Abstract
Background
Chronic Otitis Media with effusion (COME) is characterized by persistent middle ear effusions that are in most cases highly viscous, but some patients present with serous fluid. This study aimed at comprehensively characterizing the macromolecular composition of mucoid vs. serous MEEs.
Methods
MEEs from patients with COME were analyzed for proteins by Mass Spectrometry (MS) and western blot techniques, total DNA quantity, bacterial DNA (16S sequencing) and cytokine content. Proteomics datasets were studied in Ingenuity pathway analysis (IPA).
Results
Mucoid samples showed a global tendency of increased pro- inflammatory mediators. IL1-β and IL-10 were significantly more abundant in serous samples (p<0.01). Mucoid samples had higher DNA quantity (p=0.04), more likely to be positive in MUC5B protein (p=0.008) and higher peptide counts (12786 vs 2225), as well as an overall larger number of identified proteins (331 vs 177), compared to serous. IPA found the mucoid sample dataset to be related to immune cell function and epithelial remodeling whereas the serous sample dataset showed acute responses and blood related proteins. Interestingly, serous samples showed more bacterial DNA than mucoid ones, with less bacterial genera variability.
Conclusion
This study demonstrates divergent immune responses in children with COME by effusion quality.
Introduction
Otitis Media (OM) is a common middle ear condition occurring in early childhood accounting for a very high proportion of pediatric office visits and surgeries annually (1, 2) at a national health care cost estimated to be 2.88 billion dollars per year in the US (2–4). OM is a complex disease associated with different stages depending on patient symptoms and middle ear fluids characteristics. Typically, the disease is characterized by acute infectious injury and inflammation, acute OM (AOM), which can become recurrent (rAOM) or progress to chronic epithelial mucoid fluid secretion, i.e. chronic OM (5), which in turn can be itself infectious – i.e. chronic suppurative otitis media (CSOM) or mostly devoid of culturable pathogens- chronic otitis media with effusion (COME). In some cases, COME can occur spontaneously without much of a preceding infectious history. COME middle ear effusions (6) are most frequently mucoid (7–9) and viscous. It has been shown to be primarily composed of innate immunity mediators and among them from neutrophils (10, 11), such as neutrophil extracellular traps (NETs) (12, 13) along with secretory mucin glycoproteins, predominantly MUC5B (14) and a high level of pro-inflammatory cytokines especially interleukin 8 IL-8 (see review (15)). Less frequently, COME MEEs are not as viscous and are characterized as serous. Although, the biologic reasons for this are mostly unknown, studies have linked serous effusions to better medical outcomes in patients relative to those with mucoid effusions (16, 17). Matkovic et al. (18) linked the viscosity characteristics of MEEs to the duration of OM: among the 108 samples they collected, 6% of the effusions were mucoid for patients having OM less than 3 months, but 95% were found mucoid after 3 months of OM diagnosed.
Mucins are large and highly glycosylated proteins participating to the biological and physical properties of the mucus at mucosal layers. Mucins are classified by their protein backbone encoded by different mucin genes called MUC. More than 20 human MUC genes have been identified. Of these, MUC5B appears to be the predominant mucin in COME mucoid effusions (19–21). Other mucins have also been detected in lower amounts: MUC5AC, MUC2 and MUC4 (20, 22–24).
In addition to the mucins, other proteins and especially glycoproteins participate to the integrity of the mucus. Recently Radicioni et al. (25) showed that mucin proteins from tracheobronchial epithelial cells form a tight network of mucins bound to each other as well as to other glycoproteins in node like structures. It is then important to determine MEE proteins that might associate with the gel-like structure of mucoid samples. Our team recently showed neutrophilia and marked neutrophil extracellular traps (NETs) formation (11) in MEE from 50 children with COME, samples being mostly mucoid. NETs are characterized by the partial decompaction of DNA extruded in the extracellular space as long filaments carrying anti-bacterial proteins.
In order to better understand why COM patients present either mucoid or serous MEEs, we used Mass Spectrometry (MS) to identify the proteome of several mucoid and serous MEEs. These samples were also tested for inflammatory mediators, MUC5B protein, DNA quantity and bacterial DNA origin. Software analysis and correlations between different parameters were performed to characterize the potential biological pathways in each group of samples. We hypothesized that serous effusions may represent an early immune response that differs from viscous effusions in mucin and DNA content and by immune response characteristics.
Methods
Sample Collection and Preparation
The Institutional Review Board Committee of Children’s National Health System approved this study. A written consent was obtained from the legal guardian of the patients before effusions were collected. Children aged 12–150 months with COME (not CSOM) of more than 3 month duration were selected for the study as they were undergoing myringotomy with tube placement irrespective of race/ethnicity/gender. Exclusion criteria included: cleft palate or other craniofacial dysmorphic syndromes, immunosuppressive states or conditions, cystic fibrosis, immotile cilia syndrome, or prior history of skull base radiation therapy or skull base malignancy. Participants were recruited between February 2014 and February 2015. For collection, MEE was suctioned directly into a tympanostomy 7 french suction trap- and effusions from both ears were combined into one tube. Effusions were classified as mucoid or serous on the basis of presence or absence of gravitational flow of the effusion (if the effusion did not flow upon flipping the tympanostomy trap vial upside down after ten seconds, it was considered as a mucoid effusion). We found effusions from each side always equally classified. After collection, effusions were solubilized in 1ml of sterile phosphate buffered saline (PBS), aliquoted, and if not immediately used for experiments, frozen at −80°C. For DNA quantification, western blot, Luminex multiplex arrays, effusions from 39–57 patients were studied, due to limitations in sample volume. For more clarity, the raw data showing all the assays performed among the samples was added to the Supplemental Table S1). For proteomics and 16s RNA analysis, 3 of the mucoid MEEs and 3 of the serous MEEs from pediatric patients with COME were studied.
Cytokine Multiplex assay
A cytokine assay was performed to detect small proteins (less than 20kDa in size) as usually not detectable by mass spectrometry. We analyzed MEE samples for quantitative levels of IL-10, macrophage derived chemokine (MDC), IL-13, IL-17A, IL-1β, IL-6, IL-8, RANTES, TNF-α, and VEGF using a commercially available multiplex magnetic bead immunoassay (Millipore, Billerica, MA) according to the manufacturers’ instructions using provided standards and quality controls.
Western Blot Analyses
Western blot analysis was performed as previously described (14). 50 μg total proteins were loaded on a 1.0% agarose gel for electrophoresis (molecular biology grade, Life technologies, Carlsbad, CA) with a Tris-acetate-EDTA (TAE) electrophoresis buffer containing 0.1% SDS, 1mM EDTA, and 40mM Tris acetate (pH 8.0). Samples were processed in urea, SDS and β-mercaptoethanol buffer, denatured at 95°C for 10 minutes and loaded into a horizontal gel apparatus. The electrophoresis was performed at 35 V for 2 hrs, and at 15 V overnight. Then, proteins were transferred under positive pressure onto a polyvinylidine difluoride (PVDF) membrane (Milliporewith a 785 Vacuum blotter, Biorad, Richmond, CA). The membrane was exposed to a rabbit polyclonal anti-MUC5B antibody H-30 (Santa Cruz, CA) at 1:300 dilution in a 2.5% milk solution and then with a secondary antibody goat-anti rabbit coupled to HRP diluted 1:20,000 (Life Technologies). The antibody complex was revealed with SuperSignal® West Dura Extended Duration Substrate kit (Pierce, Rockland, IL) according to the manufacturer’s instructions. MUC5B signal was evaluated by counting the pixels on the films after normalizing to a saliva control using ImageJ.
DNA Quantification
200 μl of MEEs was used to extract DNA with the QiaAmp mini kit (Qiagen, Valencia, CA). DNA concentrations were assayed with the Qubit double stranded DNA kit (Life Technologies) and by optic density using a Nanodrop (Thermo Fisher Scientific, Waltham, MA) evaluating optic density at 280nm and 260nm.
Proteomic Analyses
SDS-PAGE electrophoresis followed by MS analysis were performed as previously described on 6 MEEs (14). The gel was fixed with a 50% methanol and 5% acetic acid solution, stained with Coomassie Blue and destained in water. Each sample was separated 32 to 36 pieces. In-gel digestion was performed using TrypsinGold (Promega, Madisson, WI). The solution extracted containing peptides was completely dried in a SpeedVac (ThermoScientific).
Mass Spectrometry and protein identification
MS was carried out as previously described (14). Ingenuity Pathway Analysis (IPA, version 8.5, Ingenuity Systems, Redwood City, CA) was used to generate predicted pathways activated or inhibited by the proteins in the proteomics datasets (minimum of 5 peptide counts per protein for the 6 samples). Peptide counts were normalized in every sample to their respective total peptide count, and expressed as percentages. Normalized counts from the 3 mucoid or serous samples were averaged and entered in IPA using a comparison analysis. Top pathways activated with p value (calculated by IPA using the right-tailed Fisher Exact Test) and overlap (percentage of proteins of the dataset regulated in the pathway) are presented in the result section.
16S Sequencing
DNA was purified using the QiaAmp mini kit (Qiagen, Valencia, CA). The Qubit double stranded DNA kit (Invitrogen) was used to quantify extracted DNA. All extractions yielding >50 ng of total DNA were further processed. Samples were processed and sequenced as described in (26) and sequenced for the 16S ribosomal RNA gene.
Statistical Analysis
Inflammatory mediator’s assays, DNA quantification and MUC5B expression were compared using a t test analysis or ANOVA (with SigmaPlot software). For the experiments/correlations done on the 6 samples analyzed by Proteomics, several statistical analysis methods were used, all of which were non-parametric due to the small sample size. Comparisons between two categorical parameters used Fisher’s exact or χ2 tests, between one categorical and one continuous parameter used Wilcoxon rank sum tests, and between two continuous parameters used Spearman correlations. Comparisons of peptides between serous and mucoid samples using Wilcoxon rank sum tests and were performed only when peptides were present in at least two of the three samples of each type. All p-values ≤ 0.05 were considered statistically significant. No adjustments for multiple testing were performed as our goal was not hypothesis testing, but hypothesis generating. These analyses were performed using Stata V14 (College Station, TX).
Results
Patient characteristics
Effusions from a total of 57 patients were included in the study. Out of the 57 patients, 15 were female (26.3%) and 42 were male (75.4%). Ethnicities varied with 9 Hispanic/Latino (15.7%), 25 African-American (43.8%), 13 Caucasian (22.8%), and 10 other (17.7%). Forty-five patients (79%) contained mucous type effusions, while 12 (21%) had serous effusions. The mean age for all patients was 36.4 months. Notably, patients with mucoid effusions were found to be younger than those with serous effusions (35.7 vs. 60.2 months old respectively, p=0.02). The clinical indication for tympanostomy tube placement in all cases was persistent MEE of greater than 3 months duration.
Inflammatory mediators, MUC5B and extracellular DNA composition of MEEs
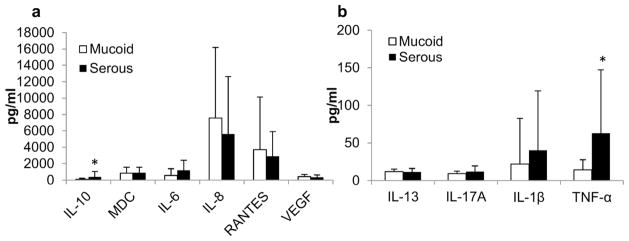
Inflammatory mediators are implicated in the pathogenesis of OM, hence a diversity of them were studied by multiplex ELISA assay in 39 MEE samples (Fig 1). Three mediators were not detected: Eotaxin, IFNγ and IL-5. Six mediators were detected with concentration >100pg/ml: IL-10, MDC, IL-6, IL-8, RANTES and VEGF (Fig 1a). Among these only IL-10 was significantly more abundant in serous samples compared to mucoid (389pg/ml vs 95pg/ml, p=0.02). There was a tendency showing a higher abundance of IL-8 (7547 pg/ml vs 5619 pg/ml, p=0.56) and RANTES (3715 pg/ml vs 2907 pg/ml, p=0.73) in mucoid samples. IL-6 was in higher concentration in serous samples compared to mucoid (1204 pg/ml vs 573 pg/ml, p=0.08) but this difference was not significant. Four mediators were detected with a concentration <100pg/ml: IL-13, IL-17A, IL1β and TNF-α (Fig 1b). Of these, TNF-α was the only mediator that showed a statistical difference, being more abundant in serous MEEs (62.7pg/ml vs 14.3pg/ml, p=0.003).
Fig 1.
Inflammatory mediator concentration in MEEs assayed by multiplex ELISA. Same volumes of MEEs were assayed for inflammatory mediators and averaged for mucoid and serous samples. a. Mediators with a concentration higher than 100pg/ml (IL-10, IL1-b, IL-8, MDC, IL-6 and RANTES). b. Mediators with a concentration lower than 100pg/ml: IL-13, IL-17A, TNF-α and VEGF).*significantly different between mucoid and serous MEEs p≤0.05
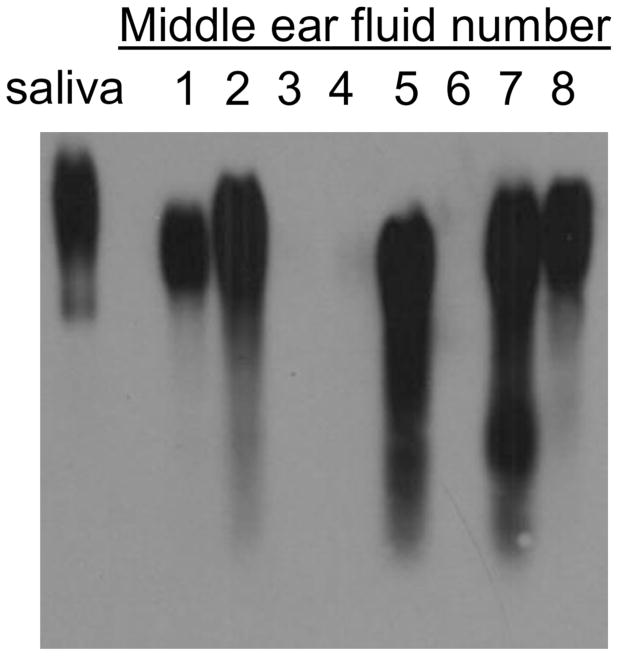
As MUC5B is the predominant mucin in mucoid COM samples (14), we evaluated its presence in these MEEs by immunoblotting (Table 1). As expected, MUC5B was highly abundant in mucoid samples as shown with the normalized pixel count. Mucoid samples had a count ranging between 0 and 8.12 with a median of 1.26 and mean of 2.77 whereas serous samples ranged from 0 to 2.87 with a median of 0.36 and mean of 0.9. This difference was not found to be significant (p=0.13). A representative Western blot for MUC5B is shown as Fig 2.
Table 1.
MUC5B protein and extracellular DNA concentration in mucoid and serous samples.
| MUC5B normalized pixelcoun | |||||
| Mean | Median | Range | p-value | ||
| Mucoid | 2.77 | 1.26 | 0–8.12 | 0.13 | n=37 |
| Serous | 0.90 | 0.36 | 0–2.87 | n=9 | |
| Qubit dsDNA | |||||
| Mean | Median | Range | p-value | ||
| Mucoid | 72.9 | 35.15 | 0.84–324 | 0.04 | n=44 |
| Serous | 18.3 | 3.24 | 0.46–106 | n=11 | |
| Nanodrop DNA | |||||
| Mean | Median | Range | p-value | ||
| Mucoid | 59.4 | 30.37 | 2.62–238.8 | 0.05 | n=44 |
| Serous | 18.8 | 6.92 | 2.36–91.01 | n=11 | |
MUC5B pixel count was normalized to the count of every positive control (10μl saliva). Double stranded DNA (dsDNA) determined by the Qubit kit was expressed as ng/μl, as well as the DNA concentration determined by Nanodrop. The number of samples (n), mean, median and ranged are detailed in the table. The p-value was obtained with an ANOVA test performed to analyze the difference between mucoid and serous datasets.
Fig 2.
MUC5B protein detected by Western Blot assay. 50μ of MEE proteins (samples 1 to 8) were separated in an agarose gel overnight and transferred on a PVDF membrane. MUC5B was detected using an antibody anti-MUC5B H-300 (Santa Cruz, CA) and revealed using a secondary antibody coupled to HRP.
As it has also been demonstrated that MEEs contain a preponderance of NETs (11) containing extracellular DNA we assayed for DNA in samples using absorbance at 260nm and 280nm for the detection of nucleic acids, as well as a double stranded DNA assay (Table 1). The two methods showed similar results: despite the variability in DNA quantity in both groups (median mucoid 35.15 for Qubit and 30.37 for Nanodrop vs median serous 3.24 for Qubit and 6.92 for Nanodrop), there was a higher abundance in mucoid MEEs that was statistically different (p=0.02).
Mass spectrometry: characteristics of the samples
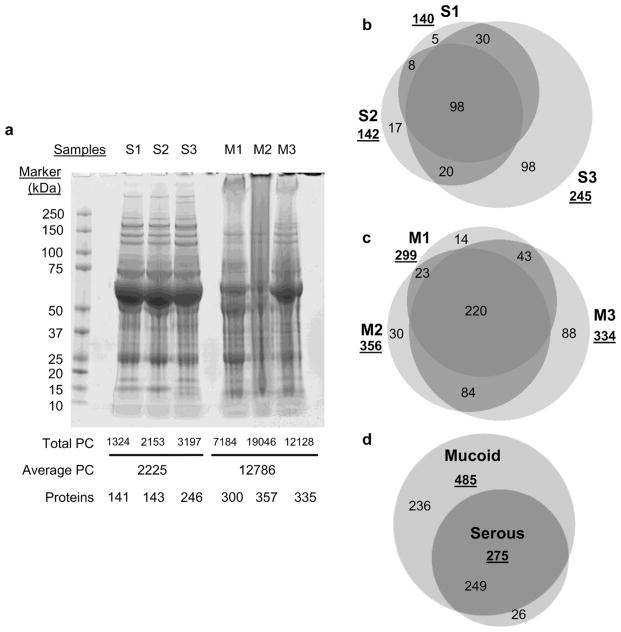
Three mucoid and serous MEEs were analyzed by MS, the demographics of these selected patients is shown in the Supplemental Table S2. The SDS-PAGE stained with Coomassie Blue is shown in Fig 3a. Serous samples had a very similar migration profile with defined bands for the larger proteins > 75kDa. Mucoid samples showed a smear-like migration, especially M2, these profiles being characteristic of viscous and DNA rich samples.
Fig 3.
MEEs protein characteristics. a. SDS-PAGE was used to separate proteins by molecular weight. Total peptide count (total PC), average peptide count and number of proteins detected are detailed below the gel picture. b. and c. Datasets were analyzed for similarity using Venn Diagrams (BioVenn http://www.cmbi.ru.nl/cdd/biovenn/). d. Proteins in common in mucoid vs serous samples were compared using Venn Diagrams.
MS detected between 1324 and 3194 peptides for serous samples (141 to 246 proteins), with a mean peptide count of 2225. Notably, more peptides and proteins were detected for mucoid samples: between 7184 and 19046 peptides (300 to 357 proteins) with a mean of peptide count of 12786, 5.7-fold higher than the mean for serous samples.
The lists of proteins identified by MS were compared to evaluate their similarities (Fig 3b, C and D). Mucoid samples shared more than 60% of proteins (220 proteins total, Fig 3b) with sample M3 containing the most proteins identified (88 more). Serous samples shared between 40% (S3, Fig 3c) and 70% of proteins (S1) or 98 proteins total. S3 differed from the others with 98 proteins detected specifically in this MEE (40% of its dataset). Most of the proteins of serous samples were detected in mucoid ones (only 26 proteins specific to this group) whereas mucoid MEEs had more proteins specific to its group (236, Fig 3d).
Analysis of the MS dataset
The whole MS dataset revealed 529 proteins total across the samples using a minimum cutoff of 5 peptide counts total for the 6 samples (Supplemental Table S3). Peptide counts were expressed as percentage of the total in order to normalize across samples. The Wilcoxon rank sum test was used to compare groups, with proteins detected in minimum 2 samples per group. Proteins statistically significant are displayed in Table 2. 26 proteins were significantly more abundant in mucoid samples and only 11 in serous ones. In mucoid samples, neutrophil related proteins (myeloperoxidase, neutrophil elastase, lactotransferrin) were found to be more abundant with fold inductions >10. MUC5B was shown to be 18 fold more abundant in mucoid effusions as well as other mucosal innate immune proteins (Catalase, BPI fold-containing family B member 1, Macrophage capping protein, Complement factor B and several immunoglobulins). Serous samples had a higher abundance of complement proteins (C4b-binding protein alpha chain, C5, C6, C7) and blood related proteins (laminogen, apolipoproteins A and C). The top 10 most abundant proteins by peptide counts (Supplemental Table S4) for mucoid and serous samples were also the ones found to be statistically significantly different between the groups (as shown in Table 2), suggesting that the more abundant proteins in each group are the ones that characterize them. Table 3 shows the most abundant (peptide count >50) proteins specific to each group. The presence of several histone 2B monomers and neutrophilic bactericidal proteins (bactericidal permeability-increasing protein and azurocidin) was specific to mucoid samples, and also protein-arginine deiminase type-4 (a marker of NETosis), and cytochrome b-245 heavy chain (a critical component of the membrane-bound oxidase of phagocytes during infection). Proteins specific to serous samples were immunoglobulin light chains (Ig kappa and lambda), extracellular matrix proteins such as extracellular matrix protein 1 and fibulin 1, and typical serum proteins: coagulation factor XII, apolipoprotein L1, hemoglobin subunit gamma-2 and hepatocyte growth factor activator.
Table 2.
Proteins detected by mass spectrometry in significantly different abundance between mucoid and serous MEEs.
| Uniprot ID | description | Serous/Mucoid | Mucoid/Serous |
|---|---|---|---|
| P35579 | Myosin-9 | 0.02 | 48.64 |
| P05164 | Myeloperoxidase | 0.03 | 37.85 |
| P13796 | Plastin-2 | 0.03 | 34.32 |
| P04040 | Catalase | 0.04 | 27.44 |
| P06744 | Glucose-6-phosphate isomerase | 0.04 | 27.33 |
| P07437 | Tubulin beta chain | 0.05 | 19.44 |
| Q9HC84 | Mucin-5B | 0.06 | 18.11 |
| P08246 | Neutrophil elastase | 0.06 | 17.54 |
| P41218 | Myeloid cell nuclear differentiation antigen | 0.06 | 16.25 |
| Q9BQE3 | Tubulin alpha-1C chain | 0.07 | 15.36 |
| Q8TDL5 | BPI fold-containing family B member 1 | 0.07 | 15.23 |
| P40199 | Carcinoembryonic antigen-related cell adhesion molecule 6 | 0.07 | 13.88 |
| P11142 | Heat shock cognate 71 kDa protein | 0.08 | 12.00 |
| P02788 | Lactotransferrin | 0.09 | 11.23 |
| P40121 | Macrophage-capping protein | 0.09 | 10.86 |
| P80188 | Neutrophil gelatinase-associated lipocalin | 0.26 | 3.89 |
| P04004 | Vitronectin | 0.32 | 3.10 |
| P05109 | Protein S100-A8 | 0.35 | 2.88 |
| P07195 | L-lactate dehydrogenase B chain | 0.37 | 2.74 |
| P30041 | Peroxiredoxin-6 | 0.40 | 2.50 |
| P04207 | Ig kappa chain V-III region CLL | 0.40 | 2.48 |
| P06396 | Gelsolin | 0.42 | 2.38 |
| P00751 | Complement factor B | 0.43 | 2.35 |
| P0CG05 | Ig lambda-2 chain C regions | 0.49 | 2.04 |
| P08185 | Corticosteroid-binding globulin | 0.49 | 2.04 |
| P02675 | Fibrinogen beta chain | 0.50 | 2.00 |
| P00747 | Plasminogen | 2.04 | 0.49 |
| P04003 | C4b-binding protein alpha chain | 2.06 | 0.48 |
| P13671 | Complement component C6 | 2.17 | 0.46 |
| P43652 | Afamin | 2.39 | 0.42 |
| P10643 | Complement component C7 | 2.96 | 0.34 |
| P55058 | Phospholipid transfer protein | 3.00 | 0.33 |
| P51884 | Lumican | 3.15 | 0.32 |
| P41222 | Prostaglandin-H2 D-isomerase | 3.67 | 0.27 |
| P01031 | Complement C5 | 4.05 | 0.25 |
| P06727 | Apolipoprotein A-IV | 5.08 | 0.20 |
| P02656 | Apolipoprotein C-III | 5.50 | 0.18 |
Peptide counts of the datasets were normalized to the total peptide count for each sample. These normalized datasets were subjected to a Wilcoxon rank sum test to compare the abundance of proteins between mucoid vs serous groups, with proteins detected in at least 2 samples per group. All the proteins listed are significantly different between mucoid and serous samples with an unadjusted p-value < 0.05. Fold inductions were calculated by dividing the average proportion in the two groups.
Table 3.
Proteins detected by mass spectrometry only present in mucoid or serous samples.
| accession | description | Sum M | Sum S |
|---|---|---|---|
| P68032 | Actin, alpha cardiac muscle 1 | 415 | 0 |
| P68133 | Actin, alpha skeletal muscle | 214 | 0 |
| P34931 | Heat shock 70 kDa protein 1-like | 194 | 0 |
| P17213 | Bactericidal permeability-increasing protein | 187 | 0 |
| P58876 | Histone H2B type 1-D | 177 | 0 |
| P23527 | Histone H2B type 1-O | 168 | 0 |
| P09960 | Leukotriene A-4 hydrolase | 128 | 0 |
| P07237 | Protein disulfide-isomerase | 121 | 0 |
| P20160 | Azurocidin | 100 | 0 |
| P68371 | Tubulin beta-4B chain | 100 | 0 |
| P80723 | Brain acid soluble protein 1 | 92 | 0 |
| P04839 | Cytochrome b-245 heavy chain | 91 | 0 |
| P30101 | Protein disulfide-isomerase A3 | 84 | 0 |
| Q00610 | Clathrin heavy chain 1 | 83 | 0 |
| Q16777 | Histone H2A type 2-C | 83 | 0 |
| P08575 | Receptor-type tyrosine-protein phosphatase C | 75 | 0 |
| P06899 | Histone H2B type 1-J | 70 | 0 |
| P01766 | Ig heavy chain V-III region BRO | 59 | 0 |
| Q9UGM3 | Deleted in malignant brain tumors 1 protein | 59 | 0 |
| P27824 | Calnexin | 58 | 0 |
| P68366 | Tubulin alpha-4A chain | 58 | 0 |
| Q13885 | Tubulin beta-2A chain | 52 | 0 |
| Q9UM07 | Protein-arginine deiminase type-4 | 52 | 0 |
| P16104 | Histone H2A.x | 51 | 0 |
| P27797 | Calreticulin | 51 | 0 |
| P20742 | Pregnancy zone protein | 0 | 60 |
| P23142 | Fibulin-1 | 0 | 45 |
| O14791 | Apolipoprotein L1 | 0 | 43 |
| P01593 | Ig kappa chain V-I region AG | 0 | 26 |
| P06314 | Ig kappa chain V-IV region B17 | 0 | 25 |
| P11277 | Spectrin beta chain, erythrocytic | 0 | 22 |
| P00748 | Coagulation factor XII | 0 | 21 |
| P07225 | Vitamin K-dependent protein S | 0 | 19 |
| P15169 | Carboxypeptidase N catalytic chain | 0 | 16 |
| P69892 | Hemoglobin subunit gamma-2 | 0 | 15 |
| P06313 | Ig kappa chain V-IV region JI | 0 | 13 |
| P01619 | Ig kappa chain V-III region B6 | 0 | 12 |
| P19320 | Vascular cell adhesion protein 1 | 0 | 11 |
| P33151 | Cadherin-5 | 0 | 11 |
| Q04756 | Hepatocyte growth factor activator | 0 | 11 |
| Q6EMK4 | Vasorin | 0 | 11 |
| P05452 | Tetranectin | 0 | 10 |
| P01612 | Ig kappa chain V-I region Mev | 0 | 8 |
| P16157 | Ankyrin-1 | 0 | 8 |
| P16403 | Histone H1.2 | 0 | 8 |
| P43251 | Biotinidase | 0 | 8 |
| Q16610 | Extracellular matrix protein 1 | 0 | 6 |
| P01034 | Cystatin-C | 0 | 5 |
| P06310 | Ig kappa chain V-II region RPMI 6410 | 0 | 5 |
| Q15063 | Periostin | 0 | 5 |
| P06317 | Ig lambda chain V-VI region SUT | 0 | 5 |
Peptide counts of proteins specific of each group were summed. The mucoid samples had 236 more proteins than serous samples but only the proteins with a sum higher than 50 peptide counts are shown. Serous samples had 26 proteins not present in mucoid samples.
IPA was used to find pathways likely to be activated in each group.. For every protein, percentages of normalized peptide counts were averaged and entered in IPA (Table 4). For mucoid samples, the remodeling of epithelial adherent functions was found to be the first pathway activated with an overlap of 26.5%. Other pathways activated were: actin cytoskeleton signaling, regulation of actin based mobility by Rho and RhoGDI signaling, all related to cell mobility. For serous samples, the actin cytoskeleton signaling was also activated but with a lower overlap: 7.8%. Glycolysis was activated as well as the complement system, the remodeling of epithelial junctions and the acute phase response signaling.
Table 4.
Predicted pathways activated by mucoid or serous protein datasets using IPA.
| Mucoid | Serous | ||||
|---|---|---|---|---|---|
| Pathway | P-value | Overlap | Pathway | P-value | Overlap |
| Remodeling of epithelial adherent junction | 9.96E-23 | 26.5% | Actin cytoskeleton signaling | 1.03E-12 | 7.8% |
| Actin cytoskeleton signaling | 2.13E-20 | 11.1% | Glycolysis | 1.01E-11 | 32% |
| Epithelial adherent junction signaling | 1.08E-17 | 13% | Complement system | 3.34E-10 | 21.6% |
| Regulation of actin based mobility by Rho | 8.72E-16 | 16.5% | Remodeling of epithelial junctions | 2.56E-9 | 13.2% |
| RhoGDI signaling | 7.01E-14 | 9.8% | Acute phase response signaling | 7.76E-9 | 7.1% |
The normalized average peptide counts for the mucoid and serous datasets were entered in IPA and a core analysis was performed. The top pathways predicted to be activated are shown with associated p values and overlap (percentage of proteins activated in the canonical pathway of IPA).
To further analyze the difference between these datasets, proteins present only in either mucoid or serous samples were entered in IPA (Table 5). Mucoid samples had 236 specific proteins that were linked to phagosome/leukocyte functions with the activation of the following pathways: phagosome maturation, antigen presentation, leukocyte extravasation, and integrin and mitochondria pathways. The 26 specific serous proteins were related to blood processes: prothrombin activation and coagulation system; and atherosclerosis related signaling and granulocyte adhesion and diapedesis.
Table 5.
Predicted pathways activated by mucoid or serous specific protein datasets using IPA.
| Mucoid Pathway | Serous | ||||
|---|---|---|---|---|---|
| Pathway | P-value | Overlap | Pathway | P-value | Overlap |
| Phagosome maturation | 2.8E-18 | 16.7% | Extrinsic prothrombin activation pathway | 9.69E-5 | 12.5% |
| Antigen presentation pathway | 3.13E-15 | 32.4% | Intrinsic prothrombin activation pathway | 3.25E-4 | 6.9% |
| Leukocyte extravasation pathway | 6.54E-11 | 8.6% | Coagulation system | 4.75E-4 | 5.7% |
| Integrin signaling | 8.3E-11 | 8.5% | Atherosclerosis signaling | 5.8E-3 | 1.6% |
| Mitochodrial dysfuntion | 6.65E-10 | 8.8% | Granulocyte adhesion and diapedesis | 1.15E-2 | 1.1% |
The normalized average peptide counts for the mucoid and serous specific datasets were entered in IPA and a core analysis was performed. The top pathways predicted to be activated are shown with associated p values and overlap (percentage of proteins activated in the canonical pathway of IPA).
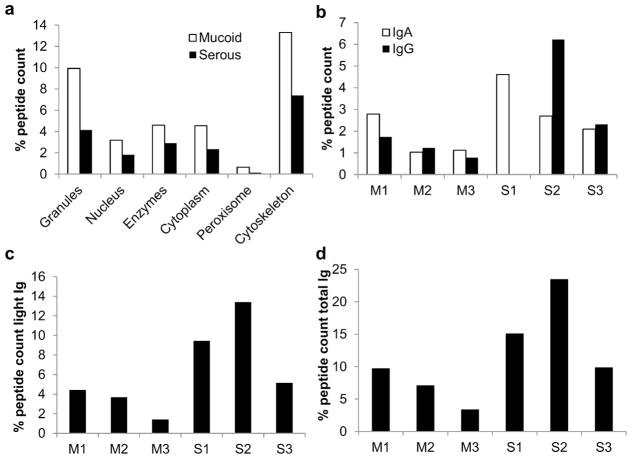
Proteins of several pathways identified by IPA were chosen and compared between groups (Table 6). The mucoid dataset showed a higher proportion of heat shock proteins, Ras/Rho/Rab, annexins, glycolysis proteins and NET proteins. Serous samples had a higher proportion of complement proteins and immunoglobulins. Fig 4 shows a more detailed analysis of NET proteins and immunoglobulins. NET proteins categorized in 6 groups were all in higher proportion for mucoid samples (granule, nucleus, enzymes, cytoplasm, peroxidases and cytoskeleton proteins: Fig 4a). Immunoglobulins IgA and IgG are shown in Fig 4b. IgG was not detected in the sample S1 likely that the MS failed to identify it. Samples M2 and M3 had the lower IgA and IgG proportions; and samples M1 and S3 seemed to have similar levels of both. S1 had two times more IgA than the others and the sample S2 had three times more IgG than the others. The same trends were seen when considering the light Ig chains kappa and lambda (Fig 4c) and the total Ig proportion (Fig 4d).
Table 6.
Abundance of proteins classified by categories among mucoid and serous samples.
| Mucoid | Serous | |||
|---|---|---|---|---|
| Peptide count | % peptide count | Peptide count | % peptide count | |
| Complement | 361 | 0.9 | 339 | 5.1 |
| Heat shock proteins | 1158 | 3.0 | 83 | 1.2 |
| Ras Rho Rab | 819 | 2.1 | 67 | 1.0 |
| Annexins | 1412 | 3.7 | 145 | 2.2 |
| Glycolysis | 2902 | 7.6 | 410 | 6.1 |
| Immunoglobulins | 2466 | 6.4 | 1022 | 15.3 |
| NET proteins | 10220 | 26.6 | 1010 | 15.1 |
Proteins from the dataset were categorized by function and the normalized total peptide count was calculated per group.
Fig 4.
Neutrophil extracellular trap (NET) related proteins and immunoglobulins. All peptide counts of NET and immunoglobulin proteins were summed and normalized to the total peptide count. Proportions of these proteins over the total counts are shown in the graphs. a. NET proteins were categorized in granule, nucleus, enzyme, cytoplasm, peroxisome and cytoskeleton proteins. Proportions were averages for mucoid and serous groups. b. Proportions of IgA and IgG. c. Proportion of light immunoglobulin chains. D. Proportion of all the immunoglobulins present in every sample.
16S sequencing of MEE DNA
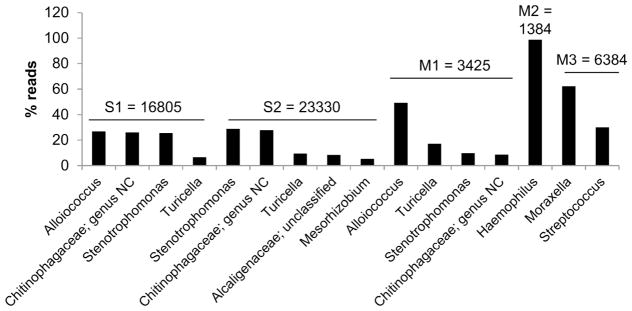
To analyze bacterial genera, we used the 16S ribosomal RNA (rRNA) gene sequencing, a well-established method for studying phylogeny and taxonomy of samples. Fig 5 shows genera with reads higher than 5% except for the sample S3 that failed the quality controls. Total reads for every sample (top of the graph) showed a higher number for serous samples (16805 for S1 and 23330 for S2 vs 3452 for M1, 1384 for M2 and 6384 for M3). S1 and M1 had similar genera: Alloicoccus, Chitinophagaceae; unclassified, Stenotrophomonas and Turacella. S2 also contained Chitinophagaceae; unclassified, Stenotrophomonas and Turacella as well as Alcaligenaceae; unclassified and Mesorhizobium. M2 was characterized by 99% of reads corresponding to Haemophilus and M3 had two genera: Moraxella and Streptococcus. In overall, S1, S2 and M1 seemed to share similarities compared to M2 and M3 that had different genera.
Fig 5.
Main genera detected by 16S rRNA sequencing. DNA was purified using a column based assay and the 16S rRNA sequencing method was used to detect the genera present in MEEs. Genera with a percentage of reads higher than 5% are shown in this graph. For each sample, the total reads is added on the top of the bars. Genus NC refers to a genus that couldn’t be characterized.
Correlations of all the parameters assayed in MEEs
All data measured in the 3 mucoid and 3 serous samples were compared in pair-wise fashion to assess any patterns. Spearman correlations were used to assess overall patterns in the relationships between parameters (Table 7, and Supplemental Table S5). Considering the small sample size, these correlations are interpreted as general tendencies, rather than appropriately powered tests of hypotheses. Of interest, MUC5B parameters (western blot, peptide count and sequence coverage) correlated positively with DNA, IL-1β, IL-8, RANTES, NET proteins and annexins. Complement proteins correlated negatively with DNA and MUC5B parameters. IgA also correlated negatively with IL-13, IL17A, TNF-α and the total peptide count and number of proteins detected by MS.
Table 7.
Protein presence correlation in 39 to 57 MEE samples.
| Mucoid/Serous | Muc5B (no/yes) | dsDNAQubit | Sample number | |
|---|---|---|---|---|
| Muc5B (no/yes) | 0.008 | 57 | ||
| dsDNA- Qubit | 0.005 (S<M) | 0.28 | 55 | |
| IL-10 | 0.58 | 0.86 | 0.573; <0.001 | 39 |
| MDC | 0.81 | 0.93 | −0.001; 0.99 | 39 |
| IL-13 | 0.86 | 0.09 | 0.165; 0.33 | 39 |
| IL-17A | 0.92 | 0.40 | 0.557; <0.001 | 39 |
| IL-1b | 0.69 | 0.31 | 0.672; <0.001 | 39 |
| IL-6 | 0.65 | 0.93 | 0.461; 0.004 | 39 |
| IL-8 | 0.13 | 0.29 | 0.705; <0.001 | 39 |
| RANTES | 0.78 | 0.37 | 0.363; 0.027 | 39 |
| TNF-α | 0.73 | 0.53 | 0.580; <0.001 | 39 |
| VEGF | 0.20 | 0.09 | 0.690; <0.001 | 39 |
| MUC5B pixel | 0.004 (S<M) | 0.17 | 0.303; 0.046 | 46 |
Single values shown are p values from either a Chi square test or Wilcoxon rank sum test. Double values are correlation coefficients and p values from Spearman correlations. S<M indicates that the serous samples had a significantly lower abundance of the parameter studied than the mucoid samples. The values in bold are p values < 0.05.
These correlations show the link between MUC5B protein detection and the viscosity of samples (MUC5B+ samples are likely mucoid, p = 0.008) and a higher abundance of MUC5B (pixel count) in mucoid samples vs serous (p = 0.004). MUC5B pixel count was also linked to the abundance of DNA (Qubit assay) in mucoid samples (p = 0.005). Several cytokines were correlated to MEE DNA abundance: IL-10, IL-17A, IL-1b, IL-8, TNF-α and VEGF. All were statistically significant with p <0.001 and IL-8 had the highest correlation coefficient (0.705), a finding relevant with the neutrophilia in OM and especially the accumulation of NETs during mucoid COM, but not during serous OM. In the case of serous samples containing more immunoglobulins and complement proteins, there was a negative correlation with MUC5B parameters, IL13, IL17A, TNF-α and MS total peptide counts and proteins detected.
Discussion
This study aimed at elucidating key macromolecular differences between mucoid vs serous MEEs from patients with COME receiving tympanostomy tubes. In our experience, most COME samples are mucoid. Typically, the high viscosity of fluids at airway mucosal membranes is mostly due to secretory mucin glycoproteins, primarily MUC5AC and MUC5B (27). In the middle ear, there has been conflicting findings, but mostly due to the fact that mucins proteins are hard to analyze. Many mucins can be found in the middle ear by mRNA analysis but only few by protein analysis. MS and western blotting show MUC5B as the predominant mucin in middle ear effusions (14). In our current analysis, MUC5B accounted for 0.4–0.6% of all peptides identified by MS in serous samples and 1.2–3.8% in mucoid samples, this higher abundance in mucoid samples being expected. Two peptides mapped to MUC18 and MUC4 were found in serous samples, but at levels considered negligible (<5 peptides detected). Higher peptide count and sequence coverage were expected in mucoid samples, especially considering their strong signal by western blot.
The viscosity of fluids is also dependent on the dynamics of mucins binding to each other and to other glycoproteins (25), underscoring the importance of other proteins. Of interest, this study identified lysozyme, the polymeric immunoglobulin receptor, LPLUNC1, DMTB1 and other proteins to be part of the mucin interactome, presumably important for the physical properties of the airway mucus. Extracellular DNA also impacts the viscosity of fluids and it is noteworthy that our samples contained a relatively high amount of DNA, especially in mucoid MEEs, with 27x more extracellular DNA compared to serous MEEs. But despite the higher abundance of NET markers in mucoid samples, 16S sequencing showed 5.4X more bacterial rRNA gene reads in serous samples compared to mucoid ones. Yet, notably, the most common otopathogens were observed in 2 of the 3 mucoid samples. The presence of these bacteria within the middle ear are likely to be involved in the promotion of mucin gene expression and over-secretion as suggested by published IN VITRO data (28–31).
The healthy middle ear epithelium is a single layer of cubical cells with a limited ability to produce mucins in the healthy state. Only the part of the epithelium close to the Eustachian tube shows characteristics of a mucocilliary epithelium to clear the middle ear (32). Patients with COM, either CSOM or COME, have a remodeled epithelium pseudostratified with ciliated cells and goblet cells, as well as the presence of mucus glands (19–21, 33–34). A remodeling of the epithelium is then needed for the production of mucoid MEEs, which can potentially indicate that patients with serous effusions would have less remodeling than patients with mucoid samples (suggested in (15)).
To better understand the pathogenesis of COM we used IPA. IPA found ‘epithelial remodeling’ as well as ‘immune cell mobility’ pathways to be predictably activated for mucoid samples. For serous samples, we found earlier response pathways, such as the ‘complement pathway’, but we also found the ‘epithelial remodeling’ pathway, albeit with a lower p value than in mucoid effusions. Proteins unique to mucoid samples were linked to leukocyte activity, while proteins specific to serous samples were related to blood pathways. It seems that the presence of MUC5B and NETs are related to each other, and both are more abundant in mucoid samples. Moreover, IL-8, IL1-β and RANTES, all MUC5B inductors, are also characteristic of mucoid samples (28, 35–37). Although it is possible that serous samples represent a distinct type of immune response, ,it is difficult on the basis of our small sample of patients to conclude on distinct profiles being representative of a continuum in the disease vs. distinct disease inflammatory entities. Based on published data demonstrating that mucoid effusions are more common in younger patients receiving tympanostomy, while serous effusions tend to be more common in older patients (38), we hypothesize that indeed these effusions represent distinct inflammatory responses. It is becoming increasingly clear that distinct mucin subtypes such as MUC5B and MUC5AC each play a dichotomous role in terms of their innate immune response, with MUC5B being required for chronic upper airway defense homeostasis (39),. Moreover, neutrophil extracellular trap formation often results in mucoid middle ear effusions, not in serous effusions as seen in our data. Finally, we posit that the effusion type is dictated by distinct inflammatory response middle ear pathophysiology and not part of a continuum from serous to chronic. On the basis of our results, it would appear that a majority of mucoid MEEs are a result of neutrophilic responses, culminating in the formation of NETs along with mucin MUC5B, these representing a direct, mostly cell mediated immune response to middle ear pathogens. Immune response resulting in serous effusions could reflect a preponderance of humoral immunity- as characterized by higher antibody levels, complement, and serum proteins, these representing either an allergic or an earlier inflammatory middle ear response.
In comparison with our study, two others investigated the composition difference of mucoid versus serous MEE looking at fewer parameters. Chung et al. (22) found more mucins, lysozyme, IgA and IL-8 in mucoid samples (n=27) compared to serous ones (n=18). However the mucin quantification approach employed (periodic acid-Schiff, PAS) was not specific to mucins. Collecting and analyzing 25 mucoid MEEs and 30 serous MEEs, Yabe et al. (40) showed 60% of the proteins of mucoid MEEs were mucins but not for serous samples. In contradistinction to our findings, their serous samples showed less IgA, IgG and lysozyme than mucoid ones.
Despite all the parameters analyzed, our study is limited by the number of samples used for proteomics (3 for each group). Notably, all inferences on the basis of our results are speculative and based on the descriptive nature of our data. Findings are correlational and not necessarily causal. However, the nature of these differences in the composition of serous and mucoid effusions do point to distinct immune responses reflected by the nature of their macromolecular components.
Conclusion
Mucoid and serous MEEs have a different composition and biological signatures. All together, the IPA analysis and the correlations performed showed that mucoid samples have a neutrophilic signature associated with MUC5B presence and extracellular DNA, confirming the implication of NETs in COME. Serous samples contained a higher amount of early innate immunity markers (complement and immunoglobulin proteins) and serum proteins. In conclusion, we hypothesize that mucoid samples are characterized by a preponderance of MUC5B, associated with an accumulation of NETs. Serous samples have an early immune response signature with a composition showing a potential transudation of serum proteins in the middle ear (MUC5B less abundant). Further studies are needed to understand why patients with COME demonstrate such distinct biologic signatures.
Supplementary Material
Acknowledgments
We thank Dr Mary C. Rose and Dr Colberg-Poley for their advice all along this project.
Footnotes
FINANCIAL DISCLOSURE: This work was supported by R01DC012377 from the NIDCD to dp
CONFLICT OF INTEREST: None to report.
References
- 1.Cherry DK, Woodwell DA. National Ambulatory Medical Care Survey: 2000 summary. Adv Data. 2002:1–32. [PubMed] [Google Scholar]
- 2.Grubb MS, Spaugh DC. Treatment failure, recurrence, and antibiotic prescription rates for different acute otitis media treatment methods. Clin Pediatr (Phila) 2010;49:970–975. doi: 10.1177/0009922810370363. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld RM, Casselbrant ML, Hannley MT. Implications of the AHRQ evidence report on acute otitis media. Otolaryngol Head Neck Surg. 2001;125:440–448. doi: 10.1067/mhn.2001.119326. discussion 439. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed S, Shapiro NL, Bhattacharyya N. Incremental health care utilization and costs for acute otitis media in children. Laryngoscope. 2014;124:301–305. doi: 10.1002/lary.24190. [DOI] [PubMed] [Google Scholar]
- 5.Brugnara L, Murillo S, Novials A, et al. Low Physical Activity and Its Association with Diabetes and Other Cardiovascular Risk Factors: A Nationwide, Population-Based Study. PLoS One. 2016;11:e0160959. doi: 10.1371/journal.pone.0160959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leidinger P, Hart M, Backes C, et al. Differential blood-based diagnosis between benign prostatic hyperplasia and prostate cancer: miRNA as source for biomarkers independent of PSA level, Gleason score, or TNM status. Tumour Biol. 2016;37:10177–10185. doi: 10.1007/s13277-016-4883-7. [DOI] [PubMed] [Google Scholar]
- 7.Lim DJ, Birck H. Ultrastructural pathology of the middle ear mucosa in serous otitis media. Ann Otol Rhinol Laryngol. 1971;80:838–853. doi: 10.1177/000348947108000611. [DOI] [PubMed] [Google Scholar]
- 8.Tos M, Bak-Pedersen K. Goblet cell population in the pathological middle ear and eustachian tube of children and adults. Ann Otol Rhinol Laryngol. 1977;86:209–218. doi: 10.1177/000348947708600212. [DOI] [PubMed] [Google Scholar]
- 9.Ryan AF, Jung TT, Juhn SK, et al. Recent advances in otitis media. 4A. Molecular biology. Ann Otol Rhinol Laryngol Suppl. 2005;194:42–49. [PubMed] [Google Scholar]
- 10.Hurst DS, Venge P. The impact of atopy on neutrophil activity in middle ear effusion from children and adults with chronic otitis media. Arch Otolaryngol Head Neck Surg. 2002;128:561–566. doi: 10.1001/archotol.128.5.561. [DOI] [PubMed] [Google Scholar]
- 11.Val S, Poley M, Brown K, et al. Proteomic Characterization of Middle Ear Fluid Confirms Neutrophil Extracellular Traps as a Predominant Innate Immune Response in Chronic Otitis Media. PLoS One. 2016;11:e0152865. doi: 10.1371/journal.pone.0152865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King LBPB, Perez AC, Reimche JL, et al. 2013 Observation of Viable Nontypeable Haemophilus Influenzae Bacteria within Neutrophil Extracellular Traps in Clinical Samples from Chronic Otitis Media. Otolaryngology. :3. [Google Scholar]
- 13.Thornton RB, Wiertsema SP, Kirkham LA, et al. Neutrophil extracellular traps and bacterial biofilms in middle ear effusion of children with recurrent acute otitis media--a potential treatment target. PLoS One. 2013;8:e53837. doi: 10.1371/journal.pone.0053837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preciado D, Goyal S, Rahimi M, et al. MUC5B Is the Predominant Mucin Glycoprotein in Chronic Otitis Media Fluid. Pediatr Res. 2010;68:231–236. doi: 10.1203/PDR.0b013e3181eb2ecc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Val S. Basic Science Concepts in Otitis Media Pathophysiology and Immunity: Role of Mucins and Inflammation. In: DP, editor. Otitis Media: State of the Art Concepts and Treatment. Springer; 2015. pp. 53–79. [Google Scholar]
- 16.Kubba H, Pearson JP, Birchall JP. The aetiology of otitis media with effusion: a review. Clin Otolaryngol Allied Sci. 2000;25:181–194. doi: 10.1046/j.1365-2273.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 17.Dodson KM, Cohen RS, Rubin BK. Middle ear fluid characteristics in pediatric otitis media with effusion. Int J Pediatr Otorhinolaryngol. 2012;76:1806–1809. doi: 10.1016/j.ijporl.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Matkovic S, Vojvodic D, Baljosevic I. Cytokine levels in groups of patients with different duration of chronic secretory otitis. Eur Arch Otorhinolaryngol. 2007;264:1283–1287. doi: 10.1007/s00405-007-0373-2. [DOI] [PubMed] [Google Scholar]
- 19.Kawano H, Paparella MM, Ho SB, et al. Identification of MUC5B mucin gene in human middle ear with chronic otitis media. Laryngoscope. 2000;110:668–673. doi: 10.1097/00005537-200004000-00024. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Tsuboi Y, Rimell F, et al. Expression of mucins in mucoid otitis media. J Assoc Res Otolaryngol. 2003;4:384–393. doi: 10.1007/s10162-002-3023-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Tsuprun V, Kawano H, et al. Characterization of mucins in human middle ear and Eustachian tube. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1157–1167. doi: 10.1152/ajplung.2001.280.6.L1157. [DOI] [PubMed] [Google Scholar]
- 22.Chung MH, Choi JY, Lee WS, et al. Compositional difference in middle ear effusion: mucous versus serous. Laryngoscope. 2002;112:152–155. doi: 10.1097/00005537-200201000-00027. [DOI] [PubMed] [Google Scholar]
- 23.Elsheikh MN, Mahfouz ME. Up-regulation of MUC5AC and MUC5B mucin genes in nasopharyngeal respiratory mucosa and selective up-regulation of MUC5B in middle ear in pediatric otitis media with effusion. Laryngoscope. 2006;116:365–369. doi: 10.1097/01.MLG.0000195290.71090.A1. [DOI] [PubMed] [Google Scholar]
- 24.Ubell ML, Kerschner JE, Wackym PA, et al. MUC2 expression in human middle ear epithelium of patients with otitis media. Arch Otolaryngol Head Neck Surg. 2008;134:39–44. doi: 10.1001/archoto.2007.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radicioni G, Cao R, Carpenter J, et al. The innate immune properties of airway mucosal surfaces are regulated by dynamic interactions between mucins and interacting proteins: the mucin interactome. Mucosal Immunol. 2016;9:1442–1454. doi: 10.1038/mi.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krueger A, Val S, Perez-Losada M, et al. Relationship of the Middle Ear Effusion Microbiome to Secretory Mucin Production in Pediatric Patients With Chronic Otitis Media. Pediatr Infect Dis J. 2017;36:635–640. doi: 10.1097/INF.0000000000001493. [DOI] [PubMed] [Google Scholar]
- 27.Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev. 2006;86:245–278. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- 28.Val S, Kwon HJ, Rose MC, et al. Middle Ear Response of Muc5ac and Muc5b Mucins to Nontypeable Haemophilus influenzae. JAMA Otolaryngol Head Neck Surg. 2015;141:997–1005. doi: 10.1001/jamaoto.2015.2338. [DOI] [PubMed] [Google Scholar]
- 29.Chen R, Lim JH, Jono H, et al. Nontypeable Haemophilus influenzae lipoprotein P6 induces MUC5AC mucin transcription via TLR2-TAK1-dependent p38 MAPK-AP1 and IKKbeta-IkappaBalpha-NF-kappaB signaling pathways. Biochem Biophys Res Commun. 2004;324:1087–1094. doi: 10.1016/j.bbrc.2004.09.157. [DOI] [PubMed] [Google Scholar]
- 30.Kim YT, Jung HH, Ko TO, et al. Up-regulation of MUC5AC mRNA expression in endotoxin-induced otitis media. Acta Otolaryngol. 2001;121:364–370. doi: 10.1080/000164801300102798. [DOI] [PubMed] [Google Scholar]
- 31.Song KS, Lee WJ, Chung KC, et al. Interleukin-1 beta and tumor necrosis factor-alpha induce MUC5AC overexpression through a mechanism involving ERK/p38 mitogen-activated protein kinases-MSK1-CREB activation in human airway epithelial cells. J Biol Chem. 2003;278:23243–23250. doi: 10.1074/jbc.M300096200. [DOI] [PubMed] [Google Scholar]
- 32.Sade J. Pathology and pathogenesis of serous otitis media. Arch Otolaryngol. 1966;84:297–305. doi: 10.1001/archotol.1966.00760030299007. [DOI] [PubMed] [Google Scholar]
- 33.Ol'shanskaia Iu V, Domracheva EV, Udovichenko AI, et al. Chromosomal aberrations in myelodysplastic syndrome. Ter Arkh. 2005;77:27–33. [PubMed] [Google Scholar]
- 34.Tos M, Caye-Thomasen P. Mucous glands in the middle ear - what is known and what is not. ORL J Otorhinolaryngol Relat Spec. 2002;64:86–94. doi: 10.1159/000057786. [DOI] [PubMed] [Google Scholar]
- 35.Fujisawa T, Chang MM, Velichko S, et al. NF-kappaB mediates IL-1beta- and IL-17A-induced MUC5B expression in airway epithelial cells. Am J Respir Cell Mol Biol. 2011;45:246–252. doi: 10.1165/rcmb.2009-0313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bautista MV, Chen Y, Ivanova VS, et al. IL-8 regulates mucin gene expression at the posttranscriptional level in lung epithelial cells. J Immunol. 2009;183:2159–2166. doi: 10.4049/jimmunol.0803022. [DOI] [PubMed] [Google Scholar]
- 37.Schousboe LP, Rasmussen LM, Ovesen T. Induction of mucin and adhesion molecules in middle ear mucosa. Acta Otolaryngol. 2001;121:596–601. doi: 10.1080/000164801316878881. [DOI] [PubMed] [Google Scholar]
- 38.Duah V, Huang Z, Val S, et al. Younger patients with COME are more likely to have mucoid middle ear fluid containing mucin MUC5B. Int J Pediatr Otorhinolaryngol. 2016;90:133–137. doi: 10.1016/j.ijporl.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy MG, Livraghi-Butrico A, Fletcher AA, et al. Muc5b is required for airway defence. Nature. 2014;505:412–416. doi: 10.1038/nature12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yabe R, Higo R, Sugita K, et al. Gel chromatographic characterization of proteins in mucous and serous middle ear effusions of patients with otitis media in comparison to serum proteins. Eur Arch Otorhinolaryngol. 2008;265:293–298. doi: 10.1007/s00405-007-0449-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.