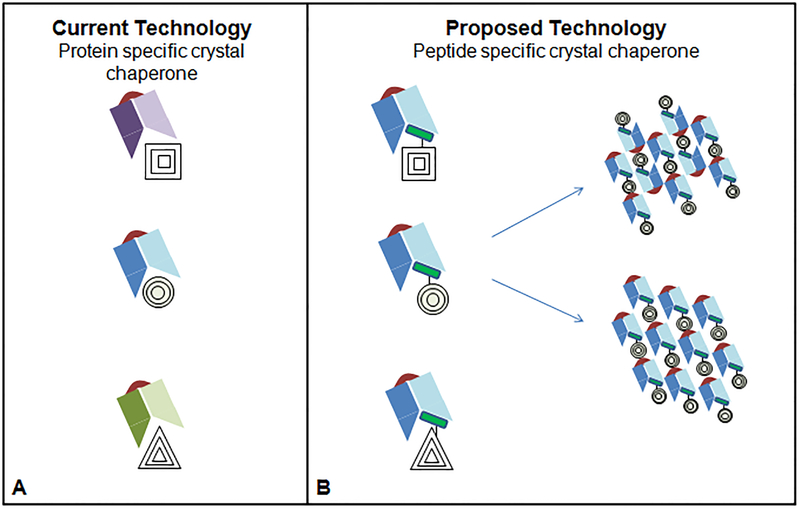
Figure 5. Comparison of the current methods to engineer non-covalent crystallization chaperones.
(a) Current hybridoma or molecular display methods are used to engineer a single chaperone to bind a single membrane protein. The chaperone cannot easily be adapted to alternative proteins, limiting the use of this method for high-throughput structural biology applications, as a new chaperone must be engineered for each additional protein. (b) An alternative is to engineer a chaperone to bind a peptide, which can be readily introduced into a construct. This would enable the use of a single chaperone for co-crystallization with many different membrane proteins.