SUMMARY
Mycoplasma pneumoniae is a common cause of human respiratory tract infections, including bronchitis and atypical pneumonia. M. pneumoniae binds glycoprotein receptors having terminal sialic acid residues via the P1 adhesin protein. Here we explored the impact of sialic acid presentation on M. pneumoniae adherence and gliding on surfaces coated with sialylated glycoproteins, or chemically functionalized with α-2,3- and α-2,6-sialyllactose ligated individually or in combination to a polymer scaffold in precisely controlled densities. In both models, gliding required a higher receptor density threshold than adherence, and receptor density influenced gliding frequency but not gliding speed. However, very high densities of α-2,3-sialyllactose actually reduced gliding frequency over peak levels observed at lower densities. Both α-2,3- and α-2,6-sialyllactose supported M. pneumoniae adherence, but gliding was only observed on the former. Finally, gliding on α-2,3-sialyllactose was inhibited on surfaces also conjugated with α-2,6-sialyllactose, suggesting that both moieties bind P1 despite the inability of the latter to support gliding. Our results indicate that the nature and density of host receptor moieties profoundly influences M. pneumoniae gliding, which could affect pathogenesis and infection outcome. Furthermore, precise functionalization of polymer scaffolds shows great promise for further analysis of sialic acid presentation and M. pneumoniae adherence and gliding.
Keywords: Mycoplasma pneumoniae, receptors, sialic acid, adherence, gliding motility
Abbreviated Summary
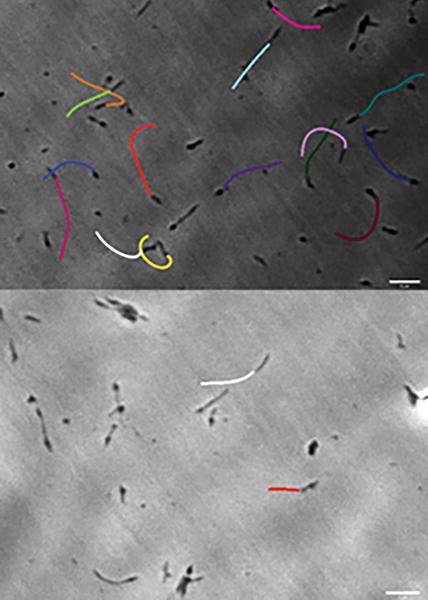
We examined the influence of sialylated receptor environment on Mycoplasma pneumoniae adherence and gliding. M. pneumoniae attached to both α-2,3- and α-2,6-sialyllactose but only glided on the former, and gliding was dependent on receptor density. Furthermore, as indicated by gliding tracks for M. pneumoniae cells attached to slides functionalized with 27% α-2,3-sialyllactose and either no α-2,6-sialyllactose (top) or 53% α-2,6-sialyllactose (bottom), the presence of α-2,6-sialyllactose reduced gliding frequency on α-2,3-sialyllactose.
INTRODUCTION
Mycoplasma pneumoniae is a small, cell wall-less bacterium with a minimal genome and limited biosynthetic capabilities (Himmelreich et al., 1996) that causes bronchitis and an atypical or “walking” pneumonia in humans. M. pneumoniae is responsible for up to 40% of community-acquired pneumonia in both adults and children (Waites and Atkinson, 2009), and infections can lead to prolonged respiratory disorders, including asthma and chronic obstructive pulmonary disease (Nisar et al., 2007; Waites et al., 2008). As a respiratory pathogen M. pneumoniae engages innate immune defenses but often avoids triggering an effective adaptive immune response (Atkinson et al., 2008), resulting in poor clearance of the organism, chronic infection, and reinfection (Waites et al., 2008).
M. pneumoniae adherence and gliding motility are essential for colonization of the airway mucosa and are mediated by a multifunctional, membrane-bound cell extension, the terminal or attachment organelle (Biberfeld and Biberfeld, 1970). The terminal organelle is complex, with eleven distinct substructures evident by electron cryotomography (Henderson and Jensen, 2006). Most relevant to the current study are the protein knobs that line the outer surface of the terminal organelle at its distal end and correspond to P1 adhesin complexes (Kawamoto et al., 2017; Krause et al., 2018; Layh-Schmitt et al., 2000; Nakane et al., 2011). Biochemical and mutant analyses indicate that these adhesin complexes include proteins P1, P40, and P90, which co-localize generally to the terminal organelle (Baseman et al., 1982; Franzoso et al., 1993; Kawamoto et al., 2017; Layh-Schmitt et al., 2000; Nakane et al., 2011). Protein P30, which localizes almost exclusively at the distal end of the terminal organelle (Baseman et al., 1987; Hasselbring et al., 2005), is essential for functional P1 adhesin complexes (Hasselbring et al., 2012; Layh-Schmitt et al., 2000). P1 operates directly in receptor binding and gliding but requires these and other accessory proteins in order to engage in adherence and gliding motility (Krause et al., 1982). Thus spontaneous loss of P40 and P90 (mutant III-4) or P30 (mutant II-3), renders P1 non-functional and M. pneumoniae non-adherent, non-motile, and avirulent (Baseman et al., 1987; Hasselbring et al., 2005; Romero-Arroyo et al., 1999; Waldo et al., 2005). The terminal organelle constitutes the gliding motor (Hasselbring and Krause, 2007), but the gliding mechanism is unique and poorly understood (Miyata, 2008). A gliding model based on electron cryotomography analysis asserts that conformational changes in the terminal organelle interior mobilize P1 adhesin complexes to treadmill on the mycoplasma surface (Henderson and Jensen, 2006; Kawamoto et al., 2017; Krause et al., 2018). Consistent with a P1 treadmill, P1-specific monoclonal antibodies detach gliding but not static mycoplasmas from an inert surface (Seto et al., 2005).
M. pneumoniae engages sialylated glycoproteins as receptors for adherence to respiratory epithelium, and pretreatment of host cells with neuraminidase to remove terminal sialic acid residues inhibits mycoplasma attachment (Baseman et al., 1982; Manchee and Taylor-Robinson, 1969; Sobeslavsky et al., 1968). Sialylated serum glycoproteins in mycoplasma growth media enable M. pneumoniae attachment to the plastic surface of cell culture flasks, and mycoplasma binding to sialylated receptors can be modeled using glycoproteins alone (Roberts et al., 1989). Sialylated glycoproteins having α-2,3 linkages, such as fetuin and laminin, but not those having α-2,6 linkages, such as human plasma fibronectin and fibrinogen, support M. pneumoniae attachment to inert surfaces (Roberts et al., 1989). Furthermore, pre-treatment with neuraminidase or competition with soluble laminin, α-2,3-sialyllactose, or other synthetic sialylated compounds having α-2,3-linked sialic acid, significantly reduces M. pneumoniae attachment and subsequent gliding on sialylated glycoproteins (Kasai et al., 2013; Roberts et al., 1989). α-2,6-sialyllactose and synthetic sialylated compounds with α-2,6-linked sialic acid also inhibit mycoplasma binding and gliding but require higher concentrations to do so, indicating a lower binding affinity (Kasai et al., 2013; Roberts et al., 1989). Thus, M. pneumoniae receptor specificity is somewhat nuanced with regard to sialic acid linkages, which is particularly significant in vivo, where diverse sialic acid linkages and other modifications are common, and in this respect M. pneumoniae recognition of sialylated receptors is similar to that of influenza virus hemagglutinin (Ji et al., 2017).
In the current study, we used two model systems to explore further the interaction of M. pneumoniae with sialylated receptor populations in vitro. In the first model, we utilized sialylated glycoproteins bound to an inert surface in order to extend the findings of Roberts et al. (1989) to include the analysis of gliding motility. The second model employed a protocol for surface-grafted glycopolymer construction (Chen et al., 2018) that allowed us to control precisely the oligosaccharide chemical presentation and surface grafting density. In both models, the surface density of sialylated residues influenced M. pneumoniae gliding frequency but not gliding speed. And while both α-2,3- and α-2,6-sialyllactose supported M. pneumoniae adherence to an inert surface, gliding motility was only observed on the former. However, when α-2,6- and α-2,3-sialyllactose were conjugated to the surface at different ratios, the former inhibited attachment and gliding on the latter. Our results demonstrate that differences in receptor environment affect gliding capability, which could influence the outcome of airway colonization in the human host. Elucidating the dynamics of mycoplasma – host receptor interactions will lead to a better understanding of pathogenesis and persistence of M. pneumoniae in the human airway.
RESULTS
M. pneumoniae binding to sialylated glycoproteins
We examined binding of M. pneumoniae to laminin and human chorionic gonadotropin (hCG) to provide a baseline for subsequent gliding analysis. We detected 16.5-19 and 3.2 mol sialic acid per mol of laminin and hCG, respectively, by high performance anion exchange chromatography coupled with pulsed amperometric detection (HPAEC-PAD) (data not shown). M. pneumoniae bound to chamber slides coated with laminin in a concentration-dependent manner, as expected, with saturation evident at 10 μg (Fig. 1A). Mycoplasma attachment to laminin at the highest levels tested was comparable to that with chamber slides pre-treated with SP4 growth medium glycoproteins, which served as a positive control. As a negative control, we used chamber slides pretreated with ovalbumin alone (Fig. 1A), for which HPAEC-PAD analysis detected only 0.5 mol sialic acid per mol (data not shown). As expected (Roberts et al., 1989), M. pneumoniae also exhibited concentration-dependent attachment to hCG (Fig. 1B), but with saturation evident at 5 μg, and attachment levels significantly lower than for the SP4 positive control or for laminin. Pre-treatment of coated chamber slides with neuraminidase to cleave terminal sialic acid residues resulted in 50-75% reductions in M. pneumoniae attachment to both laminin and SP4 (Fig. 2; P < 0.001), consistent with a binding specificity for sialic acid moieties, as expected (Roberts et al., 1989; Sobeslavsky et al., 1968).
Fig. 1.
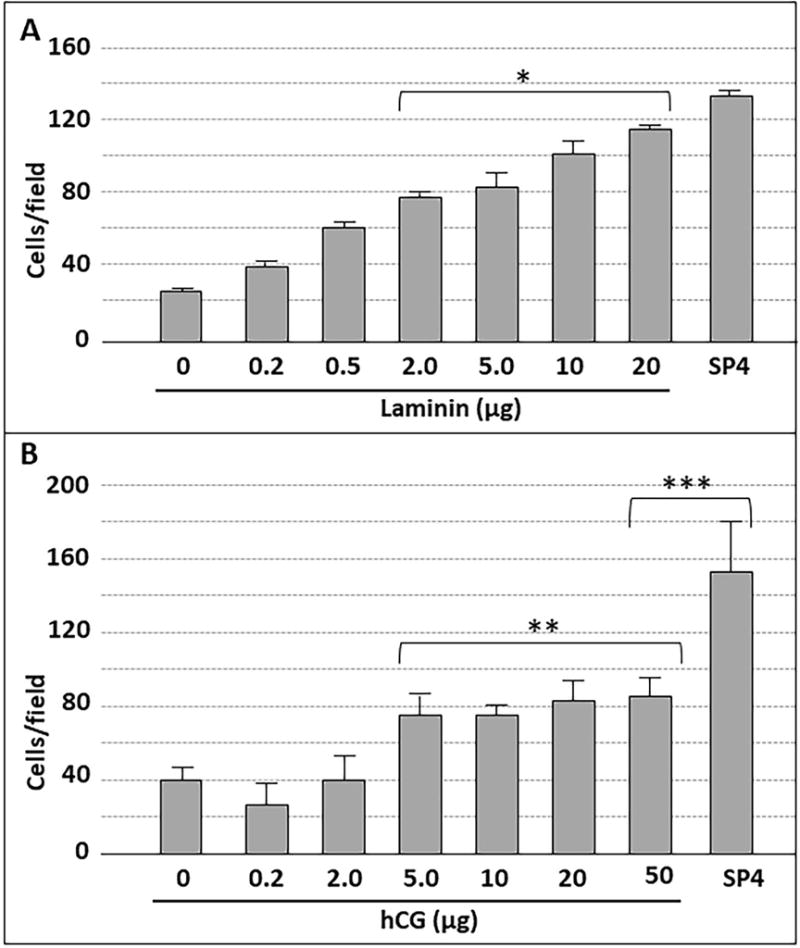
M. pneumoniae attachment to chamber slides coated with laminin (A) or hCG (B) at the indicated levels. Each bar represents the mean and positive standard error of the mean for total cell counts for three separate experiments. SP4, data for chamber slides coated with serum glycoproteins in SP4 growth medium, which served as a positive control. * P < 0.001 for 2.0, 5.0, 10, and 20 μg laminin relative to the 0 μg negative control; ** P < 0.001 for 5.0, 10, 20, and 50 μg hCG relative to the 0 μg negative control; *** P < 0.001 for 50 μg hCG relative to the SP4 control.
Fig. 2.
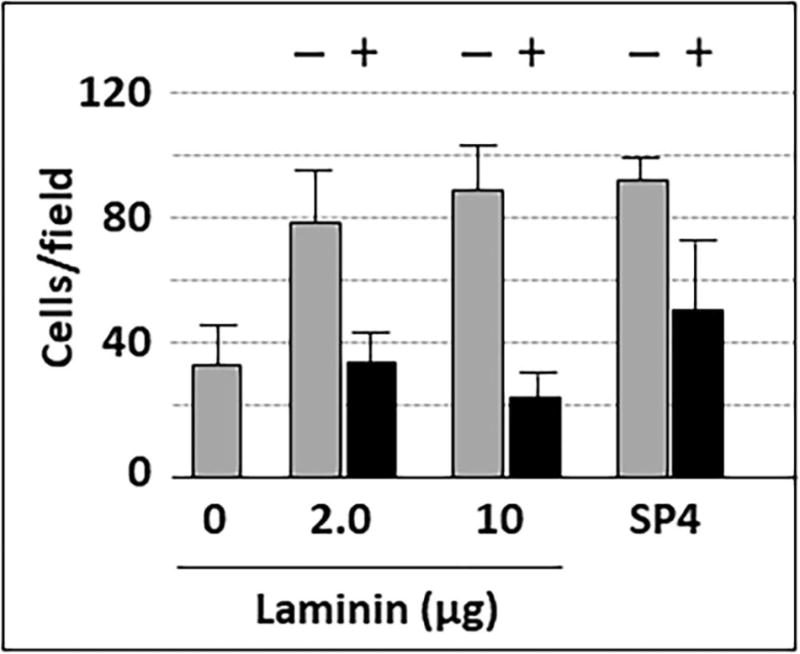
M. pneumoniae attachment to chamber slides coated with laminin, with and without pre-treatment with neuraminidase. Each bar represents the mean and positive standard error of the mean for attached cells for three separate experiments. SP4, data for chamber slides coated with serum glycoproteins in SP4 growth medium, which served as a positive control. + / −, with or without neuraminidase pre-treatment (P < 0.001).
M. pneumoniae gliding on laminin requires a receptor density threshold
We examined M. pneumoniae gliding frequency and speed on glass surfaces coated with laminin or hCG at increasing levels. We consistently observed no gliding on laminin at levels below 2 μg (Fig. 3), or on hCG at all levels tested (data not shown), despite the significant attachment noted in Fig. 1. Gliding was observed on laminin at 2 μg and higher, and gliding frequency increased with laminin from 2 to 5 μg and remained at statistically similar levels at 10 and 20 μg, yet significantly higher than the SP4 positive control (Fig. 3; P < 0.01). Finally, mean gliding speed for all cells measured was 0.32-0.34 μm/sec for all laminin concentrations where we observed gliding, as well as for the SP4 control (tracking 200 cells per concentration in triplicate).
Fig. 3.
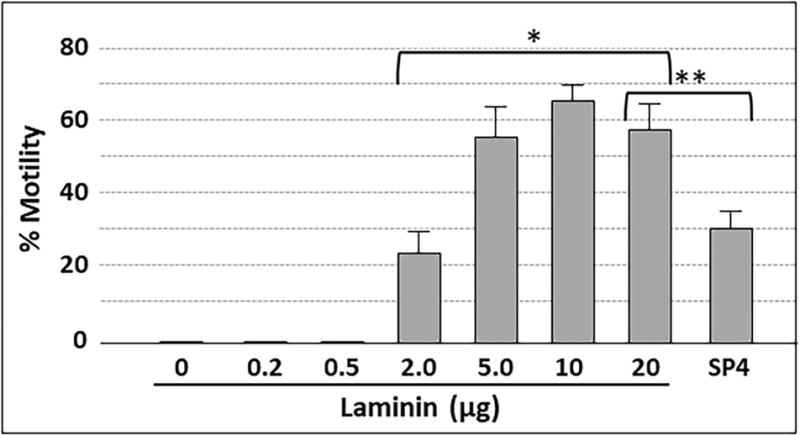
Gliding frequency for M. pneumoniae cells bound to chamber slides coated with laminin at the indicated amounts. Each bar represents the mean and positive standard error of the mean for gliding frequency for three separate experiments. SP4, data for chamber slides coated with serum glycoproteins in SP4 growth medium, which served as a positive control. * P<0.001 for 2.0, 5.0, 10, and 20 μg laminin relative to lower laminin levels; ** P < 0.001 for 20 μg laminin relative to the SP4 control.
Carbohydrate Surface Functionalization
Laminin and related sialylated glycoproteins provide a convenient model for the study of M. pneumoniae interactions with airway glycans, but variability in the extent and nature of their glycosylation, and thus the inability to control oligosaccharide density, presentation, and composition, limits their modeling value. Kasai et al. (2013) had excellent success analyzing glycan specificity based upon the inhibitory activity of synthesized oligosaccharides in solution on mycoplasma binding and gliding on an inert surface coated with serum glycoproteins. Here we sought instead to examine mycoplasma binding and gliding activity on surfaces having well defined oligosaccharide presentation. We recently described the use of hydrazide conjugation to ligate reducing sugars to poly(PFPA) (pentafluorophenylacrylate)-grafted surfaces (Chen et al., 2017). By varying the ratio of hydrazine and ethanolamine in the conjugation, it is possible to control the density of hydrazide available for ligating α-2,3- and α-2,6-sialyllactose to the poly(PFPA) scaffold (Fig. 4). We assessed the chemical modifications at each step by ellipsometry, drop shape analysis, and Fourier transform infrared spectroscopy for thickness, hydrophobicity and chemical changes of the thin polymer film, respectively (Chen et al., 2017; data not shown). On average, ~10-nm hydrophilic glycosurfaces were built on ~15-nm thick poly(PFPA) hydrophobic films. Surface analysis by atomic force microscopy demonstrated smooth and featureless topologies, with a root mean squared roughness of ~1 nm (data not shown). Table 1 shows the hydrazine/ethanolamine ratios utilized, the resulting percentages of conjugation sites occupied by sialyllactose, and the corresponding estimated sialic acid density.
Fig. 4.
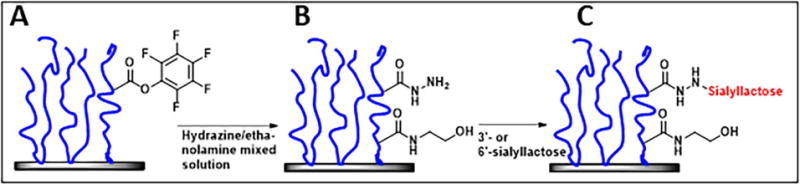
Schematic representation of poly(PFPA) brushes (blue) conjugated to glass (A), with the subsequent conjugation of hydrazine at various ratios with competing ethanolamine (B) dictating the density of hydrazide available for (C) ligation with the reducing end of sialyllactose (red).
Table 1.
Sialyllactose density conjugated to poly(PFPA) as a percentage of maximum possible at the indicated hydrazine : ethanolamine ratios.
| Hydrazine:Ethanolamine | Actual resulting % hydrazide | Final % sialyllactose | Calculated sialyllactose density (molecules/cm2) |
|---|---|---|---|
|
| |||
| 1:999 | 2% | 1.6% | 5.4 × 1013 |
| 1:199 | 10% | 8% | 2.7 × 1014 |
| 1:99 | 20% | 16% | 5.4 × 1014 |
| 1:39 | 40% | 32% | 1.1 × 1015 |
| 100:0 | 100% | 80% | 2.7 × 1015 |
M. pneumoniae attachment and gliding to sialyllactose-functionalized surfaces
We conjugated α-2,3- or α-2,6-sialyllactose at the reducing ends to poly(PFPA) at densities ranging from 1.6% to 80%. M. pneumoniae attached to α-2,3-sialyllactose in a concentration-dependent manner (Fig. 5). Attachment to α-2,3-sialyllactose at densities of 16%, 32% and 80% was statistically comparable to that seen with the SP4 growth medium control, and binding saturation was evident at 16% α-2,3-sialyllactose. Attachment to α-2,6-sialyllactose was significant but substantially lower than for α-2,3-sialyllactose and never achieved the levels observed for the SP4 control. We observed very minimal attachment (≤ 10 cells per field) to the poly(PFPA) scaffold with 100% ethanolamine (i.e. no sialyllactose; data not shown). Treatment of α-2,3-sialyllactose and α-2,6-sialyllactose with neuraminidase resulted in significantly decreased attachment, confirming specificity for the sialic acid (Fig. 6).
Fig. 5.
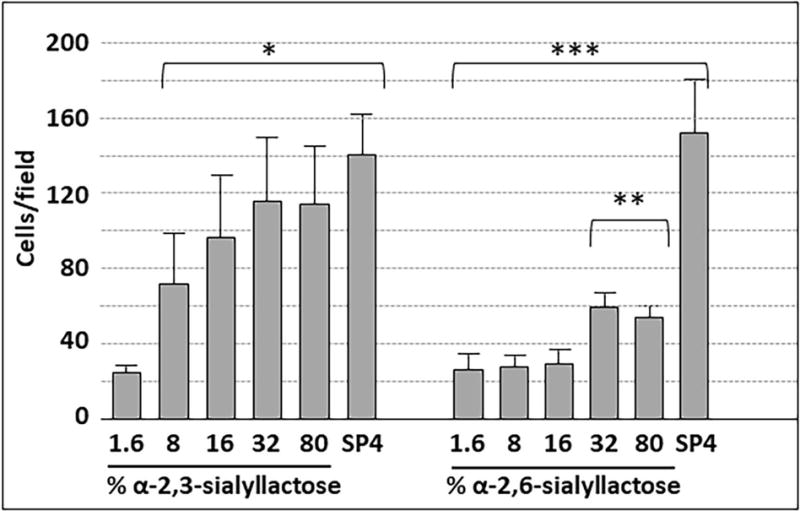
M. pneumoniae attachment to slides chemically functionalized with α-2,3- or α-2,6-sialyllactose, as indicated. Each bar represents the mean and positive standard error of the mean for total cell counts at a given sialyllactose percentage for three separate experiments. SP4, chamber slides coated with SP4 growth medium as a positive control. * P < 0.001 for cell numbers on 1.6% α-2,3-sialyllactose relative to the higher α-2,3-sialyllactose percentages; ** P < 0.001 for cell numbers on 32% and 80% α-2,6-sialyllactose relative to the lower α-2,6-sialyllactose percentages; *** P < 0.001 for cell numbers on SP4 relative to all percentages of α-2,6-sialyllactose.
Fig. 6.
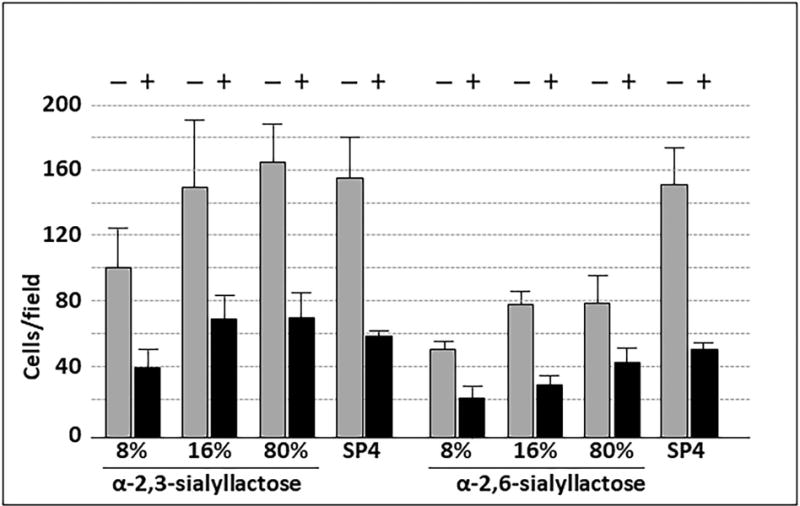
M. pneumoniae attachment to slides functionalized with sialyllactose with and without pre-treatment with neuraminidase. Each bar represents the mean and positive standard error of the mean for attached cells for three separate experiments. SP4, data for chamber slides coated with serum glycoproteins in SP4 growth medium, which served as a positive control. + / −, with or without neuraminidase pre-treatment (P < 0.001).
We observed no gliding on surfaces with a α-2,3-sialyllactose conjugation density lower than 8% (Fig. 7), despite the significant level of attachment noted in Fig. 5. Gliding was prominent on α-2,3-sialyllactose at densities of 8% and higher. Gliding frequency increased between conjugation densities of 8% and 32% but then decreased at higher conjugation densities. No gliding was observed on surfaces conjugated with α-2,6-sialyllactose at any concentration, or on α-2,3-sialyllactose following pre-treatment with neuraminidase (data not shown). Mean gliding speed for all cells measured was 0.28-0.30 μm/sec for all α-2,3-sialyllactose concentrations that supported gliding (tracking 200 cells per ratio in triplicate).
Fig. 7.
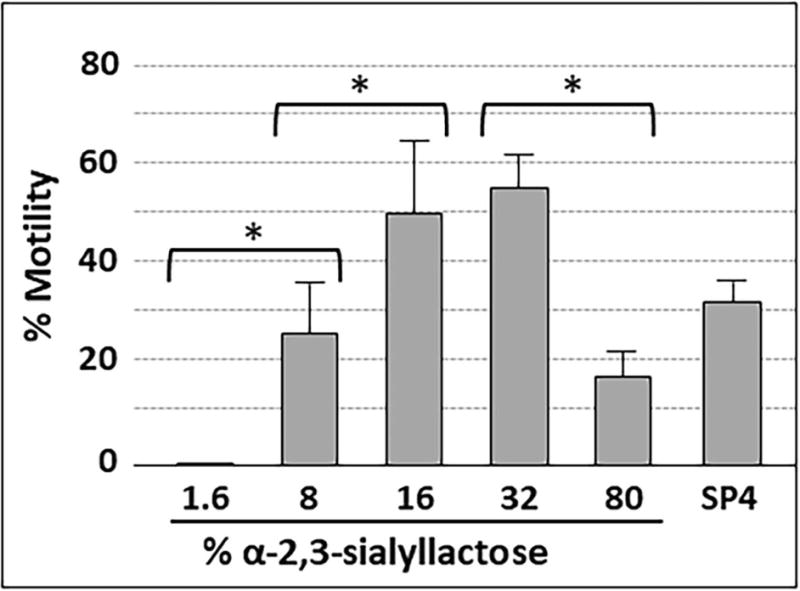
Gliding frequency for M. pneumoniae cells attached to slides chemically functionalized with α-2,3-sialyllactose. Each bar represents the mean and positive standard error of the mean for gliding frequency for three separate experiments. SP4, data for chamber slides coated with serum glycoproteins in SP4 growth medium, which served as a positive control. *P < 0.001 for the indicated sialyllactose densities.
In order to investigate whether α-2,6-sialyllactose influences attachment and gliding on α-2,3-sialyllactose, we conjugated both oligosaccharides to poly(PFPA) at different ratios (Table 2). We also prepared poly(PFPA) surfaces in parallel with competing ethanolamine, in the manner described in Table 1, to allow direct comparison on an equivalent density of α-2,3-sialyllactose alone. In each case, the conjugation of α-2,6-sialyllactose in combination with α-2,3-sialyllactose significantly reduced both the level of mycoplasma attachment (Table 2) and the gliding frequency (Table 2 and Fig. 8) below that observed on α-2,3-sialyllactose alone at the same density.
Table 2.
M. pneumoniae adherence and gliding motility on surfaces having α-2,3-sialyllactose and α-2,3-sialyllactose conjugated individually and in combination at the indicated ratios.
| Ratio of α-2,3- to α-2,6-sialyllactose for conjugation | Resulting α-2,3- : α-2,6-sialyllactose on surface (%) | Cells per field2 | Gliding Frequency (%)3 |
|---|---|---|---|
|
| |||
| NA1 | 27 : 0 | 178.7 ± 35.6 | 63% |
| 1 : 1 | 27 : 27 | 116.7 ± 20.6 | 10% |
| 1 : 2 | 27 : 53 | 92.7 ± 13.1 | 6% |
| NA | 14 : 0 | 121 ± 22.5 | 17% |
| 1 : 5 | 14 : 66 | 82.3 ± 10.8 | 1% |
| 0 : 100 | 0 : 80 | 62.9 ± 7.4 | 0% |
NA, not applicable; amount of α-2,3-sialyllactose conjugated was controlled by varying the hydrazine/ethanolamine ratio, as in Table 1, rather that its ratio to α-2,6-sialyllactose
P < 0.001 for differences between 27:0 and 27:27 or 27:53; P < 0.03 for difference between 14:0 and 14:66
P < 0.001 for differences between 27:0 and 27:27 or 27:53; P < 0.05 for difference between 14:0 and 14:66
Fig. 8.

Gliding tracks for M. pneumoniae attached to slides chemically functionalized with α-2,3- and α-2,6-sialyllactose at different ratios. (A) 1:0 α-2,3-sialyllactose and α-2,6-sialyllactose; (B) 1:1 α-2,3- and α-2,6-sialyllactose; (C) 1:2 α-2,3- and α-2,6-sialyllactose; and (D) 1:5 α-2,3- and α-2,6-sialyllactose. Gliding tracks were recorded at 1 frame/sec for 20-30 sec.
DISCUSSION
Glycoprotein receptors having terminal sialic acid residues are fundamentally important in M. pneumoniae binding to human airway epithelium and in vitro proxies thereof (Manchee and Taylor-Robinson, 1969; Sobeslavsky et al., 1968). Moreover, this requirement for sialic acid recognition encompasses mycoplasma gliding on inert surfaces (Kasai et al., 2013), and likely on epithelial surfaces, although this has only been observed indirectly (Jordan et al., 2007; Prince et al., 2014). Mycoplasma gliding motility requires repeated binding and release of receptors in order to effect sustained movement across a surface (Nagai and Miyata, 2006; Miyata, 2008; Miyata and Hamaguchi, 2016), reflecting the dynamic nature of this bacterial ligand / host receptor interaction. M. pneumoniae exhibits higher binding affinities in suspension for sialic acids having α-2,3 linkages over those linked α-2,6 (Kasai et al., 2013), but the nature of surface-bound receptor recognition in M. pneumoniae adherence and gliding activity is otherwise poorly understood and likely influenced by variability in the local receptor environment. In the current study, we utilized glass surfaces either coated by physisorption with sialylated glycoproteins, or precisely conjugated with sialyl-oligosaccharides via a polymer brush scaffold, to begin to examine the influence of receptor environment on M. pneumoniae attachment and gliding. We observed concentration-dependent and sialic acid-specific attachment of M. pneumoniae to both the heavily sialylated glycoprotein laminin, and the less sialylated glycoprotein hCG, consistent with previous studies (Roberts et al., 1989). Laminin supported mycoplasma attachment at levels that were comparable to that for serum glycoproteins in SP4 growth medium, which served as a positive control here, and significantly greater than that for hCG (Fig. 1). While laminin at concentrations of 2 μg and higher supported M. pneumoniae gliding motility (Fig. 3), M. pneumoniae was non-motile on laminin at lower concentrations, and on hCG at all concentrations, despite the significant levels of attachment observed (Fig. 1). Thus, a sialic acid receptor density threshold is required for the initiation of gliding, and that threshold is greater than the receptor density necessary to support mycoplasma attachment. Mycoplasma gliding frequency correlated with receptor density, increasing with higher concentrations of laminin. In contrast, gliding speed remained consistent for all laminin concentrations tested.
Glycoproteins adsorbed to an inert surface provide a convenient and informative model for studying M. pneumoniae – host receptor interactions. However, this approach has significant limitations, most notably that the amount of glycoprotein bound to the glass surface, and thus the actual density of sialic acid residues, is poorly defined. Moreover, glycosylation of the substrate protein is subject to variability, with no control over carbohydrates linkages, composition, or arrangement (e.g. linear vs. branched). This prompted us to pursue an approach that overcomes these limitations and yields functionalized surfaces with more precisely defined oligosaccharide presentations. To this end we recently described a methodology for ligating oligosaccharides at the reducing end to poly(PFPA)-grafted surfaces through a hydrazide linkage (Chen et al., 2017). Here we expanded upon that approach, varying the density of hydrazide linkages available by changing the ratios of hydrazine and competing ethanolamine in the conjugation to poly(PFPA) (Fig. 4), making it possible to control the density of α-2,3- or α-2,6-sialyllactose precisely in the subsequent ligation to hydrazide.
Both α-2,3- and α-2,6-sialyllactose supported M. pneumoniae attachment in a concentration-dependent manner. However, mycoplasma binding to α-2,6-sialyllactose never achieved the levels observed with α-2,3-sialyllactose or the SP4 control. This is consistent with both the lower binding affinity reported for M. pneumoniae and α-2,6-sialyllactose in suspension (Kasai et al., 2013), and the higher levels of α-2,6-sialyllactose required to inhibit M. pneumoniae attachment to laminin (Roberts et al., 1989). Neuraminidase pre-treatment of surfaces conjugated with α-2,3- or α-2,6-sialyllactose resulted in significantly decreased mycoplasma attachment (Fig. 6). Increasing the amount of neuraminidase or treatment time did not reduce attachment levels further (data not shown). The residual mycoplasma attachment to neuraminidase-treated sialyllactose probably reflects mycoplasma binding to the remaining lactose after removal of the sialic acid residues, based upon the previous observation by Krivan et al. (1989) of low levels of M. pneumoniae binding to lactosylceramide.
Only α-2,3-sialyllactose supported M. pneumoniae gliding motility (Fig. 7), and as noted above with laminin, a threshold density of α-2,3-sialyllactose existed, below which mycoplasma attachment was evident but not gliding motility. These results are consistent with the two-step model of gliding motility for M. pneumoniae (Miyata and Hamaguchi, 2016), where the “catch and release” interaction that is believed to occur between the P1 adhesin complex and sialyl receptors occurs repeatedly during gliding. The P1 adhesin complex appears to engage both α-2,3- and α-2,6- sialyl receptors, as evidenced by the ability of either in solution to competitively inhibit receptor binding by gliding M. pneumoniae, resulting in their detachment from the surface (Kasai et al., 2013), and by our attachment data here (Fig. 5 and 6). However, sialic acids linked α-2,3 but not α-2,6 may trigger a secondary recognition event, such as a conformational change in the adhesin complex, resulting in engagement of the bound adhesin complex in treadmilling, and initiation of gliding. M. pneumoniae attachment may be similar in this respect to the two-step initial and tight binding described for gliding by Mycoplasma mobile (Nagai and Miyata, 2006). If sialic acid linked α-2,3 is too sparse, insufficient P1 adhesin complexes engage to drive M. pneumoniae cell movement and gliding motility. Here no gliding occurred on chemically functionalized surfaces having α-2,3-sialyllactose densities below an estimated 0.54 residues per nm2. It should be noted that the poly(PFPA) matrix extends into three-dimensional space, but we did not consider depth in our sialyllactose density estimate for two reasons. First, we recorded thickness measurements for dehydrated matrices, and the depth of the hydrated matrix will vary with extent of conjugation. Second, the average conjugation efficiency with different oligosaccharides may vary from 80% to 90%, and it is not known if conjugation is uniform throughout the depth of the film or may reflect steric inhibition within the matrix.
Mycoplasma gliding frequency increased with α-2,3-sialyllactose density, but only to a point. At the highest α-2,3-sialyllactose densities tested, M. pneumoniae gliding frequency decreased relative to the maximum gliding frequencies observed at lower sialyllactose densities. This reduction might reflect an avidity issue, where too many adhesin complexes are engaged and therefore unavailable to sustain cell movement via treadmilling. In contrast, gliding speed did not change with receptor density, both with laminin and α-2,3-sialyllactose, and in this respect M. pneumoniae gliding behavior differed from that described for M. mobile, where gliding speed varies with sialyllactose density (Nagai and Miyata, 2006). This distinction is probably a reflection of the fundamental difference in their proposed gliding mechanisms, specifically centipede-like movement for M. mobile and inchworm-like movement with adhesin treadmilling for M. pneumoniae (Henderson and Jensen, 2006; Miyata, 2007).
Differential lectin histochemistry studies indicate that sialic acids linked both α-2,3 and α-2,6 are expressed on the surface of normal human bronchial epithelial (NHBE) cells differentiated in vitro (Kogure et al., 2006). However, the relative distribution of each linkage type reportedly varies in different regions of intact airways, again based upon lectin histochemistry (Shinya et al., 2006). In all cases, however, it is clear that the airway mucosa presents a heterogeneous array of sialylated and other oligosaccharides, with M. pneumoniae likely to encounter sialic acids in diverse linkages and relative abundance. In order to begin to explore how this diversity might affect M. pneumoniae attachment and gliding behavior we conjugated α-2,3-sialyllactose and α-2,6-sialyllactose to poly(PFPA) at varying ratios. Our results revealed reduced attachment and gliding on α-2,3-sialyllactose when α-2,6-sialyllactose was also present. We believe that the simplest interpretation of this inhibition is competition by α-2,6-sialyllactose for putative binding by P1 adhesin complexes. As α-2,6-sialyllactose fails to support gliding, this competitive inhibition could limit the number of P1 adhesin complexes engaged with α-2,3-sialyllactose and therefore contributing to cell gliding. This likely interplay between P1 adhesin complexes and sialic acids having diverse linkages and presentations on airway epithelium could have a profound impact on mycoplasma localization and mobilization in different regions of the conducting airways, depending on the relative abundance of each sialyl linkage. To illustrate, M. pneumoniae attaches to the tips of cilia on NHBE cells, migrates rapidly to the base of the cilia, spreads laterally more slowly, localizing to intercellular junctions, and invades the basolateral compartment (Prince et al., 2014; 2017). The local receptor environment, specifically the relative abundance of oligosaccharide receptors that support or are barriers to adherence and gliding, could account for this colonization pattern. In this respect a parallel can be drawn to the preferences for α-2,3- or α-2,6-sialyl linkages in receptors for human and avian influenza virus strains, respectively, and the relative abundance of each linkage in the airways of the preferred host (de Graaf and Fouchier, 2014). It is also increasingly clear that sialic acid binding by influenza virus is nuanced, extending beyond terminal sialic acid linkages to include their overall glycan topologies (Peng et al., 2017; Viswanathan et al., 2010), and the same may be true for receptor recognition by M. pneumoniae. Our strategy for conjugating oligosaccharides in a controlled manner to a poly(PFPA) matrix shows excellent promise and should allow analysis of receptor specificity beyond linkage alone to reflect the diversity that can exist in conformational flexibility and presentation. We anticipate that this approach will allow us to incorporate variations in the spacer length, as well as the composition, modifications, and combinations of oligosaccharides. As glycan analysis reveals more information about the surface environment of the airway mucosa it will be possible to define further how the receptor environment influences M. pneumoniae attachment and gliding behavior and infection outcome.
Materials and Methods
M. pneumoniae culture
We grew M. pneumoniae wild-type strain M129 (Lipman et al., 1969) for 72 h at 37°C in tissue culture flasks in SP4 medium (Tully et al., 1977) containing fetal bovine serum (FBS). When cultures reached mid-log phase (pH 6.9-7.1), the growth medium was removed and the culture vessels were washed three times with phosphate buffered saline (PBS; pH 7.2). Mycoplasmas were then scraped from the surface into PBS and harvested by centrifugation at 20,000 × g for 20 min at 4°C. The resulting pellets were washed once with PBS by centrifugation, suspended in modified SP4 medium (mSP4; no FBS or phenol red), plus 5% ovalbumin (Sigma-Aldrich), syringe-passaged ten times using a 25 gauge needle to disperse the cells, and syringed-filtered twice through a 0.45μm filter to remove cell clumps.
Preparation of glycoprotein-coated substrates
We used four-well chambered coverglasses (#1; Nunc Lab-Tek) treated with receptor preparations at varying concentrations for all binding assays. The receptor substrates tested were the sialoglycoproteins murine laminin (Invitrogen) and human chorionic gonadotropin (hCG; Sigma-Aldrich). Laminin and hCG were dissolved individually in PBS at concentrations ranging from 0.2-50 μg/ml and incubated in 500-μl volumes per chambered coverglass well for 1 h at room temperature. Thus, incubation with 10 μg protein, for example, corresponds to 5.5 μg/cm2. As a positive control for M. pneumoniae binding, chambered coverglasses were incubated with SP4 medium for 1 h at room temperature to allow FBS glycoproteins to coat the coverglass surface. Receptor solutions were then removed, all wells were washed once with PBS, and 5% ovalbumin (Sigma-Aldrich; ≥ 98% purity) was added for 1 h at room temperature to limit nonspecific binding of M. pneumoniae to the glass. As a negative control for binding, chambered coverglasses were pre-incubated with 5% ovalbumin alone for 1 h at room temperature.
Preparation of chemically functionalized surfaces and determination of sialyllactose density
All glass coverslip substrates with grafted poly(PFPA) brushes were fabricated as described previously (Chen et al., 2017). Hydrazide linkages for conjugating reducing sugars to the poly(PFPA) were generated by incubating the poly(PFPA)-grafted substrates in dimethylformamide (DMF) containing hydrazine monohydrate and trimethylamine (Chen et al., 2017). We controlled the density of hydrazide linkers by replacing various percentages of hydrazine with ethanolamine hydrochloride. These were allowed to react with the poly(PFPA) substrates for 2 h at room temperature, rinsed with DMF, and dried under a stream of nitrogen. To conjugate the sialyllactose, the disaccharide sodium salt (Carbosynth) (10 mM) and aniline (100 mM) were dissolved in 100 mM sodium acetate buffer (pH 4.5). The hydrazine / ethanolamine-modified substrates were incubated with the sialyllactose solutions in a moisture chamber for 24 h, rinsed with water, and dried under a stream of nitrogen. We carried out surface characterization as described previously (Chen et al., 2017).
To quantify the percentage of hydrazide on poly(PFPA) surfaces, coverslips prepared in parallel were incubated with p-nitrobenzaldehyde (0.03 mmol) dissolved in DMF/water (1:1 vol/vol) and then aniline (0.3 mmol) and the pH adjusted to 4.5 with 2 M HCl. After 2 h the substrates were rinsed with DMF, water, DMF again, and then dried under a stream of nitrogen. We measured the absorbance of conjugated p-nitrobenzaldehyde on each quartz substrate by UV-vis spectrometry and calculated surface hydrazide percentages by comparing the absorbance of hydrazine/ethanolamine-modified substrates with that of substrates modified with 100% hydrazine. The corresponding density was calculated by , where A is absorbance, ε is the extinction coefficient of p-nitrobenzaldehyde at λmax (330 nm), and Na is Avogadro’s number.
Analysis of M. pneumoniae binding and gliding
M. pneumoniae cell suspensions of 107-108 color-change units (Stemke and Robertson, 1982) in 600-μl volumes prepared as described above were added to each pre-treated chambered coverglass well and incubated 1 h at 37°C. The culture medium was removed, each well was washed three times with mSP4 to remove unattached mycoplasmas, and 600 μl mSP4 plus 3% gelatin was added per well. We examined coverglasses by phase contrast microscopy using a DM IRB inverted microscope (Leica Microsystems) and captured images with a digital charge-coupled-device camera (Hamamastsu Photonics K.K.). Images were analyzed using Openlab version 5.5.0 (PerkinElmer). We quantified binding by direct microscopic counts from phase contrast images. For direct microscopic counts, we analyzed 10 fields of view from three replicates, counting only individual mycoplasma cells and excluding occasional clumps of cells. Each study was repeated a minimum of three times. Neuraminidase (Sigma-Aldrich) treatments were for 1 h at 37°C, using 0.5units per well for all assays, following protein coating or oligosaccharide ligation. We assessed gliding motility as described previously (Hasselbring et al., 2005). Briefly, we captured time-lapse movies for mycoplasma cultures maintained at 37°C in an incubator chamber surrounding the microscope (Solent Scientific). We calculated gliding speed for individual cells over a minimum of 20 uninterrupted frames at a constant time interval along a collision-free cell path, with paths tracked using Openlab software. We used those measurements to calculate average gliding frequency for a minimum of 200 cells, and gliding speed for a minimum of 100 cells, per experimental treatment per experiment. Results are presented here as the means and positive standard errors of the mean for each treatment from three independent experiments.
Statistics
We analyzed data on cell attachment and gliding motility in SigmaPlot (Systat Software) by multivariate analysis of variance followed by Holm-Sidak post hoc pairwise comparisons.
Acknowledgments
This work was supported by research grant AI110098 from the National Institute of Allergy and Infectious Diseases to D.C.K. and by the NIH-funded Research Resource for Biomedical Glycomics (P41GM10349010). We thank M. Tiemeyer for technical assistance.
Footnotes
We declare no competing interests.
References
- Atkinson TP, Balish MF, Waites KB. Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol Rev. 2008;32:956–973. doi: 10.1111/j.1574-6976.2008.00129.x. [DOI] [PubMed] [Google Scholar]
- Baseman JB, Banai M, Kahane I. Sialic acid residues mediate Mycoplasma pneumoniae attachment to human and sheep erythrocytes. Infect Immun. 1982;38:389–391. doi: 10.1128/iai.38.1.389-391.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseman JB, Cole RM, Krause DC, Leith DK. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baseman JB, Morrison-Plummer J, Drouillard D, Puleo-Scheppke B, Tryon VV, Holt SC. Identification of a 32-kilodalton protein of Mycoplasma pneumoniae associated with hemadsorption. Isr J Med Sci. 1987;23:474–479. [PubMed] [Google Scholar]
- Biberfeld G, Biberfeld P. Ultrastructural features of Mycoplasma pneumoniae. J Bacteriol. 1970;102:855–861. doi: 10.1128/jb.102.3.855-861.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Leman D, Williams CR, Brooks K, Krause DC, Locklin J. A versatile methodology for glycosurfaces: direct ligation of nonderivatized reducing saccharides to poly(pentafluorophenyl acrylate) grafted surfaces via hydrazide conjugation. Langmuir. 2017;33:8821–8828. doi: 10.1021/acs.langmuir.7b00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014;33:823–841. doi: 10.1002/embj.201387442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G, Hu PC, Meloni GA, Barile MF. The immunodominant 90-kilodalton protein is localized on the terminal tip structure of Mycoplasma pneumoniae. Infect Immun. 1993;61:1523–1530. doi: 10.1128/iai.61.4.1523-1530.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbring BM, Jordan JL, Krause DC. Mutant analysis reveals a specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J Bacteriol. 2005;187:6281–6289. doi: 10.1128/JB.187.18.6281-6289.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselbring BM, Krause DC. Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol Microbiol. 2007;63:44–53. doi: 10.1111/j.1365-2958.2006.05507.x. [DOI] [PubMed] [Google Scholar]
- Hasselbring BM, Sheppard ES, Krause DC. P65 Truncation impacts P30 dynamics during Mycoplasma pneumoniae gliding motility. J Bacteriol. 2012;194:3000–3007. doi: 10.1128/JB.00091-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson GP, Jensen GJ. Three-dimensional structure of Mycoplasma pneumoniae’s attachment organelle and a model for its role in gliding motility. Mol Microbiol. 2006;60:376–385. doi: 10.1111/j.1365-2958.2006.05113.x. [DOI] [PubMed] [Google Scholar]
- Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, White YJ, Hadden JA, Grant OC, Woods RJ. New insights into influenza A specificity: an evolution of paradigms. Curr Opin Struct Biol. 2017;44:219–231. doi: 10.1016/j.sbi.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JL, Chang HY, Balish MF, Holt LS, Bose SR, Hasselbring BM, Waldo RH, 3rd, Krunkosky TM, Krause DC. Protein P200 is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infect Immun. 2007;75:518–522. doi: 10.1128/IAI.01344-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai T, Nakane D, Ishida H, Ando H, Kiso M, Miyata M. Role of binding in Mycoplasma mobile and Mycoplasma pneumoniae gliding analyzed through inhibition by synthesized sialylated compounds. J Bacteriol. 2013;195:429–435. doi: 10.1128/JB.01141-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto A, Matsuo L, Kato T, Yamamoto H, Namba K, Miyata M. Periodicity in attachment organelle revealed by electron cryotomography suggests conformational changes in gliding mechanism of Mycoplasma pneumoniae. mBio. 2016;7(2):e00243–16. doi: 10.1128/mBio.00243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure T, Suzuki T, Takahashi T, Miyamoto D, Hidari KIPJ, Guo C-T, Ito T, Kawaoka Y, Suzuki Y. Human trachea primary epithelial cells express both sialyl(α2-3)Gal receptor for human parainfluenza virus type 1 and avian influenza viruses, and sialyl(α2-6)Gal receptor for human influenza viruses. Glycoconj J. 2006;23:101–106. doi: 10.1007/s10719-006-5442-z. [DOI] [PubMed] [Google Scholar]
- Krause DC, Leith DK, Wilson RM, Baseman JB. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DC, Shi J, Chen S, Jensen AJ, Sheppard ES, Jensen GJ. Electron cryotomography of Mycoplasma pneumoniae mutants correlates terminal organelle architectural features and fucntion. Mol Microbiol. 2018 doi: 10.1111/mmi.13937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivan HC, Olson LD, Barile MF, Ginsburg V, Roberts DD. Adhesion of Mycoplasma pneumoniae to sulfated glycolipids and inhibition by dextran sulfate. J Biol Chem. 1989;264:9283–9288. [PubMed] [Google Scholar]
- Layh-Schmitt G, Podtelejnikov A, Mann M. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiol. 2000;146:741–747. doi: 10.1099/00221287-146-3-741. [DOI] [PubMed] [Google Scholar]
- Lipman RP, Clyde WA, Denny FW. Characteristics of virulent, attenuated, and avirulent Mycoplasma pneumoniae strains. J Bacteriol. 1969;100:1037–1043. doi: 10.1128/jb.100.2.1037-1043.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchee RJ, Taylor-Robinson D. Utilization of neuraminic acid receptors by mycoplasmas. J Bacteriol. 1969;98:914–919. doi: 10.1128/jb.98.3.914-919.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M. Centipede and inchworm models to explain Mycoplasma gliding. Trends Microbiol. 2007;16:6–12. doi: 10.1016/j.tim.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Miyata M. Molecular Mechanism of Mycoplasma gliding - A Novel Cell Motility System. In: Lenz P, editor. Cell motility. New York: Springer; 2008. pp. 137–175. [Google Scholar]
- Miyata M, Hamaguchi T. Integrated information and prospects for gliding mechanism of the pathogenic bacterium Mycoplasma pneumoniae. Front Microbiol. 2016;7:960. doi: 10.3389/fmicb.2016.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai R, Miyata M. Gliding motility of Mycoplasma mobile can occur by repeated binding to N-acetylneuraminyllactose (sialyllactose) fixed on solid surfaces. J Bacteriol. 2006;188:6469–6475. doi: 10.1128/JB.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakane D, Adan-Kubo J, Kenri T, Miyata M. Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J Bacteriol. 2011;193:715–722. doi: 10.1128/JB.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisar N, Guleria R, Kumar S, Chawla TC, Biswas NR. Mycoplasma pneumoniae and its role in asthma. Postgrad Med J. 2007;83:100–104. doi: 10.1136/pgmj.2006.049023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, de Vries RP, Grant OC, Thompson AJ, McBride R, Tsogtbaatar B, Lee PS, Razi N, Wilson IA, Woods RJ, Paulson JC. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe. 2017;21:23–34. doi: 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince OA, Krunkosky TM, Krause DC. In vitro spatial and temporal analysis of Mycoplasma pneumoniae colonization of human airway epithelium. Infect Immun. 2014;82:579–586. doi: 10.1128/IAI.01036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince OA, Krunkosky TM, Sheppard ES, Krause DC. In vitro modeling of persistent Mycoplasma pneumoniae infection of differentiated human airway epithelium. Cell Microbiol. 2017 doi: 10.1111/cmi.12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DD, Olson LD, Barile MF, Ginsburg V, Krivan HC. Sialic acid-dependent adhesion of Mycoplasma pneumoniae to purified glycoproteins. J Biol Chem. 1989;264:9289–93. [PubMed] [Google Scholar]
- Romero-Arroyo CE, Jordan J, Peacock SJ, Willby MJ, Farmer MA, Krause DC. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J Bacteriol. 1999;181:1079–1087. doi: 10.1128/jb.181.4.1079-1087.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Kenri T, Tomiyama T, Miyata M. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J Bacteriol. 2005;187:1875–1877. doi: 10.1128/JB.187.5.1875-1877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- Sobeslavsky O, Prescott B, Chanock RM. Adsorption of Mycoplasma pneumoniae to neuraminic acid receptors of various cells and possible role in virulence. J Bacteriol. 1968;96:695–705. doi: 10.1128/jb.96.3.695-705.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemke GW, Robertson JA. Comparison of two methods for enumeration of mycoplasmas. J Clin Microbiol. 1982;16:959–961. doi: 10.1128/jcm.16.5.959-961.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully JG, Whitcomb RF, Clark HF, Williamson DL. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science. 1977;195:892–894. doi: 10.1126/science.841314. [DOI] [PubMed] [Google Scholar]
- Viswanathan K, Chandrasekaran A, Srinivasan A, Raman R, Sasisekharan V, Sasisekharan R. Glycans as receptors for influenza pathogenesis. Glycoconj J. 2010;27:561–570. doi: 10.1007/s10719-010-9303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites KB, Atkinson TP. The role of Mycoplasma in upper respiratory infections. Curr Infect Dis Rep. 2009;11:198–206. doi: 10.1007/s11908-009-0030-6. [DOI] [PubMed] [Google Scholar]
- Waites KB, Balish MF, Atkinson TP. New insights into the pathogenesis and detection of Mycoplasma pneumoniae infections. Future Microbiol. 2008;3:635–648. doi: 10.2217/17460913.3.6.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo RH, III, Jordan JL, Krause DC. Identification and complementation of a mutation associated with loss of Mycoplasma pneumoniae virulence-specific proteins B and C. J Bacteriol. 2005;187:747–751. doi: 10.1128/JB.187.2.747-751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


