluidics. 2014;17(2):425–430. doi: 10.1007/s10404-013-1305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan CA, Tang SKY, Whitesides GM. Independent control of drop size and velocity in microfluidic flow-focusing generators using variable temperature and flow rate. Analytical Chemistry. 2009;81(6):2399–2402. doi: 10.1021/ac8026542. [DOI] [PubMed] [Google Scholar]
- Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Richman DD. Highly Precise Measurement of HIV DNA by Droplet Digital PCR. PLoS ONE. 2013;8(4):2–9. doi: 10.1371/journal.pone.0055943. Journal Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimbu K, Tavel JA. What are Biomarkers? Current Opininion in HIV and AIDS. 2011;5(6):463–466. doi: 10.1097/COH.0b013e32833ed177.What. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura S, Nakajima M, Seki M. Effect of channel structure on microchannel emulsification. Langmuir. 2002;18(15):5708–5712. doi: 10.1021/la025813a. [DOI] [Google Scholar]
- Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. BioTechniques. 1992;13(3):444–449. [PubMed] [Google Scholar]
- Syme CD, Martino C, Yusvana R, Sirimuthu NMS, Cooper JM. Quantitative characterization of individual microdroplets using surface-enhanced resonance raman scattering spectroscopy. Analytical Chemistry. 2012;84(3):1491–1495. doi: 10.1021/ac202705a. [DOI] [PubMed] [Google Scholar]
- Takahama T, Sakai K, Takeda M, Azuma K, Hida T, Hirabayashi M, Nishio K. Detection of the T790M mutation of EGFR in plasma of advanced non–small cell lung cancer patients with acquired resistance to tyrosine kinase inhibitors (West Japan oncology group 8014LTR study) inhibitors (West Japan oncology group 8014LTR study) Oncotarget. 2016;7(36):8014. doi: 10.18632/oncotarget.11303. http://doi.org/10.18632/oncotarget.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taly V, Pekin D, El Abed A, Laurent-Puig P. Detecting biomarkers with microdroplet technology. Trends in Molecular Medicine. 2012a doi: 10.1016/j.molmed.2012.05.001. [DOI] [PubMed]
- Taly V, Pekin D, El Abed A, Laurent-Puig P. Detecting biomarkers with microdroplet technology. Trends in Molecular Medicine. 2012b;18(7):405–416. doi: 10.1016/j.molmed.2012.05.001. Journal Article. [DOI] [PubMed] [Google Scholar]
- Taly V, Pekin D, Benhaim L, Kotsopoulos SK, Le Corre D, Li X, Laurent-Puig P. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clinical Chemistry. 2013;59(12):1722–1731. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- Tan SH, Nguyen NT, Yobas L, Kang TG. Formation and manipulation of ferrofluid droplets at a microfluidic T-junction. Journal of Micromechanics and Microengineering. 2010;20(4):45004. http://doi.org/Artn045004/r10.1088/0960-1317/20/4/045004. [Google Scholar]
- Tang MYH, Shum HC. One-step immunoassay of C-reactive protein using droplet microfluidics. Lab on a Chip. 2016;16(22):4359–4365. doi: 10.1039/C6LC01121G. [DOI] [PubMed] [Google Scholar]
- Tanyeri M, Perron R, Kennedy IM. Lasing droplets in a microfabricated channel. Optics Letters. 2007;32(17):2529–2531. doi: 10.1364/OL.32.002529. [DOI] [PubMed] [Google Scholar]
- Taylor GI. The Formation of Emulsions in Definable Fields of Flow. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences. 1934;146(858):501–523. doi: 10.1098/rspa.1934.0169. [DOI] [Google Scholar]
- Thorsen T, Roberts RW, Arnold FH, Quake SR. Dynamic pattern formation in a vesicle-generating microfluidic device. Physical Review Letters. 2001;86(18):4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, Barrett JC. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Tighe PJ, Ryder RR, Todd I, Fairclough LC. ELISA in the multiplex era: Potentials and pitfalls. Proteomics - Clinical Applications. 2015 doi: 10.1002/prca.201400130. [DOI] [PMC free article] [PubMed]
- Trypsteen W, Kiselinova M, Vandekerckhove L, De Spiegelaere W. Diagnostic utility of droplet digital PCR for HIV reservoir quantification. Journal of Virus Eradication. 2016;2(3):162–9. doi: 10.1016/S2055-6640(20)30460-X. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/27482456\nhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4967968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trypsteen W, Vynck M, De Neve J, Bonczkowski P, Kiselinova M, Malatinkova E, De Spiegelaere W. ddpcRquant: threshold determination for single channel droplet digital PCR experiments. Analytical and Bioanalytical Chemistry. 2015;407(19):5827–5834. doi: 10.1007/s00216-015-8773-4. [DOI] [PubMed] [Google Scholar]
- Unger MA, Chou H, Thorsen T, Scherer A, Quake SR. Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography. Science. 2000;288(5463):113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- van Ginkel JH, Huibers MMH, van Es RJJ, de Bree R, Willems SM. Droplet digital PCR for detection and quantification of circulating tumor DNA in plasma of head and neck cancer patients. BMC Cancer. 2017;17(1) doi: 10.1186/s12885-017-3424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Kawaguchi T, Isa SI, Ando M, Tamiya A, Kubo A, Koh Y. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clinical Cancer Research. 2015;21(15):3552–3560. doi: 10.1158/1078-0432.CCR-14-2151. [DOI] [PubMed] [Google Scholar]
- Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442(7101):368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- Witte AK, Fister S, Mester P, Schoder D, Rossmanith P. Evaluation of the performance of quantitative detection of the Listeria monocytogenes prfA locus with droplet digital PCR. Analytical and Bioanalytical Chemistry. 2016;408(27):7583–7593. doi: 10.1007/s00216-016-9861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Jin W, Zhou C, Han S, Yang W, Zhu Q, Mu Y. Integrated glass microdevice for nucleic acid purification, loop-mediated isothermal amplification, and online detection. Analytical Chemistry. 2011;83(9):3336–3342. doi: 10.1021/ac103129e. [DOI] [PubMed] [Google Scholar]
- Xu J, Attinger D. Drop on demand in a microfluidic chip. Journal of Micromechanics and Microengineering. 2008;18(6):65020. doi: 10.1088/0960-1317/18/6/065020. [DOI] [Google Scholar]
- Xu JH, Li SW, Tán J, Wang YJ, Luo GS. Preparation of highly monodisperse droplet in a T-junction microfluidic device. AIChE Journal. 2006;52(9):3005–3010. doi: 10.1002/aic.10924. [DOI] [Google Scholar]
- Xu P, Zheng X, Tao Y, Du W. Cross-Interface Emulsification for Generating Size-Tunable Droplets. Analytical Chemistry. 2016;88(6):3171–3177. doi: 10.1021/acs.analchem.5b04510. [DOI] [PubMed] [Google Scholar]
- Zagnoni M, Anderson J, Cooper JM. Hysteresis in multiphase microfluidics at a T-junction. Langmuir. 2010;26(12):9416–9422. doi: 10.1021/la1004243. [DOI] [PubMed] [Google Scholar]
- Zec H, Rane TD, Wang TH. Microfluidic platform for on-demand generation of spatially indexed combinatorial droplets. Lab on a Chip. 2012;12(17):3055–3062. doi: 10.1039/c2lc40399d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zec H, Shin DJ, Wang TH. Novel droplet platforms for the detection of disease biomarkers. Expert Review of Molecular Diagnostics. 2014;7159(February):1–15. doi: 10.1586/14737159.2014.945437. [DOI] [PubMed] [Google Scholar]
- Zeng S, Li B, Su X, Qin J, Lin B. Microvalve-actuated precise control of individual droplets in microfluidic devices. Lab on a Chip. 2009;9(10):1340. doi: 10.1039/b821803j. [DOI] [PubMed] [Google Scholar]
- Zhang P, Kaushik A, Hsieh K, Wang TH. Spatially encoded picoliter droplet groups for high-throughput combinatorial analysis. 2017, 19th International Conference on Solid-State Sensors, Actuators and Microsystems (TRANSDUCERS). CONF. 2017 doi: 10.1109/TRANSDUCERS.2017.7994418. [DOI]
- Zhang Y, Nguyen NT. Magnetic digital microfluidics – a review. Lab on a Chip. 2017;17(6):994–1008. doi: 10.1039/C7LC00025A. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Park S, Liu K, Tsuan J, Yang S, Wang TH. A surface topography assisted droplet manipulation platform for biomarker detection and pathogen identification. Lab on a Chip. 2011;11(3):398–406. doi: 10.1039/C0LC00296H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Park S, Yang S, Wang TH. An all-in-one microfluidic device for parallel DNA extraction and gene analysis. Biomedical Microdevices. 2010;12(6):1043–1049. doi: 10.1007/s10544-010-9458-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang TH. Full-range magnetic manipulation of droplets via surface energy traps enables complex bioassays. Advanced Materials. 2013;25(21):2903–2908. doi: 10.1002/adma.201300383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Bhattacharya S, Kotsopoulos S, Olson J, Taly V, Griffiths AD, Larson JW. Multiplex digital PCR: breaking the one target per color barrier of quantitative PCR. Lab on a Chip. 2011;11(13):2167–2174. doi: 10.1039/c1lc20126c. Journal Article. [DOI] [PubMed] [Google Scholar]
- Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, Liu X. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. Journal of Molecular Diagnostics. 2015;17(3):265–272. doi: 10.1016/j.jmoldx.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Zhu P, Wang L. Passive and active droplet generation with microfluidics: a review. Lab on a Chip. 2017;17(1):34–75. doi: 10.1039/C6LC01018K. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Fang Q. Analytical detection techniques for droplet microfluidics-A review. Analytica Chimica Acta. 2013 doi: 10.1016/j.aca.2013.04.064. [DOI] [PubMed]
- Zonta E, Garlan F, Pécuchet N, Perez-Toralla K, Caen O, Milbury C, Taly V. Multiplex Detection of Rare Mutations by Picoliter Droplet Based Digital PCR: Sensitivity and Specificity Considerations. PLoS ONE. 2016;11(7) doi: 10.1371/journal.pone.0159094. [DOI] [PMC free article] [PubMed] [Google Scholar]


