Abstract
RNA editing causes massive remodeling of the mitochondrial mRNA transcriptome in trypanosomes and related kinetoplastid protozoa. This type of editing involves the specific insertion or deletion of uridylates (U) directed by small non-coding guide RNAs (gRNAs). Because U-insertion exceeds U-deletion by a factor of ten, editing increases the nascent mRNA size by up to 55%. In T. brucei, the editing apparatus uses ~40 proteins and >1200 gRNAs to create the functional open reading frame in 12 mRNAs. Thousands of sites are specifically recognized in the pre-edited mRNAs and a myriad of partially-edited transcript intermediates accumulates in mitochondria. The control of editing is poorly understood, but past work suggests that it occurs during substrate recognition, the initiation and progression of editing, and during the life-cycle in different hosts. The growing understanding of the editing proteins offers clues about editing control. Most editing proteins reside in the “RNA free” RNA Editing Core Complex (RECC) and in the accessory RNA Editing Substrate Complex (RESC) that contains gRNA. Two accessory RNA helicases are known, including one in the RNA Editing RNA helicase 2 complex (REH2C). Both the RESC and the REH2C associate with mRNA, providing a rationale for the assembly of mRNA or its mRNPs, RESC, and the RECC enzyme. Identified variants of the canonical editing complexes further complicates the model of RNA editing. We examine specific examples of complex variants, differential effects of editing proteins on the mRNAs within and between T. brucei life stages, and possible control points in the putative holo-editosomes.
Graphical/Visual Abstract and Caption
1-. Introduction
Trypanosomes and other kinetoplastid protozoa in early-branching eukaryotic lineages utilize unique mechanisms of gene expression (Sogin 1991; Tschudi and Ullu 1994; Simpson and Maslov 1999). One example is a post-transcriptional maturation of mitochondrial mRNA transcripts by the specific insertion and deletion of uridylates (Us). mRNA primary transcripts are packed with stop codons and cannot be translated into functional proteins. The changes in sequence create functional open reading frames and can produce over 50% of the mature transcript size in some cases. This strange phenomenon was termed RNA editing and became a paradigm in RNA biology (Benne et al. 1986). RNA editing is now used broadly to indicate several post-transcriptional processes that alter the nucleotide sequence of the primary transcript and excludes changes in sequence caused by RNA splicing, 5′ capping, and 3′ tail biogenesis. Groundbreaking studies by the Stuart, Simpson, Sollner-Webb and Hajduk labs in the 1990s established that uridylate-specific editing is catalyzed by proteins, and that pre-edited mRNAs are targeted through specific hybridization with small non-coding guide RNAs (gRNAs). The pre-mRNA annealing with gRNA creates several mismatches or editing sites. The accurate “repair” of these mismatches on the mRNA side of the hybrid through concerted cycles of endonuclease cleavage, U-addition/removal and ligation results in extended sequence complementarity between the gRNA and mRNA (Blum and Simpson 1990; Seiwert et al. 1996; Rusche et al. 1997; Sabatini et al. 1998) (Fig. 1A).
Figure 1. RNA editing core complex (RECC).
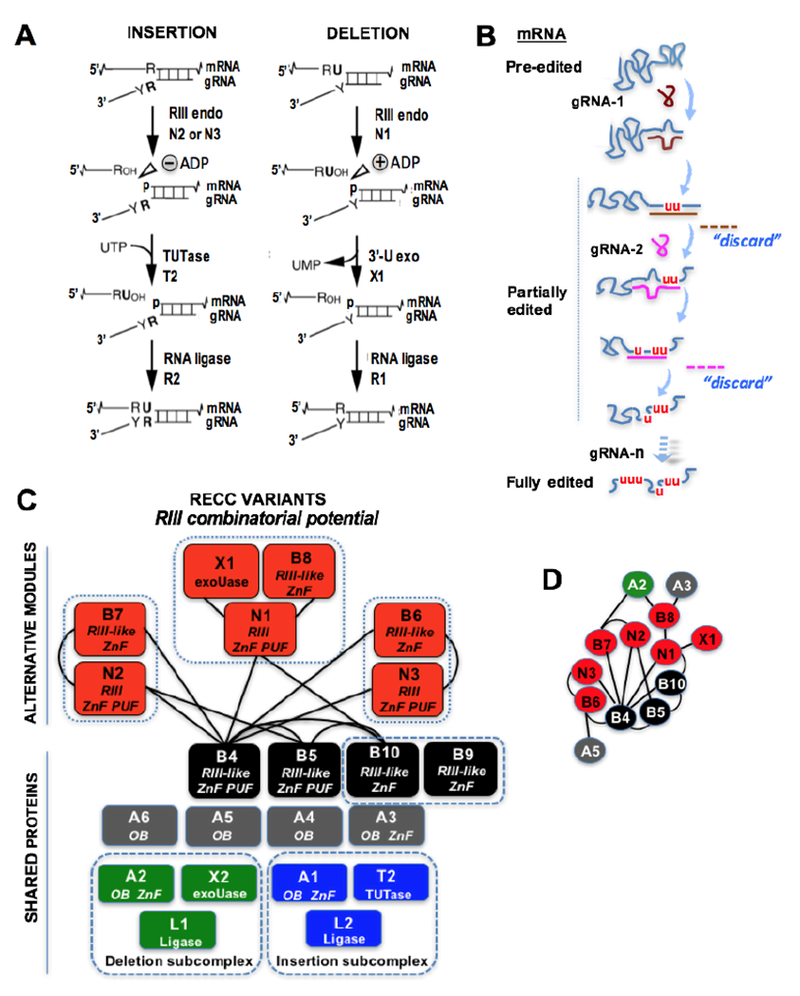
(A) Basic reaction steps at each editing site include gRNA-directed cleavage of the mRNA, followed by either a 3′-U exonuclease or a 3′ TUTase acting on the 3’-end of the cleaved upstream fragment, then resealing of the mRNA by RNA ligase. The gRNA-directed cleavage reactions at U-deletional and U-insertional editing sites require distinct endonucleases with different cleavage reaction mechanisms: the former requires an adenylate nucleotide (+), while the latter is inhibited by adenylates (−). (B) Editing by multiple gRNAs. The initiating gRNA (gRNA-1) and subsequent gRNAs (gRNA-2…gRNA-n) are depicted with pre-edited, partially edited, and fully edited mRNA. After a full block of editing is completed (depicted by a continuous duplex) the responsible gRNA is replaced by the next upstream gRNA. The exiting gRNA is thought to be degraded by a putative “discard” pathway (depicted by a broken line). (C) Variants of the RECC with shared proteins and alternative endonuclease modules for insertion and deletion editing. Note that B9 and B10 are not canonical RESC proteins because they are not regularly observed in the purifications of RECC. B9 and B10 may be only found in a subset of complexes and are depicted in a box to distinguish them from canonical RECC proteins. BS3 crosslinks between RNase III (RIII)-like proteins are indicated with lines. (D) Summary of the combinatorial potential of RIII-like proteins based on the BS3 crosslinking data in panel C. Additional potential interactions between RIII-like proteins including X1 and A-proteins are included.
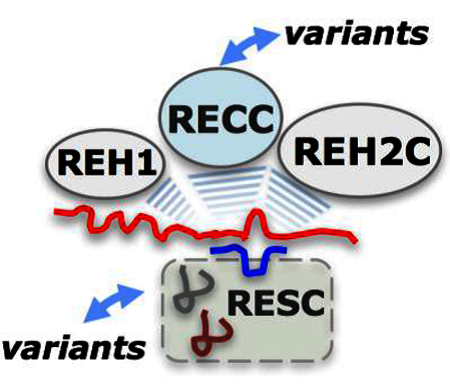
In T. brucei, most mitochondrial mRNAs are remodeled extensively in reactions directed by over 1200 gRNAs (Koslowsky et al. 2013). Pre-mRNAs are classified as pan-edited, minimally edited and never-edited. Pan-edited mRNAs use dozens of gRNAs to direct the sequence changes at hundreds of sites. gRNAs are utilized sequentially, one at the time, and a large pool of partially-edited intermediates accumulates in mitochondria (Fig. 1B). The incompletely edited transcripts may undergo proofreading cycles. This enormous task requires an editing apparatus of nearly 40 proteins that assemble around mRNA transcripts. Most of these proteins are located in macromolecular trans factors: the catalytic RNA Editing Core Complex (RECC, also termed the 20S editosome) and two auxiliary assemblies: RNA Editing Substrate Complex (RESC) and RNA Editing Helicase 2 Complex (REH2C). The composition and general function of these complexes have been described in recent reviews (Read et al. 2016; Aphasizheva and Aphasizhev 2016; Cruz-Reyes et al. 2016) but additional components and possible roles are rapidly emerging. The RECC enzyme is RNA-free (Rusche et al. 1997; Golas et al. 2009; Aphasizheva et al. 2014) while RESC and REH2C carry RNA substrates and products (Weng et al. 2008; Madina et al. 2014; Aphasizheva et al. 2014). The RECC enzyme and its auxiliary subcomplexes associate with each other via RNA interactions (Fig. 2). Outstanding central questions in the field include how the core and auxiliary components are assembled around mRNA transcripts, how the RECC enzyme is directed to each editing site, how the RECC enzyme traverses across multiple gRNAs enabling processive editing, and how each of these steps may be controlled. Several layers of control are likely during the editing process. The parasite exhibits vital adaptations in the bloodstream of the mammalian host and in the insect transmission vector, the tsetse fly (Vickerman 1985). These developmental stages exhibit differences in the editing profiles of mRNAs that correlate with changes in mitochondrial function and metabolism between the two hosts (Stuart et al. 1997). Also, specificity controls must be in place to avoid “off-targets” in editing including abundant tRNA and rRNA species. Bloodstream-form (BF) and insect-infecting procyclic-form (PF) trypanosomes grow optimally at different temperatures that could impact the structure of the RNA substrates or their RNPs during editing (Koslowsky et al. 1996; Reifur et al. 2010). Studies of the RECC enzyme and its auxiliary components suggest several potential sources of editing control. These include the identification of three variants of RECC with differing site-specificity (Carnes et al. 2011). Also, recent discoveries showed that some RECC subunits exhibit differential roles in BF and PF trypanosomes (McDermott et al. 2015b) and suggested that specialized heterodimers in RECC may modulate editing site recognition. The RECC enzyme will be discussed in section 2. In addition, accessory RNP variants and potential assembly factors are emerging, and two RNA helicases are known, including one in an RNP with mRNA. The accessory components will be discussed in section 3. Thus, both core and accessory components of the putative holo-editosomes may contribute to the control of this remarkable phenomenon. Several observations from various laboratories discussed below offer interesting insights on possible control points in trypanosome RNA editing.
Figure 2. Accessory editing components of the RECC enzyme.
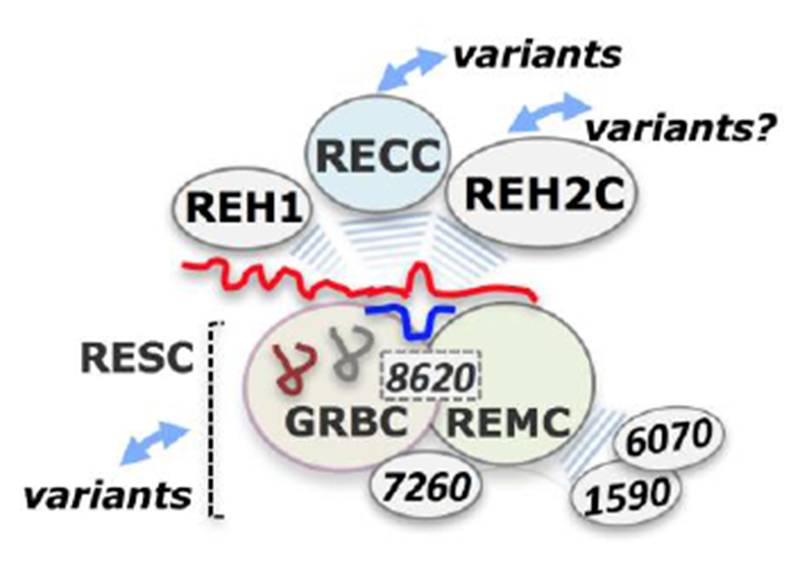
Core and accessory components of the editing apparatus assemble around the mRNA substrates. These include the gRNA-bound RESC subcomplex, the REH2C helicase subcomplex, a MRB1590/6070-containing subcomplex, and REH1 helicase. The core RECC enzyme and its accessory editing factors have been found to associate with each other via transient RNA-mediated interactions. RESC is divided into two functionally distinct modules, the GRBC and REMC. Several RESC proteins may be critical for the integrity and organization of RESC. MRB8620, a canonical GRBC protein, may control the assembly of the GRBC/REMC modules in RESC (MRB8620 is depicted as a dotted box bridging the two modules). MRB7260 binds weakly with RESC proteins and is not a canonical protein. It may be an assembly factor of RESC. Purifications of RESC and REH2C proteins were shown to contain mRNA substrates and products of editing. This supports a current model of the RNA editing machinery in which RESC-mediated targeting of mRNA or its mRNPs creates suitable mRNA-gRNA hybrid substrates for the RECC and RNA helicase enzymes and other factors during concerted phases of editing: substrate recognition, editing initiation and progression, complex organization and co-complex dynamics. Identified variants of RECC, RESC and potentially other mRNA trans factors may provide additional layers of complexity and control points in editing.
2-. Core editing enzyme
2.1-. Multi-protein RECC enzyme variants
The initiation step in processes such as those catalyzed by replisomes, ribosomes, splicesomes and transcription complexes is typically subject to strict control. Trypanosome RNA editing begins with the recognition of a suitable mRNA/gRNA bi-molecular substrate and the ensuing endonucleolytic cleavage at the first (3′ most) editing site on the mRNA strand. Subsequent steps of U-specific addition or removal and RNA ligase-mediated resealing of the cleaved strand complete a full editing cycle at that site (Fig. 1A). This basic three-step catalytic cycle repeats at individual sites as editing progresses in a general 3′-to-5′ direction (Seiwert et al. 1996; Cruz-Reyes and Sollner-Webb 1996; Kable et al. 1996; Carnes et al. 2017). Together, substrate recognition and mRNA cleavage may mark the physical and functional engagement of the editing apparatus. Therefore, these events may be key check-points in the control of RNA editing.
The first reported purification of the RECC enzyme identified seven protein subunits (Rusche et al. 1997), but subsequent studies nearly tripled this number (Panigrahi et al. 2003a; Panigrahi et al. 2003b; Stuart et al. 2005). A key for current protein terminology is shown in Table 1. Three of these proteins, REN1 (N1), REN2 (N2) and REN3 (N3), each have a functional RNase III-type endonuclease domain. Typical RNase III-type nucleases form a homodimer in which both monomers contribute to the creation of a dsRNA-binding surface. The dimer makes a double-stranded RNA cleavage with each monomer cleaving one of the two strands in the RNA duplex (Gan et al. 2008). In contrast, purifications of RECC enzyme are only thought to cleave the mRNA strand in mRNA-gRNA duplexes (Seiwert et al. 1996; Rusche et al. 1997; Carnes et al. 2005; Trotter et al. 2005; Hernandez et al. 2008). However, all in vitro editing assays have used labeled mRNA and unlabeled gRNA. Also, a recombinant version of REN1 cleaved the mRNA strand in an in vitro assay (Kang et al. 2006).
Table 1.
Proteins of the RECC subcomolex variants
| Name | Synonyms | T. brucei Gene ID | ||
|---|---|---|---|---|
| RECC | ||||
| KREPA1 | A1 | TbMP81 | Tb927.2.2470 | |
| KREPA2 | A2 | TbMP63 | Tb927.10.8210 | |
| KREPA3 | A2 | TbMP42 | Tb927.8.620 | |
| KREPA4 | A4 | TbMP24 | Tb927.10.5110 | |
| KREPA5 | A5 | TbMP19 | Tb927.8.680 | |
| KREPA6 | A6 | TbMP18 | Tb927.10.5120 | |
| KREPB4 | B4 | TbMP46 | Tb927.11.2990 | |
| KREPB5 | B5 | TbMP44 | Tb927.11.940 | |
| KREPB6 | B6 | TbMP49 | Tb927.3.3990 | |
| KREPB7 | B7 | TbMP47 | Tb927.9.5630 | |
| KREPB8 | B8 | TbMP41 | Tb927.8.5690 | |
| KREPB9 | B9 | Tb927.9.4440 | ||
| KREPB10 | B10 | Tb927.8.5700 | ||
| KREN1 | N1 | REN1 | TbMP90 | Tb927.1.1690 |
| KREN2 | N2 | REN2 | TbMP67 | Tb927.10.5440 |
| KREN3 | N3 | REN3 | TbMP61 | Tb927.10.5320 |
| KRET2 | T2 | RET2 | TbMP57 | Tb927.7.1550 |
| KREX1 | X1 | REX1 | TbMP100 | Tb927.7.1070 |
| KREX2 | X2 | REX2 | TbMP99 | Tb927.10.3570 |
| KREL1 | L1 | REL1 | TbMP52 | Tb927.9.4360 |
| KREL2 | L2 | REL2 | TbMP48 | Tb927.1.3030 |
RECC proteins in T. brucei
Early in vitro studies of gRNA-directed cleavage using mitochondrial extracts and native purified RECC enzyme identified basic requirements for the endonuclease reaction, including a robust stimulation by adenosine nucleotides (ADP or ATP) at deletion sites and their converse inhibition at insertion sites (Cruz-Reyes et al. 1998a; Cruz-Reyes et al. 1998b) (Fig. 1A). This early observation revealed a presumed allosteric control of the editing endonucleases and represents the earliest indication of differential control of the cleavage step at sites for insertion and deletion. Nucleotides immediately adjacent to a scissile phosphodiester bond can be manipulated to enhance overall efficiency or artificially convert deletion sites to insertion sites, and vice versa (Cruz-Reyes et al. 1998a; Cruz-Reyes et al. 2001; Cifuentes-Rojas et al. 2005). A-form dsRNA is required at, or near, the editing site and a 2′ hydroxyl is essential at the scissile bond for cleavage (Cifuentes-Rojas et al. 2007). Also, the cleavage activity minimally requires a single helical turn of RNA in the anchor region (Cifuentes-Rojas et al. 2007; Hernandez et al. 2008). While basic requirements for efficient endonucleolytic cleavage in vitro were identified, the discrimination of bona fide editing sites in the mitochondrial milieu leading to a productive cleavage most likely faces additional structural constraints and challenges. The observations above raise central questions, including: how the unique single-stranded cleavage activity of editing nucleases is established, how editing sites are precisely recognized, and how the key editing nuclease step is controlled in vivo.
Genetic studies of the REN proteins in trypanosomes led to important insights regarding these questions. Purification of each tagged endonuclease revealed three variants or isoforms of the RECC enzyme: RECC1, RECC2, and RECC3 (Fig. 1C). These variant complexes share a common set of proteins and are distinguished by specific sets of 3 or 2 proteins, N1/B8/X1, N2/B7, or N3/B6, respectively. Reciprocal purifications of tagged B8, B7, and B6 confirmed the typifying proteins in each RECC variant (Carnes et al. 2011). Interestingly, B8, B7, and B6 carry one degenerate RNase-like domain that lacks critical catalytic amino acids. Also, the RECC variants exhibit different substrate specificity. Such specificity has been studied in vitro and in vivo (Carnes et al. 2011; Carnes et al. 2017). RECC1 primarily cleaves at deletion sites, RECC2 primarily cleaves at insertion sites, and RECC3 primarily cleaves at insertion sites of the mRNA COII. However, the in vivo studies suggest some flexibility in the specificity of these enzymes at insertion and deletion sites. A current hypothesis is that the single-stranded cleavage activity of the RECC variants is due to heterodimerization of each REN enzyme with a non-catalytic RNase III protein partner. Consistent with this model, recent studies were unable to find evidence for homodimerization of the REN proteins (Carnes et al. 2011).
Interestingly, four of the shared proteins in the RECC variants, B4, B5, B9, and B10, also carry degenerate RNase III domains (McDermott et al. 2016; Carnes et al. 2018). So, each RECC variant includes one catalytic REN endonuclease and up to five RNase-III like proteins that are catalytically inert. However, B9 and B10 are infrequently observed in purifications of RECC, and a pulldown of B9 led to a purification of a partial complex lacking endonucleases (Lerch et al. 2012). It is possible that B9/B10 are only found in a subset of complexes, or that they are transiently or weakly associated with RECC. Thus, the possible binary partnerships may include one REN nuclease and one degenerate RNase III protein, or two degenerate RNase III-like proteins. Such combinatorial potential of the RNase III protein network may provide fine-tuning while expanding the recognition of editing substrates. The editing machinery must discriminate thousands of editing sites in mitochondria and cleave them efficiently despite the frequent changes in covalent structure of the mRNA and global conformation of the bi-molecular mRNA-gRNA structure which changes to adjust to the variable uridine composition of the mRNA as editing progresses (Reifur et al. 2010). The flexibility and ability of the editing machinery to recognize such highly variable substrates could also contribute to the control of differential editing during the life cycle of T. brucei.
Recent studies of the RECC variants in procyclic trypanosomes used chemical cross-linking and mass spectrometry to determine proximities between protein subunits in these complexes (McDermott et al. 2015a; McDermott et al. 2016). A large network of possible interactions was established using this powerful approach. This review specifically focuses on the catalytic and degenerate RNase III proteins. Importantly, REN1, REN2, and REN3 formed no detectable crosslinks with each other (Figs. 1C-D). This observation is consistent with the exclusive nature of the REN enzymes in the RECC variants (Carnes et al. 2011). However, inter-crosslinks were detected for the typifying pairs N1/B8, N2/B7 and N3/B6, in RECC1, RECC2 and RECC3, respectively. The RNase III-like protein B4, which is shared in the RECC variants, exhibited the largest number of proximities including with all three REN enzymes and the RNase III-like proteins B5, B6, B7, and B10. The network of detected crosslinks between RNase-like proteins is summarized in Fig. 1D. The B4 protein showed similar tripartite subsets with N2/B7, N3/B6, and N1/B10 (McDermott and Stuart 2017). Also, B4 is in a tripartite subset with N2 and B5. Besides the crosslink network between RNase-like proteins, there were many crosslinks between N1 and X1 in the same endonuclease module.
Finally, some RNase-like proteins (B6, B7 and B8) crosslinked with A proteins (A2, A3, A5). This result suggested possible interaction surfaces between the nuclease modules and the shared core structure in RECC. No inter-protein crosslinks were detected involving B9. However, as it was mentioned before B9 and B10 are not considered conical RECC proteins. A lack of crosslinks may be due to the substoichimetric level of B9 in the examined complexes or may represent a limitation of the BS3 cross-linker used in these studies. This cross-linker has a linker arm of 11.4 Å, which can react with two lysine residues whose alpha carbon atoms are up to 30 Å apart (McDermott et al. 2016). Thus, it is conceivable that bona fide binding surfaces in some proteins lack pairs of lysines separated by appropriate distances and locations for inter-crosslinking. Conversely, proximal crosslinks do not necessarily reflect true protein contacts. Valuable insights about the architecture of RECC complexes were generated. However, the combinatorial potential of the editing RNase III proteins will require validation using complementary approaches, including the use of isolated recombinant proteins. In summary, three alternative RECC variants, alongside their potential binary combinations of three catalytic and seven degenerate RNase III proteins, may control key initiating steps in the discrimination and cleavage of editing sites.
2.2-. Differential effects of the RECC proteins B4, B5, and A3 in two trypanosome life stages
Genetic studies of B4, B5 and A3 showed that these proteins, which are common to the RECC variants, exhibit differential behaviors in BF and PF cells. As described above, B4 and B5 have degenerate non-catalytic RNase III domains and could potentially dimerize with catalytic or other non-catalytic RNase III editing proteins. A3 also lacks catalytic motifs but may interact with the RNase-III interaction network via B8 (Fig. 1D). However, all three proteins, B4, B5 and A3, carry domains that suggest functional interactions with RNA or proteins (Fig. 1C). Initial RNAi-based genetic knockdowns in BF cells showed that B5 and A3 affect the integrity of RECC complexes. Analysis of A3 in PF cells shows a partial effect in PF cells (Law et al. 2008; Guo et al. 2010; McDermott et al. 2015b). More detailed characterizations using conditional null cell lines also showed that the lack of B5 eliminated the RECC complexes in BF cells but only partially disrupted them in PF cells. The same results are also observed for the B4 conditional null (McDermott et al. 2015b; McDermott and Stuart 2017). Also, the lack of A3 eliminated the RECC complexes in BF cells but only slightly decreased their sedimentation density in PF cells. However, both B5 and A3 are required for RNA editing and cell growth. Furthermore, the substitution of amino acids at selected sites within these proteins differentially affected cell growth, the integrity of the RECC complexes, and RNA editing in the two life stages (McDermott et al. 2015b). Some mutations in various domains of B5 and A3 affected PF cells, BF cells, or both. The focus in that study was on the B5 PUF and RNase-III domains, predicted to bind RNA, and on the A3 zinc-fingers predicted to bind RNA or protein.
In a complementary study, a random mutagenesis of the functional domains of B5 identified eight amino acid substitutions that are lethal in BF cells but not in PF cells. Most of these positions were in the degenerate RNase III-like domain, consistent with the idea that this type of domain controls editing (McDermott et al. 2015a). The exception was a mutation of a conserved glycine in B5 thought to be essential for RNase III dimerization, and consequently inhibited editing and growth in both BF and PF cells. In the case of B4, a recent study showed that this protein is similar to B5 in that mutations in their degenerate RNase III domain more severely impact the integrity of the RECC complexes in BF than in PF cells. In this study of B4, mutations in the RNase III domain strongly inhibited BF and PF growth and editing. Also, the equivalent conserved glycine in B4, thought to mediate RNase III dimerization, was critical in BF and PF growth and editing (McDermott and Stuart 2017).
Originally, the RECC complexes were thought to largely provide the basic catalytic center in the holo-editosomes. However, the above mutagenic studies provided the first evidence of differential relevance for some subunits of these complexes in BF and PF trypanosomes. So, the RECC complexes may also play a role in the control of stage-specific RNA editing. The above observations also showed that functional studies in one stage are not necessarily valid in another stage. A number of possible mechanisms may account for the differential roles of specific subunits of the RECC complexes including differences in protein modification, conformation, or interactions with other proteins or RNA. Interestingly, the A3 protein is a proposed structural core component of RECC complexes. In PF cells, A3 showed a number of crosslinks with specialized components of both the U insertion and the U deletion pathways (McDermott et al. 2015a). It will be interesting to compare how the inter-crosslink network of A3, B4 and B5 differ in purified complexes from both PF and BF cells. B4 seems also particularly interesting because it generated in PF cells the largest number of crosslinks with other RNase III-type proteins. Yet another RNase III protein, like B5, or another core protein besides A3 (i.e., also shared in the RECC variants), may be a major interaction point in BF cells. The RECC complexes in BF and PF cells most likely have the same protein composition. However, the low expression of tagged proteins in BF have prevented isolation of sufficient RECC complexes for analysis in this life cycle stage (Carnes et al. 2011). Even if RECC complexes are compositionally identical in BF and PF cells, the dramatic differences caused by the mutations described above suggest that the architecture of the RECC complexes, or their protein-protein and protein-RNA interactions, differ substantially between the two stages.
3-. Accessory components in the RNA editing apparatus
Besides the RECC proteins many non-RECC proteins promote efficient editing in vivo. Of the ~40 proteins in the editing apparatus that have been identified in T. brucei mitochondria, nearly half of these proteins are found in accessory subcomplexes: RESC, REH2C, and a possible MRB6070/1590-containing subcomplex (Read et al. 2016; Shaw et al. 2015; Aphasizheva and Aphasizhev 2016; Cruz-Reyes et al. 2016) Fig. 2; Table 2). Another RNA helicase, REH1, may be largely free or in a small assembly (Fig. 2) (Li et al. 2011). The core enzyme (RECC) and its accessory editing trans factors are linked in an interaction network mediated by RNA. A large study of RESC proteins proposed two separate functional modules: the gRNA-Binding Complex (GRBC) and the more loosely defined RNA Editing Mediator Complex (REMC) (Aphasizheva et al. 2014). This is consistent with extensive analyses of Y2H-predicted interactions between accessory proteins (Ammerman et al. 2012) and additional studies summarized in (Read et al. 2016; Aphasizheva and Aphasizhev 2016). Two GRBC protein paralogs GAP1 and GAP2 (alias GRBC2 and GRBC1, respectively) were shown to form a stable α2β2 heterotetramer that binds and stabilizes gRNA (Weng et al. 2008). Purifications of canonical RESC proteins contain mRNA (Madina et al. 2014; Aphasizheva et al. 2014). This was confirmed in more recent studies (Huang et al. 2015; McAdams et al. 2018). Also, purifications of the REH2C subcomplex from either normal or gRNA-free mitochondria contain mRNA. Thus, REH2C is an mRNP (Kumar et al. 2016). The presence of gRNA and mRNA in purifications of accessory editing proteins supported the current model whereby mRNA, or its mRNPs, bind to gRNA-loaded RESC. Thus, the RNA-free core editing enzyme may catalyze efficient and processive editing through productive interactions with the RNA substrate-loaded accessory apparatus (Fig. 2).
Table 2.
Proteins of RESC and REH2C, and additional factors
| Name | Synonym | Function (6) | Motifs | T. brucei Gene ID | |
|---|---|---|---|---|---|
| GRBC | GRBC1 | GAP2 | gRNA binding, gRNA stability | Tb927.7.2570 | |
| GRBC2 | GAP1 | gRNA binding, gRNA stability | Tb927.2.3800 | ||
| GRBC3 | MRB8620 | RESC organization | Tb927.11.16860 | ||
| GRBC4 | MRB5390 | Tb11.02.5390 | |||
| GRBC5 | MRB11870 | RESC organization | Pentein | Tb927.10.11870 | |
| GRBC6 | MRB3010 | Tb927.5.3010 | |||
| GRBC7 | MRB0880 | Tb927.11.9140 | |||
| MRB7260 | Editing progression. gRNA exchange/utilization. RESC organization | PhyH | Tb927.9.7260 | ||
| REMC | REMC1 (1) | MRB10130 | ARM/HEAT | Tb927.10.10130 | |
| REMC2 | MRB1860 | Tb927.2.1860 | |||
| REMC3 | MRB800 | Tb927.7.800 | |||
| REMC4 | MRB8180 | Editing progression. RNA binding | Tb927.8.8180 | ||
| REMC5 (2) | MRB4160 | Editing progression. RNA binding | Tb927.4.4160 | ||
| REMC5A (2) | MRB8170 | Editing progression. RNA binding | Tb927.8.8170 | ||
| RGG2 | TbRGG2 | Editing progression. RNA binding, annealing, unwinding (4) | RRM, RGG | Tb927.10.10830 | |
| REH2C | REH2 | RNA helicase. RNA binding. | dsRBD, DEAH/RHA, HA2, OB | Tb927.4.1500 | |
| H2F1 | REH2 adaptor, REH2 stability | C2H2 zinc fingers | Tb927.6.1680 | ||
| H2F2 | RNA binding (5) | Tb927.6.2140 | |||
| Other characterized proteins not assigned to the subcomplexes above | |||||
| REH1 | Mhel61 | RNA helicase. gRNA exchange | DEAD-box, RNA helicase | Tb927.11.8870 | |
| MRB1590 (3) | RNA binding, ATPase activity | ABC-ATPase | Tb927.3.1590 | ||
| MRB6070 (3) | RanBPs zinc-fingers | Tb927.2.6070 | |||
RNAi depletion caused a full loss of TbRGG2. It also decreased editing in all tested mRNA substrates
Paralogs with 73.3% translated sequence identity
RNase-resistant association between MRB1590 and MRB6070. These two proteins may form a separate subcomplex.
Unwinding acitivity was determined using an E. coli reporter assa
Kumar et al., 2016; Kumar et al., unpublished data.
Citations of functional studies of these proteins are summarized in Read et al, 2015; Aphasizheva & Aphasizhev, 2016; and Cruz-Reyes et al, 2016. More recent functional studies are also discussed in this review.
The current placement of the known editing proteins in the accessory subcomplexes was largely guided by RNase-resistant or RNA-independent association, cross-tagging, yeast two-hybrids (Y2H), recombinant proteins, and editing phenotypes after RNAi-based depletion of individual proteins (Table 2). These studies were discussed in recent reviews (Read et al. 2016; Aphasizheva and Aphasizhev 2016; Cruz-Reyes et al. 2016) and more recent work described below. The GRBC and REMC modules are intimately interconnected through an intricate network of protein-protein contacts. Some of these contacts were affected by RNase treatment, suggesting synergy between protein and RNA-mediated interactions. Besides the presumed “intact” RESC modules, variations in RESC organization are emerging. Also, some studies of GRBC and REMC, and new experimental approaches, are revealing additional functional layers within each module. The first part of this section discusses examples of natural GRBC and REMC variants and examples of genetically-induced changes in complex organization, including both protein and RNA components. It addresses possible roles of the alternative assemblies in editing control. Interestingly, a common theme in the examples discussed is the variable interaction between the GAP1/2 tetramer and MRB3010 (GRBC6). An interesting alternative assembly that includes GRBC proteins but that seems to function outside of RNA editing is also included. The second part of this section discusses the RNA editing helicases REH1 and REH2, and examines possible roles of these accessory enzymes in editing control.
3.1-. Natural variants of GRBC and REMC
A study of MRB6070 interactions may provide the earliest example of natural GRBC isoforms that exhibit a variable stoichiometry of canonical GRBC proteins in vivo (Ammerman et al. 2012). MRB6070 is not a canonical protein of RESC but associates with RESC via RNA (Fig. 2). Ammerman et al performed a tandem-affinity purification of PTP-tagged MRB6070 and western blots showing that the eluates contained the canonical REMC proteins TbRGG2 and MRB8170 (REMC5A) and the canonical GRBC proteins GAP1/GAP2. However, the GRBC proteins MRB3010 and MRB11870 (GRBC5) were not detected in the pulldown. These proteins exhibited comparable levels in cell extracts. This suggested the presence of a native GRBC-related variant in which the GAP1/GAP2 tetramer has a reduced interaction with at least MRB3010 and MRB11870. An equivalent purification of PTP-tagged MRB6070 but from an RNase-treated cell lysate contained TbRGG2 and MRB8170, albeit at a lower level, but not GAP1/GAP2. Consistent with these observations, mass spectrometry of the anti-protein C elution from RNase-treated PTP-MRB6070 extracts revealed a higher coverage for GAP1/GAP2, TbRGG2 and MRB8170, while only 1 and 2 unique peptides of MRB3010 and MRB11870 were detected, respectively (Ammerman et al. 2012). The authors also showed that the association of MRB6070 with RESC proteins is entirely mediated by RNA. Ammerman et al suggested that the stoichiometry between the GAP1/GAP2 tetramer and other GRBC proteins can change in native GRBC variants.
The role of the GRBC variant associated with MRB6070 is unknown, but its ability to support editing activity should be compromised without MRB3010 and MRB11870 present (Ammerman et al. 2012; Ammerman et al. 2013). However, the gRNA-loaded GAP1/GAP2 tetramer could be used to “mark” or recruit specific mRNAs via hybridization. The Ammerman study also suggested that the GAP1/GAP2 tetramer may assemble with REMC components such as TbRGG2 in the absence of other GRBC proteins. Section 3.2 below further discusses the association of GAP1/GAP2 tetramer with other components of the editing apparatus, including TbRGG2, upon the induced knockdown of specific proteins. MRB6070 could participate in early stages of the GRBC/REMC modular assembly (Ammerman et al. 2012). MRB6070 was found to associate with MRB1590 in an RNase-resistant manner. MRB6070 and MRB1590 are known to bind RESC via RNA (Shaw et al. 2015). Also, both MRB6070 and MRB1590 were first detected in a purification of the RNA helicase REH2, but their co-purification with REH2 was RNase sensitive (Hernandez et al. 2010). Shaw et al (2015) found that a MRB1590 knockdown specifically decreased the level of edited A6 mRNA, although this effect was modest as shown by qRT-PCR. These authors further showed that this A6-specific effect may be due to a reduced editing progression (i.e., increased pausing) within a GC-rich region. Finally, Shaw et al, also found that MRB1590 exhibits ATPase and RNA-binding activities, suggesting a possible mechanism for how A6 editing may pause at the G-C rich region. Thus, while MRB6070 and MRB1590 are not being considered canonical RESC proteins, these two proteins may form a separate subcomplex (Fig. 2). It will be interesting to determine the possible effect of MRB6070 on A6 mRNA and other transcripts. The MRB6070-MRB1590 containing subcomplex could serve as a chaperon or assembly factor in RESC formation with mRNA-specific or preferential effects.
A study by Madina et al provided another example of natural GRBC isoforms that exhibit differing stoichiometry in at least one canonical protein component in vivo (Madina et al. 2014). In a study of the RNA editing helicase 2 (REH2) the authors examined immunoprecipitations (IPs) of endogenous REH2 and MRB3010 in mitochondrial extracts of procyclic trypanosomes. The IPs used affinity-purified peptide polysera (Madina et al. 2014). As described above, REH2 is part of the REH2C subcomplex which associates with RESC and RECC via RNA linkers (Hernandez et al. 2010; Madina et al. 2014; Kumar et al. 2016). Both endogenous REH2 and MRB3010 pulldowns contain gRNA and the examined canonical proteins in western blots of GRBC, REMC and RECC, including: GAP1, GAP2, TbRGG2, A1 and A2. Editing ligases in both IPs were detected in adenylation assays (Hernandez et al. 2010; Madina et al. 2014; Madina et al. 2015; Kumar et al. 2016)(Kumar et al., unpublished data). However, MRB3010 was not detected or was barely visible in western blots of the REH2 IPs (Madina et al. 2014; Kumar et al. 2016). The presence of GAP1, GAP2 and gRNA in the pulldowns of endogenous REH2 implied that the REH2-associated GRBC variant includes a gRNA-loaded GAP1/2 tetramer. MRB3010 is an essential GRBC protein that is required in early editing (Ammerman et al. 2011). Consistent with the differential content in MRB3010, qRT-PCR analyses of the first editing block in two examined mRNAs in IPs of REH2 and MRB3010 showed a higher level of 3’ early editing in mRNAs from the MRB3010 IPs. Also, a small-scale RNA-seq experiment of gRNA content in these IPs indicated a relative enrichment of some initiating gRNAs in the MRB3010 IPs. A reversible assembly of the gRNA-loaded GAP1/2 tetramer with critical proteins such as MRB3010 could serve as a toggle switch at specific phases of the editing process. A momentary pause or slowdown in editing activity would facilitate REH2C function, presumably in RNP remodeling, before MRB3010 is re-instated, and the process can then continue with higher efficiency.
Several studies of REMC have provided evidence that the organization of REMC proteins vary substantially (Madina et al. 2011; Kafkova et al. 2012; Dixit et al. 2017; Simpson et al. 2017). One study directly compared the canonical REMC protein paralogs MRB8170 (REMC5A) and MRB4160 (REMC5), which share 77.3% amino acid sequence identity. In this case, affinity purification of each paralog under the same conditions showed similar, but not identical, sets of associated canonical proteins of REMC and GRBC (Kafkova et al. 2012). The purification of each tagged paralog contained an under-represented number of unique peptides from the other paralog, and an RNase-treated purification of MRB4160 eliminated the co-purification of MRB8170. Sedimentation studies also indicated distinct protein associations between MRB8170 or MRB4160 because a genetic knockdown of MRB4160 did not affect the stability or sedimentation of MRB8170. The paralogs MRB8170 and MRB4160 are mutually exclusive and form distinct REMC variants (Kafkova et al. 2012). Besides the stable interactions of MRB8170 and MRB4160 with canonical RESC proteins, the paralogs MRB8170 and MRB4160 also exhibited RNase-resistant interactions with TbRGG1, MRP1 and Nudix hydrolase. The latter three proteins affect the RNA stability of several mitochondrial transcripts (Aphasizheva et al. 2014; Vondruskova et al. 2005; Fisk et al. 2009; Hashimi et al. 2009). MRP1, in a complex with MRP2, is also proposed to specifically control editing of Cyb mRNA (Vondruskova Lukes 2005; Fisk 2009). Available studies of the possible impact of TbRGG1 RNAi on editing are conflicting, with one study finding no effect (Aphasizheva et al. 2014) and another study suggesting a modest decrease in edited mRNAs (Hashimi et al. 2008). Thus, canonical REMC proteins may be found in natural subcomplex variants with roles outside RNA editing. A more recent study also indicated the existence of multiple RESC-related subcomplexes of variable composition (McAdams et al. 2018). Also, Simpson et al (2017) reported different levels of some REMC proteins. Namely, MRB8180 is 15-fold less abundant than MRB8170 and 30-fold less abundant than TbRGG2, making it unlikely that all REMC variants contain MRB818880. A differential contribution of MRB8170, MRB4160 and other RESC proteins to the editing process is discussed below (section 3.2).
An interesting study of TbRGG3 (Tb927.3.1820) revealed an unexpected case of a natural TbRGG3-associated subcomplex that contains GAP1 but no other examined canonical GRBC proteins. TbRGG3 is mitochondrial but dispensable for RNA editing (McAdams et al. 2015). Although, this review focuses on potential control points in the editing process, TbRGG3 provides an important precedent of natural GRBC-related assemblies that could play roles outside the editing apparatus. TbRGG3 was originally found in double-affinity purifications of each of these proteins: TbRGG1, GAP1 and GAP2 (Hashimi et al. 2008). Purifications of native RECC and native RNA helicase REH2 also detected TbRGG3, but only one and two representing unique peptides of TbRGG3 were found, respectively. However, the TbRGG3 association with RECC and REH2 was sensitive to RNase (Hernandez et al. 2010). TbRGG3 exhibited strong Y2H-interactions with GAP1 but weak interactions with MRB3010 and other GRBC proteins (Ammerman et al. 2012). TbRGG3 is arginine-glycine rich, highly methylated at the arginines, and exhibits RNA UV-crosslinking and annealing activity with synthetic RNA transcripts (Fisk et al. 2013; McAdams et al. 2015). McAdams et al showed that TbRGG3 may function in the stabilization of specific pre-edited mRNAs. These authors performed pulldowns of a myc-tagged TbRGG3 from cell extracts and confirmed that TbRGG3 interacts with GAP1 and TbRGG1 in an RNase-resistant manner. However, the TbRGG3 pulldown lacked MRB3010 or TbRGG2. It will be interesting to determine if the TbRGG3-associated complex includes GAP2 or gRNA. However, the GAP proteins may be largely found associated in GAP1/2 tetramers in vivo because the GAP1 stability was compromised upon RNAi silencing of GAP2 and vice versa, and overexpression of one GAP protein did not cause its accumulation in a free form (Weng et al. 2008; Hashimi et al. 2008). The presence of TbRGG1 in the TbRGG3 pulldowns also indicated that the TbRGG3-purified subcomplex is important in mRNA stability. Other proteins that control the stability or 3’ processing of RNAs including editing substrates, have been characterized and recently reviewed (Aphasizheva and Aphasizhev 2016). If the TbRGG3-purified subcomplex indeed includes gRNA-loaded GAP1/2 tetramers, it could potentially target specific mRNA transcripts via gRNA hybridization. The gRNA-selected mRNA targets could be directed to a RNA “discard pathway” for degradation.
Other interesting REMC-related variants include a number of assemblies that involve TbRGG2. This canonical REMC protein is essential in editing, as it was discussed above, but it may also function outside of RNA editing. TbRGG2 exhibited RNA binding and RNA annealing activities with synthetic RNA transcripts and was able to melt RNA secondary structure in an E. coli reporter system (Ammerman et al. 2010). Thus, TbRGG2 may serve as a chaperone factor in the control of RNA structure during RNA editing. However, the genetic analysis of TbRGG2 has added complications because this factor is also present in a few identified alternative assemblies (Sprehe et al. 2010; Zimmer et al. 2011; Madina et al. 2011). A TbRGG2 interaction with p22 is particularly interesting because RNAi of p22 decreased the steady-state level of edited mRNA CO2, the only substrate using a cis-acting gRNA. The level of pre-edited mRNA CO2 was not affected in the p22 knockdown. p22 also exhibited an RNase-resistant interaction with the RECC enzyme (Sprehe et al. 2010). Because RNAi of TbRGG2 decreases the editing of pan-edited mRNAs but not minimally-edited mRNA CO2 (Ammerman et al. 2010) the possible function of TbRGG2 in the p22-TbRGG2 assembly is intriguing. It is feasible that p22 and TbRGG2, serving as a chaperone, work in concert to stabilize the edited mRNA CO2. TbRGG2 also appeared to be in small MRB-7260 containing complexes identified in a sedimentation analysis (McAdams et al. 2018). Further studies of this and other non-canonical TbRGG2-containing assemblies will be interesting.
3.2-. Induced changes in RESC organization
Some studies have proposed that specific protein components could serve as organizers of RESC, including proteins that exhibit strong Y2H-predicted interactions with canonical proteins both in GRBC and REMC (Read et al. 2016). This section examines specific observations in detailed studies that suggest possible control points in RESC.
A study by Ammerman et al characterized MRB11870, a canonical GRBC protein that is required in early editing (Ammerman et al. 2013). MRB11870 affects the editing of virtually all substrates as it has been found for several canonical GRBC proteins (Ammerman et al. 2011; Weng et al. 2008; Aphasizheva et al. 2014; Hashimi et al. 2009). This is consistent with a model in which the GRBC supplies all trans-acting gRNA in the editing pathway, and findings that the canonical GRBC is a stable assembly held by RNA-independent interactions. Ammerman et al established a procyclic cell line with an MRB11870-RNAi construct and an endogenously PTP-tagged MRB3010 allele. This cell line allowed them to examine the impact of MRB11870 depletion on the interaction of MRB3010 with examined canonical proteins in GRBC and REMC in cell extracts. MRB3010 is known to exhibit strong Y2H-interactions with GAP1 and MRB11870 (Ammerman et al. 2012). In the MRB11870 knockdown, the MRB3010 pulldown contained a reduced level of GAP1, MRB11870 and TbRGG2. Analysis of gRNA steady-state in the cell extract suggested that the stability of GAP1/2 tetramer was not affected by the MRB11870 knockdown. Thus, the coupling of GAP1/2 tetramer with MRB3010 and other components seems to require MRB11870. It will be interesting to determine whether the MRB11870 ablation reduced all protein interactions with MRB3010 or only a specific subset. It is possible that MRB11870 occupies a critical site in GRBC that facilitates bonding of the GAP1/2 tetramer with MRB3010 and other canonical GRBC proteins. An impact on the GRBC organization would account for the reduced level of TbRGG2 (a REMC protein) in the MRB3010 pulldowns. If a canonical protein resides on the surface of GRBC its ablation would be expected to inhibit all RNA editing but not the assembly of GAP1/2 tetramer with other GRBC proteins. Although MRB11870 is a core subunit of GRBC it could offer a control point in editing. That is, a modulation in MRB11870 interactions could control the engagement of the GAP1/2 tetramer in GRBC and thereby the gRNA entry into the pathway. This would stall the editing apparatus at initiation.
A study by Huang et al examined how the RNAi of the canonical GRBC proteins MRB8620 (GRBC3) or GAP1 affect the assembly of MRB3010 with other RESC proteins (Huang et al. 2015). The authors established a procyclic cell line that contained an endogenously V5-tagged MRB3010 allele and an RNAi construct of either MRB8620 or GAP1. After the depletion of MRB8620, pulldowns of tagged MRB3010 were examined in western blots of the canonical proteins GAP1 and MRB11870 (in GRBC) and TbRGG2 and MRB8180 (in REMC). The MRB8620 knockdown reduced all examined proteins in the pulldown. The steady-state level of the examined proteins was normal in the MRB8620-depleted mitochondrial extract. Thus, the MRB8620 knockdown affected the integrity of MRB3010-associated RESC. However, analysis of total gRNA in the extract suggested that the GAP1/2 tetramer is normal in the MRB8620 knockdown. Using the same approach but this time in a GAP1 knockdown background, Huang et al (2015) showed that the tagged MRB3010 pulldown contained all examined RESC proteins in the absence of GAP1. This implied that GRBC-related assemblies that lack GAP1/2 are stable. The GAP1/2 tetramer is not a pre-requisite for the assembly of MRB3010 with MRB11870, presumably with other canonical GRBC proteins, and with REMC proteins. Huang et al proposed that that GRBC includes the GAP1/2 tetramer and a “GRBCcore” (alias MRB1 core) that could contain the remaining GRBC proteins.
It will be interesting to determine if the proposed GRBCcore and the GAP1/2 tetramer modules are natural variants of GRBC and can be found separately in normal cells. The intact GRBC is known to carry mRNA (pre-edited, partially edited and fully edited) (Madina et al. 2014; Aphasizheva et al. 2014; Huang et al. 2015; McAdams et al. 2018). During their studies, Huang et al (2015) also showed that the mRNA content of RESC changed in the MRB8620 knockdown. Using antibody IPs of native GAP1, the authors found that the MRB8620 ablation reduced the RESC association with the RECC editing enzyme. Consequently, this caused an increase in the level of RESC-associated pre-edited or partially edited mRNA. Thus, the Huang study suggested different points of possible editing control involving the organization of RESC and its interactions. The docking of the GAP1/2 tetramer with the proposed GRBCcore, and an MRB8620-mediated bridging between the GRBCcore and REMC proteins could be used to control the amount of active RESC. Also, an MRB8620-mediated modulation of RECC enzyme association and mRNA substrate accumulation in RESC could control the efficiency of editing catalysis. Thus, MRB8620 may serve as an assembly factor (Fig. 2).
A recent study by McAdams et al (2018) characterized MRB7260. This interesting protein exhibits weak Y2H-predicted interactions with some RESC proteins, however, it is not considered a canonical protein of REMC or GRBC (Ammerman et al. 2012; McAdams et al. 2018). MRB7260 has a PhyH domain typically found in glycosomal proteins. However, MRB7260 is mitochondrial and its PhyH domain seems to be catalytically inactive (McAdams et al. 2018; Guther et al. 2014). This study by McAdams et al showed that MRB7260 is required for RNA editing and exhibits differential effects on protein and RNA interactions within RESC (see section 3.3). Relevant to the discussion in this section are the observations in the McAdams study (2018) examining the effects of MRB7260 on RESC organization. The authors used a procyclic cell line containing both an MRB7260 RNAi construct and a constitutive HTM-tagged TbRGG2 allele. Pulldowns of tagged TbRGG2 were utilized to assess the impact of MRB7260 ablation on the RESC organization. Western blots showed that the level of MRB8180 and MRB8170 (REMC proteins) and GAP1 in the GRBC were normal in the TbRGG2 pulldowns. However, the level of the canonical GRBC protein MRB3010 decreased in the pulldown upon MRB7260 ablation. The total gRNA was normal in these cells and a prior purification of myc-tagged TbRGG2 contained both GAP1 and GAP2 (Ammerman et al. 2012). This indicated that the GAP1/2 tetramer was not affected by the MRB7260 depletion. McAdams et al (2018) compared the above pulldowns of tagged TbRGG2 with reciprocal purifications using a PTP-tagged MRB3010 allele. In this case, the authors found that the MRB7260 ablation reduced the canonical REMC proteins TbRGG2, MRB8170 and MRB8180 in the pulldowns of MRB3010. However, the GRBC proteins GAP1 and MRB11870 were not affected in the same MRB3010 pulldowns. The contrasting relative levels of GAP1 and MRB3010 (i.e., altered and normal, respectively) in the above pulldowns of TbRGG2 and MRB3010 from MRB7260-RNAi extracts may be interpreted in different ways. One possibility is that the MRB3010-purifications largely include fully assembled RESC, and maybe other RESC variants, in which the interaction of MRB3010 with the GAP1/2 tetramer and other RESC proteins is not affected by the MRB7260 RNAi. However, TbRGG2-purifications may include multiple TbRGG2-containing complexes including some in which MRB7260 RNAi can affect the content of MRB3010 but no other examined proteins: GAP1 in GRBC, or MRB8170 and MRB8180 in REMC. This may reflect a dynamic composition and function of TbRGG2-purified complexes and of MRB7260-containing complexes that are discussed below in section 3.3. Consistent with a stable association of MRB3010 with the GAP1/2 tetramer in the presumed fully assembled RESC, a GAP1 knockdown decreased the level of MRB3010 (Kumar et al. 2016). However, RNAi of MRB3010 did not seem to affect the level of GAP1/2 (Ammerman et al. 2011). A variable association of MRB3010 with the GAP/1/2 tetramer in at least some complex variants is expected because glycerol gradients of cell extracts, and of some pulldowns (e.g., MRB7260-IPs), exhibit different sedimentation patterns of GAP1 and MRB3010, including fractions with only one of these proteins and fractions with both (Ammerman et al. 2012; McAdams et al. 2018). This is also reminiscent of the REH2C subcomplex association with GRBC variants that differ in MRB3010 content (Madina et al. 2014; Madina et al. 2015; Kumar et al. 2016). The McAdams study suggests that the coupling of GAP1/2 tetramer with MRB3010 is affected by MRB7260. The authors also found evidence that MRB7260 and GAP1 associate in heterodispersed RESC-related complexes of unclear function. As noted above, MRB7260 exhibits weak Y2H-interactions with some RESC proteins but it is not a canonical GRBC or a REMC protein. Overall, these studies of MRB7260 suggest that this protein may be part of a specialized assembly factor of RESC (Fig. 2).
3.3. Differential effects of accessory editing proteins on mitochondrial mRNAs
Studies of the accessory editing proteins seeking to dissect basic control mechanisms of RNA editing remain challenging. Most of the accessory proteins lack conserved sequence motifs that can provide clues about their function. Also, the dynamic nature of the accessory subcomplexes may enable individual proteins to adopt a range of activity levels or even different functional roles depending on their associated partners in the subcomplex variants. Nonetheless, several accessory proteins exhibit differential phenotypes that are being examined. This section analyzes specific observations by several labs that offer a glimpse of the current questions being addressed, the approaches being taken and complex problems ahead. The focus is on specific or preferential effects of the examined proteins on different mitochondrial mRNAs. Some of these studies discussed below used combinations of RNAi-based silencing, protein tagging, high-throughput RNA sequencing and CLIP methodologies. These studies provided important insights into the differential control of the editing process.
An extensive study of proteins that stably co-purify with the gRNA-loaded GAP1/2 tetramer by the Aphasizhev lab used RNAi-based depletion to compare the impact of the examined proteins on editing (Aphasizheva et al. 2014). This RNAi study of the examined proteins revealed intricate differential editing patterns and associations involving protein and RNA. One of their approaches examined the sedimentation distribution and steady-state level of canonical markers in GRBC and REMC: the GAP1/2 tetramer and TbRGG2, respectively. While glycerol gradients provide relatively limited resolution, downregulation of some proteins caused dramatic changes in the sedimentation or stability of the canonical markers. However, RNAi of other proteins exhibited a moderate or minimal effect in sedimentation, despite causing an editing phenotype. This Aphasizheva study (2014) found that depletion of MRB10130 (REMC1) caused a full loss of TbRGG2, and knockdowns of MRB5390 (GRBC4) and MRB0880 (GRBC7) caused a partial loss of TbRGG2 in some fractions. Also, RNAi of MRB8180 caused a partial loss of TbRGG2. However, another study reported no loss of TbRGG2 in a MRB8180 knockdown (Simpson et al. 2017). Importantly, observed protein destabilization in the RNAi studies usually reflect predicted Y2H-interactions in RESC (Ammerman et al. 2012). Surprisingly, RNAi of MRB0880 eliminated GAP2 but only slightly impacted the level or mobility of GAP1 in the Aphasizheva study. However, other studies showed that a knockdown of GAP1 destabilized GAP2, and vice versa, indicating that these protein paralogs stabilize each other in the natural tetramer (Weng et al. 2008; Hashimi et al. 2009). It will be interesting to determine how GAP1 is stabilized in the MRB0880 knockdown because this finding implied that some natural GRBC variants could potentially carry only one of the GAP paralogs.
Other interesting points of contrast in the Aphasizheva study (2014) include the observation that the depletion of the GAP1/2 tetramer minimally affected the sedimentation of TbRGG2 (see also (Huang et al. 2015)), whereas RNAi of either TbRGG2 or MRB0880 increased the sedimentation of the GAP1/2 tetramer. Simpson et al (2017) reported that RNAi of TbRGG2 also decreases the level of MRB8170 and MRB8180. There are differing results about the impact of the simultaneous knockdown of the paralogs MRB4160/8170 on TbRGG2. That is, some authors reported a partial decrease in TbRGG2 whereas others did not find a loss of TbRGG2 upon the double MRB4160/8170 RNAi (Kafkova et al. 2012; Aphasizheva et al. 2014; Simpson et al. 2017). The RNAi studies revealed complex editing phenotypes in a large panel of mRNA substrates. However, a common theme of the GRBC proteins was their impact on all editing directed by trans-acting gRNA. REMC proteins mostly affected pan-edited mRNAs and exhibited differential effects on the mRNA types. This has been observed in several studies (Kafkova et al. 2012; Simpson et al. 2017; Fisk et al. 2008; Aphasizheva et al. 2014). However, MRB8180 had little or no effect on edited ND7 and ND8 (Aphasizheva et al. 2014; Simpson et al. 2017). Some studies have differed on the impact of knockdowns of MRB8180, TbRGG2 and the paralogs MRB4160/8170 on the minimally edited mRNAs CYb and CO2 (Fisk et al. 2008; Kafkova et al. 2012; Aphasizheva et al. 2014). The placement of MRB10130 in the RESC modules is complicated because RNAi of this protein inhibited all editing but also caused a full loss of TbRGG2, as mentioned above (Aphasizheva et al. 2014). Thus, GRBC or REMC proteins are generally distinguished. However, the intimate association of the protein components may complicate the interpretation of some of the data, particularly if a knockdown of one protein destabilizes another. Although “intact” GRBC and REMC modules have not been isolated from each other, the cumulative data from several labs indicates a physical and functional segregation of the modules. Thus, the GRBC and REMC are proposed to participate in distinct phases of RNA editing (Read et al. 2016; Aphasizheva and Aphasizhev 2016).
The Read lab used a novel high-throughput sequencing platform named the Trypanosome RNA Editing Alignment Tool (TREAT) to examine the impact of RNAi of individual RESC proteins on the editing of the mRNAs RPS12 and ND7 (Simpson et al. 2016; Simpson et al. 2017; McAdams et al. 2018). Despite the existence of REMC variants and stabilizing protein-protein contacts discussed above these RNA-seq studies have clearly indicated a differential contribution of the examined REMC proteins in editing progression. RNAi of MRB8180 or TbRGG2 substantially increased editing pausing at many sites on RPS12 but only affected a few sites on ND7. This finding may reflect previous reports indicating that RNAi of MRB8180 has little or no effect on the pan-edited mRNA ND7 (Simpson et al. 2017; Aphasizheva et al. 2014). In contrast, RNAi of TbRGG2 caused a robust loss of edited RPS12 (Fisk et al. 2008; Aphasizheva et al. 2014). Simpson et al (2017) found that upon TbRGG2 RNAi, RPS12 mRNA editing initiates normally, but progression is stalled, due to apparent problems with gRNA-mRNA alignment. In this case, the loss of RPS12 editing in the TbRGG2 knockdown may largely reflect the many predicted Y2H interactions of TbRGG2 with RESC proteins (Ammerman et al. 2012). As it was mentioned above, TbRGG2 RNAi decreased the level of both MRB8170 and MRB8180 (Simpson et al. 2017). In contrast to the above effects of MRB8180 and TbRGG2, a double knockdown of the paralogs MRB4160/8170 caused the opposite effect. That is, it increased pausing at many sites in ND7 but only at a few sites in RPS12 in the RNA-seq studies.
The same RNA-seq approach was also used to examine a knockdown of MRB7260 (McAdams et al. 2018). MRB7260 is unusual in that it was originally found in only a subset of pulldowns of RESC proteins (Ammerman et al. 2012; Aphasizheva et al. 2014). The MRB7260 knockdown did not destabilize canonical RESC proteins. Looking at the pausing profiles, McAdams et al proposed that the MRB7260 knockdown reduces gRNA exchange that occurs as editing progresses from one gRNA-directed block to the next (Fig. 1B). MRB7260 appeared to have an effect on gRNA-mRNA positioning. The initiating gRNA was not affected by MRB7260, so editing initiation is controlled by other proteins. Although, only mRNA RPS12 was examined by RNA-seq, analyses of editing phenotypes by qRT-PCR implied that MRB7260 could similarly affect other pan-edited mRNAs. The McAdams study (2018) also found that MRB7260 may modulate additional steps in editing. gRNA utilization normally involves specific mRNA matching with a cognate gRNA. The authors noted that the RNAi of MRB7260 increased the use a “non-cognate” gRNA that directed non-canonical editing. Thus, MRB7260 may contribute to the proofreading of the mRNA-gRNA pairs. Moreover, McAdams et al (2018) reported differential RNA-mediated effects during the co-purification of MHT-tagged MRB7260 with other RESC proteins. The relative level of GAP1 and MRB81870 in the MRB7260 pulldown was reduced by treatment of the cell lysates with RNase. However, levels of TbRGG2, MRB8180, MRB10130 and MRB3010 were increased in the MRB7260 pulldown of lysates treated with RNase than of untreated lysates. The RNAi of MRB7260 also differentially affected the content of mRNA and gRNA in the RESC. The MRB7260 depletion increased the content of mRNA in pulldowns of HTM-tagged TbRGG2 but decreased a few-fold the gRNA content in pulldowns of PTP-tagged MRB3010. Furthermore, glycerol gradients of a MRB7260 pulldown showed a broad distribution of MRB7260-associated complexes including at ~5S with GAP1 and TbRGG2.
Together the McAdams studies suggest an intricate network of protein- and RNA-mediated contacts in RESC that may be modulated by MRB7260. It is unclear if the GAP1/2 tetramer or only GAP1 was present in the ~5S particles. Such small particles could participate in the assembly of the RESC variants. Finally, RNAi of MRB7260 induced a robust growth defect in procyclic trypanosomes but only had a modest effect in the bloodstream stage, suggesting that MRB7260 may have a differential role(s) in the T. brucei life cycle. The use of high-throughput sequencing will be informative to examine the differential role of other accessory editing proteins, including non-canonical protein components such as MRB1590 which may exhibit a mRNA-specific differential effect on the editing of the mRNA A6 (Shaw et al. 2015).
Several proteins in RESC are known to UV-crosslink with RNA including TbRGG2, MRB8180 and the paralogs MRB4160/8170 (Simpson et al. 2017; Dixit et al. 2017). A study by the Lukes lab applied iCLIP techniques to examine the RNA-binding specificity of the paralogs MRB4160/8170 in procyclic trypanosomes (Dixit et al. 2017). Nucleotide-resolution analyses of the targets in this study showed that both paralogs bind the mRNA CO3 but not the mRNA ND3. Consistent with this finding, qRT-PCR analyses of edited mRNA showed that a double-knockdown of MRB4160/8170 decreased the level of edited CO3 but had no effect on edited ND3 (Aphasizheva et al. 2014). Another study that quantitated both edited and pre-edited mRNA CO3 found that both edited and pre-edited mRNA CO3 were downregulated in the MRB4160/8170 double-knockdown (Kafkova et al. 2012). This effect was also seen in the single MRB8170 knockdown but not in the single MRB4160 knockdown (Kafkova et al. 2012). Thus, the two paralogs bind CO3 with differential effects on editing but neither seems to bind ND3. The interaction of MRB8170 with mRNA CO3 could involve a synergy with MRB4160, together modulating the stability of this mRNA. The Dixit study (2017) reported binding of both paralogs with other mitochondrial mRNAs either pre-edited, fully-edited and never-edited. However, binding to never-edited mRNA was minimal. The possible basis of CO3 selective binding by MRB4160/8170 was not examined. However, the paralogs could use a different binding strategy to control editing rather than stability of mRNAs. These include mRNAs A6, ND7 and RPS12 where the MRB4160/8170 double-knockdown decreased the level of edited substrate without evidently affecting the level of pre-edited substrate (Kafkova et al. 2012). Thus, the MRB4160/8170 paralogs could use alternative binding strategies to direct different mRNA targets to either decay or editing pathways.
3.4-. RNA editing helicases as potential control points
Two of the ~40 proteins in the editing apparatus are RNA helicases: the DEAD-box helicase REH1 (alias mHel61p) and the DEAH/RHA helicase subfamily REH2 (Kruse et al. 2013; Cruz-Reyes et al. 2016) (Fig. 2)(Table 2). RNA helicases are expected to modulate many RNA-RNA and RNA-protein interactions during the editing process. Given the number of proteins, mRNAs, and gRNAs in trypanosome RNA editing, the use of only two RNA helicases in this process is surprising. In comparison, mRNA splicing uses over 100 proteins largely in the form of RNPs including small non-coding RNAs that are assembled around pre-mRNAs (Nguyen et al. 2016; Yan et al. 2015). This process uses several DEAD- and DExH-box RNA helicases. Ribosomal RNA biogenesis requires an even a larger number of RNA helicases (Jankowsky 2011; Jarmoskaite and Russell 2014). REH1 and REH2 are members of the superfamily 2 (SF2). SF2-type RNA helicases are monomeric and can be found in all eukaryotes. Their motif II sequence is found as DEAD, DEAH or DExH and provides the names of the subgroups in SF2 (Jankowsky 2011). DExH-proteins including REH2 are also distinguished by an oligonucleotide-binding (OB-fold) domain that is thought to be regulatory (Walbott et al. 2010). The observations below provide insights into the involvement of REH1 and REH2 in RNA helicase-catalyzed remodeling of the RNAs involved in editing or their RNPs. These RNA helicases are potential control points in this process.
3.4.1-. REH1
The RNA helicase REH1 is one of the first editing proteins characterized in trypanosomes. Most REH1 in mitochondria may be free or in small assemblies that sediment in the 5–10S region (Missel et al. 1997; Li et al. 2011). REH1 RNAi caused a strong editing inhibition in two pan-edited mRNAs, A6 and CR3. Other examined mRNAs, pan-edited or minimally edited, exhibited a moderate inhibition upon REH1 depletion. In vitro, REH1 (~60 kDa) is an ATP-requiring RNA helicase with both 5’ → 3’ and 3’ → 5’ specificity. In vivo, REH1 appears to promote the relay of gRNAs as editing progresses from one block of editing sites to the next along the mRNA, according to analyses of two tested mRNAs, pan-edited A6 and minimally edited Cyb (Li et al. 2011) (Fig. 1B). That is, ablation of REH1 did not affect the processing in the first editing block of the tested mRNAs. However, REH1 was required for the editing of two or more blocks (Li et al. 2011). Thus, REH1 plays a critical role in gRNA utilization or exchange in the examined mRNAs. However, no binding protein partner that may serve as regulator has been identified. Also, it will be interesting to determine if REH1 affects the gRNA relay between all editing blocks or only the examined blocks by Li et al. Recombinant REH1 is active but only a small fraction of the native REH1 co-sediments with unwinding activity in mitochondrial extracts (Missel et al. 1997; Hernandez et al. 2010). However, Li et al found that a portion of ectopically expressed TAP-tagged REH1 in the 20–25S region, co-sediments with RECC, and in the >25S region in higher-order complexes. Their REH1-TAP purification without RNase contained a substoichiometric level of REL1/2 ligases (RECC proteins). Also, REH1 was present in biochemical purifications of RECC, and in antibody pulldowns of the RNA helicase REH2 without RNase (Panigrahi et al. 2003b; Panigrahi et al. 2003a; Hernandez et al. 2010). REH1 may directly interact with each gRNA-mRNA pair. The REH1 activity could vary between the hundreds of possible mRNA-gRNA duplex structures. Also, REH1 must be signaled to specifically act at the completion of an editing block. The molecular basis of this signal or trigger to act at the end of an editing block is unknown. These properties of REH1 may exhibit mRNA substrate preferences and could be controlled during the life cycle in trypanosomes.
3.4.2-. REH2
In contrast with the proposed role REH1 in gRNA exchange that is needed by the second gRNA and subsequent gRNAs as editing progresses, the much larger REH2 (~240 kDa) is required for editing within the first “initiating” block, as well as for editing at upstream blocks (Hashimi et al. 2009; Madina et al. 2015). Thus, REH2 may be required for editing at most if not all blocks. Purifications of native REH2 carry an ATP-dependent unwinding activity. This activity exhibits a 3’ → 5’ direction and requires conserved motifs in REH2 for ATP binding and dsRNA binding (RecAI and dsRBD, respectively) (Hernandez et al. 2010). Also, recombinant REH2 is catalytically active in vitro (Kumar et al, unpublished data). REH2 is part of the REH2C subcomplex which also includes the REH2-associated factor 1 (H2F1) and H2F2 (Kumar et al. 2016). As mentioned above, REH2 carries a putative regulatory OB fold in its C-terminus. H2F1 is a multi-zinc finger protein that stabilizes REH2 and serves as an adaptor directing REH2 to the editing apparatus. The 15S REH2C subcomplex was identified in RNA-free mitochondria after ablation of the single mitochondrial RNA polymerase. Another purification of REH2 after a GAP1 knockdown (i.e., without gRNA-loaded GAP1/2 tetramer) showed that REH2C is an mRNP that contains pre-edited, partially-edited and fully-edited mRNAs (Madina et al. 2015; Kumar et al. 2016). REH2 exhibits UV-crosslinking with synthetic RNA (Hernandez et al. 2010; Madina et al. 2014). The REH2C mRNP associates via RNA linkers with RESC and RECC, but the REH2C association with RESC seems more stable that with the RECC enzyme (Hernandez et al. 2010; Madina et al. 2014; Madina et al. 2015; Kumar et al. 2016)(Fig. 2). RNAi of REH2 caused a shift of GAP1, a canonical RESC protein, to lighter sedimentation fractions (Hashimi et al. 2009). These observations are consistent with the view that the RECC enzyme and its accessory factors are associated via RNA-mediated contacts. Also, RESC may recruit mRNPs, rather than mRNA “naked” transcripts, including the REH2C mRNP. Kumar et al proposed that the REH2C mRNP potentially hybridizes with gRNA-loaded GRBC (Kumar et al. 2016). In support of this model, inactivating mutations in RecAI (ATP binding) and dsRBD2 dissociated REH2 from mRNA, gRNA and GAP1 (Madina et al. 2015). As it was previously described in section 3.1 on natural GRBC variants, the REH2C exhibits stable and weak interactions with GRBC variants of relatively different MRB3010 content, respectively (alias GRBC* and GBRC). As indicated before, the MRB3010-bound GRBC variant seems particularly active in editing. Notably, a REH2 knockdown revealed important trans effects on this GRBC variant. These trans effects included a reduction of examined pre-edited mRNAs, and an increase in pausing in partially edited molecules. Similarly, a knockdown of H2F1, which binds directly to REH2 in the REH2C caused a reduction of examined pre-edited mRNAs in MRB3010-bound GRBC (Madina et al. 2015; Kumar et al. 2016). In contrast to REH1 which facilitates gRNA exchange and may be largely found free or in small assemblies, the RNA helicase REH2 may join the editing apparatus bound to mRNA in an mRNP. This REH2 mRNP may be required in substrate loading and editing progression including at the first block in RESC-associated mRNAs. An ongoing high-throughput experiment may provide additional information on the role of REH2 at individual sites. Like REH1, the helicase REH2 activity may be controlled in a substrate or cell cycle specific manner.
Conclusion
Highly dynamic molecular machines such as mRNA spliceosomes, ribosomes, and the RNA holo-editosomes discussed here assemble several trans factors around mRNA. These molecular machines recognize thousands of different substrates and require numerous control points. The nuclear splicing machinery uses over 100 proteins including five multi-subunit snRNPs plus additional protein factors. In comparison, the mitochondrial RNA editing machinery in T. brucei uses nearly 40 proteins including the multi-subunit core enzyme and its accessory components. The latter include at least two multi-subunit RNPs and additional protein factors. As other multi-step RNA processes, RNA editing may be controlled during complex assembly, substrate recognition, initiation and progression, and during development. Studies of editing control in T. brucei remain challenging. However, a combination of genetic, biochemical, high-throughput RNA sequencing and CLIP approaches are providing important insights. Besides the canonical editing apparatus, emerging variants or isoforms of both core and accessory subcomplexes add potential layers of regulation. These complex variants may provide the necessary flexibility and fine-tuning for the specific recognition of thousands of editing sites in pre-edited substrates and a myriad of intermediates in the mitochondrial milieu. The mRNA cleavage step by the RECC enzyme provides a key checkpoint in the basic catalytic cycle of RNA editing. Suitable mRNA-gRNA substrates for RECC cleavage are assembled and enabled by auxiliary mRNA trans factors discussed in this review, including RESC, REH2C and REH1. The available cross-linking analyses of purified RECC suggests an intriguing combinatorial potential of its catalytic and non-catalytic RNase III proteins. The identification of bona fide partners in proposed RNase III heterodimers and the roles of these heterodimers will be exciting. The pre-assembly or stepwise assembly of core and accessory trans factors on the mRNA transcripts may respond to the metabolic needs of the growing parasites or their adaptation to insect and mammal hosts.
Several questions regarding basic editing mechanisms and their control, include: the formation of the gRNA-bound GAP1/2 tetramer, the tetramer docking into a modular RESC (i.e., including GRBC and REMC), the general mRNA-targeting by GRBC, the differential effects of REMC, the REH2C helicase mRNP and the REH1 helicase in RNA remodeling, coordination of core and accessory co-complex interactions, the role of complex variants, the transition between sites and gRNAs as editing progresses, potential “discard” pathways in gRNA exchange or mRNA quality control. This review focused on a few described examples of core and accessory complex variants, natural or induced, and observed differential effects in the growing parasites or in different life cycle stages. However, other control mechanisms not discussed here may specifically target initiating gRNAs which are relatively rare (Koslowsky et al. 2013), regulate alternative editing including a differential use of start codons and more dramatic changes in coding capacity (Ochsenreiter et al. 2008; Koslowsky et al. 2013; Madina et al. 2014; Simpson et al. 2016; Kirby and Koslowsky 2017). Multimeric versions of the editing apparatus could integrate subcomplex variants. Powerful structural approaches recently identified tetrameric supraspliceosomes (Sperling and Sperling 2017). Also, the orchestration of higher order interactions of holo-editosomes with stability factors and translation factors may be controlled in mitochondrial RNA metabolism (Hernandez et al. 2010; Aphasizheva et al. 2011; Read et al. 2011). Trypanosome RNA editing remains an enigmatic phenomenon in RNA biology. However, the rapidly evolving model of the holo-editosome organization offers interesting paths moving forward.
Acknowledgments
We thank Suzanne McDermott for reading the manuscript and her expertise particularly on recent RECC studies. This work was supported by the National Science Foundation (Collaborative Research 1616865 to J.C.-R. and 1616845 to B.H.M.M.); AgriLife at TAMU (to J.C.-R.); and Presbyterian Health Foundation (to B.H.M.M.). Work by BHMM was supported in part by NIH P20GM103640 (to Ann West). We apologize to colleagues whose work was not cited due to space limits.
Contributor Information
*Jorge Cruz-Reyes, Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843, cruzrey@tamu.edu.
Blaine H.M. Mooers, Department of Biochemistry & Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, OK 73104, Blaine-Mooers@ouhsc.edu.
Pawan K. Doharey, Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843, pawan_doharey@tamu.edu.
Joshua Meehan, Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843, joshua.meehan1@outlook.com.
Shelly Gulati, Department of Biochemistry & Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, OK 73104, shelly-gulati@ouhsc.edu.
References
- Ammerman ML, Downey KM, Hashimi H, Fisk JC, Tomasello DL, Faktorova D, Kafkova L, King T, Lukes J, Read LK (2012) Architecture of the trypanosome RNA editing accessory complex, MRB1. Nucleic acids research 40 (12):5637–5650. doi: 10.1093/nar/gks211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Hashimi H, Novotna L, Cicova Z, McEvoy SM, Lukes J, Read LK (2011) MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA 17 (5):865–877. doi: 10.1261/rna.2446311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Presnyak V, Fisk JC, Foda BM, Read LK (2010) TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA 16 (11):2239–2251. doi: 10.1261/rna.2285510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman ML, Tomasello DL, Faktorova D, Kafkova L, Hashimi H, Lukes J, Read LK (2013) A core MRB1 complex component is indispensable for RNA editing in insect and human infective stages of Trypanosoma brucei. PloS one 8 (10):e78015. doi: 10.1371/journal.pone.0078015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Aphasizhev R (2016) U-Insertion/Deletion mRNA-Editing Holoenzyme: Definition in Sight. Trends Parasitol 32 (2):144–156. doi: 10.1016/j.pt.2015.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Maslov D, Wang X, Huang L, Aphasizhev R (2011) Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell 42 (1):106–117. doi: 10.1016/j.molcel.2011.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizheva I, Zhang L, Wang X, Kaake RM, Huang L, Monti S, Aphasizhev R (2014) RNA binding and core complexes constitute the U-insertion/deletion editosome. Mol Cell Biol 34 (23):4329–4342. doi: 10.1128/MCB.01075-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Van den Burg J, Brakenhoff JP, Sloof P, Van Boom JH, Tromp MC (1986) Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46 (6):819–826 [DOI] [PubMed] [Google Scholar]
- Blum B, Simpson L (1990) Guide RNAs in kinetoplastid mitochondria have a nonencoded 3’ oligo(U) tail involved in recognition of the preedited region. Cell 62 (2):391–397 [DOI] [PubMed] [Google Scholar]
- Carnes J, McDermott S, Anupama A, Oliver BG, Sather DN, Stuart K (2017) In vivo cleavage specificity of Trypanosoma brucei editosome endonucleases. Nucleic acids research 45 (8):4667–4686. doi: 10.1093/nar/gkx116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, McDermott SM, Stuart K (2018) RNase III Domain of KREPB9 and KREPB10 Association with Editosomes in Trypanosoma brucei. mSphere 3 (1). doi: 10.1128/mSphereDirect.00585-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Soares CZ, Wickham C, Stuart K (2011) Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J Biol Chem 286 (22):19320–19330. doi: 10.1074/jbc.M111.228965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J, Trotter JR, Ernst NL, Steinberg A, Stuart K (2005) An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc Natl Acad Sci U S A 102 (46):16614–16619. doi: 10.1073/pnas.0506133102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Halbig K, Sacharidou A, De Nova-Ocampo M, Cruz-Reyes J (2005) Minimal pre-mRNA substrates with natural and converted sites for full-round U insertion and U deletion RNA editing in trypanosomes. Nucleic acids research 33 (20):6610–6620. doi: 10.1093/nar/gki943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Pavia P, Hernandez A, Osterwisch D, Puerta C, Cruz-Reyes J (2007) Substrate determinants for RNA editing and editing complex interactions at a site for full-round U insertion. J Biol Chem 282 (7):4265–4276. doi: 10.1074/jbc.M605554200 [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J, Mooers BH, Abu-Adas Z, Kumar V, Gulati S (2016) DEAH-RHA helicase•Znf cofactor systems in kinetoplastid RNA editing and evolutionarily distant RNA processes. RNA Dis 3 (2):e1336. doi: 10.14800/rd.1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Rusche LN, Piller KJ, Sollner-Webb B (1998a) T. brucei RNA editing: adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol Cell 1 (3):401–409 [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J, Rusche LN, Sollner-Webb B (1998b) Trypanosoma brucei U insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic acids research 26 (16):3634–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Sollner-Webb B (1996) Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc Natl Acad Sci U S A 93 (17):8901–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes J, Zhelonkina A, Rusche L, Sollner-Webb B (2001) Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol Cell Biol 21 (3):884–892. doi: 10.1128/MCB.21.3.884-892.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit S, Muller-McNicoll M, David V, Zarnack K, Ule J, Hashimi H, Lukes J (2017) Differential Binding of Mitochondrial Transcripts by MRB8170 and MRB4160 Regulates Distinct Editing Fates of Mitochondrial mRNA in Trypanosomes. MBio 8 (1). doi: 10.1128/mBio.02288-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Ammerman ML, Presnyak V, Read LK (2008) TbRGG2, an essential RNA editing accessory factor in two Trypanosoma brucei life cycle stages. J Biol Chem 283 (34):23016–23025. doi: 10.1074/jbc.M801021200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Li J, Wang H, Aletta JM, Qu J, Read LK (2013) Proteomic analysis reveals diverse classes of arginine methylproteins in mitochondria of trypanosomes. Molecular & cellular proteomics : MCP 12 (2):302–311. doi: 10.1074/mcp.M112.022533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk JC, Presnyak V, Ammerman ML, Read LK (2009) Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol Cell Biol 29 (19):5214–5225. doi: 10.1128/MCB.00520-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan J, Shaw G, Tropea JE, Waugh DS, Court DL, Ji X (2008) A stepwise model for double-stranded RNA processing by ribonuclease III. Molecular microbiology 67 (1):143–154. doi: 10.1111/j.1365-2958.2007.06032.x [DOI] [PubMed] [Google Scholar]
- Golas MM, Bohm C, Sander B, Effenberger K, Brecht M, Stark H, Goringer HU (2009) Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J 28 (6):766–778. doi: 10.1038/emboj.2009.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Ernst NL, Carnes J, Stuart KD (2010) The zinc-fingers of KREPA3 are essential for the complete editing of mitochondrial mRNAs in Trypanosoma brucei. PloS one 5 (1):e8913. doi: 10.1371/journal.pone.0008913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guther ML, Urbaniak MD, Tavendale A, Prescott A, Ferguson MA (2014) High-confidence glycosome proteome for procyclic form Trypanosoma brucei by epitope-tag organelle enrichment and SILAC proteomics. J Proteome Res 13 (6):2796–2806. doi: 10.1021/pr401209w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Cicova Z, Novotna L, Wen YZ, Lukes J (2009) Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA 15 (4):588–599. doi: 10.1261/rna.1411809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H, Zikova A, Panigrahi AK, Stuart KD, Lukes J (2008) TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA 14 (5):970–980. doi: 10.1261/rna.888808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Madina BR, Ro K, Wohlschlegel JA, Willard B, Kinter MT, Cruz-Reyes J (2010) REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem 285 (2):1220–1228. doi: 10.1074/jbc.M109.051862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A, Panigrahi A, Cifuentes-Rojas C, Sacharidou A, Stuart K, Cruz-Reyes J (2008) Determinants for association and guide RNA-directed endonuclease cleavage by purified RNA editing complexes from Trypanosoma brucei. J Mol Biol 381 (1):35–48. doi: 10.1016/j.jmb.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Faktorova D, Krizova A, Kafkova L, Read LK, Lukes J, Hashimi H (2015) Integrity of the core mitochondrial RNA-binding complex 1 is vital for trypanosome RNA editing. RNA. doi: 10.1261/rna.052340.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E (2011) RNA helicases at work: binding and rearranging. Trends in biochemical sciences 36 (1):19–29. doi: 10.1016/j.tibs.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R (2014) RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem 83:697–725. doi: 10.1146/annurev-biochem-060713-035546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kable ML, Seiwert SD, Heidmann S, Stuart K (1996) RNA editing: a mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273 (5279):1189–1195 [DOI] [PubMed] [Google Scholar]
- Kafkova L, Ammerman ML, Faktorova D, Fisk JC, Zimmer SL, Sobotka R, Read LK, Lukes J, Hashimi H (2012) Functional characterization of two paralogs that are novel RNA binding proteins influencing mitochondrial transcripts of Trypanosoma brucei. RNA 18 (10):1846–1861. doi: 10.1261/rna.033852.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Gao G, Rogers K, Falick AM, Zhou S, Simpson L (2006) Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc Natl Acad Sci U S A 103 (38):13944–13949. doi: 10.1073/pnas.0604476103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby LE, Koslowsky DJ (2017) Mitochondrial dual-coding genes in Trypanosoma brucei. PLoS Negl Trop Dis 11 (10):e0005989. doi: 10.1371/journal.pntd.0005989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowsky DJ, Kutas SM, Stuart K (1996) Distinct differences in the requirements for ribonucleoprotein complex formation on differentially regulated pre-edited mRNAs in Trypanosoma brucei. Molecular and biochemical parasitology 80 (1):1–14 [DOI] [PubMed] [Google Scholar]
- Koslowsky DJ, Sun Y, Hindenach J, Theisen T, Lucas J (2013) The insect-phase gRNA transcriptome in Trypanosoma brucei. Nucleic acids research. doi: 10.1093/nar/gkt973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse E, Voigt C, Leeder WM, Goringer HU (2013) RNA helicases involved in U-insertion/deletion-type RNA editing. Biochimica et biophysica acta 1829 (8):835–841. doi: 10.1016/j.bbagrm.2013.04.003 [DOI] [PubMed] [Google Scholar]
- Kumar V, Madina BR, Gulati S, Vashisht AA, Kanyumbu C, Pieters B, Shakir A, Wohlschlegel JA, Read LK, Mooers BH, Cruz-Reyes J (2016) REH2C Helicase and GRBC Subcomplexes May Base Pair through mRNA and Small Guide RNA in Kinetoplastid Editosomes. J Biol Chem 291 (11):5753–5764. doi: 10.1074/jbc.M115.708164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, O’Hearn SF, Sollner-Webb B (2008) Trypanosoma brucei RNA editing protein TbMP42 (band VI) is crucial for the endonucleolytic cleavages but not the subsequent steps of U-deletion and U-insertion. RNA 14 (6):1187–1200. doi: 10.1261/rna.899508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch M, Carnes J, Acestor N, Guo X, Schnaufer A, Stuart K (2012) Editosome accessory factors KREPB9 and KREPB10 in Trypanosoma brucei. Eukaryotic cell 11 (7):832–843. doi: 10.1128/EC.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Herrera J, Zhou S, Maslov DA, Simpson L (2011) Trypanosome REH1 is an RNA helicase involved with the 3’−5’ polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc Natl Acad Sci U S A 108 (9):3542–3547. doi: 10.1073/pnas.1014152108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madina BR, Kumar V, Metz R, Mooers BH, Bundschuh R, Cruz-Reyes J (2014) Native mitochondrial RNA-binding complexes in kinetoplastid RNA editing differ in guide RNA composition. RNA 20 (7):1142–1152. doi: 10.1261/rna.044495.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madina BR, Kumar V, Mooers BH, Cruz-Reyes J (2015) Native Variants of the MRB1 Complex Exhibit Specialized Functions in Kinetoplastid RNA Editing. PloS one 10 (4):e0123441. doi: 10.1371/journal.pone.0123441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madina BR, Kuppan G, Vashisht AA, Liang YH, Downey KM, Wohlschlegel JA, Ji X, Sze SH, Sacchettini JC, Read LK, Cruz-Reyes J (2011) Guide RNA biogenesis involves a novel RNase III family endoribonuclease in Trypanosoma brucei. RNA 17 (10):1821–1830. doi: 10.1261/rna.2815911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams NM, Ammerman ML, Nanduri J, Lott K, Fisk JC, Read LK (2015) An arginine-glycine-rich RNA binding protein impacts the abundance of specific mRNAs in the mitochondria of Trypanosoma brucei. Eukaryotic cell 14 (2):149–157. doi: 10.1128/EC.00232-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams NM, Simpson RM, Chen R, Sun Y, Read LK (2018) MRB7260 is essential for productive protein-RNA interactions within the RNA editing substrate binding complex during trypanosome RNA editing. RNA 24 (4):540–556. doi: 10.1261/rna.065169.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Carnes J, Stuart K (2015a) Identification by Random Mutagenesis of Functional Domains in KREPB5 That Differentially Affect RNA Editing between Life Cycle Stages of Trypanosoma brucei. Mol Cell Biol 35 (23):3945–3961. doi: 10.1128/MCB.00790-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Guo X, Carnes J, Stuart K (2015b) Differential Editosome Protein Function between Life Cycle Stages of Trypanosoma brucei. J Biol Chem 290 (41):24914–24931. doi: 10.1074/jbc.M115.669432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Luo J, Carnes J, Ranish JA, Stuart K (2016) The Architecture of Trypanosoma brucei editosomes. Proc Natl Acad Sci U S A 113 (42):E6476–E6485. doi: 10.1073/pnas.1610177113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott SM, Stuart K (2017) The essential functions of KREPB4 are developmentally distinct and required for endonuclease association with editosomes. RNA. doi: 10.1261/rna.062786.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missel A, Souza AE, Norskau G, Goringer HU (1997) Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol Cell Biol 17 (9):4895–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SH, Nagai K (2016) Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 A resolution. Nature 530 (7590):298–302. doi: 10.1038/nature16940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreiter T, Cipriano M, Hajduk SL (2008) Alternative mRNA editing in trypanosomes is extensive and may contribute to mitochondrial protein diversity. PloS one 3 (2):e1566. doi: 10.1371/journal.pone.0001566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP (2003a) Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. Journal of the American Society for Mass Spectrometry 14 (7):728–735. doi: 10.1016/S1044-0305(03)00126-0 [DOI] [PubMed] [Google Scholar]
- Panigrahi AK, Schnaufer A, Ernst NL, Wang B, Carmean N, Salavati R, Stuart K (2003b) Identification of novel components of Trypanosoma brucei editosomes. RNA 9 (4):484–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read LK, Lukes J, Hashimi H (2016) Trypanosome RNA editing: the complexity of getting U in and taking U out. Wiley Interdiscip Rev RNA 7 (1):33–51. doi: 10.1002/wrna.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read LK, Zimmer SL, Ammerman ML (2011) Marked for translation: long A/U tails as an interface between completion of RNA editing and ribosome recruitment. Mol Cell 42 (1):6–8. doi: 10.1016/j.molcel.2011.03.013 [DOI] [PubMed] [Google Scholar]
- Reifur L, Yu LE, Cruz-Reyes J, Vanhartesvelt M, Koslowsky DJ (2010) The impact of mRNA structure on guide RNA targeting in kinetoplastid RNA editing. PloS one 5 (8):e12235. doi: 10.1371/journal.pone.0012235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Cruz-Reyes J, Piller KJ, Sollner-Webb B (1997) Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J 16 (13):4069–4081. doi: 10.1093/emboj/16.13.4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini RS, Adler BK, Madison-Antenucci S, McManus MT, Hajduk SL (1998) Biochemical methods for analysis of kinetoplastid RNA editing. Methods 15 (1):15–26. doi: 10.1006/meth.1998.0602 [DOI] [PubMed] [Google Scholar]
- Seiwert SD, Heidmann S, Stuart K (1996) Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84 (6):831–841 [DOI] [PubMed] [Google Scholar]
- Shaw PL, McAdams NM, Hast MA, Ammerman ML, Read LK, Schumacher MA (2015) Structures of the T. brucei kRNA editing factor MRB1590 reveal unique RNA-binding pore motif contained within an ABC-ATPase fold. Nucleic acids research 43 (14):7096–7109. doi: 10.1093/nar/gkv647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L, Maslov DA (1999) Evolution of the U-insertion/deletion RNA editing in mitochondria of kinetoplastid protozoa. Annals of the New York Academy of Sciences 870:190–205 [DOI] [PubMed] [Google Scholar]
- Simpson RM, Bruno AE, Bard JE, Buck MJ, Read LK (2016) High-throughput sequencing of partially edited trypanosome mRNAs reveals barriers to editing progression and evidence for alternative editing. RNA 22 (5):677–695. doi: 10.1261/rna.055160.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RM, Bruno AE, Chen R, Lott K, Tylec BL, Bard JE, Sun Y, Buck MJ, Read LK (2017) Trypanosome RNA Editing Mediator Complex proteins have distinct functions in gRNA utilization. Nucleic acids research 45 (13):7965–7983. doi: 10.1093/nar/gkx458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin ML (1991) Early evolution and the origin of eukaryotes. Current opinion in genetics & development 1 (4):457–463 [DOI] [PubMed] [Google Scholar]
- Sperling J, Sperling R (2017) Structural studies of the endogenous spliceosome - The supraspliceosome. Methods 125:70–83. doi: 10.1016/j.ymeth.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprehe M, Fisk JC, McEvoy SM, Read LK, Schumacher MA (2010) Structure of the Trypanosoma brucei p22 protein, a cytochrome oxidase subunit II-specific RNA-editing accessory factor. J Biol Chem 285 (24):18899–18908. doi: 10.1074/jbc.M109.066597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K, Allen TE, Heidmann S, Seiwert SD (1997) RNA editing in kinetoplastid protozoa. Microbiology and molecular biology reviews : MMBR 61 (1):105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K, Schnaufer A, Ernst NL, Panigrahi AK (2005) Complex management: RNA editing in trypanosomes. Trends in biochemical sciences 30 (2):97–105. doi: 10.1016/j.tibs.2004.12.006 [DOI] [PubMed] [Google Scholar]
- Trotter JR, Ernst NL, Carnes J, Panicucci B, Stuart K (2005) A deletion site editing endonuclease in Trypanosoma brucei. Mol Cell 20 (3):403–412. doi: 10.1016/j.molcel.2005.09.016 [DOI] [PubMed] [Google Scholar]
- Tschudi C, Ullu E (1994) Trypanosomatid protozoa provide paradigms of eukaryotic biology. Infectious agents and disease 3 (4):181–186 [PubMed] [Google Scholar]
- Vickerman K (1985) Developmental cycles and biology of pathogenic trypanosomes. British medical bulletin 41 (2):105–114 [DOI] [PubMed] [Google Scholar]
- Vondruskova E, van den Burg J, Zikova A, Ernst NL, Stuart K, Benne R, Lukes J (2005) RNA interference analyses suggest a transcript-specific regulatory role for mitochondrial RNA-binding proteins MRP1 and MRP2 in RNA editing and other RNA processing in Trypanosoma brucei. J Biol Chem 280 (4):2429–2438. doi: 10.1074/jbc.M405933200 [DOI] [PubMed] [Google Scholar]
- Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N (2010) Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J 29 (13):2194–2204. doi: 10.1038/emboj.2010.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Aphasizheva I, Etheridge RD, Huang L, Wang X, Falick AM, Aphasizhev R (2008) Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell 32 (2):198–209. doi: 10.1016/j.molcel.2008.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y (2015) Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349 (6253):1182–1191. doi: 10.1126/science.aac7629 [DOI] [PubMed] [Google Scholar]
- Zimmer SL, McEvoy SM, Li J, Qu J, Read LK (2011) A novel member of the RNase D exoribonuclease family functions in mitochondrial guide RNA metabolism in Trypanosoma brucei. J Biol Chem 286 (12):10329–10340. doi: 10.1074/jbc.M110.152439 [DOI] [PMC free article] [PubMed] [Google Scholar]


