INTRODUCTION
A major obstacle to an HIV cure is a persistent subset of quiescent, infected cells, known as the latent reservoir [1–3]. Proviruses in their quiescent state remain undetected by the immune system and impervious to antiretroviral therapy (ART) [4, 5]. Upon ART interruption, these proviruses can quickly resume viral replication [6–9]. One unmet need is a means to safely and effectively unmask these infected cells in the setting of ART. Multiple latency reversing agents have been investigated for this purpose, but none has yet demonstrated the ability to significantly reduce the size of the latent reservoir, and most have significant safety concerns [10–14].
We recently reported that clinical vaccines administered to people living with HIV can induce cellular HIV RNA expression during virally suppressive ART [15]. In that study, vaccination was associated with increased immune activation and enhanced HIV-specific responses. However, it remains unknown how vaccine-specific immune responses correlate with HIV activation and whether standard vaccines induce HIV expression selectively from a small pool of antigen-specific, activated HIV-infected cells, or non-selectively, i.e. from a broad pool of bystander activated HIV infected cells.
Here, we used deep sequencing to characterize HIV reactivation following a standard influenza vaccination in a group of seven people living with HIV who were virally suppressed with ART.
RESULTS
Study Population
Seven participants who had a median duration of HIV infection of 21 years [interquartile range (IQR): 17–31] were included. Their median age was 59 years (IQR: 55–64). All started ART during chronic infection and achieved sustained viral suppression below 20 copies/mL for at least the last 6 months, and were virally suppressed at time of vaccine administration. Their median CD4+ T cell count was 613 cells/mm3 (IQR= 327–721). Population characteristics are summarized in SDC Table S2. Upon enrollment, participants provided a baseline blood sample and then received a 0.5mL intramuscular injection of a standard influenza vaccine (Fluarix®, GSK). Blood samples were collected at baseline and days 2, 4, 7, 14 and 28 following vaccination.
HIV RNA Transcription and Immune Activation Following Vaccination
We first measured cell-associated HIV RNA encoding for gag at each sampled time point, in duplicate by digital droplet polymerase chain reaction (ddPCR), to determine which showed increased HIV transcription following vaccination. Participants K2, K3 and K4 had increased copy numbers of gag RNA (median increase 256 copies/106 cells, Range: 177–924) in the week following vaccination, relative to their baseline measures; there was a median of 852 copies of HIV gag DNA (Range: 143–5769) and 590 copies of HIV gag RNA (Range: 79–3481) per 106 cells in these participants (SDC Figures S1a and S1b).
We next analyzed HLA-DR and CD38 expression (markers of cellular activation) by flow cytometry and found that CD4+ T cells from three of the seven participants (K3, K4 and K5) showed increasing percentages of HLA-DR and CD38 expression during the four weeks after vaccination compared with baseline (SDC Figure S1c). We also compared the specificity of immune responsiveness of the participants by measuring influenza-specific Immunoglobulin G (IgG) in the serum at baseline and at days 7, 14 and 28. Participants K3, K4 and K7 exhibited the greatest increases in influenza-specific IgG (SDC Figure S1d).
Source of HIV Transcription
To better understand the source of detectable cell-associated HIV RNA, we deeply sequenced HIV DNA and RNA populations in blood from participants K2, K3 and K4 at all collected time points. Deep sequencing of the HIV gag p24, pol reverse transcriptase, and env C2-V3 coding regions was performed. After quality filtering, over 50 million reads were analyzed with a median of 320,452 reads/region/sample (IQR: 198,053–1,494,661). A median of 6 (IQR: 3–11) and 14 (IQR: 8–23) haplotypes per HIV RNA and DNA sample were generated and analyzed. Comparing sequence diversity within the three coding regions via pairwise Tamura-Nei 93 distances between reads with at least 100 overlapping base pairs [16] showed no significant changes in sequence diversity in any sequenced HIV coding region between pre-vaccination samples and samples collected one month after vaccination (SDC Figure S2). We next assessed viral compartmentalization between HIV DNA and RNA sequences for participants K2, K3 and K4 using a distance-based FST test on collapsed and re-expanded haplotypes and a tree-based Slatkin-Maddison test [17] (SDC Table S3). We found no evidence of viral compartmentalization between paired HIV DNA and RNA samples via either method. Overall, maximum likelihood phylogenetic trees of the HIV DNA and RNA sequences, and tree topologies exhibited extensive intermingling of sequences between HIV RNA and DNA populations at each time point (Figure 1), especially compared with those from D0 and D28 in the other 4 participants (SDC Figure S3). Together, these results suggested that the vaccine activated HIV DNA populations for transcription non-selectively.
Figure 1. Maximum likelihood phylogenetic reconstruction of sequences generated from longitudinally collected HIV-1 RNA and DNA Populations Following Influenza Vaccination.
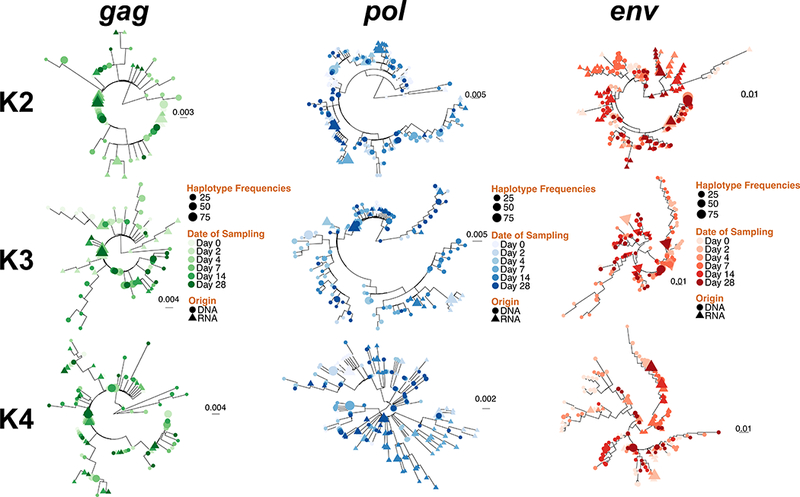
HIV DNA and RNA haplotypes above a minimal frequency threshold of 0.01 were extracted from reads covering the gag (panel A), pol (panel B) and env (panel C) regions for individual K2, K3 and K4 and were used to construct maximum likelihood phylogenies using FastTree (Price et al., 2009). HIV DNA and RNA haplotypes are depicted in circles and triangles respectively. Time point in days (0 to 28 days from vaccination) is indicated for each variant. Scale bars are in substitutions/site.
DISCUSSION
Many current HIV curative strategies have focused on developing methods that induce expression of the virus from infected cells during ART, so that viral proteins are revealed, and cellular reservoirs can be cleared by the host immune response, while ART prevents new cells from being infected [18, 19]. Prophylactic vaccination represents a potential means of transiently activating the immune system, and has been associated with increased levels of cell-free HIV RNA after vaccinations for influenza [20–29], pneumococcus [30–32], tetanus [28, 33], hepatitis B [34] and cholera [35]. Our group recently published data generated from a randomized clinical trial which showed that standard vaccination can increase HIV transcription comparably to what has been observed with the HDACi, Vorinostat [15].
Out of our seven study participants, K2, K3 and K4 most clearly spiked in their HIV gag RNA expression following vaccination (SDC Figure S1a,b). Such variable responsiveness to standard influenza [36–39]. In these three participants, HIV DNA population diversity did not change during the four weeks following vaccination, suggesting that sampling of infected circulating cells did not change appreciably after vaccination (SDC Figure S3). Maximum likelihood phylogenetic reconstructions of HIV gag, pol and env sequences showed extensive overlap between HIV DNA and RNA sequences at all time points (Figures 1, S4), suggesting that cell-associated HIV RNA sequences were likely derived from a broad, non-selective pool of cellular reservoirs of HIV DNA. This is consistent with a study of the HDACi Panobinostat, which showed panmixis of sequences generated from HIV DNA and RNA populations in circulating PBMC [40].
Our study has several limitations. First, our sample size cannot allow broad generalization to others who have HIV reactivation after influenza vaccination. Since this was a pilot study and not a clinical trial, some people may have selective activation of HIV reservoirs following vaccination. Along these lines, increased cell-associated HIV RNA after vaccination may be due to factors other than vaccination. To explore this further, we chose the three participants with the best evidence for increased HIV expression following vaccination (SDC Figure S1a,b). In our previous randomized clinical trial of a larger cohort of people receiving a schedule of vaccines, we were able to detect significant increases in copy numbers of cell-associated, unspliced gag following vaccination [15]. Next, our number of sequence reads suggest high levels of PCR amplification, which can introduce primer biases. However, if template selection based on non-ideal annealing temperatures occurred, we would expect our data to underestimate the true number of haplotypes and bias in amplification. An under-estimation or biased amplification of templates between HIV RNA and DNA populations would likely increase the observation of compartmentalization and thus selective activation, which we did not detect. Along these lines, we did not barcode HIV templates before amplification. While this decreased amplification bias, it precluded us from measuring clonal expansion of HIV DNA or large bursts of HIV RNA from a single provirus in our samples; however, such conditions would be expected to also increase the potential for observation of compartmentalization, which we did not detect. Lastly, our compartmentalization analyses may have been confounded by the number of HIV DNA and RNA haplotypes analyzed. To reduce the likelihood of sampling bias, we grouped all time points for each participant. Compartmentalization was detected in some samples when we analyzed each time point individually (data not shown), but this was likely due to limited HIV RNA haplotypes available for analysis at each time point.
The hunt is on for strategies to provoke HIV expression from quiescent cellular reservoirs and then eradicate the exposed, infected cells. Earlier studies examining the effects of clinical vaccinations on people living with HIV suggested that vaccines can potently activate cellular reservoirs of HIV. We demonstrated here HIV RNA was broadly expressed from a phylogenetically representative pool of circulating cellular reservoirs following vaccination. The ability to reactivate a diverse pool of cellular HIV reservoirs will be critical to HIV eradication approaches. Our findings are a proof-of-concept that standard clinical vaccines can broadly re-activate latent HIV from reservoirs suppressed with ART. Standard influenza vaccination will not cure anyone of HIV. However - after further investigation to determine the mechanisms of HIV activation and improve their potency - vaccinations could become a relatively safe addition to various cure efforts.
Supplementary Material
Acknowledgments
Funding Support:
This work was supported by the National Institute of Health grants K24 AI100665, R01 AI118422, R01 AI120009, and P30 AI036214. Aaron Christensen-Quick is supported as a trainee with institutional training grants from the National Institute of Allergy and Infectious Diseases 5T32AI00738. Fabio Zanini is supported by a Long-Term Fellowship from the European Molecular Biology Organization (EMBO), n. ALTF 269–2016.
References
- 1.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323(5919):1304–7. [DOI] [PubMed] [Google Scholar]
- 2.Trono D, Van Lint C, Rouzioux C, Verdin E, Barre-Sinoussi F, Chun TW, et al. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329(5988):174–80. [DOI] [PubMed] [Google Scholar]
- 3.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–300. [DOI] [PubMed] [Google Scholar]
- 4.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi RT, Bosch RJ, Aga E, Albrecht M, Demeter LM, Dykes C, et al. No evidence for decay of the latent reservoir in HIV-1-infected patients receiving intensive enfuvirtide-containing antiretroviral therapy. J Infect Dis. 2010;201(2):293–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruiz L, Martinez-Picado J, Romeu J, Paredes R, Zayat MK, Marfil S, et al. Structured treatment interruption in chronically HIV-1 infected patients after long-term viral suppression. Aids. 2000;14(4):397–403. [DOI] [PubMed] [Google Scholar]
- 7.Papasavvas E, Ortiz GM, Gross R, Sun J, Moore EC, Heymann JJ, et al. Enhancement of human immunodeficiency virus type 1-specific CD4 and CD8 T cell responses in chronically infected persons after temporary treatment interruption. J Infect Dis. 2000;182(3):766–75. [DOI] [PubMed] [Google Scholar]
- 8.Orenstein JM, Bhat N, Yoder C, Fox C, Polis MA, Metcalf JA, et al. Rapid activation of lymph nodes and mononuclear cell HIV expression upon interrupting highly active antiretroviral therapy in patients after prolonged viral suppression. Aids. 2000;14(12):1709–15. [DOI] [PubMed] [Google Scholar]
- 9.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–5. [DOI] [PubMed] [Google Scholar]
- 10.Sung JA, Pickeral J, Liu L, Stanfield-Oakley SA, Lam CY, Garrido C, et al. Dual-Affinity Re-Targeting proteins direct T cell-mediated cytolysis of latently HIV-infected cells. J Clin Invest. 2015;125(11):4077–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai P, Wu G, Baker CE, Thayer WO, Spagnuolo RA, Sanchez R, et al. In vivo analysis of the effect of panobinostat on cell-associated HIV RNA and DNA levels and latent HIV infection. Retrovirology. 2016;13(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banga R, Procopio FA, Cavassini M, Perreau M. In Vitro Reactivation of Replication-Competent and Infectious HIV-1 by Histone Deacetylase Inhibitors. J Virol. 2015;90(4):1858–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS pathogens. 2014;10(10):e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang G, Dandekar S. Targeting NF-kappaB signaling with protein kinase C agonists as an emerging strategy for combating HIV latency. AIDS Res Hum Retroviruses. 2015;31(1):4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yek C, Gianella S, Plana M, Castro P, Scheffler K, Garcia F, et al. Standard vaccines increase HIV-1 transcription during antiretroviral therapy. Aids. 2016;30(15):2289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular biology and evolution. 1993;10(3):512–26. [DOI] [PubMed] [Google Scholar]
- 17.Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123(3):603–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deeks SG. HIV: Shock and kill. Nature. 2012;487(7408):439–40. [DOI] [PubMed] [Google Scholar]
- 19.Margolis DM. How Might We Cure HIV? Current infectious disease reports. 2014;16(3):392. [DOI] [PubMed] [Google Scholar]
- 20.Ho DD. HIV-1 viraemia and influenza. Lancet. 1992;339(8808):1549. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien WA, Grovit-Ferbas K, Namazi A, Ovcak-Derzic S, Wang HJ, Park J, et al. Human immunodeficiency virus-type 1 replication can be increased in peripheral blood of seropositive patients after influenza vaccination. Blood. 1995;86(3):1082–9. [PubMed] [Google Scholar]
- 22.Staprans SI, Hamilton BL, Follansbee SE, Elbeik T, Barbosa P, Grant RM, et al. Activation of virus replication after vaccination of HIV-1-infected individuals. J Exp Med. 1995;182(6):1727–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosok B, Voltersvik P, Bjerknes R, Axelsson M, Haaheim LR, Asjo B. Dynamics of HIV-1 replication following influenza vaccination of HIV+ individuals. Clin Exp Immunol. 1996;104(2):203–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tasker SA, O’Brien WA, Treanor JJ, Weiss PJ, Olson PE, Kaplan AH, et al. Effects of influenza vaccination in HIV-infected adults: a double-blind, placebo-controlled trial. Vaccine. 1998;16(9–10):1039–42. [DOI] [PubMed] [Google Scholar]
- 25.Gunthard HF, Wong JK, Spina CA, Ignacio C, Kwok S, Christopherson C, et al. Effect of influenza vaccination on viral replication and immune response in persons infected with human immunodeficiency virus receiving potent antiretroviral therapy. J Infect Dis. 2000;181(2):522–31. [DOI] [PubMed] [Google Scholar]
- 26.Kolber MA, Gabr AH, De La Rosa A, Glock JA, Jayaweera D, Miller N, et al. Genotypic analysis of plasma HIV-1 RNA after influenza vaccination of patients with previously undetectable viral loads. AIDS. 2002;16(4):537–42. [DOI] [PubMed] [Google Scholar]
- 27.Vigano A, Bricalli D, Trabattoni D, Salvaggio A, Ruzzante S, Barbi M, et al. Immunization with both T cell-dependent and T cell-independent vaccines augments HIV viral load secondarily to stimulation of tumor necrosis factor alpha. AIDS Res Hum Retroviruses. 1998;14(9):727–34. [DOI] [PubMed] [Google Scholar]
- 28.Ostrowski MA, Krakauer DC, Li Y, Justement SJ, Learn G, Ehler LA, et al. Effect of immune activation on the dynamics of human immunodeficiency virus replication and on the distribution of viral quasispecies. J Virol. 1998;72(10):7772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramilo O, Hicks PJ, Borvak J, Gross LM, Zhong D, Squires JE, et al. T cell activation and human immunodeficiency virus replication after influenza immunization of infected children. The Pediatric infectious disease journal. 1996;15(3):197–203. [DOI] [PubMed] [Google Scholar]
- 30.Calmy A, Bel M, Nguyen A, Combescure C, Delhumeau C, Meier S, et al. Strong serological responses and HIV RNA increase following AS03-adjuvanted pandemic immunization in HIV-infected patients. HIV Med. 2012;13(4):207–18. [DOI] [PubMed] [Google Scholar]
- 31.Brichacek B, Swindells S, Janoff EN, Pirruccello S, Stevenson M. Increased plasma human immunodeficiency virus type 1 burden following antigenic challenge with pneumococcal vaccine. J Infect Dis. 1996;174(6):1191–9. [DOI] [PubMed] [Google Scholar]
- 32.Negredo E, Domingo P, Sambeat MA, Rabella N, Vazquez G. Effect of pneumococcal vaccine on plasma HIV-1 RNA of stable patients undergoing effective highly active antiretroviral therapy. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2001;20(4):287–8. [DOI] [PubMed] [Google Scholar]
- 33.Stanley SK, Ostrowski MA, Justement JS, Gantt K, Hedayati S, Mannix M, et al. Effect of immunization with a common recall antigen on viral expression in patients infected with human immunodeficiency virus type 1. N Engl J Med. 1996;334(19):1222–30. [DOI] [PubMed] [Google Scholar]
- 34.Rey D, Krantz V, Partisani M, Schmitt MP, Meyer P, Libbrecht E, et al. Increasing the number of hepatitis B vaccine injections augments anti-HBs response rate in HIV-infected patients. Effects on HIV-1 viral load. Vaccine. 2000;18(13):1161–5. [DOI] [PubMed] [Google Scholar]
- 35.Ortigao-de-Sampaio MB, Shattock RJ, Hayes P, Griffin GE, Linhares-de-Carvalho MI, Ponce de Leon A, et al. Increase in plasma viral load after oral cholera immunization of HIV-infected subjects. AIDS. 1998;12(14):F145–50. [DOI] [PubMed] [Google Scholar]
- 36.Atashili J, Kalilani L, Adimora AA. Efficacy and clinical effectiveness of influenza vaccines in HIV-infected individuals: a meta-analysis. BMC Infect Dis. 2006;6:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talbot HK, Griffin MR, Chen Q, Zhu Y, Williams JV, Edwards KM. Effectiveness of seasonal vaccine in preventing confirmed influenza-associated hospitalizations in community dwelling older adults. J Infect Dis. 2011;203(4):500–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talbot HK, Zhu Y, Chen Q, Williams JV, Thompson MG, Griffin MR. Effectiveness of influenza vaccine for preventing laboratory-confirmed influenza hospitalizations in adults, 2011–2012 influenza season. Clin Infect Dis. 2013;56(12):1774–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Havers F, Sokolow L, Shay DK, Farley MM, Monroe M, Meek J, et al. Case-Control Study of Vaccine Effectiveness in Preventing Laboratory-Confirmed Influenza Hospitalizations in Older Adults, United States, 2010–2011. Clin Infect Dis. 2016;63(10):1304–11. [DOI] [PubMed] [Google Scholar]
- 40.Barton K, Hiener B, Winckelmann A, Rasmussen TA, Shao W, Byth K, et al. Broad activation of latent HIV-1 in vivo. Nat Commun. 2016;7:12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


