Abstract
One of the newest frontiers of physical therapy is the field of epigenetics, which examines how pervasive environmental factors such as exercise regulate the expression of genes. The epigenome may be one of the most powerful systems through which exercise exerts its beneficial effects on health and longevity. Large epidemiology studies show that individuals who regularly exercise demonstrate a lower “epigenetic age,” experience fewer metabolic diseases, and enjoy greater longevity. However, the dose, mode, intensity, and duration of exercise required to achieve a healthy epigenetic profile is unknown. As experts in exercise prescription, physical therapists are ideally suited to contribute to the discovery of this dose-response relationship. This perspective makes a case for the genesis of “precision physical therapy,” which capitalizes on epigenetic discoveries to optimize exercise-based interventions. Summarized here is the emerging body of knowledge supporting epigenetic adaptations to exercise in humans, including the intriguing possibility that these environmentally modified traits could be passed down to offspring. In the future, it is likely that epigenetic data will enhance our understanding of individual disease risk and individual response to prescribed exercise. The profession of physical therapy must be alert to new epigenetic knowledge that can enhance the specificity and efficacy of movement-based treatments.
Physical therapists prescribe physical activity/exercise to improve cell, tissue, organ, and system function to advance the health and well-being of individuals. Our mechanistic understanding of the ways in which our interventions elicit the desired adaptations evolved with our advancement of our foundational science. For example, we know that when our patient with C6 quadriplegia develops a strong tenodesis grip, she is benefiting from a collection of adaptive cellular responses. In her brain, sensory and motor networks that once coordinated her hand muscles “re-map” to serve her wrist extensors, increasing the neural substrate available to these functionally essential muscles.1 Through advances in neuroscience, we understand the chemical signaling cascades and the changes in gene expression that make this adaptation possible. From research conducted with animal models, we are beginning to understand the ways that these cellular adaptations depend upon the dose and type of exercise. Although physical therapists usually only observe the broad functional outcomes of our interventions, the day is coming when we may understand the intricate cellular adaptations upon which every functional gain is predicated. It will then be easy to see that all physical therapy occurs at a cellular level.
As this knowledge accumulates, it will not be necessary for physical therapists to become biochemists or molecular geneticists. Physical therapists will enhance the care they provide, however, if they can use key new cell biology discoveries to guide and direct their powerful exercise-based interventions. This expansion of the discipline is already underway, as all accredited physical therapy education programs now include genetics as a component of entry-level curriculum.2 For clinicians without this background, a review of the genetic basis for the heath-promoting effects of exercise may be instructive. Moreover, the newest frontier of physical therapy lies within the field of epigenetics,3 the study of how regular exposures to environmental factors (exercise, nutrition, emotional stress, etc) promote the expression or repression of certain genes. The epigenome may be one of the most powerful systems through which exercise exerts its pervasive beneficial effects. As experts in exercise, physical therapists are ideally suited to contribute to the burgeoning knowledge base of this field. As importantly, physical therapy clinicians have much to gain by capitalizing on new epigenetic knowledge flowing outward from the basic sciences. This perspective paper will make a case for the genesis of “precision physical therapy,” which capitalizes on epigenetic discoveries to optimize exercise-based interventions. This paper will highlight a few key epigenetic principles that can inform and strengthen the contemporary scientific rationale for physical therapy interventions. By remaining abreast of this rapidly expanding field, physical therapists can expertly tailor novel movement interventions for patients of all ages, activity levels, and health states.
Introduction to Epigenetics
In the nucleus of each cell are chromosomes, genes, and DNA that contain the protein-coding sequence of nucleotides for the entire genome. Through a process called transcription, the DNA nucleotide code is converted to messenger RNA (mRNA) molecules that contain the “blueprint” for assembling complex proteins. During the process of translation, cellular structures called ribosomes use the information contained within mRNA to direct the construction of proteins from individual amino acids. These proteins include enzymes, hormones, structural proteins, cellular messengers, and the many other classes of biomolecules from which every cell is constructed. The coordination of protein production from the genetic code is the chief means by which an organism executes growth, adaptation, and repair.
In the inactive state, DNA is tightly wound around histone proteins to form chromatin. Chromatin strands further twist and condense to form chromosomes. When signaled to create new proteins, chromatin relaxes and the portion of DNA coding for a protein unwinds. Enzymes called DNA helicases unzip the double strand of DNA to expose the protein-coding DNA region (gene) to the molecules that will carry out transcription. An enzyme called RNA polymerase binds to a “promoter region” of DNA that serves as a uniform starting point for transcription of the gene. RNA polymerase synthesizes an mRNA strand “complement” of the exposed DNA (Fig. 1). This mRNA is akin to a photo negative of the DNA, containing nucleotide base pairs that exactly oppose the original DNA sequence. Each nucleotide triplet of the protein-coding region of mRNA encodes a particular amino acid. mRNA leaves the nucleus and travels to the ribosomes, which lie immediately adjacent to the nucleus in a structure called the rough endoplasmic reticulum. Ribosomes oversee the translation of information encoded in mRNA into complex, three-dimensional proteins. The ribosomes read the mRNA “photo negative” and assemble amino acids into long strands called polypeptides. Through a complex series of chemical interactions and modifications, the polypeptides twist and fold into completed proteins.
Figure 1.
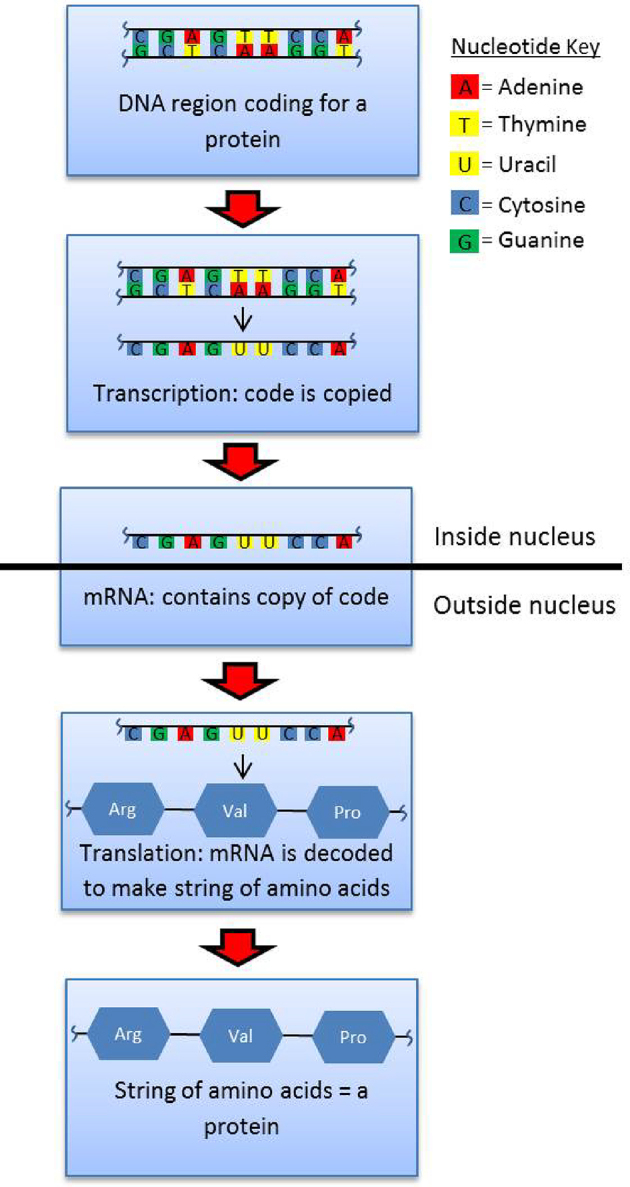
Simplified illustration of the steps involved in transcription and translation. Arg, Val, and Pro are arginine, valine, and proline amino acids, respectively.
“Epigenetics” means “above the genome” and refers to processes that guide and direct the expression of genes contained in the genome. Epigenetic changes do not cause alterations to the DNA nucleotide base pairs themselves (mutation), but rather consist of reversible “tags” that can be applied to the DNA or to the histone molecules that bind DNA into chromatin. DNA methylation and histone acetylation are the epigenetic modifications most studied in relation to physical activity4-8 and thus will be the two modifications emphasized in this perspective. Even though they do not cause changes to the genetic code, epigenetic modifications are “stably heritable,” meaning they can be passed down during cell division, either through mitosis or meiosis.9 Although some epigenetic traits can be handed down to an organism's progeny, most of the present discussion will center around epigenetic inheritance of traits across successive cell generations in the lifespan of a single organism. While an organism or cell's genotype (DNA code) is relatively stable over its lifespan (barring DNA damage/mutation), the transcription/translation of the genome into proteins may be strongly influenced by epigenetic modifications, resulting in a phenotype that reflects the individual or cell's pervasive environmental influences.5
DNA methylation is carried out by a protein called DNA methyltransferase (DNMT). The process can occur at many DNA nucleotide base-pair locations, but occurs most readily where cytosine nucleotides lie adjacent to guanine nucleotides (CpG methylation).8 Depending on the gene region to which the methylation “tag” is applied, transcription of the methylated gene may be enhanced or decreased. If methylation occurs at a gene's promoter region or at a gene “enhancer region,” transcription of the gene into mRNA will be blocked.8 This would result in decreased production of the protein encoded by the methylated gene. If the methylation occurs beyond a gene's promoter or enhancer regions, in the gene region containing the specific instructions for protein assembly, the influence of epigenetic tagging is less clear. Researchers generally believe that intragenic methylation enhances gene transcription,5,8 which would result in increased production of the protein encoded by the methylated gene.
Epigenetic modifications may also alter the interaction between DNA and the histone molecules around which the DNA strand coils (Fig. 2). Histone molecules include a “tail” region that can accept an acetyl molecule tag via the action of an enzyme called histone acetyl transferase (HAT). This process of acetylation relaxes the DNA-histone association, which makes the DNA accessible for transcription. In this way, acetylation of histone tails increases gene expression.5 In an opposing process, an enzyme called histone deacetylase (HDAC) removes acetyl groups from the histone tails, which causes DNA to associate more tightly with the histone core, thus inhibiting gene transcription and protein translation.5
Figure 2.
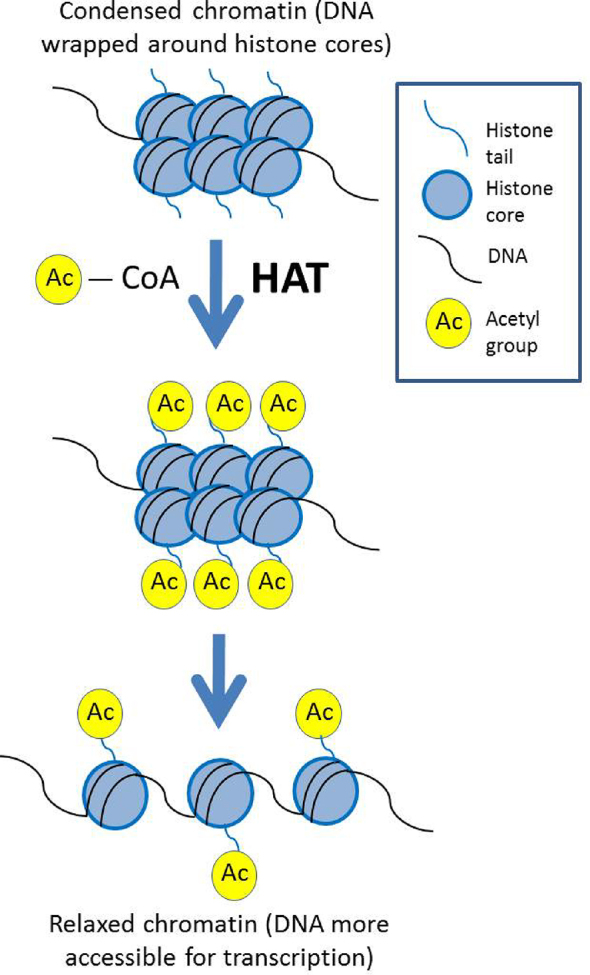
Histone acetylation. Histone acetyl transferase (HAT) transfers an acetyl group from acetyl-CoA to histone tails. Acetylation relaxes the DNA-histone arrangement, which makes the DNA accessible for transcription, increasing gene expression.
The ability to quantify epigenetic adaptations is rapidly evolving, and researchers are continually developing new techniques to measure DNA methylation and histone acetylation, both within cells and for an entire organism.6,10-18 As our ability to measure epigenetic modifications continually improves, so does our understanding of which genes may be epigenetically modified in response to exercise.4,5,19 The intersection of epigenetic discoveries with the science of exercise is an important new frontier for physical therapy.3
Exercise as a Stimulus for Epigenetic Adaptations
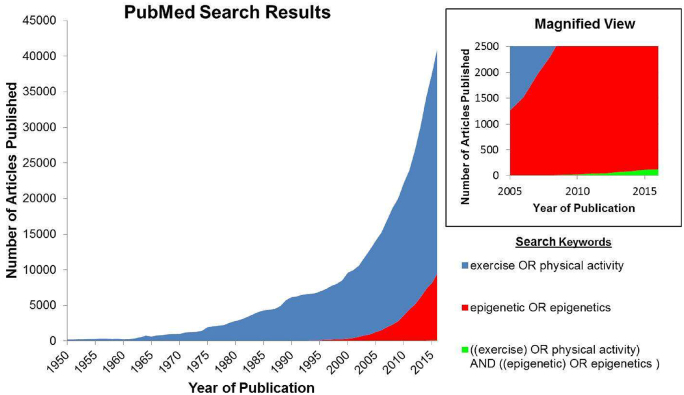
Intuitive links between exercise and health have existed for millennia, but scientific study of this relationship began in earnest only a few decades ago. The number of scientific studies of exercise began to rapidly increase in the 1970s, culminating in over 400,000 exercise-related publications today (Fig. 3). Through this work, we know a great deal about the inverse relationship between physical activity and metabolic disease20,21 and about the added morbidity faced by people with disability, who often do not or cannot engage in regular exercise.22 The field of epigenetics is poised to experience the same type of knowledge growth that exercise experienced in past decades. The current number of epigenetics articles being published per year is comparable to the number of exercise articles that were published in 2000 (Fig. 3). However, fewer than 600 articles to date have explored the interrelationships between exercise and epigenetics, with only a few dozen using human participants.23 These studies are critically important for physical therapists because the majority of our movement inventions are designed to be implemented for a long duration, precisely the type of pervasive “environmental” stimulus that can cause stable epigenetic adaptations. Physical therapy is a discipline that may have a tremendous amount to gain by understanding the complex interactions of exercise and epigenetic modifications.
Figure 3.
PubMed search results regarding exercise and epigenetics. Through 2016, over 400,000 articles with “exercise” or “physical activity” as a keyword had been published. Articles with “epigenetic” or “epigenetics” as a keyword totaled ∼60,000. Fewer than 600 articles include both sets of keywords (inset panel).
Large-scale epidemiology studies have begun to reveal links between exercise-mediated epigenetic changes (DNA methylation) and human longevity. Researchers quantified methylation levels at 485,577 specific CpG nucleotide pairs in the blood cells of a multi-ethnic, international cohort of > 4500 participants, predominantly women.24 This approach yielded a “DNA methylation age” that served as a biomarker for epigenetic aging. This estimate of epigenetic aging has been shown to predict longevity25 and all-cause mortality,18,26,27 and to correlate with a number of diseases.24,28,29 Researchers computed the difference between the participants’ chronological age and their “epigenetic age,” as indicated by DNA methylation status, then examined relationships between this difference and diet, exercise, and many clinical markers of metabolic health. Participants with low epigenetic age tended to consume more fruits, vegetables, fish, and poultry, to be moderate alcohol consumers, to have higher educational attainment and income, to engage in physical activity, and to have superior clinical markers of metabolic health. The statistical relationships between epigenetic age and physical activity were tenuous in this study, likely because participants were merely classified as “active” or “sedentary.” A similar study of elders living in Sweden stratified physical activity into 4 tiers and noted a significant, linear association between lifetime exercise dose and epigenetic aging (blood cell methylation status).30 This association remained significant even after adjusting for BMI, smoking, and a range of metabolic markers, supporting that exercise independently exerts a limiting effect on epigenetic aging. These studies provide some of the first direct evidence that exercise, working in partnership with diet to protect metabolic health, correlates with epigenetic adaptations that predict longevity. These studies add an important piece to our understanding of how exercise exerts its potent, long-lasting effects on overall health.
Other studies have administered exercise interventions over a finite time span to more prospectively examine the epigenetic adaptations by which exercise may protect health and limit aging. In sedentary men, 6 months of a cycling and aerobics intervention (1.8 sessions per week, 1 hour per session) led to several significant global metabolic changes (reduced waist/hip ratio, increased VO2max, reduced diastolic blood pressure, and increased HDL).31 In adipose cell biopsies from these participants, CpG site methylation significantly changed in 18 obesity genes and 21 candidate genes for type 2 diabetes. These methylation changes were accompanied by mRNA changes, supporting that the observed epigenetic changes did indeed lead to gene expression changes after exercise training. Adipose tissue is a key endocrine regulator of metabolism and glucose homeostasis,32 suggesting intriguing downstream effects for the epigenetic adaptations observed for obesity and diabetes genes. The study offers initial support that epigenetic mechanisms underlie the wide-ranging regulatory effects of adipose tissue upon clinical markers of metabolism.
Exercise is a systemic stressor that may elicit different epigenetic adaptations in different tissues. Adaptations in myocytes are of particular interest because of their central role in exercise and metabolic regulation. Epigenetic adaptations have been observed in response to a single bout of endurance exercise: immediately after 60 minutes of cycling at about 76% V̇O2max, participants in one study experienced significant changes in histone acetylation in the vastus lateralis muscle.6 Other researchers found that a long protocol (6 months) of endurance training (cycling and aerobics) modified methylation of 134 genes in the vastus lateralis, many of which had functions relating to type 2 diabetes.33 In participants who performed unilateral knee extension exercise for 3 months (a within-subject control design that mitigates many sources of variability), DNA methylation and mRNA expression changed for over 4000 gene loci in vastus lateralis muscle.34 A pathway analysis revealed that changes in gene networks regulating carbohydrate metabolism and structural remodeling were especially prevalent, suggesting a “coordinated transcriptional and epigenetic response” to exercise. The authors suggested that the regulation and maintenance of endurance exercise-based adaptations may, to a large degree, be predicated upon epigenetic changes.
Other researchers are working to establish a dose-response relationship for epigenetic adaptations to exercise. Barrès and colleagues found that 35 minutes of high-intensity exercise (80% maximal aerobic capacity) decreased the promoter-region methylation of genes involved in oxidative metabolism, thereby increasing expression of oxidative genes (Table).4 Lower-intensity exercise (40% V̇O2max) did not elicit significant demethylation, leading the authors to conclude that only high-intensity exercise yields epigenetic changes that foster oxidative metabolism. However, other studies suggest that long-term exposure to low-intensity exercise may indeed be a sufficient stimulus to elicit epigenetic modifications. In a pilot study, Ren and colleagues examined DNA methylation at 60 CpG sites in salivary DNA from participants who either did or did not practice Tai Chi.35 Despite Tai Chi being a low-intensity activity (1.5 to 3.0 METs36), researchers found significantly different methylation at six CpG sites, even after controlling for various possible confounders.35 Although a dose-response relationship between exercise intensity and epigenetic change is likely to exist, too little is currently known about this relationship to form conclusive guidelines. Many studies rely upon participant recall of physical activity intensity and duration, making the estimation of lifetime exercise dose very difficult. A well-controlled prospective study of epigenetic adaptations to low-intensity exercise is urgently needed.
Table.
| Gene | Metabolic Function62 |
|---|---|
| Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPARGC1A/PGC-1α)a,b | Mitochondrial biogenesis (multiplication) and determination of muscle fiber type; critical “transcription factor” gene that initiates extensive downstream gene expression changes. |
| Mitochondrial transcription factor A (TFAM)a | Mitochondrial transcription factor that governs mitochondrial DNA replication and repair. |
| Myocyte enhancer factor 2A (MEF2A)a | Regulates muscle development and neuronal differentiation. |
| Pyruvate dehydrogenase kinase, isozyme 4 (PDK4)a | Regulates glucose metabolism. |
| Myogenin (MYF4)b | Induces myogenesis. |
| Muscle creatine kinase (CKM)b | Crucial for energy transduction. |
| Glucose transporter 4 (SLC2A4/GLUT4)b | Transports glucose across the cell membrane; regulated by insulin. |
| Hexokinase 2 (HK2)b | Catalyzes first step of glucose metabolism. |
| ATP synthase beta (ATPB)b | Catalyzes ATP synthesis. |
| Carnitine palmitoyltransferase I (CPT1)b | Controls rate of long-chain fatty acid beta-oxidation. |
aSample of genes governing hypertrophy or oxidative metabolism that showed epigenetic adaptations (demethylation) after high-intensity exercise.
bSample of genes governing hypertrophy or oxidative metabolism that showed transcriptional adaptations (mRNA expression change) after high-intensity exercise.
An additional knowledge gap in the study of epigenetic response to exercise pertains to non-endurance, strength-based activity. Only one study to date has examined this mode of exercise in humans, via a supervised 8-week strength training program in healthy volunteers.37 Changes in leukocyte DNA methylation occurred in many of the same gene pathways previously observed to change in response to aerobic exercise. However, strength training differed from aerobic exercise by triggering methylation changes in pathways regulating anabolic growth factors. Because leukocytes play a role in inflammation and repair, these adaptations may reflect a generalized response to muscle cell damage. However, the study provides initial support that strength training may trigger epigenetic adaptations that foster muscle cell growth and repair. Studying epigenetic adaptations directly in skeletal muscle tissue would be an important next avenue for research.
Despite uncertainty regarding the optimal exercise mode or intensity for beneficial epigenetic modifications, we know that inactivity is epigenetically disadvantageous. Nine days of bedrest induced a metabolic state of insulin resistance in healthy participants, accompanied by widespread deleterious changes in gene pathways that control oxidative metabolism.38 These transcriptional changes were paired with a trend toward increased methylation of the promoter region of PPARGC1A (also known as PGC-1α), one of the most important genes regulating mitochondrial function and oxidative metabolism. The presence of these epigenetic modifications, particularly at the PGC-1α gene, may help explain why 4 weeks of retraining did not restore oxidative gene expression levels to baseline levels. This study illustrates the potential long-lasting epigenetic consequences of inactivity. Unfortunately, injury and disease processes relegate many of our patients to protracted periods—even a lifetime—without access to regular exercise. Patients who experience trauma, rheumatic disease, obesity, and any number of neuromuscular disorders may be unable to exercise a large percentage of their muscle mass. They may also be unable to achieve the intensity or duration of exercise shown to elicit clear-cut epigenetic benefits. Blanket recommendations for exercise dosage made for healthy populations4,5,39 may be unrealistic for these individuals. This further underscores the need for prospective studies of epigenetic adaptations to low-intensity exercise, which may be the most attainable form of regular activity for people living with disease or disability.
As mentioned previously, exercise is a systemic stressor that may trigger different epigenetic adaptations in different tissues. Skeletal muscle and adipose tissue have received a great deal of attention because of their clear relatedness to exercise and to chronic diseases like diabetes. These tissues are also easily accessed (via biopsy) in human participants. However, physical therapists understand that exercise is a cognitive and behavioral challenge that can be facilitated or impeded by factors such as attention and mood. The possibility of exercise-mediated epigenetic regulation of brain and nervous tissues therefore warrants consideration by physical therapists. Because of the inaccessibility of these tissues in human participants, all studies to date have employed rat and mouse models.40 Aerobic tasks such as treadmill and wheel running trigger epigenetic modifications in the hippocampus, a brain region that is critical for learning, memory, and mood.41,42 Exercise decreases hippocampal expression of DNMT, the protein that carries out DNA methylation.13 It also decreases methylation at the promoter region of the brain-derived neurotrophic factor (BDNF) gene.15 BDNF plays essential roles in synaptic plasticity, learning, and memory formation as well as in mood and attention.43,44 Although much work remains to be done in this area, epigenetic adaptations that favor BDNF production may play a role in the cognition-enhancing and mood-regulating effects of exercise.40 In physical therapy, the cognitive and mood sequelae of conditions such as multiple sclerosis,45 spinal cord injury,46,47 and obesity48,49 strongly affect patients’ capacity to engage in therapeutic activities. The ability to epigenetically modulate these factors via aerobic exercise is an exciting prospect for an adjunctive treatment to accompany targeted functional interventions.
Epigenetic Heritability
For physical therapists, the heritability of epigenetic traits is most germane in the context of cell replication within a single organism. Healthful adaptations induced by exercise may be passed down during mitotic cell division that occurs during tissue growth and repair. With sufficient exposure to exercise, the accumulation of epigenetically adapted cells and tissues may yield systemic changes in an organism that enhance health and longevity. However, in accordance with the Institute for Healthcare Improvement's “Triple Aim,”50 the physical therapy profession also bears a responsibility to address population health and promote the health of populations.51-53 This raises the provocative question of whether epigenetic adaptations induced by exercise could echo across generations in humans. As importantly, physical therapy clinicians must ask whether epigenetic adaptations arising from the lack of exercise could potentially play a role in the population-wide epidemic of obesity and metabolic disease.54
Some researchers postulate that because humans evolved in a calorie-restrictive environment, our epigenome is primed to adapt in ways that favor increased body weight and fat mass.5 In our present world of caloric excess, this predilection for energy conservation is maladaptive, leading to high rates of obesity and type 2 diabetes in the industrialized world. While a clear cause-and-effect relationship in humans has not yet been discovered, there is evidence for epigenetic differences in families with diabetes. Compared to individuals with no family history of diabetes, at least 65 genes in individuals with a positive family history of diabetes are differentially methylated, suggesting epigenetic heritability as a possible contributing factor.33
Animal studies provide abundant evidence that exercise-mediated epigenetic changes may be inherited by offspring. It is important to remember that three generations can simultaneously encounter a single epigenetic stressor: the mother, the fetus, and the (female) fetal gametes. Gestational exposure to toxins has been shown to affect the methylation status of rodent offspring out to the fourth generation,55 implicating changes in fetal gametes. It is theoretically possible, but not yet proven, that exercise-based epigenetic changes could yield similar germline adaptations.54 Paternal and maternal exercise both appear to alter the epigenetic status of offspring in rodent models. Paternal obesity transmitted an increased risk of metabolic syndrome to female offspring, but this disadvantageous inheritance could be corrected by offspring exercise.56 Another study demonstrated that a maternal high-fat diet caused hypermethylation of the PGC-1α promoter region in offspring, accompanied by metabolic disease that persisted through adulthood (12 months).57 In offspring of overfed dams who also exercised, both the hypermethylation and metabolic syndrome observed in offspring waned by 9 months of age. In humans, beneficial epigenetic inheritance of exercise-mediated traits has not yet been conclusively demonstrated. DNA methylation of certain genes has been shown to differ in infants born to mothers at different self-reported activity levels58 and body weights,59 but the biological significance of these adaptations is unknown. No prospective studies of exercise-based epigenetic adaptations in human parents and their children have yet been completed.
Summary: Epigenetic Application to Physical Therapy
Physical therapy, along with all other health care disciplines, will be affected by the rapid progression of precision medicine. Each year, more examples emerge of ways that individual patient genotyping can be used to optimize pharmaceutical treatment or to avoid dangerous drug sensitivities.60 Tailoring exercise recommendations to individual patient genotypes will be a future application for precision medicine.61 Complete genome sequencing can be done rapidly and at low cost (a few hundred dollars), making this a feasible component for inclusion in a patient's medical history in the coming decades. Health care systems are developing the massive data infrastructure needed to integrate genomic data into electronic medical records, and most importantly, to provide clinicians with the genomic data they need to prescribe optimized treatments.60 As knowledge advances about the genetic determinants of individual response to exercise, physical therapy exercise prescription will also capitalize on precision genomic information.
It will soon be possible to rapidly and affordably characterize the epigenome in the same way that the genome can currently be analyzed. We predict that epigenetic modification of key genes will be used to identify patients at an elevated risk for obesity, diabetes, cognitive decline, and other pathologies. Epigenetic markers will also be used as signposts to help contextualize the after-effects of periods of inactivity or immobility, especially with regard to muscle atrophy and metabolic dysregulation. Physical therapists will intervene to prescribe exercise-based treatments to mitigate these inherited or acquired epigenetically modulated traits. Physical therapy clinicians, as the health care team's experts in exercise dose prescription, will be at the forefront of research to uncover the optimal modes, intensities, and durations of exercise for preventing or reversing disease in patients with unfavorable epigenetic status. Much contemporary physical therapy research uses short exercise-based interventions (lasting a few weeks to a few months) to elicit desired adaptations. This practice will need to be re-examined if future epigenetic research indicates that stable epigenetic changes require a longer time course to consolidate. Fortunately, physical therapy clinical interventions are designed to be delivered over a prolonged period of time—ideally culminating in a lifestyle of patient-initiated exercise—providing the type of lengthy environmental exposure that favors healthful epigenetic adaptation.
Conclusion
Continued research examining the relationships between epigenetic modifications and exercise will enhance exercise prescription guidelines for healthy individuals and for patients living with disease or disability. Key factors to be considered in the future application of epigenetic knowledge to rehabilitation include differences in sampled tissues, differences in exercise dose and mode, and uncertainty about the heritability of exercise-modulated epigenetic traits. In the future, it is likely that epigenetic data will enhance our understanding of individual responses to prescribed exercise. The profession of physical therapy must be alert to new epigenetic knowledge that can enhance the specificity and efficacy of movement-based treatments.
Author Contributions
Concept/idea/research design: R.K. Shields
Writing: J.R. Woelfel, S. Dudley-Javoroski, R.K. Shields
Data collection: J.R. Woelfel, R.K. Shields
Data analysis: J.R. Woelfel, R.K. Shields
Project management: R.K. Shields
Fund procurement: R.K. Shields
Providing facilities/equipment: R.K. Shields
Providing institutional liaisons: R.K. Shields
Clerical/secretarial support: R.K. Shields
Consultation (including review of manuscript before submitting): R.K. Shields
Funding
The authors’ time to develop this manuscript was supported by funding from National Institutes of Health grants R01-HD082109 and R01-HD084645. The funders played no role in the writing of this perspective.
Disclosures
The authors completed the ICJME Form for Disclosure of Potential Conflicts of Interest. They reported no conflicts of interest.
References
- 1. Freund P, Rothwell J, Craggs M, Thompson AJ, Bestmann S. Corticomotor representation to a human forearm muscle changes following cervical spinal cord injury. Eur J Neurosci. 2011;34:1839–1846. [DOI] [PubMed] [Google Scholar]
- 2. Commission on Accreditation in Physical Therapy Education Standards and Required Elements for Accreditation of Physical Therapist Education Programs. Alexandria, VA: 2016. [Google Scholar]
- 3. Shields RK. 48th Mary McMillan lecture: Turning over the hourglass. Phys Ther. 2017;97:949–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrès R, Yan J, Egan B et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–411. [DOI] [PubMed] [Google Scholar]
- 5. Kirchner H, Osler ME, Krook A, Zierath JR. Epigenetic flexibility in metabolic regulation: disease cause and prevention? Trends Cell Biol. 2013;23:203–209. [DOI] [PubMed] [Google Scholar]
- 6. McGee SL, Fairlie E, Garnham AP, Hargreaves M. Exercise-induced histone modifications in human skeletal muscle. J Physiol. 2009;587:5951–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rasmussen M, Zierath JR, Barrès R. Dynamic epigenetic responses to muscle contraction. Drug Discov Today. 2014;232;19:1010–1014. [DOI] [PubMed] [Google Scholar]
- 8. Denham J, Marques FZ, O’Brien BJ, Charchar FJ. Exercise: putting action into our epigenome. Sports Med. 2014;44:189–209. [DOI] [PubMed] [Google Scholar]
- 9. Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eads CA, Danenberg KD, Kawakami K et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chatterjee A, Rodger EJ, Morison IM, Eccles MR, Stockwell PA. Tools and strategies for analysis of genome-wide and gene-specific DNA methylation patterns. Methods Mol Biol. 2017;1537:249–277. [DOI] [PubMed] [Google Scholar]
- 12. Fazzari MJ, Greally JM. Introduction to epigenomics and epigenome-wide analysis. Methods Mol Biol. 2010;620:243–265. [DOI] [PubMed] [Google Scholar]
- 13. Abel JL, Rissman EF. Running-induced epigenetic and gene expression changes in the adolescent brain. Int J Dev Neurosci. 2013;31:382–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gomez-Pinilla F, Zhuang Y, Feng J, Ying Z, Fan G. Exercise impacts brain-derived neurotrophic factor plasticity by engaging mechanisms of epigenetic regulation. Eur J Neurosci. 2011;33:383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wheatley KE, Nogueira LM, Perkins SN, Hursting SD. Differential effects of calorie restriction and exercise on the adipose transcriptome in diet-induced obese mice. J Obes. 2011;2011:265417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Narayan PJ, Dragunow M. High content analysis of histone acetylation in human cells and tissues. J Neurosci Methods. 2010;193:54–61. [DOI] [PubMed] [Google Scholar]
- 18. Christiansen L, Lenart A, Tan Q et al. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell. 2016;15:149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brown WM. Exercise-associated DNA methylation change in skeletal muscle and the importance of imprinted genes: a bioinformatics meta-analysis. Br J Sports Med. 2015;49:1567–1578. [DOI] [PubMed] [Google Scholar]
- 20. American College of Sports Medicine Position Stand The recommended quantity and quality of exercise for developing and maintaining cardiorespiratory and muscular fitness, and flexibility in healthy adults. Med Sci Sports Exerc. 1998;30:975–991. [DOI] [PubMed] [Google Scholar]
- 21. Carlström S, Karlefors T. Studies on fatty acid metabolism in diabetics during exercise. IV. Plasma free fatty acid concentrations and hemodynamics in juvenile diabetics during exercise before and after insulin treatment. Acta Med Scand. 1967;181:747–757. [PubMed] [Google Scholar]
- 22. Carroll DD, Courtney-Long EA, Stevens AC et al. Vital signs: disability and physical activity—United States, 2009–2012. MMWR Morb Mortal Wkly Rep. 2014;63:407–413. [PMC free article] [PubMed] [Google Scholar]
- 23. Voisin S, Eynon N, Yan X, Bishop DJ. Exercise training and DNA methylation in humans. Acta Physiol (Oxf). 2015;213:39–59. [DOI] [PubMed] [Google Scholar]
- 24. Quach A, Levine ME, Tanaka T et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging. 2017;9:419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horvath S, Pirazzini C, Bacalini MG et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging. 2015;7:1159–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Marioni RE, Shah S, McRae AF et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raina A, Zhao X, Grove ML et al. Cerebral white matter hyperintensities on MRI and acceleration of epigenetic aging: the Atherosclerosis Risk in Communities Study. Clin Epigenetics. 2017;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ambatipudi S, Horvath S, Perrier F et al. DNA methylome analysis identifies accelerated epigenetic ageing associated with postmenopausal breast cancer susceptibility. Eur J Cancer. 2017;75:299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luttropp K, Nordfors L, Ekstrom TJ, Lind L. Physical activity is associated with decreased global DNA methylation in Swedish older individuals. Scand J Clin Lab Invest. 2013;73:184–185. [DOI] [PubMed] [Google Scholar]
- 31. Ronn T, Volkov P, Davegardh C et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet. 2013;9:e1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ronti T, Lupattelli G, Mannarino E. The endocrine function of adipose tissue: an update. Clin Endocrinol (Oxf). 2006;64:355–365. [DOI] [PubMed] [Google Scholar]
- 33. Nitert MD, Dayeh T, Volkov P et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61:3322–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindholm ME, Marabita F, Gomez-Cabrero D et al. An integrative analysis reveals coordinated reprogramming of the epigenome and the transcriptome in human skeletal muscle after training. Epigenetics. 2014;9:1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ren H, Collins V, Clarke SJ et al. Epigenetic changes in response to tai chi practice: a pilot investigation of DNA methylation marks. Evid Based Complement Alternat Med. 2012;2012:841810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ainsworth BE, Haskell WL, Herrmann SD et al. 2011 Compendium of physical activities: a second update of codes and MET values. Med Sci Sports Exerc. 2011;43:1575–1581. [DOI] [PubMed] [Google Scholar]
- 37. Denham J, Marques FZ, Bruns EL, O’Brien BJ, Charchar FJ. Epigenetic changes in leukocytes after 8 weeks of resistance exercise training. Eur J Appl Physiol. 2016;116:1245–1253. [DOI] [PubMed] [Google Scholar]
- 38. Alibegovic AC, Sonne MP, Hojbjerre L et al. Insulin resistance induced by physical inactivity is associated with multiple transcriptional changes in skeletal muscle in young men. Am J Physiol Endocrinol Metab. 2010;299:E752–E763. [DOI] [PubMed] [Google Scholar]
- 39. Garber CE, Blissmer B, Deschenes MR et al. American College of Sports Medicine position stand. Quantity and quality of exercise for developing and maintaining cardiorespiratory, musculoskeletal, and neuromotor fitness in apparently healthy adults: guidance for prescribing exercise. Med Sci Sports Exerc. 2011;43:1334–1359. [DOI] [PubMed] [Google Scholar]
- 40. Fernandes J, Arida RM, Gomez-Pinilla F. Physical exercise as an epigenetic modulator of brain plasticity and cognition. Neurosci Biobehav Rev. 2017;80:443–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ieraci A, Mallei A, Musazzi L, Popoli M. Physical exercise and acute restraint stress differentially modulate hippocampal brain-derived neurotrophic factor transcripts and epigenetic mechanisms in mice. Hippocampus. 2015;25:1380–1392. [DOI] [PubMed] [Google Scholar]
- 42. Patki G, Solanki N, Atrooz F et al. Novel mechanistic insights into treadmill exercise based rescue of social defeat-induced anxiety-like behavior and memory impairment in rats. Physiol Behav. 2014;130:135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuipers SD, Bramham CR. Brain-derived neurotrophic factor mechanisms and function in adult synaptic plasticity: new insights and implications for therapy. Curr Opin Drug Discov Devel. 2006;9:580–586. [PubMed] [Google Scholar]
- 44. Hwang J, Castelli DM, Gonzalez-Lima F. The positive cognitive impact of aerobic fitness is associated with peripheral inflammatory and brain-derived neurotrophic biomarkers in young adults. Physiol Behav. 2017;179:75–89. [DOI] [PubMed] [Google Scholar]
- 45. Wynia K, Middel B, van Dijk JP, De Keyser JH, Reijneveld SA. The impact of disabilities on quality of life in people with multiple sclerosis. Mult Scler. 2008;14:972–980. [DOI] [PubMed] [Google Scholar]
- 46. Bloch A, Tamir D, Vakil E, Zeilig G. Specific deficit in implicit motor sequence learning following spinal cord injury. PLoS One. 2016;11:e0158396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Craig A, Guest R, Tran Y, Middleton J. Cognitive impairment and mood states after spinal cord injury. J Neurotrauma. 2017;34:1156–1163. [DOI] [PubMed] [Google Scholar]
- 48. Capuron L, Poitou C, Machaux-Tholliez D et al. Relationship between adiposity, emotional status and eating behaviour in obese women: role of inflammation. Psychol Med. 2011;41:1517–1528. [DOI] [PubMed] [Google Scholar]
- 49. Kerwin DR, Zhang Y, Kotchen JM et al. The cross-sectional relationship between body mass index, waist-hip ratio, and cognitive performance in postmenopausal women enrolled in the Women's Health Initiative. J Am Geriatr Soc. 2010;58:1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Institute for Healthcare Improvement. IHI Triple Aim Initiative http://www.ihi.org/engage/initiatives/tripleaim/Pages/default.aspx. Accessed June 13, 2018. [Google Scholar]
- 51. Jette AM. A bold vision for physical therapy. Phys Ther. 2017;97:946–947. [DOI] [PubMed] [Google Scholar]
- 52. Jensen GM, Nordstrom T, Mostrom E, Hack LM, Gwyer J. National study of excellence and innovation in physical therapist education: part 2—a call to reform. Phys Ther. 2017;97:875–888. [DOI] [PubMed] [Google Scholar]
- 53. Jette AM. 43rd Mary McMillan Lecture. Face into the storm. Phys Ther. 2012;92:1221–1229. [DOI] [PubMed] [Google Scholar]
- 54. Denham J. Exercise and epigenetic inheritance of disease risk. Acta Physiol (Oxf). 2018;e12881. [DOI] [PubMed] [Google Scholar]
- 55. Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McPherson NO, Owens JA, Fullston T, Lane M. Preconception diet or exercise intervention in obese fathers normalizes sperm microRNA profile and metabolic syndrome in female offspring. Am J Physiol Endocrinol Metab. 2015;308:E805–E821. [DOI] [PubMed] [Google Scholar]
- 57. Laker RC, Lillard TS, Okutsu M et al. Exercise prevents maternal high-fat diet-induced hypermethylation of the Pgc-1alpha gene and age-dependent metabolic dysfunction in the offspring. Diabetes. 2014;63:1605–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McCullough LE, Mendez MA, Miller EE, Murtha AP, Murphy SK, Hoyo C. Associations between prenatal physical activity, birth weight, and DNA methylation at genomically imprinted domains in a multiethnic newborn cohort. Epigenetics. 2015;10:597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soubry A, Murphy SK, Wang F et al. Newborns of obese parents have altered DNA methylation patterns at imprinted genes. Int J Obes (Lond). 2015;39:650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aronson SJ, Rehm HL. Building the foundation for genomics in precision medicine. Nature. 2015;526:336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ramirez-Velez R, Lobelo F, Izquierdo M. Exercise for disease prevention and management: a precision medicine approach. J Am Med Dir Assoc. 2017;18:633–634. [DOI] [PubMed] [Google Scholar]
- 62. Weizmann Institute of Science Gene Cards: The Human Gene Database. http://www.genecards.org/. Accessed September 14, 2017. [Google Scholar]