Abstract
Through analysis of the Drosophila ionotropic receptors (IRs), a family of variant ionotropic glutamate receptors, we reveal that most IRs are expressed in peripheral neuron populations in diverse gustatory organs in larvae and adults. We characterise IR56d, which defines two anatomically-distinct neuron classes in the proboscis: one responds to carbonated solutions and fatty acids while the other represents a subset of sugar- and fatty acid-sensing cells. Mutational analysis indicates that IR56d, together with the broadly-expressed co-receptors IR25a and IR76b, is essential for physiological responses to carbonation and fatty acids, but not sugars. We further demonstrate that carbonation and fatty acids both promote IR56d-dependent attraction of flies, but through different behavioural outputs. Our work provides a toolkit for investigating taste functions of IRs, defines a subset of these receptors required for carbonation sensing, and illustrates how the gustatory system uses combinatorial expression of sensory molecules in distinct neurons to coordinate behaviour.
Subject terms: Genetics, Neuroscience
Little is known about the role of variant ionotropic glutamate receptors (IRs) in insect taste. Here the authors characterise the expression pattern of IRs in the Drosophila gustatory system and highlight the role of one receptor, IR56d, in the detection of carbonation
Introduction
Classic models of gustatory perception in mammals highlight the existence of a small number of taste classes signalling nutritive content (e.g. sugars and amino acids) or toxicity (e.g. bitter) that determine—through activation of hard-wired neural circuits—behavioural acceptance or rejection of food1,2. Different classes of tastants are recognised by discrete sensory channels that express distinct, and relatively small, receptor families. For example, detection of all sugars depends upon a single heterodimeric G protein-coupled receptor (GPCR) complex, T1R2/T1R3, while bitter cells—which detect an enormous diversity of noxious compounds—co-express a few dozen GPCRs of the T2R family1,2.
Such models have been pervasive in interpreting how gustatory perception occurs in other animals, including insects, where analogous segregated sensory pathways for sweet and bitter compounds have been defined3–6. However, in contrast to mammals, where taste—mediated by lingual taste buds—informs only feeding decisions, insect gustation occurs in multiple sensory appendages, including the proboscis, legs, wings and sexual organs, and controls diverse behaviours, such as foraging, feeding, sexual/social recognition and oviposition3–6. In addition to stereotyped appetitive and aversive feeding responses to sweet and bitter compounds, respectively, insects display behavioural reactions to many other types of chemicals, including salt7, water8, carbonation (i.e. aqueous CO2)9, organic and inorganic acids10,11, and pheromonal cuticular hydrocarbons12.
The wide-ranging roles of the insect gustatory system are reflected in the molecular receptors that mediate peripheral sensory detection. The best-characterised taste receptor repertoire is the Gustatory Receptor (GR) family, which are a divergent set of presumed heptahelical ion channels that function in the detection of sugars, bitter compounds and certain sex pheromones3,13. A second large repertoire of receptors implicated in insect gustation is the Ionotropic Receptor (IR) family, which are ligand-gated ion channels that have derived from synaptic ionotropic glutamate receptors (iGluRs)14–17. Unlike iGluRs, IRs display enormous diversification both in the size of the repertoire across insects (ranging from tens to several hundreds15,16,18), and in their protein sequences (with as little as 10% amino acid identity between pairs of receptors). IRs are best-characterised in the vinegar fly, Drosophila melanogaster, which possesses 60 intact Ir genes. Of these, the most thoroughly understood are the 17 receptors expressed in the adult antenna. Thirteen of these are expressed in discrete populations of sensory neurons, and function as olfactory receptors for volatile acids, aldehydes and amines16,19,20 or in humidity detection21–24. The remaining four (IR8a, IR25a, IR76b and IR93a) are expressed in multiple, distinct neuron populations and function, in various combinations, as co-receptors with the selectively-expressed tuning IRs21,22,25.
By contrast, little is known about the sensory functions of the remaining, large majority of non-antennal IRs. Previous analyses described the expression of transgenic reporters for subsets of these receptors in small groups of gustatory sensory neurons (GSNs) in several different contact chemosensory structures15,26–28. While these observations strongly implicate these genes as having gustatory functions, the evidence linking specific taste ligands to particular receptors, neurons and behaviours remains sparse. For example, IR52c and IR52d are expressed in sexually-dimorphic populations of leg neurons and implicated in male courtship behaviours26, although their ligands are unknown. Reporters for IR60b, IR94f and IR94h are co-expressed in pharyngeal GSNs that respond to sucrose, which may limit overfeeding29 or monitor the state of externally digested food30. IR62a is essential for behavioural avoidance of high Ca2+ concentrations, but the precise neuronal expression of this receptor is unclear31. As in the olfactory system, these selectively-expressed IRs are likely to function with the IR25a and/or IR76b co-receptors, which are broadly-expressed in contact chemosensory organs, and required for detection of multiple types of tastants, including polyamines32, inorganic, carboxylic and amino acids28,33–35, and Ca2+31.
Here we describe a set of transgenic reporters for the entire Ir repertoire. We use these to survey the expression of this receptor family in both larval and adult stages. Using this molecular map, we identify IR56d as a selectively-expressed receptor that acts with IR25a and IR76b to mediate physiological and attractive behavioural responses to carbonation, a previously orphan taste class9. Furthermore, we extend recent studies33,36,37 to show that IR56d is also required in sugar-sensing GR neurons to mediate distinct behavioural responses to fatty acids.
Results
A toolkit of transgenic reporters for IRs
We generated transgenic reporters for all non-antennal IRs, comprising 5’ genomic regions of individual Ir loci placed upstream of Gal4 (Methods and Supplementary Table 1). Although the location of relevant gene regulatory sequences is unknown, this strategy has yielded faithful reporters of endogenous expression patterns for essentially all antennal Irs14,20,21,38,39. These constructs were integrated into identical sites in the genome to avoid positional effects on transgene expression. Such reporters of receptor expression provide greater sensitivity and spatial resolution than is permitted by RNA fluorescent in situ hybridization (FISH), which is inadequate to reliably detect Ir transcripts outside the antenna14. Moreover, when used to drive the membrane-targeted mCD8:GFP effector, they allow tracing of the innervation of labelled neurons in the brain and ventral nerve cord.
IR neuronal expression and projections in larvae and adults
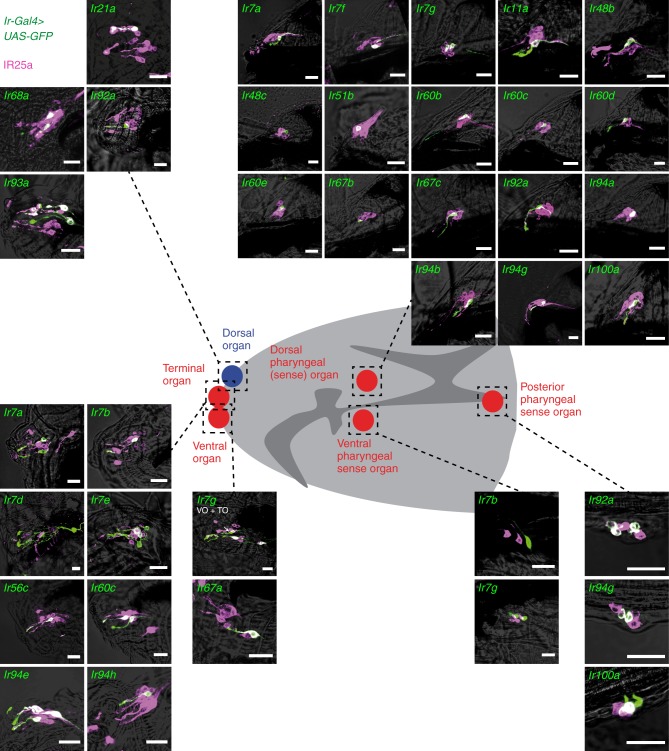
We first analysed Ir-Gal4 driven mCD8:GFP expression in third instar larvae (Figs. 1, 2 and Supplementary Fig. 1). In this analysis, we also included Ir-Gal4 lines that are expressed in the adult antennae20, and incorporated our previous observations on a small subset of the non-antennal IR reporters15,28. The larva contains a bilaterally-symmetric olfactory organ (dorsal organ) and several distinct gustatory organs located on the surface of the head and the internal lining of the pharynx (Figs. 1, 2)40. As described previously27,28, the drivers for the co-receptors IR25a and IR76b (but not IR8a) are broadly expressed in all of these chemosensory organs (Figs. 1, 2). Expression of Gal4 drivers for only four other antennal IRs was detected in the dorsal organ: IR21a and IR93a, which act (with IR25a) in cool temperature-sensing21,41, IR68a, which functions (with IR25a and IR93a) in moist air sensing22,24 and IR92a, which mediates olfactory sensitivity to ammonia14,42. These observations suggest that the larval dorsal organ, like the adult antenna, has olfactory, thermosensory and hygrosensory roles.
Fig. 1.
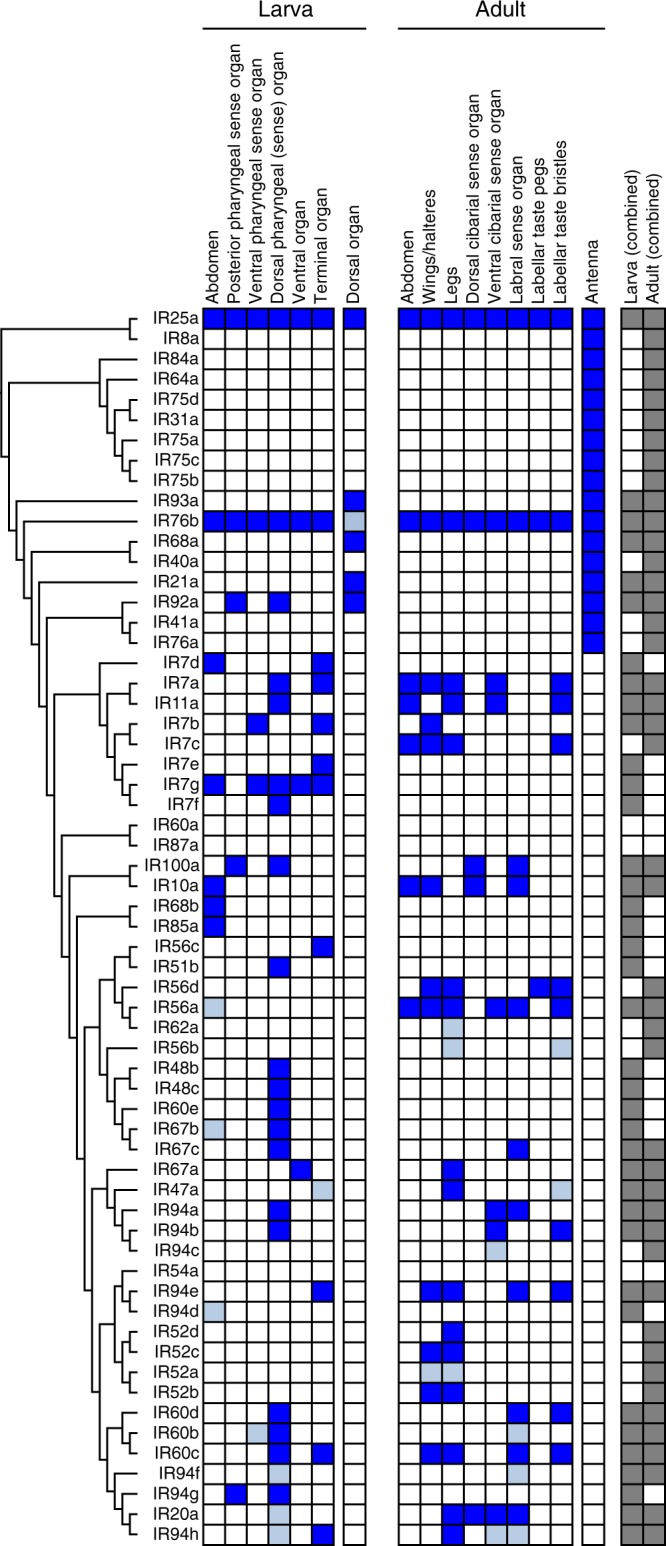
Ir-Gal4 expression patterns and IR phylogeny. Summary of the expression (indicated by dark blue shading) of Gal4 drivers for the entire D. melanogaster IR repertoire in peripheral chemosensory organs in third instar larvae and adult flies (see also Figs 2 and 3). Some lines, in particular antennal-expressed drivers, were previously described15,20–22,25,28,39,41,74,75. Light blue shading indicates additional expression reported for independently-generated Ir-Gal4 drivers26,27. We did not distinguish expression in the dorsal pharyngeal organ and dorsal pharyngeal sense organ. The tree on the left shows a cladogram representing the phylogenetic relationships between D. melanogaster IRs. Protein sequences were aligned with MUSCLE, and the tree was made with RaxML under the WAG model of substitution, with 1000 bootstrap replicates. The columns on the right highlight drivers exhibiting common or stage-specific expression in larvae and adults
Fig. 2.
Expression of Ir-Gal4 drivers in the peripheral nervous system of larval Drosophila. Immunofluorescence with anti-GFP (green) and anti-IR25a (magenta), overlaid on bright-field images, on whole-mount tissue of third instar larvae, revealing the expression of Ir-Gal4 lines in different chemosensory organs (as schematised in the cartoon of the larval head in the centre). Ir7g-Gal4 is expressed in both terminal organ (TO) and ventral organ (VO) neurons; the VO neuron is indicated with an arrow. Images for Ir7b, Ir7e, Ir7g, Ir56c, Ir60c and Ir94e drivers are adapted from28. Genotypes are of the form: w;UAS-mCD8:GFP;IrX-Gal4. Scale bars: 20 μm
Most reporters (27/44) of the remaining non-antennal IR repertoire are detected in bilaterally-symmetric populations of ∼1–3 neurons in one or more larval gustatory organs, including head sensory neurons in the terminal and ventral organs, and internal neurons in the dorsal, ventral and posterior pharyngeal sense organs (Figs 1 and 2). Commensurate with these different peripheral expression patterns, the labelled neurons display diverse projection patterns in the primary gustatory centre, the subesophageal zone (SEZ) (Supplementary Fig. 1). Several reporters, for IR7d, IR7g, IR10a, IR68b and IR85a, are also detected in neurons in each segment of the abdomen, which project to the ventral nerve cord (VNC) (Fig. 1 and Supplementary Fig. 1).
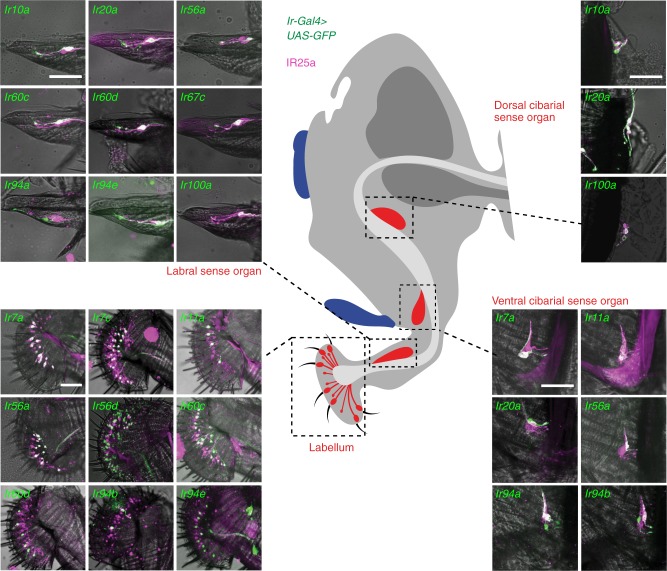
In adults, analysis of the new Ir-Gal4 drivers did not identify any additional antennal-expressed IRs (Fig. 1). However, 21 reporters were detected within one or more populations of sensory neurons in external taste organs, including the taste bristles that project from the surface of the labellum, the labellar taste pegs, and the pharyngeal taste organs (Fig. 1 and Fig. 3). Furthermore, from examination of the central projections of these neurons to the SEZ and VNC, we surmised their expression in a variety of other taste organs, including the legs, wings, as well as neurons that may originate in the abdomen (Fig. 1 and Supplementary Fig. 2). We noted sexually-dimorphic projection patterns in only two reporters: Ir52c-Gal4 (similar to that previously described26) and Ir94e-Gal4 (Supplementary Fig. 2); the latter driver also displays expression in a few soma within the SEZ (Supplementary Fig. 2).
Fig. 3.
Expression of Ir-Gal4 drivers in the proboscis of adult Drosophila. Immunofluorescence with anti-GFP (green) and anti-IR25a (magenta), overlaid on bright-field images, on whole-mount proboscides revealing the expression of Ir-Gal4 lines in different adult gustatory organs (as schematised in the cartoon in the centre). Genotypes are of the form: w;UAS-mCD8:GFP;IrX-Gal4. Scale bars: 50 μm
Relating receptor phylogeny, expression and life stage
We combined these results with information on additional sites of expression revealed by a distinct set of reporters for a subset of IRs (the IR20a clade26, which were built using 5’ genomic regions of slightly different lengths as well as 3’ sequences) to produce a global picture of Ir expression (Fig. 1). These data were organised by IR phylogeny, to examine the relationship between receptor protein sequences and spatiotemporal expression patterns. For the 44 non-antennal IRs, 32 reporters were expressed in larvae and 27 in adults, of which 17 were common to these life stages. Stage-specific receptors were found throughout the phylogeny (Fig. 1), rather than being confined to a single clade. Of the larval-specific IRs, nothing is currently known about their function; the adult-specific repertoire includes the Ir52a-d clade, some members of which control male mating behaviours26.
In both life stages, drivers for some IRs that are closely-related in sequence (and often—but not always—encoded by tandemly-arrayed genes) are expressed in the same contact chemosensory organ (e.g. IR48b, IR48c, IR60e, IR67b and IR67c). This observation suggests that these more recently duplicated receptor genes retain similar cis-regulatory elements. However, this relationship is not strictly-held, as reporters for other, recently-diverged receptors can have quite different expression patterns (e.g. IR10a and IR100a).
IR56d is expressed in labellar taste peg and bristle neurons
To analyse the gustatory function of the non-antennal IRs, we focussed on IR56d, due to its unique expression: Ir56d-Gal4 is the only reporter—apart from the broadly-expressed Ir25a-Gal4 and Ir76b-Gal4—detected in neurons housed in the taste pegs, a class of short sensory hairs that lie between cuticular grooves (pseudotracheae) on the inner medial surface of the labellum (Fig. 4a, b). The driver is also expressed in neurons innervating taste bristles, which project from the external surface of the labellum (Fig. 4a, b). As we were unable to validate the expression of the Ir56d-Gal4 transgene by RNA FISH, we used CRISPR/Cas9 genome editing to replace the endogenous Ir56d locus with Gal4 to generate an independent driver line (Ir56dGal4) in which all relevant genomic regulatory regions should be present (Supplementary Fig. 3a). When combined with UAS-mCD8:GFP, Ir56dGal4 displayed a highly similar expression pattern to the Ir56d-Gal4 transgene (Supplementary Fig. 3b), indicating that the original promoter reporter faithfully recapitulates endogenous gene expression.
Fig. 4.
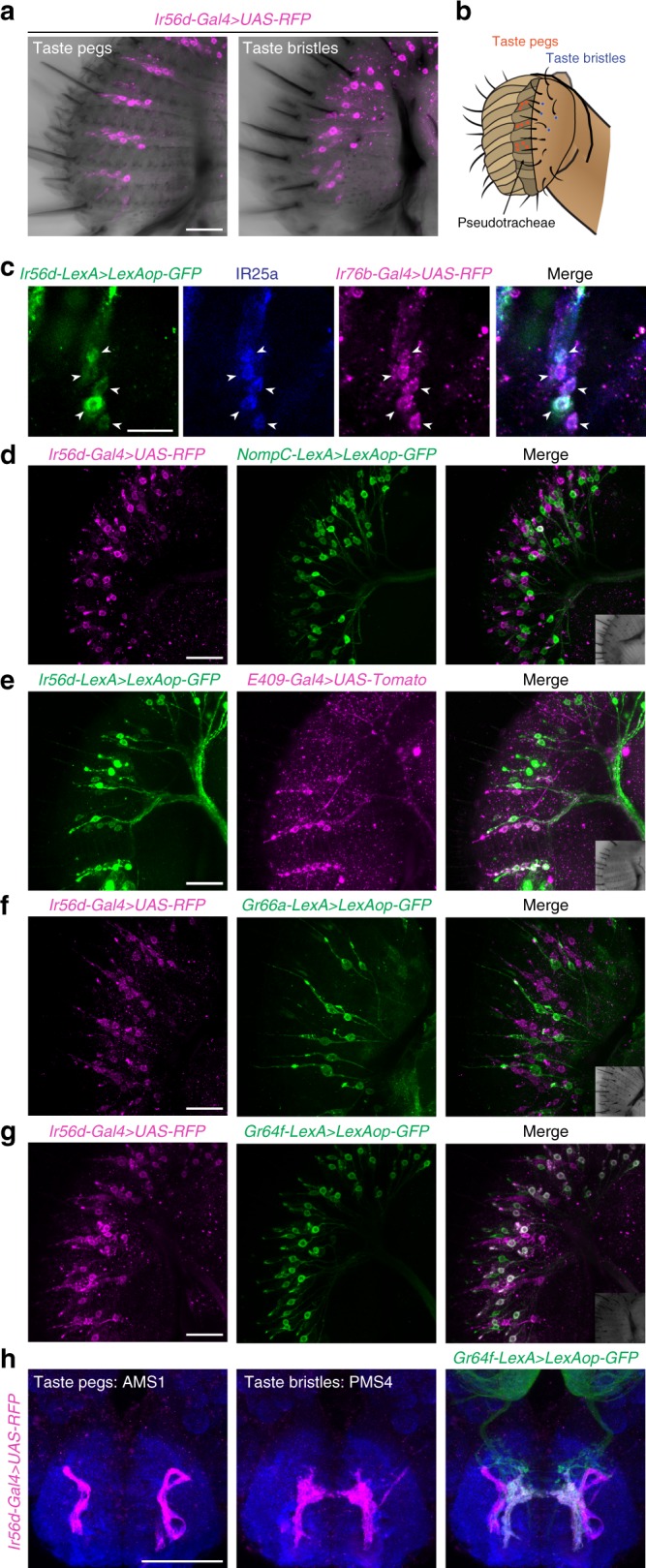
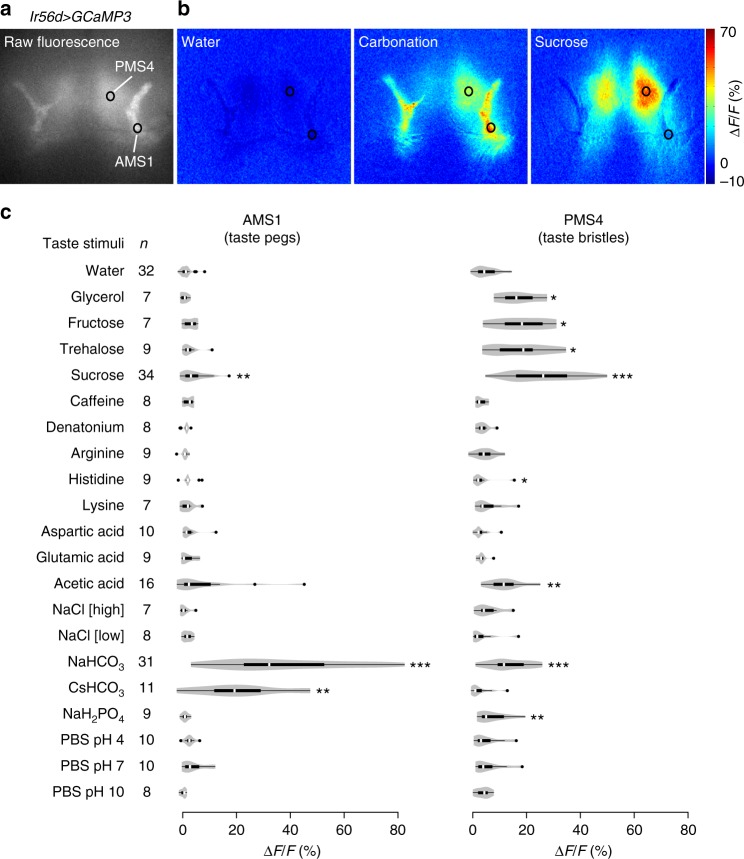
IR56d is expressed in two populations of neurons in the labellum. a Immunofluorescence with anti-RFP (magenta), overlaid on bright-field images, on a whole-mount proboscis of a w;UAS-mCD8:RFP;Ir56d-Gal4 animal. The left image corresponds to the maximal projection of the inner face of one labellar palp, and the right image corresponds to the surface of one labellar palp. Scale bar: 25 μm. b Schematic representing the anatomical location of the taste peg neurons (orange) and taste bristle neurons (blue) in the labellum. c Immunofluorescence with anti-GFP (green), anti-IR25a (blue) and anti-RFP (magenta) on a whole-mount proboscis of a w;LexAop-mCD8:GFP-2A-mCD8:GFP/UAS-mCD8:RFP;Ir56d-LexA/Ir76b-Gal4 animal. The images show a close-up of taste peg neurons (arrowheads) to visualise the co-localisation of the three markers. Scale bar: 25 μm. d Immunofluorescence with anti-RFP (magenta) and anti-GFP (green) on a whole-mount proboscis of a w;LexAop-mCD8:GFP-2A-mCD8:GFP/UAS-mCD8:RFP;NompC-LexA/Ir56d-Gal4 animal. The inset in the merged image shows a bright-field view of the imaged tissue (here and in the following panels). Scale bar: 25 μm. e Immunofluorescence with anti-RFP (magenta) and anti-GFP (green) on a whole-mount proboscis of a w;LexAop-mCD8:GFP-2A-mCD8:GFP/E409-Gal4;Ir56d-LexA/UASCD4:tdTomato animal. Scale bar: 25 μm. f Immunofluorescence with anti-RFP (magenta) and anti-GFP (green) on a whole-mount proboscis of a Gr66a-LexA/+; LexAop-rCD2:GFP/UAS-mCD8:RFP;Ir56d-Gal4/(TM6B or TM2) animal. Scale bar: 25 μm. g Immunofluorescence with anti-RFP (magenta) and anti-GFP (green) on a whole-mount proboscis of a w;LexAop-mCD8:GFP-2A-mCD8:GFP/UAS-mCD8:RFP;Gr64f-LexA/Ir56d-Gal4 animal. Scale bar: 25 μm. h Immunofluorescence with anti-RFP (magenta), anti-GFP (green) and nc82 (blue) on a whole-mount brain of a w;LexAop-mCD8:GFP-2A-mCD8:GFP/UAS-mCD8:RFP;Gr64f-LexA/Ir56d-Gal4 animal. Both left and middle panels show the expression of only the Ir56d-Gal4 driver. The left panel shows the maximal projection of the anterior SEZ; the middle panel shows the maximal projection of the most posterior optical slices of the SEZ. The right panel shows the overlay of the Ir56d-Gal4- and Gr64f-LexA-expressing populations. AMS1 anterior maxillary sensory zone 1, PMS4 posterior maxillary sensory zone 4. Scale bar: 50 μm
To characterise the identity of the IR56d neurons, we combined the Ir56d-Gal4 driver (or an equivalent Ir56d-LexA transgene; see Methods) with reporters for other populations of labellar neurons. We first confirmed that IR56d neurons express IR25a and IR76b (Fig. 4c), suggesting that IR56d may function with one or both of these co-receptors. Morphological studies have shown that taste pegs contain one presumed mechanosensory and one chemosensory neuron43. The mechanosensory neuron can be visualised with a promoter reporter for the NOMPC mechanoreceptor (NompC-LexA)44,45. We observed that NompC-LexA-labelled neurons paired, but did not overlap, with Ir56d-Gal4-positive taste peg neurons (Fig. 4d). By contrast, Ir56d-Gal4-expressing cells in the taste pegs co-localised with those labelled by the E409-Gal4 enhancer trap, which labels at least a subset of the peg chemosensory neurons9 (Fig. 4e). Taste bristles house two to four gustatory neurons, including those tuned to sweet and bitter stimuli, labelled by reporters for Gr64f and Gr66a, respectively3,6. Ir56d-Gal4 taste bristle neurons were completely distinct from Gr66a-positive cells, but overlapped with a subset of the Gr64f-expressing neurons (Fig. 4f, g).
Consistent with the expression in pegs and bristles, Ir56d-Gal4 neurons project to two distinct regions of the SEZ: the anterior maxillary sensory zone 1 (AMS1), and the posterior maxillary sensory zone 4 (PMS4) (Fig. 4h)46. Co-labelling of these neurons with the Gr64f reporter demonstrated that the taste bristle subpopulation innervates PMS4, indicating that the taste peg neurons project to AMS1 (Fig. 4h), consistent with previous observations9,46.
IR56d taste peg neurons are gustatory carbonation sensors
To determine the physiological specificity of IR56d neurons, we expressed the fluorescent calcium indicator GCaMP3 under the control of Ir56d-Gal4 (Fig. 5a), and measured changes in fluorescence in their axon termini in the SEZ upon presentation to the proboscis of a panel of diverse taste stimuli, including sugars, bitter compounds, amino and organic acids, high and low NaCl concentrations, carbonated solutions and buffers of different pH (Fig. 5b, c). We separately quantified GCaMP3 fluorescence changes in the AMS1 and PMS4 projections, reflecting activity of taste peg and taste bristle subpopulations, respectively. AMS1-innervating neurons responded strongly to carbonated solutions (Fig. 5c), but not to other tastants in this panel. These data—together with our co-expression analysis (Fig. 4e)—identify the Ir56d taste peg neurons as the carbonation-sensing cells that were previously recognised by their expression of the E409-Gal4 enhancer trap9.
Fig. 5.
IR56d neurons respond to carbonation and sugars. a Raw fluorescence image of IR56d neuron axons innervating the SEZ in a w;UAS-GCaMP3;Ir56d-Gal4 animal. The black circles indicate the approximate position of the regions-of-interest (ROIs) in the AMS1 and PMS4 used for the quantifications in c. b Colour-coded images of ∆F/F (reflecting the maximal GCaMP3 fluorescence intensity changes; scale bar on the far-right) of IR56d neuron responses in the same animal as in a to the application of distilled water, a carbonated solution or sucrose to the labellum. c Quantification of changes in ∆F/F in the ROIs in AMS1 and PMS4 upon application of the indicated taste stimuli to the labellum of w;UAS-GCaMP3;Ir56d-Gal4 animals. Violin plots (in this and all following figures) show the density of the data points extending from the minimum to the maximum value. Internal boxplots show the median and the interquartile range of the distribution of responses (IQR; first and third quartile). Whiskers represent 1.5x IQR. Black dots represent outliers. n denotes the number of replicates for each stimulus. Concentrations of each of the taste stimuli are listed in Supplementary Table 2. For the statistical analysis the response data for each stimulus were compared with water; only significant differences are shown: *P < 0.05, **P < 0.01, ***P < 0.001 (Wilcoxon rank sum test)
PMS4-innervating neurons displayed a broader response profile, showing the largest GCaMP3 fluorescence changes upon stimulation with sucrose and other sugars, consistent with these neurons representing a subset of the Gr64f-expressing sweet-sensing neurons housed in taste bristles (Fig. 4g). We also detected weaker responses to glycerol, acetic acid, and, somewhat variably, to carbonated solutions (Fig. 5c). These observations indicate that Ir56d subpopulations are both anatomically and physiologically distinct.
Ir56d-Gal4 is additionally expressed in leg neurons (Fig. 1 and Supplementary Fig. 2); consistent with previous observations33, these cells are also labelled with a Gr64f reporter (Supplementary Fig. 4a). By calcium imaging in leg tarsi, we found these cells respond to sucrose but not carbonated solutions (Supplementary Fig. 4b-c). These results suggest that the legs contain only one type of IR56d neuron, which is equivalent to that housed in labellar taste bristles.
IR56d, IR25a and IR76b are required for carbonation sensing
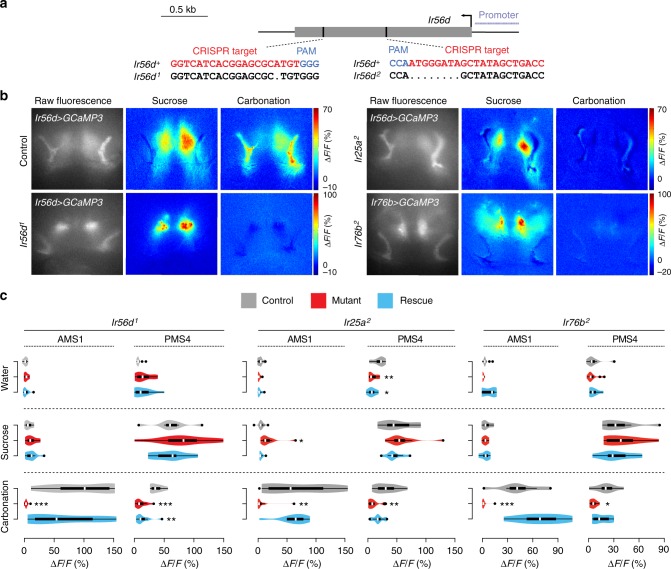
To address the contribution of IR56d to the sensory responses of the neurons in which it is expressed, we used CRISPR/Cas9 genome editing to generate two Ir56d mutant alleles; these contain frame-shift generating deletions predicted to truncate the protein within the presumed ligand-binding domain (Ir56d1) or before the ion channel domain (Ir56d2) (Fig. 6a). We performed calcium imaging in IR56d neurons in Ir56d mutant flies using sucrose and carbonation stimuli, which were the strongest agonists for the taste bristle (PMS4) and taste peg (AMS1) subpopulations, respectively (Fig. 5c). While responses of the mutants to sucrose were unaffected compared to control animals, responses to carbonation were abolished in Ir56d mutants (Fig. 6b, c). The defect in sensitivity to carbonation was restored upon selective expression of a wild-type Ir56d cDNA in these neurons (Fig. 6c).
Fig. 6.
IR56d, IR25a and IR76b are required for sensory responses to carbonation but not sucrose. a Schematic of the Ir56d locus (single exon in grey), showing the position of the CRISPR target and the sequence of the Ir56d mutant alleles in these regions. PAM protospacer adjacent motif. b Raw fluorescence of GCaMP3 expressed in IR56d neurons, and colour-coded images (reflecting the maximal GCaMP3 fluorescence intensity changes; scale bars for each genotype on the right) in control, Ir56d1, Ir25a2 and Ir76b2 mutant flies stimulated with 1 M sucrose and a carbonated solution (as in Fig. 5c). For genotypes, see c. c Quantification of changes in ∆F/F in the ROIs as shown in Fig. 5a, b upon application of the indicated chemicals to the proboscis of the indicated genotypes: IR56d: Control: w;Bl/+; UAS-GCaMP3,Ir56d-Gal4/+ (n = 8); Mutant: w;Ir56d1/Ir56d1; UAS-GCaMP3,Ir56d-Gal4/+ (n = 11); Rescue: w;Ir56d1,UAS-Ir56d/Ir56d1;UAS-GCaMP3,Ir56d-Gal4/+ (n = 10). IR25a: Control: w;Bl/+; UAS-GCaMP3,Ir56d-Gal4/+ (n = 11); Mutant: w;Ir25a2/Ir25a2;UAS-GCaMP3,Ir56d-Gal4/+ (n = 11); Rescue: w;Ir25a2,UAS-Ir25a/Ir25a2; UAS-GCaMP3,Ir56d-Gal4/+ (n = 9). IR76b: Control: w;Ir76b-Gal4/CyO;UAS-GCaMP3/TM6B (n = 8); Mutant: w;Ir76b-Gal4/+; UAS-GCaMP3,Ir76b2/Ir76b2 (n = 10); Rescue: w;Ir76b-Gal4,UAS-Ir76b/+;UAS-GCaMP3,Ir76b2/Ir76b2 (n = 7). We used Ir76b-Gal4 in the rescue experiments of the Ir76b mutant due to constraints of the chromosomal location of the relevant transgenes; although Ir76b-Gal4 is more broadly-expressed than Ir56d-Gal4, the AMS1 and PMS4 projections are still easily recognisable. For the statistical analysis the response data for each stimulus were compared with water; only significant differences are shown: *P < 0.05, **P < 0.01, ***P < 0.001 (Wilcoxon rank sum test with Bonferroni correction for multiple comparisons)
We next tested the contribution of the two other IRs expressed in IR56d neurons, IR25a and IR76b. Mutations in each of these genes produced phenotypes that were very similar to those of Ir56d mutants: sucrose responses in the PMS4 were unaffected, while responses to carbonation were completely lost (Fig. 6b, c). Sensitivity to carbonation could be rescued by expression of wild-type cDNA transgenes in the corresponding mutant background (Fig. 6b, c). Together these data suggest that the carbonation sensor comprises, at least in part, a complex of IR56d with the co-receptors IR25a and IR76b. The persistent sucrose responses in Ir56d taste bristle neurons lacking these IRs are consistent with the well-established role of GRs in sugar sensing in these cells3,6.
Carbonation induces Ir56d-dependent attraction
Previous analysis of flies’ behavioural responses to carbonation indicated that this stimulus mediates taste-acceptance behaviour9. However, the requirement of E409-Gal4-positive labellar neurons was difficult to determine because the E409-Gal4 enhancer trap is expressed in many central neurons in addition to the taste pegs9, limiting its usefulness for neuronal manipulation experiments. With our characterisation of IR56d, we were better positioned to examine more precisely the sensory basis of carbonation-evoked behaviours.
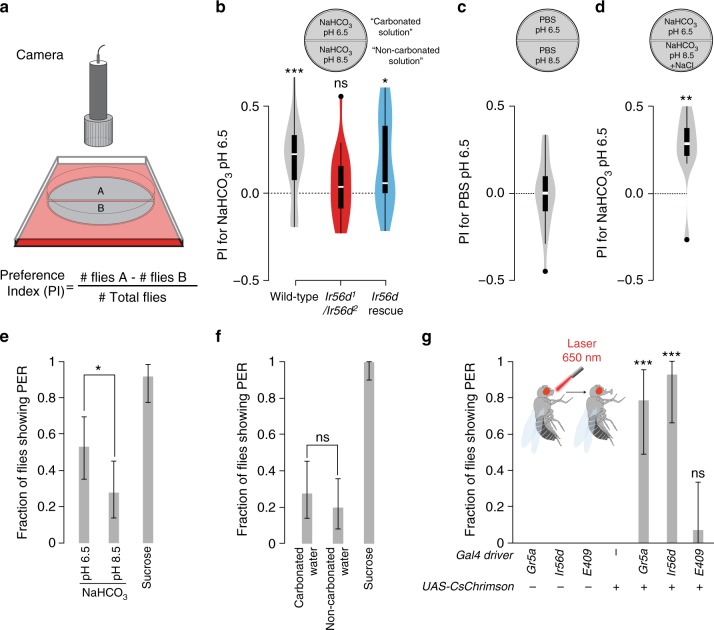
We first established a two-choice assay in which flies could freely explore a circular arena containing separate semicircles of filter paper soaked in carbonated or non-carbonated solutions (100 mM NaHCO3 pH 6.5 and 100 mM NaHCO3 pH 8.5, respectively; these ensure a long-lasting source of carbonation9). After 90 min, we observed the position of flies in the arena and calculated a preference index (Fig. 7a). Wild-type flies showed a clear preference for the carbonated solution (Fig. 7b). This preference was not due to the pH difference of the solutions as flies did not show preference for phosphate buffered saline (PBS) pH 6.5 over PBS pH 8.5 (Fig. 7c); similarly, the slightly different salt concentrations in the carbonated and non-carbonated solutions (see Methods) could not account for the preference observed (Fig. 7d). These observations are consistent with those made using a different positional-preference assay9, confirming that carbonation (a product of microbial fermentation) is a modestly attractive stimulus for Drosophila. Importantly, this preference was completely abolished in Ir56d mutant flies (Fig. 7b) and restored, albeit not to wild-type levels, by expression of Ir56d cDNA in IR56d neurons (Fig. 7b).
Fig. 7.
Carbonation promotes IR56d-dependent feeding behaviours: a Schematic of the two-choice positional preference arena assay. Flies can choose to feed from two substrates comprising stacks of blotting paper soaked in different tastant solutions on opposite sides of a 94 mm diameter Petri dish. Fly position was quantified automatically and used to calculate a Preference Index (PI) as indicated below the scheme. b Two-choice assay showing the preference of the indicated genotypes for a carbonated solution (100 mM NaHCO3 pH 6.5) over a control non-carbonated solution (100 mM NaHCO3 pH 8.5). Genotypes: w1118 (n = 21 arenas; 70–80 flies per arena); Ir56d mutant: w;Ir56d1/Ir56d2 (n = 21 arenas); Rescue: w;Ir56d1,UAS-Ir56d/Ir56d2;Ir56d-Gal4/+ (n = 14 arenas). ns non-significant, *P < 0.05, **P < 0.01, ***P < 0.001 (Wilcoxon signed rank test (H0 = 0)). c Two-choice assay showing the preference of w1118 flies (n = 24 arenas) for a PBS pH 6.5 solution vs. a PBS pH 8.5 solution. d Two-choice assay showing the preference of w1118 flies (n = 10 arenas) for a carbonated solution (100 mM NaHCO3 pH 6.5) over a non-carbonated solution (100 mM NaHCO3 pH 8.5) that was supplemented with NaCl to achieve a final [Na+] of 150 mM. **P < 0.01 (Wilcoxon signed rank test (H0 = 0)). e Fraction of w1118 flies (n = 36) showing the proboscis extension reflex (PER) to the tastants indicated (100 mM NaHCO3 at pH 6.5 or pH 8.5, 100 mM sucrose). Error bars represent the ±95% binomial confidence intervals; *P < 0.05 (Fisher exact test). f Fraction of w1118 flies (n = 36) showing PER to commercial carbonated water, non-carbonated water and 100 mM sucrose. Error bars represent the ±95% binomial confidence intervals; ns non-significant (Fisher exact test). g Fraction of flies of the indicated genotypes (n = 14 for all) showing PER upon optogenetic stimulation using red light. Genotypes (left-to-right along the x-axis): (i) w;Bl/CyO;Gr5a-Gal4/TM6B (ii) w;Bl/CyO;Ir56d-Gal4/TM6B (iii) w;Bl/CyO;E409-Gal4/TM6B (iv) w;UAS-CsChrimson/CyO;TM2/TM6B (v) w;UAS-CsChrimson/CyO;Gr5a-Gal4/TM6B (vi) w;UAS-CsChrimson/CyO;Ir56d-Gal4/TM6B (vii) w;UAS-CsChrimson/CyO;E409-Gal4/TM6B. Error bars represent the ±95% binomial confidence intervals; ns non-significant, ***P < 0.001 (Fisher exact test)
To investigate why flies display positional preference for carbonation, we performed additional behavioural assays. The best-established response of insects to attractive gustatory stimuli is the proboscis extension reflex (PER), which promotes contact of the feeding organ with the substrate. While PER is robustly triggered by sucrose (Fig. 7e), the carbonated solution used in the two-choice assay (100 mM NaHCO3 pH 6.5) triggered a small PER response that was only slightly higher than the control non-carbonated solution (100 mM NaHCO3 pH 8.5) (Fig. 7e). To eliminate any contribution of salt-evoked PER, we also performed PER assays with fresh commercial carbonated and non-carbonated water, which have only trace levels of minerals (Supplementary Table 2). Here, both stimuli induced similarly low levels of PER (Fig. 7f). Finally, we examined whether PER can be triggered by optogenetic activation of taste peg neurons using the red-light sensitive channelrhodopsin CsChrimson (Fig. 7g). In positive control animals, in which CsChrimson was expressed under the control of a Gr5a driver or our Ir56d driver (which is expressed in both taste pegs and sugar-sensing neurons in taste bristles), exposure of the labellum to red light induced, as expected, robust PER (Fig. 7g). By contrast, selective activation of the taste peg neurons (using the E409-Gal4 driver) did not (Fig. 7g). Together these results suggest that the carbonation-sensing taste peg neurons do not activate the PER motor circuit.
During the positional preference assay, flies might also taste the substrate with their legs. However, direct carbonation stimulation of legs does not evoke PER (Supplementary Fig. 4d), which is consistent with the lack of physiological sensitivity of tarsal neurons to this stimulus (Supplementary Fig. 4b-c). These observations argue that the IR56d leg neurons are unlikely to contribute to the behavioural responses to carbonation, and that labellar taste peg neurons are the principal (and potentially only) carbonation sensors in the animal.
Finally, we asked whether carbonation influences food ingestion using Expresso, an automated feeding assay that can measure the number and volume of individual meal-bouts47. When comparing feeding of wild-type flies on carbonated and control solutions, we found no difference in any of the parameters measured (Supplementary Fig. 5a). However, we noted that these stimuli were very poor inducers of feeding, with fewer than half the flies consuming very low volumes of solutions. We reasoned this was due to the lack of a nutritious substance, and repeated the assays in the presence of a low concentration of sucrose (5 mM), which is moderately attractive to Drosophila48. This sugar supplement greatly increased consumption by the flies, but we again did not observe any enhancement of feeding by carbonation (Supplementary Fig. 5b-c). Thus, the attractiveness of carbonation to flies does not appear to be related to an ability to induce increased consumption, at least in this assay and with the tested conditions.
Ir56d-dependent hexanoic acid sensing in taste bristles
In the course of completion of our study, the taste bristle neurons that co-express Ir56d and sweet-sensing Grs in the labellum and legs were found to mediate physiological and behavioural responses to medium chain fatty acids33,36. We confirmed these observations by showing that hexanoic acid activates IR56d neurons, noting that the strongest responses occur in taste peg neurons (Supplementary Fig. 6a-b). Hexanoic acid responses were abolished in Ir56d mutants (Supplementary Fig. 6a-b), suggesting that IR56d functions both in carbonation and fatty acid detection. In contrast to carbonation, however, fatty acids evoke PER, and this behaviour is abolished in Ir56d mutants (Supplementary Fig. 6c). As taste peg neuron activation does not trigger PER (Fig. 7e–g), these observations suggest that hexanoic acid-evoked activity in taste bristles is responsible for this behaviour, as proposed previously36. Consistent with this hypothesis, RNAi of Ir56d specifically in the sweet-sensing Gr neuron subpopulation eliminates fatty acid-induced PER33.
Discussion
This work describes the first family-wide survey of the expression of IRs in Drosophila, revealing remarkable diversity in the neuronal expression patterns of members of this repertoire across all known chemosensory organs in both larvae and adults. These observations reinforce previous conclusions from analyses of subsets of these genes15,26–28 that the non-antennal IRs function to detect a myriad of chemical stimuli to evoke a variety of behavioural responses. Such properties presumably apply to the vast, divergent IR repertoires of other insect species15, for example, the 455 family members in the German cockroach Blatella germanica18, or the 135 IRs in the mosquito Aedes aegypti49. Within Drosophila we did not detect obvious relationships between IR phylogeny and stage- or organ-specific expression patterns. Phylogenetic proximity may therefore be the most indicative of functional relationships between IRs, as is the case for those expressed in the antenna20. If this hypothesis is correct, the expression data presented here suggest that functionally-related clades of receptors act in several types of chemosensory organ.
An important caveat to the transgenic approach we used to reveal expression is the faithfulness of these reporters to the endogenous expression pattern of Ir genes. Although this strategy has been widely (and successfully) used for antennal IRs and other chemosensory receptor families, it is impossible to determine reporter fidelity without a complementary tool (e.g. receptor-specific antibodies or tagging of the endogenous genomic locus). We note discrepancies between the expression of some of our Ir-Gal4 lines and those described previously26; many of these probably reflect differences in the length of regulatory regions used to create these distinct transgenes. Precise comparison of independently-constructed transgenic constructs may in fact be useful in informing the location of enhancer elements directing particular temporal or spatial expression patterns. Moreover, transgenic reporters provide powerful genetic tools for visualisation and manipulation of specific neuronal populations. The reagents generated here should therefore provide a valuable resource for further exploration of the IRs in insect gustation.
Using our atlas, we identified IR56d—together with the broadly-expressed co-receptors IR25a and IR76b—as essential for responses of labellar taste peg neurons to carbonation. Such observations implicate IR56d as the previously unknown tuning receptor for this stimulus9. However, these IRs do not appear to be sufficient for carbonation detection, as their misexpression in other neurons failed to confer sensitivity to carbonated stimuli (Supplementary Fig. 7). This observation suggests that additional molecules or cellular specialisations are required. Such a factor may be rather specific to taste pegs, given the minimal/absent responses of Ir56d-expressing taste bristle/leg neurons to carbonation, but does not appear to be another IR, as we have not identified other IR reporters expressed in this population of cells.
While precise mechanistic insights into carbonation sensing will require the ability to reconstitute IR56d-dependent carbonation responses in heterologous systems, it is interesting to compare how insects and mammals detect this stimulus. The main mammalian gustatory carbonation sensor, the carbonic anhydrase Car450 is an enzyme tethered to the extracellular surface of sour (acid) taste receptor cells in lingual taste buds, where it is thought to catalyse the conversion of aqueous CO2 into hydrogencarbonate (bicarbonate) ions (HCO3−) and protons (H+). The resulting free protons, but not hydrogencarbonate ions, provide a relevant signal for the sour-sensing cells50. By contrast, IR56d neurons are not responsive to low pH, suggesting a different chemical mechanism of carbonation detection. Our observation that IR56d is also essential for sensitivity to hexanoic acid suggests that IR56d could recognise the common carboxyl group of hydrogencarbonate and fatty acid ligands. However, IR56d neurons are not responsive to all organic acids, indicating that this cannot be the only determinant of ligand recognition.
Our characterisation of IR56d neurons extends previous reports33,36,37 to reveal an unexpected complexity in the molecular and neuronal basis by which attractive taste stimuli are encoded. The taste bristle population of IR56d neurons represents a subset of sugar-sensing cells that are also responsive to fatty acids, glycerol and, minimally, to carbonation. Although activation of these neurons promotes PER, we find that carbonation-evoked stimulation is insufficient to trigger this behaviour, which suggests that taste bristles are not a relevant sensory channel for this stimulus. While members of a specific clade of GRs are well-established to mediate responses to sugars and glycerol3,6,37,51, the detection mechanisms of fatty acids appear to be more complex. Earlier work demonstrated an important role of a phospholipase C homologue (encoded by norpA) in labellar fatty acid responses10. More recently, GR64e was implicated as a key transducer of fatty acid-dependent signals, but suggested to act downstream of NorpA, rather than as a direct fatty acid receptor37. By contrast, an independent study of the legs showed that all sugar-sensing Gr genes (including Gr64e) were dispensable for fatty acid detection, and provided evidence instead for an important role of IR25a and IR76b in these responses33. Analysis of our Ir56d mutants indicates an IR-dependent fatty acid-detection mechanism also exists in the labellum; future work will be needed to relate this to the roles of GR64e and NorpA.
The IR56d taste peg population is, by contrast, sensitive to carbonation and fatty acids (but not sugars or glycerol), and these responses can be ascribed to IR56d (a Gr64eLexA reporter is not expressed in taste peg neurons52). Although these neurons mediate taste-acceptance behaviour, they do not appear to promote proboscis extension or food ingestion. Recent work using optogenetic neuronal silencing experiments provided evidence that taste peg neuron activity is important for sustaining, rather than initiating, feeding on yeast, by controlling the number of sips an animal makes after proboscis extension53. These observations are concordant with the internal location of taste pegs on the labellum, as they will not come into contact with food until the proboscis has been extended, and could explain the positional preference for carbonated substrates that we observed. We have attempted to determine whether carbonation can influence sipping behaviour using flyPAD54. Although these experiments did not reveal a statistically-significant effect (Supplementary Fig. 8), interpretation is complicated by the difficulty of providing and maintaining carbonation stimuli in the solid medium used in flyPAD assays. Future development of other approaches to provide this stimulus in feeding assays will be necessary. Nevertheless, our data strengthen the view that carbonation, a non-nutritious microbial fermentation product, regulates—via activation of IR56d taste peg neurons—a distinct motor programme to PER as part of a multicomponent behavioural response.
Methods
Transgene generation
Ir-Gal4 lines were constructed with sequences from the Oregon R strain (OR) using standard methods15,28 (Supplementary Table 1) and inserted into the attP2 landing site55, by normal transformation procedures (Genetic Services, Inc.). Ir56d-LexA was made by subcloning the same genomic sequence as in Ir56d-Gal4 upstream of LexA:VP16-SV4056 in pattB57 and transformation into attP2. UAS-Ir56d was made by PCR amplification of the Ir56d (single-exon) ORF from w1118 genomic DNA, which was T:A cloned into pGEM-T Easy and sequenced, before subcloning into pUASTattB57, and transformation into attP4055. UAS-Ir56dmut contains a deletion of a single nucleotide at position 1010 of the Ir56d ORF; this frameshift mutation is predicted to cause premature translation termination and a non-functional receptor fragment.
Drosophila strains
Drosophila stocks were maintained on a standard corn flour, yeast and agar medium under a 12 h light:12 h dark cycle at 25 °C; different culture conditions for certain behavioural assays are described below. The wild-type strain was w1118. Other mutant and transgenic strains were: Ir25a214, Ir76b27, Ir25a-Gal425, Ir76b-Gal4 (insertions on chromosome 2 or 3)20, Gr5a-Gal458, Gr64f-LexA52, Gr66a-LexA59, Gr66a-Gal448, NompC-LexA44, E409-Gal49, UAS-Ir25a25, UAS-Ir76b7, UAS-GCaMP360, UAS-mCD8:GFP61, UAS-CD4:tdTomato62, UAS-mCD8:RFP63, LexAop-mCD8:GFP-2A-mCD8:GFP56, LexAop-rCD2:GFP56, UAS-CsChrimson64.
CRISPR/Cas9-based genome editing
Ir56d1 and Ir56d2: we identified two CRISPR target sequences within the Ir56d locus using ZiFiT (zifit.partners.org/ZiFiT/)65 that are both unique within the genome and which contain an adjacent 3’ protospacer adjacent motif (PAM) (Fig. 6a). We generated DNA templates for synthetic guide RNA synthesis by PCR using standard procedures66 using the following oligonucleotides: CRISPRsgR with either CRISPRsgF-Ir56d1 or CRISPRsgF-Ir56d2 (Supplementary Table 3). The template was transcribed in vitro with T7 polymerase, RNA was microinjected into vas-Cas9 flies (expressing Cas9 specifically in the germline67) and mutations in the target sequence region screened by Genetic Services, Inc. After establishment of homozygous mutant lines, mutations were reconfirmed by Sanger sequencing.
Ir56dGal4: the Gal4 knock-in allele was generated via CRISPR/Cas9 mediated homologous recombination. Two sgRNAs targeting the Ir56d locus were cloned into pCFD568 by Gibson Assembly to generate pCFD5-Ir56dsgRNAs. Homology arms for the Ir56d locus were fused to the Gal4-hsp70-3’UTR by PCR amplification using genomic DNA and pGal4attB15 as templates, respectively. The product was ligated into pHD-DsRed-attP67 after digestion with SapI and AarI (to generate the donor vector pHD-Ir56d-Gal4-DsRed-attP). pCFD5-Ir56dsgRNAs and pHD-Ir56d-Gal4-DsRed-attP were co-injected into Act5C-Cas9,lig4[169] flies69 following standard protocols. Successful integration events were identified by screening for DsRed expression and diagnostic PCR. Subsequently, the DsRed marker was removed by injection of Cre recombinase. The oligonucleotides used are listed in Supplementary Table 3 and Supplementary Fig. 3a depicts a schematic of the Ir56dGal4 allele before and after DsRed removal.
Histology
Immunofluorescence on peripheral and central tissues from larvae and adult flies was performed following standard procedures28,45. Primary antibodies: rabbit anti-IR25a (1:500)14, guinea pig anti-IR25a (1:200)21, mouse anti-GFP (1:500; Invitrogen), chicken anti-GFP (1:500; Abcam), rabbit anti-RFP (1:500; Abcam) and mouse monoclonal nc82 (1:10; Developmental Studies Hybridoma Bank). Secondary antibodies (all diluted 1:100–200): goat anti-mouse Alexa 488 (Invitrogen), goat anti-rabbit Cy3 (Milan Analytica, AG), goat anti-chicken Alexa488 (Abcam), goat anti-guinea pig Cy5 (Abcam) and goat anti-mouse Cy5 (Jackson ImmunoResearch). Images were collected with a Zeiss LSM 710 inverted laser scanning confocal microscope (Zeiss, Oberkochen, Germany), and processed with ImageJ and Fiji.
Optical imaging
Subesophageal zone imaging: Imaging was performed adapting previous protocols70,71. In brief, a 1–3 week-old fly was cold-anaesthetised and inserted into a plastic holder glued to a custom Plexiglas chamber. The head and proboscis of the animal were separated by a plastic barrier that prevents contact between the buffer solution applied to the brain, and the tastant solution. The proboscis was extended using a blunted syringe needle (30 g Blunt, Warner Instruments #SN-30) connected to a vacuum pump (KNF Laboport #N86KN.18) and kept extended using UV curing glue (Tetric EvoFlow, A1, Ivoclar Vivadent) solidified using a UV lamp (Bluphase C8, Ivoclar vivadent). Heads were fixed using the same UV glue and covered with Adult Haemolymph like-Saline buffer (in mM: 108 NaCl, 5 KCl, 2 CaCl2, 8.2 MgCl2, 4 NaHCO3, 1 NaH2PO4, 15 Ribose, 5 HEPES; pH 7.5; 265 mOsm). Brains were exposed by removing the cuticle using a microsurgical knife (Sharpoint, Surgical Specialties #72-1501). Complete exposure of the subesophageal zone required the removal of the oesophagus. Delivery of the tastants was performed manually upon the emission of an acoustic signal at frame 20 after the onset of the recording, using a blunted 30 g syringe needle place on a 1 ml syringe containing the solution (BD Plastipak #300013) and mounted on a micromanipulator (Narishige).
Images were acquired with a CCD camera (CoolSNAP-HQ2 Digital CameraSystem) mounted on a fluorescence microscope (upright fixed stage Carl Zeiss Axio Examiner D1) equipped with a 40x water-immersion objective (W Plan-Apochromat 40× /1,0 VIS-IR DIC). Excitation light of 470 nm was produced with an LED light (Cool LED pE-100, VisiChrome). Binned image size was 1000 × 700 pixels on the chip, corresponding to 250 × 175 μm in the preparation. Exposure time was 100 ms. Twenty-second films were recorded with an acquisition rate of 4 Hz. Metafluor software (Visitron) was used to control the camera, light, and data acquisition.
Data were processed using NIH ImageJ and custom programmes in Matlab (v9.0). Time-series images corresponding to one experiment were first aligned using StackReg/TurboReg (bigwww.epfl.ch/thevenaz/stackreg/) in NIH ImageJ. Raw images were then segmented into individual 80-frame measurements. Each measurement was bleach-corrected by fitting a double-exponential function to the relative mean fluorescence in the ROI over time, excluding the frames covering 12.5 s after stimulus onset. We then calculated the relative change in fluorescence (ΔF/F) for each frame of each measurement as (ΔFi − F0)/F0 × 100, where F0 is the mean fluorescence value of frames 10–15 (before tastant presentation at frame 20), and Fi is the fluorescence value for the ith frame of the measurement. A circular ROI (diameter 7 pixels) was used for quantification of all measurements from the same animal. The maximal ΔF/F between frames 20 and 60 for each stimulus was used for data representation and statistical analysis.
Foreleg calcium imaging: imaging was performed adapting previous protocols33. A custom-made bottom-glass imaging chamber was built by drilling a 10 mm hole in a 35 mm Petri dish (Falcon #351008) onto which an 18 × 18 mm coverslip (Menzel-Gläser #631-1331) was glued. A 1–3 week-old fly was cold-anaesthetised and the forelegs cut with a razor blade between the femur and the tibia. UV-curing glue was used to seal the cut end of the leg, which was then mounted laterally on the glass surface of the imaging chamber. 1% low melting point agarose (Peqlab #35–2010) was used to cover the leg leaving exposed only the fourth and fifth tarsal segments. The preparation was covered with 150 µl of milliQ filtered water. For the stimulations, 150 µl of the desired chemical were added manually to the preparation. The sample was subsequently washed five times using milliQ filtered water and left with 150 µl of water for equilibration during 3 min before the next stimulation. Imaging was performed using an inverted confocal microscope (Zeiss LSM 710) using an oil immersion 40x objective (Plan Neofluar 40x Oil immersion DIC objective with a 1.3 NA). 40 frames were taken per stimulation at 250 ms/frame. Different focal planes were used to image the 5b and 5s sensilla neurons. Images were analysed as indicated in the previous section.
Behaviour
Two-choice positional preference assay: assays were performed in 94 mm Petri dishes (Greinier-bio-one #632180; 94 × 16 mm), divided into two halves (A and B) by placing two stacks of three-layered semicircles of blotting paper (Macherey-Nagel #742113) separated by a 3–5 mm gap. Prior to the start of the experiment each semicircle stack was soaked with 3 ml of the desired test solution (see below and Supplementary Table 2). Up to 16 arenas were placed on a methacrylate panel (1.5 cm thickness) elevated 5.5 cm from the light source, which consisted of a 60 × 60 cm LED Panel (Ultraslim LED Panel, 360 Nichia LEDs, Lumitronix) covered with red film (106 Primary Red, Showtec). 70–80 flies (mixed sexes; 2–3 days old, starved for 24 h in glass culture tubes with a Kimwipe (Kimtech #7552) soaked with 2 ml of tap water; cold anaesthetised) were introduced into the centre of each arena and the lids replaced. When all flies had recovered mobility, the assay was started. Pictures were taken (using a USB 3.0 100 CMOS Monochrome Camera 2048 × 2048 Pixel and a CCTV Lens for 2/300f:16 mm (iDS)) every 10 min up to 90 min using a custom Matlab code. The distribution of animals in the arena at 90 min (excluding the rare flies that were non-motile or that died during the assay) was quantified using a custom macro in Fiji (code available upon request). Preference indices were calculated as: (# flies in A − # flies in B)/total # flies. For the experiments in Fig. 7b, different genotypes were run in parallel, and randomised with respect to arena position.
For carbonation preference tests, in order to ensure a slow but constant production of CO2 over the course of the assay, we used solutions of freshly-prepared 100 mM NaHCO3 that were adjusted to pH 6.5 (with 5 M NaH2PO4; ∼1–1.5 ml/100 ml) for the carbonated side and pH 8.5 (with NaOH; <50 μl/100 ml) for the non-carbonated side9. To test for preference due to pH, we use phosphate buffered saline (7.8 mM NaH2PO4, 12.2 mM Na2HPO4, 153.8 mM NaCl) solutions at pH 6.5 or 8.5 (Fig. 7c). To eliminate the possibility that preference differences were due to Na+ imbalance (due to a larger volume of 5 M NaH2PO4 required to set the pH of NaHCO3 at pH 6.5 than NaOH to set the pH to 8.5), we supplemented the NaHCO3 pH 8.5 solution with NaCl to achieve an ∼150 mM [Na+] in both test solutions; flies retained the preference for the carbonated solution (Fig. 7d).
Proboscis extension reflex (PER) assay: PER in response to labellar stimulation was assessed following a standard protocol72. Individual flies (mixed sexes; 3–5 days old, starved for 24 h) were introduced into yellow pipette tips (Starlabs #S1111.0706), whose narrow end was cut in order that only the fly’s head could protrude from the opening, leaving the rest of the body, including legs, constrained inside the tip. Tastants (Supplementary Table 2) were delivered using a piece of Kimwipe. Each fly was first tested with water; where this caused PER, water was offered ad libitum, and the animal tested again. Only flies that showed negative PER for water were assayed with the other stimuli. Up to six flies were prepared simultaneously and tastants were randomised across trials.
For leg stimulation-evoked PER we adapted a published protocol33. Groups of six cold anesthetized flies (mixed sexes; 3–5 days old, starved for 24 h) were glued on their back on top of 76 × 26 mm microscope slides (Menzel-Gläser; #631–0649) using UV curing glue (Tetric EvoFlow, A1, Ivoclar Vivadent) solidified using a UV lamp (Bluphase C8, Ivoclar vivadent). Up to five groups of six flies were prepared at the same time and allowed to recover for 30 min in a humidified chamber. Only one group of six flies were tested at a time for the whole set of stimuli presented using a piece of Kimwipe72. Before the beginning of the stimulations, flies were allowed to drink water ad libitum. Each stimulus was presented once allowing the flies to touch the Kimwipe with all the legs for 5 s. Only full extensions of the proboscis were considered as positive responses. Between stimuli, flies were allowed to drink water ad libitum and the legs washed with water.
Optogenetic induction of PER: flies were grown on standard food; prior to the experiment 3–5 days old flies were starved for 24 h in tubes containing a Kimwipe soaked in 2 mM all-trans-retinal (Sigma #R2500) in tap water. Flies were cold-anaesthetised and glued on their backs to the external side of a 94 × 16 mm plastic plate using UV curing glue (see above). Groups of 6–8 flies of the same genotype were prepared in a row and tested for PER to stimulation by a 650 nm laser diode (1 mW, Adafruit Industries #1054) aimed at the proboscis with an intensity of 2–2.5 μW/mm2. Only full proboscis extensions were considered as positive.
Expresso food ingestion measurements and analysis: flies were maintained on conventional cornmeal-agar-sucrose medium at 23–25 °C and 60–70% relative humidity (RH), under a 12 h light:12 h dark cycle (lights on at 6 am). Carbonated and non-carbonated control solutions were prepared as described above (either in water or with 5 mM sucrose). Food ingestion was measured in the Expresso device:47 individual flies (2–5 days old male w1118 flies, starved 24 h) were placed in the behavioural chamber with the doors in the closed position to prevent access to the liquid food in the calibrated glass capillaries. Expresso data acquisition software was started at which point all doors were opened giving flies synchronised access to liquid food. Each trial lasted ~33 min, and 10 flies were tested in parallel in two Expresso sensor banks. For each condition, 20–30 flies were tested. The measurements were performed at Zeitgeber Time 6–10. The Expresso food ingestion data were analysed using a custom programme in Python (available upon request). The change points in the Expresso signal that denote a meal bout and the amount of food ingested were detected using the Pruned Exact Linear Time algorithm. Total ingestion was calculated as the total volume ingested per fly per trial. The latency was calculated as the time before the first meal after door opening. When a fly did not consume any food, the total meal bout volume was scored as 0 and latency to first meal bout was scored as the total time of the assay (i.e. 33 min). All data were analysed in R statistical software.
flyPAD: we assayed mated w1118 female flies that were reared at 25 °C, 70% RH on a 12 h light:12 h dark cycle. Flies were reared at standard density and were matched for age and husbandry conditions. The fly medium contained, per litre, 80 g cane molasses, 22 g sugar beet syrup, 8 g agar, 80 g corn flour, 10 g soya flour, 18 g yeast extract, 8 ml propionic acid, and 12 ml nipagin (15% in ethanol). The day before the assay the fully fed flies were flipped into new vials to ensure a fully fed metabolic state. The starvation state was induced by transferring flies for 24 h before the assay into vials containing a tissue soaked with water.
flyPAD assays53,54 were performed using only one well of the arena per assay. The well was filled with a solid substrate comprising 20 mM sucrose made with either non-carbonated or carbonated water (Supplementary Table 2) in 1% agarose. Flies were individually transferred to flyPAD arenas by mouth aspiration and allowed to feed at 25 °C and 70% RH for 60 min. flyPAD data were acquired using the Bonsai framework73, and analysed in MATLAB using custom-written software54.
Statistics
Sample size was determined based upon preliminary experiments. Data were analysed and plotted using R (v1.0.153; R Foundation for Statistical Computing, Vienna, Austria, 2005; R-project-org) (code available upon request). Except for PER and flyPAD experiments, quantitative data are represented showing their distribution by superimposing a boxplot on top of a violin plot. The violin plot shows the kernel density estimate; for the boxplots the whiskers are calculated as follows: the upper whisker equals the third quartile plus 1.5× the interquartile range (IQR) and the lower whisker equals the first quartile minus 1.5× the IQR. Any data points above the superior or below the inferior whisker values are considered as outliers. The outliers were included in the statistical comparisons as we performed non-parametric rank tests. Data were analysed statistically using different variants of the Wilcoxon test, except where indicated. For comparisons between distributions, the Wilcoxon rank sum test was used. When P value correction for multiple comparisons was required, the Bonferroni method was used. For the experiments in Fig. 7b–d, we performed a Wilcoxon Signed Rank Test with the null hypothesis that the median of sampled values differs from zero. For PER results we used the Fisher exact test. For Expresso assay data, pairwise comparisons using the Tukey and Kramer (Nemenyi) test with Tukey-Dist approximation for independent samples were performed. flyPAD results were compared using the Wilcoxon rank-sum test.
Electronic supplementary material
Acknowledgements
We thank Carolina Gomez-Diaz for assistance with CRISPR primer design, and acknowledge the Bloomington Drosophila Stock Centre (NIH P40OD018537) and the Developmental Studies Hybridoma Bank (NICHD of the NIH, University of Iowa) for reagents. We thank members of the Benton lab for discussions and comments on the manuscript. We thank Célia Baltazar for assistance in performing the flyPAD assays. J.A.S.-A. was supported by a Federation of European Biochemical Societies Long Term Fellowship, an EMBO Long Term Fellowship and a Human Frontier Science Programme Long-term Fellowship. V.C. was supported by a Boehringer Ingelheim Foundation Fellowship. T.O.A. was supported by a Human Frontier Science Programme Long-term Fellowship. D.M. was supported by a DFG Research Fellowship (MU 4116/1-1). Research in S.G.S.’s laboratory was supported by a European Research Council Starting Independent Research Grant (309832) and the Swiss National Science Foundation (31003A_149499). Research in N.Y.’s laboratory was supported by a Cornell University Nancy and Peter Meinig Family Investigatorship Programme, a Pew Biomedical Scholar Award, and the Alfred P. Sloan Foundation Award. Research in C.R.’s laboratory was supported by the BIAL Foundation grant (279/16) and the Champalimaud Foundation. Research in R.B.’s laboratory was supported by the University of Lausanne, ERC Starting Independent Researcher and Consolidator Grants (205202 and 615094) and an SNSF Sinergia Grant (CRSII3_136307).
Author contributions
The authors have made the following declarations about their contributions: J.A.S.-A., A.F.S., V.C., A.K.S., S.Y.S., D.M., K.S., T.O.A., G.L.N.-M., S.G.S., N.Y., C.R., and R.B. conceived and designed the experiments. J.A.S.-A. (Fig. 4, Figs 6 and 7; Supplementary Fig. 3, 4, 6 and 7), A.F.S. (Fig. 5), V.C. (Figs 1 and 2; Supplementary Fig. 1), G.Z. (Fig. 7a–d), A.K.S. (Fig. 1, Fig. 3, Supplementary Fig. 2), S.Y.S. (Supplementary Fig. 5), T.O.A. (Supplementary Fig. 3), S.C. (Fig. 3), G.L.N.-M. (Fig. 2), R.B. (Fig. 6a), K.S. and D.M. (Supplementary Fig. 8) performed the experiments. J.A.S.-A., A.F.S., V.C., A.K.S., S.Y.S., D.M., K.S., T.O.A., G.L.N.-M., N.Y., C.R. and R.B. analysed the data. J.A.S.-A., A.F.S., V.C., L.A., T.O.A. and R.B. contributed reagents/materials/analysis tools. J.A.S.-A. and R.B. wrote the paper, with input from all authors.
Data availability
All relevant data supporting the findings of this study are available from the corresponding author on request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ana Florencia Silbering, Vincent Croset, Giovanna Zappia.
Change history
2/25/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41467-018-06453-1.
References
- 1.Liman ER, Zhang YV, Montell C. Peripheral coding of taste. Neuron. 2014;81:984–1000. doi: 10.1016/j.neuron.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yarmolinsky DA, Zuker CS, Ryba NJ. Common sense about taste: from mammals to insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freeman EG, Dahanukar A. Molecular neurobiology of Drosophila taste. Curr. Opin. Neurobiol. 2015;34:140–148. doi: 10.1016/j.conb.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright GA. To feed or not to feed: circuits involved in the control of feeding in insects. Curr. Opin. Neurobiol. 2016;41:87–91. doi: 10.1016/j.conb.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Benton R. The neurobiology of gustation in insect disease vectors: progress and potential. Curr. Opin. Insect Sci. 2017;20:19–27. doi: 10.1016/j.cois.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Scott K. Gustatory processing in Drosophila melanogaster. Annu. Rev. Entomol. 2018;63:15–30. doi: 10.1146/annurev-ento-020117-043331. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cameron P, Hiroi M, Ngai J, Scott K. The molecular basis for water taste in Drosophila. Nature. 2010;465:91–95. doi: 10.1038/nature09011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischler W, Kong P, Marella S, Scott K. The detection of carbonation by the Drosophila gustatory system. Nature. 2007;448:1054–1057. doi: 10.1038/nature06101. [DOI] [PubMed] [Google Scholar]
- 10.Masek P, Keene AC. Drosophila fatty acid taste signals through the PLC pathway in sugar-sensing neurons. PLoS Genet. 2013;9:e1003710. doi: 10.1371/journal.pgen.1003710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlu S, Wisotsky Z, Medina A, Dahanukar A. Acid sensing by sweet and bitter taste neurons in Drosophila melanogaster. Nat. Commun. 2013;4:2042. doi: 10.1038/ncomms3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingleby FC. Insect cuticular hydrocarbons as dynamic traits in sexual communication. Insects. 2015;6:732–742. doi: 10.3390/insects6030732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joseph RM, Carlson JR. Drosophila Chemoreceptors:a molecular interface between the chemical world and the brain. Trends Genet. 2015;31:683–695. doi: 10.1016/j.tig.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croset V, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rytz R, Croset V, Benton R. Ionotropic receptors (IRs): chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 2013;43:888–897. doi: 10.1016/j.ibmb.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 17.van Giesen L, Garrity PA. More than meets the IR: the expanding roles of variant ionotropic glutamate receptors in sensing odor, taste, temperature and moisture. F1000Res. 2017;6:1753. doi: 10.12688/f1000research.12013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison MC, et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2018;2:557–566. doi: 10.1038/s41559-017-0459-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai M, et al. Acid sensing by the Drosophila olfactory system. Nature. 2010;468:691–695. doi: 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silbering AF, et al. Complementary function and integrated wiring of the evolutionarily distinct Drosophila. J. Neurosci. 2011;31:13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knecht ZA, et al. Distinct combinations of variant ionotropic glutamate receptors mediate thermosensation and hygrosensation in Drosophila. eLife. 2016;5:e17879. doi: 10.7554/eLife.17879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knecht ZA, et al. Ionotropic receptor-dependent moist and dry cells control hygrosensation in Drosophila. eLife. 2017;6:e26654. doi: 10.7554/eLife.26654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enjin A, et al. Humidity sensing in Drosophila. Curr. Biol. 2016;26:1352–1358. doi: 10.1016/j.cub.2016.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank DD, et al. Early integration of temperature and humidity stimuli in the Drosophila brain. Curr. Biol. 2017;27:2381–2388 e2384. doi: 10.1016/j.cub.2017.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abuin L, et al. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koh TW, et al. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. 2014;83:850–865. doi: 10.1016/j.neuron.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart S, Koh TW, Ghosh AC, Carlson JR. Candidate ionotropic taste receptors in the Drosophila larva. Proc. Natl Acad. Sci. USA. 2015;112:4195–4201. doi: 10.1073/pnas.1503292112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Croset V, Schleyer M, Arguello JR, Gerber B, Benton R. A molecular and neuronal basis for amino acid sensing in the Drosophila larva. Sci. Rep. 2016;6:34871. doi: 10.1038/srep34871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joseph RM, Sun JS, Tam E, Carlson JR. A receptor and neuron that activate a circuit limiting sucrose consumption. eLife. 2017;6:e24992. doi: 10.7554/eLife.24992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szyszka P, Galizia CG. The role of the sucrose-responsive IR60b neuron for Drosophila melanogaster: A hypothesis. Chem. Senses. 2018;43:311–312. doi: 10.1093/chemse/bjy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Poudel S, Kim Y, Thakur D, Montell C. Calcium taste avoidance in Drosophila. Neuron. 2018;97:67–74 e64. doi: 10.1016/j.neuron.2017.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hussain A, et al. Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 2016;14:e1002454. doi: 10.1371/journal.pbio.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahn JE, Chen Y, Amrein HO. Molecular basis of fatty acid taste in Drosophila. eLife. 2017;6:e30115. doi: 10.7554/eLife.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganguly A, et al. A molecular and cellular context-dependent role for Ir76b in detection of amino acid taste. Cell Rep. 2017;18:737–750. doi: 10.1016/j.celrep.2016.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Amrein H. Ionotropic receptors mediate Drosophila oviposition preference through sour gustatory receptor neurons. Curr. Biol. 2017;27:2741–2750 e2744. doi: 10.1016/j.cub.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tauber JM, et al. A subset of sweet-sensing neurons identified by IR56d are necessary and sufficient for fatty acid taste. PLoS Genet. 2017;13:e1007059. doi: 10.1371/journal.pgen.1007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H, et al. Drosophila Gr64e mediates fatty acid sensing via the phospholipase C pathway. PLoS Genet. 2018;14:e1007229. doi: 10.1371/journal.pgen.1007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grosjean Y, et al. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478:236–240. doi: 10.1038/nature10428. [DOI] [PubMed] [Google Scholar]
- 39.Prieto-Godino LL, et al. Evolution of acid-sensing olfactory circuits in drosophilids. Neuron. 2017;93:661–676 e666. doi: 10.1016/j.neuron.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 40.Vosshall LB, Stocker RF. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 41.Ni L, et al. The ionotropic receptors IR21a and IR25a mediate cool sensing in Drosophila. eLife. 2016;5:e13254. doi: 10.7554/eLife.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min S, Ai M, Shin SA, Suh GS. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc. Natl Acad. Sci. USA. 2013;110:E1321–E1329. doi: 10.1073/pnas.1215680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nayak SV, Singh RN. Sensilla on the tarsal segments and mouthparts of adult Drosophila melanogaster Meigen (Diptera: Drosophilidae) Int. J. Insect Morph. Embryol. 1983;12:273–291. [Google Scholar]
- 44.Shearin HK, Dvarishkis AR, Kozeluh CD, Stowers RS. Expansion of the gateway multisite recombination cloning toolkit. PLoS ONE. 2013;8:e77724. doi: 10.1371/journal.pone.0077724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez-Alcaniz JA, Zappia G, Marion-Poll F, Benton R. A mechanosensory receptor required for food texture detection in Drosophila. Nat. Commun. 2017;8:14192. doi: 10.1038/ncomms14192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyazaki T, Ito K. Neural architecture of the primary gustatory center of Drosophila melanogaster visualized with GAL4 and LexA enhancer-trap systems. J. Comp. Neurol. 2010;518:4147–4181. doi: 10.1002/cne.22433. [DOI] [PubMed] [Google Scholar]
- 47.Yapici N, Cohn R, Schusterreiter C, Ruta V, Vosshall LB. A taste circuit that regulates ingestion by integrating food and hunger signals. Cell. 2016;165:715–729. doi: 10.1016/j.cell.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117:981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 49.Matthews, B. J., et al. Improved Aedes aegypti mosquito reference genome assembly enables biological discovery and vector control. Pre-print at https://www.biorxiv.org/content/early/2017/12/29/240747 (2017).
- 50.Chandrashekar J, et al. The taste of carbonation. Science. 2009;326:443–445. doi: 10.1126/science.1174601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wisotsky Z, Medina A, Freeman E, Dahanukar A. Evolutionary differences in food preference rely on Gr64e, a receptor for glycerol. Nat. Neurosci. 2011;14:1534–1541. doi: 10.1038/nn.2944. [DOI] [PubMed] [Google Scholar]
- 52.Fujii S, et al. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr. Biol. 2015;25:621–627. doi: 10.1016/j.cub.2014.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steck K, et al. Internal amino acid state modulates yeast taste neurons to support protein homeostasis in Drosophila. eLife. 2018;7:e31625. doi: 10.7554/eLife.31625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Itskov PM, et al. Automated monitoring and quantitative analysis of feeding behaviour in Drosophila. Nat. Commun. 2014;5:4560. doi: 10.1038/ncomms5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nat. Neurosci. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 57.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chyb S, Dahanukar A, Wickens A, Carlson JR. Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc. Natl Acad. Sci. USA. 2003;100(Suppl 2):14526–14530. doi: 10.1073/pnas.2135339100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell. 2012;149:1140–1151. doi: 10.1016/j.cell.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 62.Han C, Jan LY, Jan YN. Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc. Natl Acad. Sci. USA. 2011;108:9673–9678. doi: 10.1073/pnas.1106386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sander JD, et al. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gratz SJ, et al. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Port F, Bullock SL. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods. 2016;13:852–854. doi: 10.1038/nmeth.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Koolhaas WH, Schnorrer F. A versatile two-step CRISPR- and RMCE-based strategy for efficient genome engineering in Drosophila. G3. 2014;4:2409–2418. doi: 10.1534/g3.114.013979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silbering AF, Bell R, Galizia CG, Benton R. Calcium imaging of odor-evoked responses in the Drosophila antennal lobe. J. Vis. Exp. 2012;61:2976. doi: 10.3791/2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Flood TF, et al. A single pair of interneurons commands the Drosophila feeding motor program. Nature. 2013;499:83–87. doi: 10.1038/nature12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shiraiwa T, Carlson JR. Proboscis extension response (PER) assay in Drosophila. J. Vis. Exp. 2007;3:193. doi: 10.3791/193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lopes G, et al. Bonsai: an event-based framework for processing and controlling data streams. Front. Neuroinform. 2015;9:7. doi: 10.3389/fninf.2015.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prieto-Godino LL, et al. Olfactory receptor pseudo-pseudogenes. Nature. 2016;539:93–97. doi: 10.1038/nature19824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silbering AF, et al. Ir40a neurons are not DEET detectors. Nature. 2016;534:E5–E7. doi: 10.1038/nature18321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data supporting the findings of this study are available from the corresponding author on request.