Abstract
ATP-binding cassette (ABC) transporters are of major importance for the restricted access of toxins and drugs to the human body. At the body's barrier tissues like the blood–brain barrier, these transporters are highly represented. Especially, ABCB1 (P-glycoprotein) has been a priority target of pharmaceutical research, for instance, to aid chemotherapy of cancers, therapy resistant epilepsy, and lately even neurodegenerative diseases. To improve translational research, the humanization of mouse genes has become a popular tool although, like recently seen for Abcb1, not all approaches were successful. Here, we report the characterization of another unsuccessful commercially available ABCB1 humanized mouse strain. In vivo assessment of transporter activity using positron emission tomography imaging revealed a severe reduction of ABCB1 function in the brain of these mice. Analyses of brain mRNA and protein expression showed that the murine Abcb1a gene is still expressed in homozygous humanized animals while expression of the human gene is minimal. Promoter region analyses underpinned that the introduced human gene might dysregulate normal expression and provided insights into the regulation of both transcription and translation of Abcb1a. We conclude that insertion of the human coding DNA sequence (CDS) into exon 3 instead of exon 2 most probably represents a more promising strategy for Abcb1a humanization.
Keywords: ABCB1, P-gp, ABC transporter, PET imaging, mouse models, humanization
Introduction
Although most of the ATP-binding cassette (ABC) transporter family members are not linked to multidrug resistance, the probably most prominent one, ABCB1 (P-gp), is mainly known for its role in therapy resistant cancer. However, during the past 30 years, it became evident that it is not only of major importance for controlling access of cancer therapies to tumors, but that ABCB1(1) has a pronounced impact on drug distribution, excretion, and drug–drug interactions [1–3]; (2) is a stem cell marker [4–6]; (3) is linked to therapy resistant epilepsy and depression [7]; (4) is involved in the pathogenesis of neurodegenerative diseases [8–11]; and (5) is essential in a number of signaling pathways [5, 12–14]. In mice, the Abcb1 gene underwent a duplication resulting in the two isoforms Abcb1a and Abcb1b. The proteins share 87.3% and 80.5% identical amino acids, respectively, to human ABCB1, and several substrate specific differences were identified regarding affinity and specificity [15–17]. Although in vitro assays exist to assess the interaction of drugs with ABCB1 and predict certain in vivo characteristics of a substance, most drug development pipelines include mouse in vivo studies to assess pharmacokinetics, metabolism, toxicity, and side effects of new drugs [18]. To eliminate uncertainties of inter-species differences, humanized mouse models are being deployed recently [19]. Dallas et al. [20] reported the successful humanization of the Abcg2 gene in mice by exchanging the 107 kbp mouse gene for the 141 kbp human gene. The approach may suffer from the unpredictability of disrupting long-range transcriptional elements or possible incompatibilities between mouse and human binding sites. This, most probably, is the reason for the reported reduced protein expression in major organs. However, the model seems clearly useful for drug development and drug–drug interaction studies [20]. Sadiq et al. [21] published data on a humanized ABCB1 mouse model from Taconic using a fusion of the human ABCB1 coding DNA sequence (CDS) directly behind the start codon of both the Abcb1a and Abcb1b genes, instead of full humanization of the respective gene loci. Exhaustive protein expression analyses revealed that the human protein was expressed at negligible levels at the blood–brain barrier (BBB) while other proteins were not influenced. Interestingly, the murine ABCB1A protein was detected at only 10% of the wild-type level, but still about 7-fold higher than the human protein. Accordingly, the humanized ABCB1 mice rather resembled Abcb1a/b knockout mice than wild-type animals when assessing the brain distribution of several ABCB1 substrates [21]. Due to the increasing importance of ABCB1 in Alzheimer's disease (AD) and the increasing number of failed AD treatment trials, we set out to find another ABCB1 humanized mouse model that could serve as a basis for new AD mouse models. Here, we report the characterization of a second commercially available ABCB1 humanized mouse model that has been developed by genOway (Lyon, France) almost in parallel to the one reported by Sadiq et al. [21].
Materials and Methods
Mouse Models
Humanized ABCB1 mice in C57BL/6 background (hABCB1) have been purchased from genOway S.A. (Lyon, France) and from TaconicArtemis GmbH (Cologne, Germany) [21]. C57BL/6J wild-type and B6.C-Tg(CMV-cre)1Cgn/J (Cre-deleter) mice have been purchased from The Jackson Laboratory (Bar Harbor, USA). DNA sequencing data acquired during this study revealed that the human ABCB1 coding sequence in hABCB1 mice had been flanked by loxP sites. To induce hABCB1 knockout, we crossbred hABCB1 to Cre-deleter mice. The resulting homozygous knockout mice are referred to as human ABCB1–/– mice. All strains have been bred and housed under SOPF (specific and opportunistic pathogen-free) conditions at 21 ± 1 °C, 12 h/12 h light/dark cycle with food (PM3, Special Diet Services) and acidified water ad libitum. Genotyping of mice was done on ear biopsies employing a three-primer PCR that differentiated wild-type, knock-in, and knock-out mice (primers: humB1fw: 5′-GGCGTAGATTGAGCATGCTA-3′, humB1rc: 5′-AAAACACCGTCCTGAAAGCT-3′, humB1ko: 5′-AACAGCATATGGCTCAGGTG-3′). All protocols involving the breeding and use of animals were approved by the Landesverwaltungsamt Sachsen-Anhalt, Halle, Germany; the Norwegian Food Safety Authority (Mattilsynet); and the Austrian authority (Amt der Niederösterreichischen Landesregierung). All study procedures were performed in accordance with the European Communities Council Directive of September 22, 2010 (2010/63/EU).
Positron Emission Tomography (PET) Imaging
General
Tariquidar dimesylate was obtained from Xenova Ltd. (Slough, UK). For PET experiments, tariquidar was freshly dissolved prior to each administration in 2.5% (w/v) aqueous (aq.) dextrose solution and injected i.v. at a volume of 4 mL/kg body weight. (R)-[11C]verapamil was synthesized as described before [22] and formulated for i.v. injection in physiological saline solution (0.9%, w/v) containing 1% (v/v) Tween 80 at an approximate concentration of 370 MBq/mL. Specific activity at end of synthesis was 137 ± 34 GBq/μmol, and radiochemical purity was >98%.
Imaging Procedures
Groups of female C57BL/6N mice (age, 14–16 weeks), hABCB1 mice (genOway), and hABCB1 mice (TaconicArtemis GmbH) were either i.v. injected with vehicle solution (2.5% [w/v] aq. dextrose solution) or 15 mg/kg body weight tariquidar at 2 h before start of PET imaging (n = 3–6 per group). For injection, mice were pre-anesthetized in an induction chamber using isoflurane (2.5–3.5% in oxygen) and placed on a heated animal bed (38 °C), and the lateral tail vein was cannulated. Animal respiratory rate and body temperature were constantly monitored (SA Instruments Inc., Stony Brook, NY, USA), and the isoflurane level was adjusted (1.5–2% in oxygen) to achieve a constant and sufficient depth of anesthesia. Anesthesia was maintained for the whole pre-treatment and imaging period. Following administration, the animal bed was transferred into the gantry of a microPET scanner (Focus 220, Siemens Medical Solutions, Knoxville, TN, USA) and a 10-min transmission scan using a 57Co point source was recorded. Subsequently, at 2 h after tariquidar or vehicle injection, (R)-[11C] verapamil (32 ± 9 MBq, <0.5 nmol, 0.1 mL) was administered as an i.v. bolus over 1 min, and a 60 min dynamic PET scan (energy window, 250–750 keV; timing window, 6 ns) was initiated at the start of radiotracer injection. After completion of the imaging procedure, a terminal blood sample was withdrawn under isoflurane anesthesia from the retro-orbital sinus vein and the animals were sacrificed by cervical dislocation. Blood was measured for radioactivity in a gamma counter (Wizard 1470, Perkin-Elmer, Wellesley, MA, USA).
PET Data Analysis
The 60 min dynamic emission PET data were sorted into 23 frames, which incrementally increased in time length from 5 s to 10 min. Images were reconstructed using Fourier rebinning of the 3D sonograms followed by two-dimensional filtered back projection with a ramp filter, resulting in an image voxel size of 0.4 × 0.4 × 0.796 mm. A standard data correction protocol (normalization, attenuation and decay correction) was applied to the PET data. The PET units were converted into units of radioactivity concentration by applying a calibration factor derived from imaging of a cylindrical phantom with a known 11C-radioactivity concentration. Using the image analysis software Amide [23], whole brain was manually outlined on the PET images, and time-activity curves, expressed in standardized uptake value ((radioactivity per g/injected radioactivity) × body weight), were derived. Individual brain-to-blood concentration ratios (Kb,brain) were calculated by dividing the brain radioactivity concentration in the last PET frame (50 to 60 min after radiotracer injection) by the corresponding blood radioactivity concentration as determined by the gamma counter measurements.
Quantitative PCR
Mice were sacrificed by cervical dislocation. After quick intracardial perfusion with 10 mL ice-cold PBS, one hemisphere of each brain was snap-frozen in liquid nitrogen within 3 min after death. Brains of 100-day- and 200-day-old wild-type male mice (3 and 4, respectively) and hABCB1 male mice (5 each) were preserved using Allprotect® Tissue Reagent (Qiagen, Germany). Total RNA was isolated using the RNeasy® Mini Kit (Qiagen, Germany). RNA concentrations were assessed photometrically. cDNA was synthesized using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, USA). Gene expression was analyzed with qRT-PCR using TaqMan Hybridization Probes. Primer sets with according TaqMan probes for Abcb1a (Mm00440761_m1), human ABCB1 (Hs00184500_m1), and mouse Actb (Mm02619580_g1) genes were purchased from Thermo Fisher Scientific Inc. (USA). Gene expression was normalized to the housekeeping gene actin. All 17 cDNA samples were tested with each primer (Abcb1a, human ABCB1, and Actb), including a non-template control. Reactions were composed according to manufacturer's instructions with a final volume of 20 μL and 30 ng of template. PCR amplification conditions were 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. For comparison of expression values between strains, the unpaired Student's t test has been used. For analyses of transcript variants, RNA of a new set of brains was prepared from female wild-type, hABCB1, and human ABCB1–/– mice (5 each) and assayed using the mentioned assay and conditions plus assays Mm00440745_m1 and Mm00440751_m1.
Protein Quantification (QTAP)
Reagents
HBSS, HEPES, Tris-HCl, and sodium phosphate (Na2HPO4 and NaH2PO4) were provided by Sigma-Aldrich (Saint Quentin Fallavier, France). The reagents used for plasma membrane isolation and protein digestion, NaCl, MgCl2, KCl, sucrose, EDTA, guanidine–HCl, DTT, iodoacetamide, and urea also came from Sigma-Aldrich (Saint Quentin Fallavier, France). The complete Mini (EDTA-free) Protease Inhibitor Cocktail tablets were provided by Roche (Bâle, Switzerland). Chloroform (HiPerSolv, Chromanorm for high-performance liquid chromatography [HPLC]) was supplied by VWR (Strasbourg, France). HPLC-grade acetonitrile and methanol were purchased at Merck (Nogent-sur-Marne, France). Formic acid (99% w/w), HPLC grade, was supplied by Fischer Scientific (Illkirch, France). All the water was prepared with a Milli-Q water purification system (Millipore, Molsheim, France). Sequencing grade modified trypsin, mass spectrometry grade rLys-C, and ProteaseMAX surfactant were from Promega (Charbonnières-les-Bains, France). The measurement of protein concentration was carried out by using the Micro BCA Protein Assay Kit (Thermo Scientific, Illkirch, France). The standard solutions of peptides were provided by UMR 8638, Chimie Organique Médicinale et Extractive — Toxicologie Expérimentale or by Pepscan (Lelystad, The Netherlands).
Plasma Membrane Protein (PMP) Fractionation
PMP fractions of mouse brains were obtained as previously described with minor modifications [20, 24]. The frozen brains were thawed at 4 °C, and all the following steps were performed on ice. The samples were washed at least twice by using an isotonic buffer solution (10 mM phosphate buffer pH 7.4, 0.1 M KCl) supplemented with Protein Inhibitor Cocktail, minced with scalpels until obtaining 1 mm pieces and homogenized with an Ultraturrax® (IKA®-Werke GmbH & Co. KG, Staufen, Germany) for 2 min. The homogenates were centrifuged (15 min at 10,000g); the supernatants were transferred to ultracentrifuge tubes and centrifuged (60 min at 100,000g). The pellet, corresponding to the total membrane fraction, was suspended in 250 mM sucrose buffer (with 20 mM Tris pH 7.4 and 5.4 mM EDTA) and deposited on top of a 38% (w/v) sucrose solution. After ultracentrifugation (30 min at 100,000), the turbid layer at the interface was collected, suspended in 250 mM sucrose buffer and ultracentrifuged (30 min at 100,000). The pellet corresponding to the plasma membrane fraction was recovered in 250 mM sucrose buffer containing protease inhibitor cocktail.
Protein Digestion
The plasma membrane fractions were digested as described previously without modifications [25–27]. Briefly, 50 µg of proteins was solubilized in denaturing buffer (7 M guanidine hydrochloride, 10 mM EDTA, 500 mM Tris pH 8.5), reduced by DTT and alkylated by iodoacetamide. The alkylated proteins were precipitated with methanol–chloroform–water and resolubilized in 1.2 M urea and 0.1 M Tris, pH 8.5. Samples were first digested by using rLysC endoprotease (enzyme:protein ratio = 1:50) for 3 h at room temperature. Then trypsin (enzyme–protein ratio = 1:100) and 0.05% (w/v) ProteaseMAX were added, and samples were incubated at 37 °C overnight. The reaction was stopped by adding formic acid (3 µL at 99%, v/v). The stable isotope-labeled peptide mixture (750 fmol of each labeled peptide) was added in tryptic digest before ultrahigh-performance liquid chromatography–tandem mass spectrometry (UHPLC–MS/MS) analysis.
Protein Quantification by UHPLC–MS/MS
The proteins were quantified by the determination of the peptide concentration by using UHPLC–MS/MS with multiplexed selected reaction monitoring (SRM) method. Each peptide analyzed was specific to each protein and released after protein digestion by trypsin. The selected peptides for Abcb1a (LANDAAQVK), Abcb1a/b (IATEAIENFR), ABCB1 (FYDPLAGK), and GFAP (LDQLTANSAR); common peptide for Na+/K+ATPase α1/α2/α3 (AAVPDAVGK); and specific peptides for Na+/K+ATPase a1 (IVEIPFNSTNK), Na+/K+ATPase a2 (GIVIATGDR), and Na+/K+ATPase a3 (GVVVATGDR) were already reported by Hoshi et al. [25], Kamiie et al. [28], and Chaves et al. [29]. The samples were injected into an Acquity UPLC® system (Waters, Manchester, UK), equipped with an Acquity UPLC BEH® C18 column (Peptide BEH® C18 Column, 300Å, 1.7 µm, 2.1 mm × 100 mm) supplied by Waters (Guyancourt, France). The mobile phase consisted of a mixture of water (formic acid 0.1%, v/v) and ACN. It was operated with a flow rate of 0.5 mL/min in gradient mode, at a temperature of 30 °C. The total duration of analysis was 30 min.
Data were recorded with a Waters Xevo® TQ-S mass spectrometer (Waters, Manchester, UK). Measurements were performed by using positive electrospray ionization (ESI) with ion spray capillary voltage at 2.80 kV. Drying gas temperature was set to 650 °C at a flow rate of 800 L/h. Detection was performed in multiplexed SRM mode by using three transitions per native or labeled peptide. Skyline software [30, 31] was used for the optimization of the specific transition parameters (i.e., collision energy [CE] and peak integration). The area ratios light to labeled peptide were exported from Skyline, and the quantification was performed from calibration curves. Each sample contained two hemi cortices from different mice. It was digested in triplicate, and each triplicate was injected once. Therefore, the calculated concentrations (Table 1) take into account the experimental and biological variability.
Table 1.
Protein quantitation
| Protein | Mouse ABCB1A/B | Mouse ABCB1A | Human ABCB1 | Mouse ATP1A1 | Mouse ATP1A2 | Mouse ATP1A3 | Mouse GFAP |
|---|---|---|---|---|---|---|---|
| Peptide | IATEAIENFR | LANDAAQVK | FYDPLAGK | IVEIPFNSTNK | GIVIATGDR | GVVVATGDR | LDQLTANSAR |
| Strain | Concentration [fmol/µg total protein] (CV [%]) | ||||||
| LoD/LoQ | 0.05/0.16 | 0.08/0.23 | 0.23/0.70 | 0.58/1.73 | 0.04/0.12 | 0.07/0.25 | 0.19/0.28 |
| hABCB1 | <LoD | <LoD | <LoQ | 131.5 (34.9) | 86.0 (19.2) | 435.2 (26.9) | 1.0 (14.0) |
| hABCB1–/– | 0.44 (30.6) | 0.36 (14.4) | <LoD | 106.8 (17.0) | 78.8 (13.8) | 371.6 (11.0) | 1.0 (32.0) |
| WT | 0.72 (13.7) | 0.64 (12.1) | <LoD | 119.9 (16.6) | 80.9 (12.6) | 392.2 (15.6) | 0.8 (25.1) |
- Neurons, astrocytes, endothelial cells express ATP1A1.
- Both astrocytes and endothelial cells express ATP1A2, but endothelial cells in very low amounts.
- Neurons express ATP1A3.
- Astrocytes express GFAP.
The concentrations of different markers are not different between the samples and thus show the homogeneity of their cell composition. Therefore, the concentrations of ABCB1 proteins in the samples can be compared with each other. The QuaSAR plugin (Mani et al. [42]) was used to calculate the limit of detection and limit of quantitation.
CV (%) – biological and experimental variability, LoQ – lower limit of quantitation, LoD – lower limit of detection
Western Blot
Mice were sacrificed by cervical dislocation. After quick intracardial perfusion with 10 mL ice-cold PBS, one hemisphere of each brain was snap-frozen in liquid nitrogen within 3 min after death. Hemispheres of three 100-day-old male mice of each genotype were preserved using Allprotect® Tissue Reagent (Qiagen, Germany). After removal of Allprotect® reagent, brains were homogenized using a bead homogenizer (SpeedMill, AnalytikJena, Germany). Twenty millgrams of homogenate was mixed with homogenization buffer (20 mM EDTA, 140 mM NaCl, 5% SDS, cOmplete® mini protease inhibitor [Roche]) and incubated at 50 °C for 1 h [32]. After centrifugation at 13,000 rpm (30 min, 20 °C), protein concentration was determined using a BCA protein assay kit (ThermoFisher Scientific, USA). Samples were diluted to a protein concentration of 3 µg/µL in Laemmli buffer and denaturated at 42 °C for 30 min. Sixty milligrams of protein was loaded onto 12.5% TGX gels (Biorad, Germany) for electrophoresis and transferred onto 0.22 µm PVDF membranes using a TransBlot Turbo (Biorad, Germany). Membranes were blocked with 1.5% non-fat dry milk powder solubilized in PBS (0.01% Tween20) for 1 h at room temperature. Primary antibody was incubated over night at 4 °C, and secondary antibody incubation was 1 h at room temperature. Antibodies used were anti-ABCB1 (1:100, D-11, SantaCruz Biotechnologies), anti-β-Tubulin (1:10.000, SDL.3D10, Novus Biologicals), and anti-mouse HRP (1:10.000, NordicBiosite). Bands were detected using Clarity® Western ECL substrate (Biorad, Germany) and the Octoplus QPLEX imager (NHDyeAgnostics, Germany). Data analyzes were performed using Image Studio Lite (LI-COR Biosciences, Germany) and Microsoft Excel.
Sequencing
Genomic DNA of a hABCB1 mouse was purified using an innuPREP DNA Mini Kit (AnalytikJena AG, Germany). The humanized genomic locus was amplified in 25 µL reactions using TaKaRa LA TaqDNA Polymerase (TaKaRa Bio USA Inc.) according to manufacturer recommendations using 0.4 µM of each primer and an annealing temperature of 60 °C, resulting to single-banded PCR products. After amplification, the remaining primers and dNTPs were digested. The amplified fragments were sequenced using the Sanger process utilizing a primer walking strategy. For the sequencing of exons 3 and 4, PCR reactions were run with 12.5 ng template DNA in a total volume of 12.5 µL. PCR reactions including 0.3 µM of each PCR primer, 200 µM dNTPs, 1.5 mM MgCl2 and 0.02 U/µL KAPA2G Robust polymerase. 35 cycles were run for each PCR with an initial denaturation step of 180 s at 95 °C followed by 35 cycles with a denaturation step for 20 s at 95 °C, an annealing step at 61 °C for 20 s, an elongation step for 100 s at 72 °C and a final elongation step at 72 ° for 45 s. Excess dNTPs and primers were removed from PCR reactions and Sanger sequencing was performed. All primers used are listed in Supplementary Table 1. Amplification, primer design, and sequencing were realized by Microsynth AG (Switzerland).
Supplementary Table S1.
Primers used for amplification and primer walking
| Primer | Sequence | Purpose |
|---|---|---|
| ABCB1_Fw | ACTGGGGACTGTCCTCTTTC | Amplification |
| ABCB1_Rev | CCTCCCCACCATTAAAGTTC | Amplification |
| forward1 | ACT GTC CTC TTT CTG GTT TG | Primer walking |
| forward2 | ACT CAG AGC CGC TTC TTC | Primer walking |
| forward4 | GGA ACT TTG GCT GCC ATC | Primer walking |
| forward5 | GTA GCT GAA GAG GTC TTG | Primer walking |
| forward6 | TGG ACA GGA TAT TAG GAC | Primer walking |
| forward7 | AAG ATC CAG TCT AAT AAG | Primer walking |
| forward8 | TAC TCT TAG CAA TTG TAC | Primer walking |
| forward9 | AGA CGC TGG CTC TGG TG | Primer walking |
| forward10 | TCA ATG GTC AGT GTC CAG | Primer walking |
| 1_reverse1 | AGC TGA TAA CTT CGT ATA G | Primer walking |
| 1_reverse2 | GTG GGA ATG GTT AAG CAG | Primer walking |
| forward3.1 | AGA AAC CAA CTG TCA GTG | Primer walking |
| reverse2 | TAA TAG GCA TAC CTG GTC | Primer walking |
| 1_reverse3 | AGT GGA GAG AAA TCA TAG | Primer walking |
| Abcb1a_Ex02_Fw | TCCTGGCTCTCATCGGTAAC | Amplification |
| Abcb1a_Ex02_Rev | AGCATATTGTTCCAGGCGAA | Amplification |
| Abcb1a_Ex03_Fw | GGGCAGCTTAGACTAGTAGCT | Amplification |
| Abcb1a_Ex03_Rev | CACCCACCACATAGACCACT | Amplification |
DNA Accessibility
Hotspots for DNA accessibility have been extracted from the ENCORE database. DNAse-seq data sets used were as follows: ENCSR337EDG, ENCSR179PIH, ENCSR358ESL, ENCSR469VGZ, ENCSR292QBA, and ENCSR767AJS. ATAC-seq data from ENCFF386CJN. Chromatin immunoprecipitation sequencing (ChIP-seq) data were extracted from ENCBS139ENC.
Data Availability
The data sets generated during and analyzed during the current study are available on reasonable request. Correspondence and requests for materials should be addressed to M.K. (markus.krohn@medisin.uio.no), J.P. (jens. pahnke@medisin.uio.no), W.L. (wolfgang.loescher@tihohannover.de), or O.L. (oliver.langer@ait.ac.at). All ENCODE data sets are publicly available at www.encodeproject.org.
Results
The humanized ABCB1 (hABCB1) strain under investigation was an unsolicited development by genOway (Lyon, France). The company used a strategy it claimed to have successfully employed earlier to introduce a canine ABCB1 gene into the murine Abcb1a locus [33, 34]. To generate the hABCB1 strain, the mouse Abcb1a gene was humanized by introduction of the human ABCB1 CDS into the gene's ATG start codon in exon 2. The mouse Abcb1b gene was partially deleted by insertion of a hygromycin cassette replacing exons 3 and 4 to ensure that only the human gene would be expressed. At purchase from genOway, the promising information available was that the human ABCB1 mRNA is expressed at similar levels as is Abcb1a in wild-type animals in the tissues tested (brain, liver, intestine, kidney). Correct integration of the transgene had been checked using Southern blot. However, the knockout capability genOway aimed to introduce by gene floxing was reported to be non-functional and no protein expression data had been ascertained either. Thus, we agreed to verify the usefulness of this model in more detail and analyze ABCB1 transport activity/functionality and expression in the brain.
Positron Emission Tomography (PET) Imaging
The most important measure of validity for humanized ABC transporter mouse models is the functionality of the transporters in vivo. As transporter functionality is not assessable through mRNA or protein expression levels, we assessed ABCB1 transport activity at the BBB by PET imaging using the validated ABCB1 substrate radiotracer (R)-[11C]verapamil in combination with administration of the ABCB1 inhibitor Tariquidar as described before [35, 36]. The hABCB1 strain from Taconic was included here, because it was extensively investigated before and shown to be insufficient for studies regarding ABCB1 function [21]. In vehicle-treated animals, brain uptake of (R)-[11C]verapamil expressed as the brain-to-blood radioactivity concentration ratio at 60 min after radiotracer injection (Kb,brain) was significantly higher in hABCB1 mice from genOway (Kb,brain = 3.26 ± 0.13) and Taconic (Kb,brain = 2.76 ± 0.56) as compared with C57BL/6 mice (Kb,brain = 1.22 ± 0.06, p < 0.001, 2-way ANOVA with Bonferroni post-hoc test, Figure 1). Tariquidar treatment significantly increased (p < 0.05, Student's t test) Kb,brain in C57BL/6 mice by 3.6-fold (Kb,brain = 4.35 ± 0.45), whereas increases were only 1.4-fold in hABCB1 mice from both genOway (Kb,brain = 4.40 ± 0.35) and Taconic (Kb,brain = 3.90 ± 0.21), respectively. For comparison, previously published Kb,brain in vehicle-treated Abcb1a/b knockout mice (Abcb1a/b(–/–) mice) in C57BL/6 background was 4.81 ± 0.51 [36]. Although both humanized strains show some remaining transport capacity, as seen by the increase in Kb,brain after tariquidar treatment, we concluded that the hABCB1 mice from genOway display the same lack of functionality as does the Taconic strain.
Figure 1.
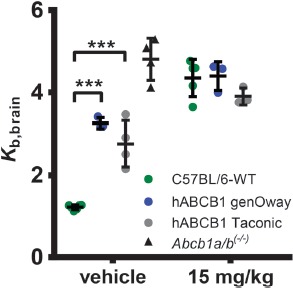
ABCB1 transport activity at the BBB measured with (R)-[11C]verapamil PET imaging. Brain uptake of (R)-[11C]verapamil expressed as brain-to-blood radioactivity concentration ratio (Kb,brain) at 60 min after radiotracer injection in female C57BL/6 mice (veh: n = 6, tariquidar: n = 5), hABCB1 mice (genOway; veh: n = 3, tariquidar: n = 3), and hABCB1 mice (Taconic; veh: n = 4, tariquidar: n = 3) treated with vehicle solution (2.5% [w/v] aq. dextrose solution) or tariquidar (15 mg/kg body weight) at 2 h before radiotracer injection. For comparison, Kb,brain in vehicle-treated Abcb1a/b(–/–) mice (n = 4) is also shown [36]. Data are mean ± standard deviation. Statistical significance was determined by 2-way ANOVA with Bonferroni post-hoc test. **p < 0.01; ***p < 0.001
mRNA Expression Analyses
To reveal the cause for this lack of functionality, we rechecked human ABCB1 and Abcb1a brain expression in the mice developed by genOway. The cycle threshold (ct) values of the amplification plots did not show a difference between mice of different age and were combined into one group for each strain. Target amplification could be verified for Actb (as endogenous reference) and Abcb1a mRNA in each sample, whereas ABCB1 mRNA expression was only detected in humanized mice. Unexpectedly, mRNA expression of Abcb1a was still apparent at wild-type levels (5.0 × 10–3 ± 2.4 × 10–3) in hABCB1 mice (4.0 × 10–3 ± 9.9 × 10–4) (Figure 2). At the same time, Abcb1a mRNA levels in ABCB1 humanized mouse brains were 35.9 times higher than human ABCB1 expression (4.0 × 10–3 ± 9.9 × 10–4 vs. 1.1 × 10–4 ± 7.7 × 10–5, one-way ANOVA, Tukey's honest significant difference post-hoc test, p < 0.0001, Figure 2).
Figure 2.
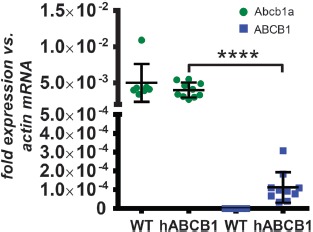
Relative mRNA expression of Abcb1a and ABCB1. Human ABCB1 mRNA is expressed at very low levels in hABCB1 mice only. No decrease of Abcb1a mRNA expression was detected in hABCB1 mice compared to wild-type animals. Shown are the relative expression of mouse Abcb1a (green) and human ABCB1 (blue) mRNA in male wild-type (n = 7) and hABCB1 (n = 10) mice based on the comparison of ct values. Ct values were normalized to mouse Actb mRNA expression using 2(–Δct) calculation. Data are mean ± standard deviation. Statistical significance was determined by 1-way ANOVA with Tukey's honest significant difference post-hoc test. ****p < 0.0001
mRNA expression analysis is sensitive to the position of primers and probes chosen. In this case, the primer set chosen for the Abcb1a mRNA was located over exon boundary 20–21, far distal to the introduced gene, because it covers all known splice variants of the gene. Thus, the qPCR outcome could result from the amplification of a greatly truncated Abcb1a mRNA that does not give rise to a functional protein. We performed an in silico translation starting from the first base of exon 3 to find possible open reading frames and see whether any truncated versions of ABCB1A proteins could be expressed in the humanized mice. As shown in Supplementary Figure 1, in frame 2 of exon 3, the last codon is an ATG which could initiate an open reading frame for a protein of 1239 amino acids length which would be fully identical to ABCB1A except for an N-terminal truncation of 37 amino acids. While this protein might still be functional, probably none of other 24 possible variants would (Supplementary Figure 1). Thus, we analyzed Abcb1a mRNA expression using two other assays located at exon boundaries 3–4 and 9–10, respectively, to verify the abundance of longer Abcb1a transcript variants. Figure 3 shows that the level of expression for all analyzed positions is similar between wild-type and hABCB1 mice. Resequencing of the engineered Abcb1a locus (see following section) confirmed the presence of valid loxP sites flanking the human ABCB1 CDS. We sought to assess if these are not functional as has been stated by genOway. To do so, we crossed hABCB1 mice to a Cre-deleter mouse strain (CMV-driven Cre recombinase, see Section 0 for details) to check Cre recombination. Genotyping PCR confirmed that this crossing resulted in recombination of the locus and generation of genomic ABCB1 knockout mice (referred to as human ABCB1–/– mice). However, as can be seen in Figure 3, Cre recombination had no effect on the Abcb1a mRNA expression level as assessed by any of the assays when compared to hABCB1 and wild-type mice. mRNA of the hABCB1 transgene was undetectable in these ABCB1–/– mice (not shown). Upon further communication, genOway revealed that only human ABCB1 mRNA expression had been analyzed in-house, but not mouse gene expression. This fact might explain why the lack of functionality has not been detected earlier.
Supplementary Figure 1. Full exonic sequence of Abcb1a.


Capital letters indicate the coding sequence, lower case indicate untranslated bases. ATG start codons that could establish an open reading frame are indicated green. Exon junctions are indicated by an |. Pink letters indicate the central base of the probes of the TaqMan assays used (according manufactures information).
Figure 3.
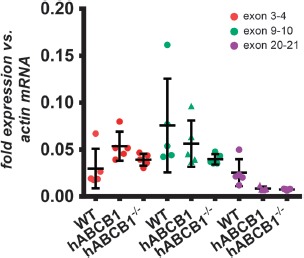
Exon junction-specific mRNA expression of Abcb1a. Abundance of Abcb1a mRNAs with different lengths was assessed using assays covering the indicated exon junctions in female wild-type mice (n = 5), hABCB1 mice (n = 5), as well as in hABCB1 mice (n = 5) crossed to a Cre-deleter mouse strain (human ABCB1–/–). No differential expression between strains was found within each assay location. Data are mean ± standard deviation. Statistical significance was determined by 1-way ANOVA with Tukey's honest significant difference post-hoc test, significance level p < 0.05
Protein Expression Analyses
Due to the unavailability of species-specific antibodies that allow differentiation between human and mouse ABCB1 proteins, we investigated protein expression of mouse ABCB1A/B and human ABCB1 using quantitative targeted absolute proteomics (QTAP) [26]. By detection of tryptic peptides specific for either the mouse or human protein variant, this method allows quantification of the transporters in whole brain homogenates. Table 1 shows that in wild-type mice both the peptide specific for mouse ABCB1A and the mouse ABCB1A/B-co-specific peptide are readily detectable. Although no ABCB1B-specific peptide has been analyzed, it can be assumed that most peptides found originate from ABCB1A proteins only, since both mouse specific peptides were found at similar quantities. In contrast, in hABCB1 mouse brains, no ABCB1A/B could be detected. Moreover, the human ABCB1 protein amount was below the lower limit of quantification (LoQ). These data are well in line with functional PET imaging data but seem to contradict qPCR results. However, it is known that mRNA expression data do often not correlate well with protein expression data. Most interestingly, human ABCB1–/– mice displayed partly rescued ABCB1A protein expression. However, also with this method, we were not able to determine the extent of the N-terminal truncation that was inflicted by Cre recombination of the Abcb1a locus. Western blot analyses using a C-terminal targeting antibody additionally supported our PET imaging data; however, no obvious protein truncation could be detected (Supplementary Figure 2).
Supplementary Figure 2. Western blot analysis of ABCB1 expression.
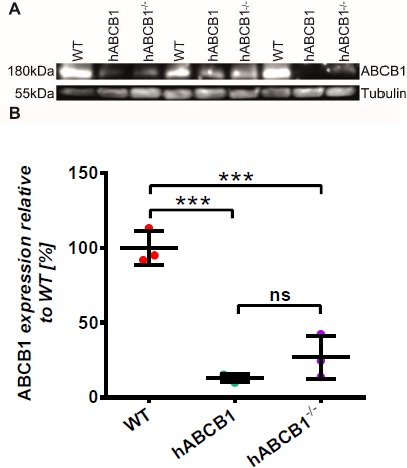
Western blot against ABCB1 confirmed PET imaging findings in hABCB1 animals and shows drastically reduced protein expression of any ABCB1 variant in hABCB1 as well as hABCB1–/– mice. Data are mean ± standard deviation. Statistical significance was determined by 1-way ANOVA with Tukey's Honest Significant Difference post-hoc test, significance level p<0.05, *** p<0.001.
Genomic DNA Sequencing
We sought to find possible explanations for the revealed shortcomings and to gather knowledge for a more successful knock-in strategy by sequencing of the humanized genomic locus. The sequenced region comprised 6089 bp including segments of mouse genomic DNA at both the 5′-and 3′-ends, suggesting that the engineered locus has been covered in full. As indicated in Figure 4, the human CDS including its 3′-untranslated region (UTR), but without the 5′-UTR, was successfully fused to the start codon of the Abcb1a gene located in exon 2. Exon 2 of Abcb1a was deleted except for its untranslated base pairs -1 to -6 from transcription start site (TSS). At TSS +4598 bp, the unaltered mouse genomic DNA sequence starts again with the first base pair aligning to intronic base pair TSS +267 of the mouse Abcb1a gene in wild-type mice (Supplementary Figure 3). Thus, exon 2 (TSS 0 to +65) and the first 202 bp of intron 2 have been deleted by the human CDS construct, in total 268 bp. Additional sequencing of exons 3 and 4 revealed that neither exon 3 nor exon 4 has been deleted nor altered during gene targeting, a fact later confirmed by genOway (Supplementary Figure 4). These data corroborate the mRNA expression analyses and again imply the possibility of expression of a truncated ABCB1A protein that is, however, most probably dysfunctional.
Figure 4.
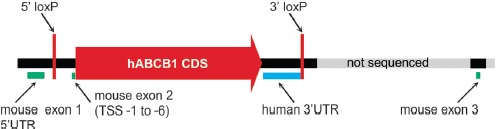
Depiction of the humanized Abcb1a gene locus. Shown are the sequenced region (6089 bp, black) and the downstream adjacent mouse sequence not included in the sequencing (light grey). The human ABCB1 CDS was fused into the TSS, which left the untranslated base pairs TSS -1 to -6 of exon 2 behind (green). ABCB1 CDS has been introduced with its 3′UTR and additional 45 bp of human genomic downstream sequence. At the 3′ transition towards the mouse genomic sequence, a loxP site has been introduced which is flanked by altogether 99 bp of unknown origin. The same is true for the 5′ situated loxP site, which is flanked by additional 37 bp, respectively, and inserted into intron 1 of mouse Abcb1a. The picture is not proportional to the genomic arrangement.
Supplementary Figure 3. In hABCB1 mice 266 bp of Abcb1a are deleted.

Alignment of hABCB1 sequence starting at the first base pair downstream of the loxP insertion site with Abcb1a starting at TSS.
Supplementary Figure 4. Abcb1a exons 3 and 4 are unaltered.

Alignment of exon 3 (a) and exon 4 (b) as sequenced from hABCB1 mice with Abcb1a reference sequence (Gene ID: 18671). Bold letters indicate exons.
Unexpectedly, in addition to the loxP sites flanking the hABCB1 gene, we found DNA of unknown origin flanking both sites. In case of the 5′-loxP site, there are 20 bp upstream and 17 bp downstream of the actual Cre recombinase recognition motif. The 3′-loxP site is flanked by 76 bp and 23 bp, respectively. BLAST results strongly suggest remnants of cloning vectors containing loxP sites. The alien DNA containing the 5′-loxP site is situated –413 bp to –492 bp from TSS, thereby disrupting intron 1 of Abcb1a and possibly interfering with promoter elements.
Thus, we searched the ENCODE database for information about DNA accessibility of this region [37, 38]. Several DN-Ase-seq (DNase I hypersensitive sites sequencing) data sets (deposited after generation of this mouse model) derived from adult mouse brains revealed hotspots in the region of interest. The combined hotspot region (covered by at least 5 data sets) reaches from about 200 bp upstream of exon 1 until about 160 bp downstream of exon 2 (Figure 5). ATAC-seq (Assay for Transposase-Accessible Chromatin sequencing) data generated from postnatal mouse forebrains revealed a hotspot of about 690 bp reaching until exon 2 (Figure 5). Furthermore, ChIP-seq experiments targeting tri-methylated histone H3 (H3K4me3) found a similar hotspot in cortical plate samples of adult mice (Figure 5). Due to the repeated occurrence of high DNA accessibility using different methods, we conclude that a region of high regulatory importance includes at least intron 1 of Abcb1a but most likely exon 2 as well.
Figure 5.
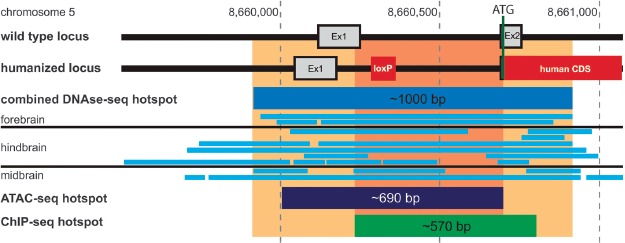
Abcb1a promoter region analysis. Shown are the Abcb1a loci of wild-type and hABCB1 mice in comparison. ENCODE database searches revealed the indicated hotspots of open/accessible chromatin found in mouse brains using DNAse-seq, ATAC-seq, and ChIP-seq methods. The area of highest density of overlapping hotspots (orange/light orange) covers more than 200 bp indicating the importance of this region for Abcb1a gene expression.
Discussion
With the study presented here, we set out to assess the functionality and expression of human ABCB1 protein in the brain of an Abcb1a humanized, Abcb1b knockout mouse model [21] developed by genOway. Sequence analyses of the humanized locus confirmed proper insertion and targeting of the mouse Abcb1a gene including the deletion of mouse exon 2. Nevertheless, mRNA expression data revealed a very low expression of ABCB1 mRNA at a level less than 3% of that of Abcb1a in wild-type mice (Figure 2). Accordingly, human ABCB1 protein did not reach the lower limit of quantitation and functional PET analyses revealed a knockout phenotype of hABCB1 mice. Whereas (R)-[11C]verapamil PET imaging without tariquidar treatment led to significantly higher radioactivity uptake in the brains of hABCB1 mice as compared to wild-type mice, ABCB1 inhibition with tariquidar induced only a minor increase in radioactivity uptake in hABCB1 animals. Hence, the genOway hABCB1 mice display a functional phenotype very similar to that of hABCB1 mice introduced by Taconic (see also Sadiq et al. [21]).
Intriguingly, murine Abcb1a mRNA is still present at wild-type levels in the humanized mice's brain (Figure 2) indicating the expression of a truncated Abcb1a gene product starting downstream of exon 2. In silico translation analysis of the Abcb1a coding sequence and qPCR revealed the possibility of a protein being expressed that lacks at least the first 37 amino acids and thus the cytoplasmic N-terminus up to transmembrane helix 1 (Figure 3). However, mouse ABCB1A/B proteins were not detectable at all in hABCB1 mice (Table 1).
Since the human CDS has been inserted correctly and this strategy was employed successfully to introduce a canine ABCB1 earlier [33, 34], the main question is: what caused the lack of expression in both (Taconic and genOway) humanized mouse models? A review of the canine ABCB1 mouse data points to an answer. The major difference to both humanized strains is the insertion of a mutated ABCB1 CDS in the canine model. Especially, dogs of the Collie lineage often suffer from severe adverse responses to ivermectin, which is a commonly used anthelmintic [39]. It is also that substrate of ABCB1 and genetic analyses of ivermectin-sensitive Collies revealed an exonic deletion of 4 bps leading to a frameshift and premature termination of translation [40]. The canine ABCB1 mouse strain was developed as an alternative model to Collies in the search for anthelmintic drugs that do not pose a life danger to such dogs. The canine ABCB1 mouse strain, therefore, is functionally an ABCB1-knockout model in which a lack of expression of the canine ABCB1 protein cannot be detected using drug-induced phenotypes or PET measurements. Thus, it is very likely that the canine ABCB1 mice exhibit a similar lack of protein expression as observed in the human ABCB1 strains developed by Taconic and genOway (see Table 2 for comparison).
Table 2.
ABCB1 knock-in mouse model comparison
| Model | Producer | Insertion site | Insertion | Special features | Ref |
|---|---|---|---|---|---|
| Canine ABCB1 | genOway | Abcb1a ATP-start codon | Canine cDNA (~4300 bp) | Intended 4 bp deletion with frame shift at pos. TSS +294 | [33] |
| Humanized ABCB1 | Taconic | Abcb1a ATP-start codon | Human cDNA (~4300 bp) | Abcb1b knockout | [21] |
| Humanized ABCB1 | genOway | Abcb1a ATP-start codon | Human cDNA (5106 bp) | loxP insertion TSS –492 and TSS +4628 | This work |
Overview of ABCB1 knock-in mouse models published to date with insertions site of the alien DNA and distinctive features. None of the models express functional ABCB1 proteins.
Notably, all strains have been designed using insertion of ABCB1 CDS into the ATG start codon of Abcb1a. Sadiq et al. [21] found the human ABCB1 protein expression in brain capillaries of hABCB1 mice from Taconic to be about 1.5% of that of ABCB1A in wild-type mice. Interestingly, the Taconic hABCB1 mice still expressed mouse ABCB1A protein, though at a level of only about 10% of that of wild-type mice [21]. In the genOway strain, ABCB1A/B protein levels in hABCB1 mice did not reach the lower limit of detection. This difference is most probably due to the different modes of tissue preparation used in both studies. While Sadiq et al. analyzed capillary-enriched brain preparations only, we used whole brain homogenates. Although detection of any ABCB1 protein in whole brain homogenates is hampered by a low enrichment of microvessels, ABCB1A was readily detectable in wild-type mice (Table 1). Unfortunately, the LoQ of human ABCB1 was rather high compared to the murine variants (0.7 vs. 0.23 fmol/µg protein). Nevertheless, human ABCB1 protein was at least detectable in hABCB1 mice whereas ABCB1A was not despite its much lower LoD (0.23 vs. 0.08 fmol/µg protein). We therefore conclude that the residual increase of Kb,brain in hABCB1 mice seen after tariquidar treatment is the result of a very low expression of human ABCB1 only.
More importantly, the combined data from our study and from Sadiq et al. show that especially the disruption of exon 2 of Abcb1a diminishes expression of the inserted gene/protein. Only the ABCB1 humanized mice developed by genOway possess a known alteration of intron 1, but neither the hABCB1 mice from Taconic nor the canine ABCB1 mice, suggesting that the structural integrity of exon 2 is of paramount importance since all models lack protein expression. The overlap of several hotspots of DNA accessibility extracted from ENCODE data sets within the region of CDS insertion reinforces its importance in gene regulation. Interestingly, the lack of two-thirds of intron 1 and all of exon 2 in human ABCB1–/– mice restored mouse ABCB1A protein expression to about 50% of wild-type levels according to QTAP results. However, Western blot results corroborate PET imaging data and QTAP results but, confusingly, do not reveal truncated protein variants. The lack of a visible shift in protein size might be due to the extremely weak signals though and a lack of resolution of the chosen gel matrix.
One could speculate that recombination of the locus restores some regulatory features partly located in intron 2 that have been disrupted by insertion of the CDS. More likely, however, it seems that alternative splicing and/or alternative polyadenylation occurs, which leads to truncated/unstable mRNAs and low translation yields. Nevertheless, the somewhat sloppy integration of the loxP sites might have had an additional effect on gene expression.
In conclusion, we think a humanization of the Abcb1a gene would have a higher likelihood of success when the human CDS is being inserted into exon 3 while intron 1/exon 2 is left untouched or introduced changes kept to a minimum. Exon 2 encodes for the first 21 aa of ABCB1A which share only 52.2% identity to human ABCB1. Hence, insertion of the human CDS into exon 3 without adapting exon 2 would generate a chimeric hABCB1 model. The same strategy was recently successfully employed to develop humanized ABCC1 and humanized ABCA7 mice (Krohn et al., unpublished). On the other hand, humanization of the first 21 aa could be achieved by exchanging 13 of the 68 bp of exon 2 plus an additional insertion of one codon. Thus, this decision might become a trade-off between protein expression and protein function. Although the functional relevance of this very N-terminus remains to be determined, it seems unlikely to have much impact on substrate affinities which is the most important issue. To our knowledge, in this region, no single nucleotide polymorphisms (SNPs) or mutations of clinical relevance are known for any ABC transporter. In addition, the use of a codon usage optimized CDS should aid expression levels. We therefore suggest that the approach of choice should be a chimeric hABCB1 mouse strain that will most likely retain the original level and pattern of protein expression.
Acknowledgments
The authors thank Sandra Noack, Ivan Eiriz Delgado, David Gomez-Zepeda, Meryam Taghi, Wang-Qing Liu, Cerina Chhuon, Michel Vidal, and Ida-Chiara Guerrera for their assistance. They also thank genOway S.A. for their openness in discussing the findings and all members of the ENCODE Consortium and Portal for providing the valuable data sets.
Footnotes
Funding Sources
This work was supported by grants from the Deutsche Forschungsgesellschaft (DFG) to J.P. (grant number: DFG PA930/9-1) and W.L. (grant number: DFG LO274/16-1), the Austrian Science Fund (FWF) (grant number: I 1609-B24 to O.L.), and the Lower Austria Corporation for Research and Education (NFB) (grant number: LS14-008 [T. Wanek]; LS15-003 [O. Langer]). The work of J.P. was also financed by the following grants: Deutsche Forschungsgemeinschaft/Germany (DFG PA930/12-1); Leibniz Society/Germany (SAW-2015-IPB-2); HelseSØ/Norway (2016062); Norsk forskningsrådet/Norway (246392, 247179 [NeuroGeM], 248772, 251290, 260786 [PROP-AD]); Horizon 2020/European Union (643417 [PROP-AD]). NeuroGeM is an EU Joint Programme — Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organisations under the aegis of JPND — www.jpnd.eu (CIHR — Canada, BMBF — Germany, NRF no. 247179 — Norway, ZonMW — The Netherlands). PROP-AD is an EU Joint Programme — Neurodegenerative Disease Research (JPND) project. The project is supported through the following funding organisations under the aegis of JPND — www.jpnd.eu (AKA no. 301228 — Finland, BMBF no. 01ED1605 — Germany, CSO-MOH no. 30000-12631 — Israel, NFR no. 260786 — Norway, SRC no. 2015-06795 — Sweden). This project has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement no. 643417 (JPco-fuND).
Authors’ Contributions
M.K., O.L., W.L., and J.P. planned the study. M.K. and A.N. performed the qPCR. T.W. and O.L. performed the PET analyses and prepared Figure 1. M.-C.M. and X.D. performed the QTAP. M.K. analyzed the sequencing and ENCODE data. M.K. wrote the article and prepared the figures. All authors discussed the results and commented on the article.
Conflict of Interest
The authors have no conflict of interest to report.
References
- 1.Kis O, Robillard K, Chan GN, Bendayan R. The complexities of antiretroviral drug–drug interactions: role of ABC and SLC transporters. Trends Pharmacol Sci. 2010;31:22–35. [DOI] [PubMed] [Google Scholar]
- 2.Marchetti S, Mazzanti R, Beijnen JH, Schellens JH. Concise review: clinical relevance of drug drug and herb drug interactions mediated by the ABC transporter ABCB1 (MDR1, P-glycoprotein). Oncologist. 2007;12:927–41. [DOI] [PubMed] [Google Scholar]
- 3.The International Transporter Consortium Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schumacher T, Krohn M, Hofrichter J, Lange C, Stenzel J, Steffen J, et al. ABC transporters B1, C1 and G2 differentially regulate neuroregeneration in mice. PLoS One. 2012;7:e35613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alison MR. Tissue-based stem cells: ABC transporter proteins take centre stage. J Pathol. 2003;200:547–50. [DOI] [PubMed] [Google Scholar]
- 6.Islam MO, Kanemura Y, Tajria J, Mori H, Kobayashi S, Shofuda T, et al. Characterization of ABC transporter ABCB1 expressed in human neural stem/progenitor cells. FEBS Lett. 2005;579:3473–80. [DOI] [PubMed] [Google Scholar]
- 7.Löscher W, Potschka H. Drug resistance in brain diseases and the role of drug efflux transporters. Nature reviews Neuroscience. 2005;6:591–602. [DOI] [PubMed] [Google Scholar]
- 8.Cirrito JR, Deane R, Fagan AM, Spinner ML, Parsadanian M, Finn MB, et al. P-glycoprotein deficiency at the blood–brain barrier increases amyloid-beta deposition in an Alzheimer disease mouse model. J Clin Invest. 2005;115:3285–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krohn M, Bracke A, Avchalumov Y, Schumacher T, Hofrichter J, Paarmann K, et al. Accumulation of murine amyloid-beta mimics early Alzheimer's disease. Brain. 2015;138:2370–82. [DOI] [PubMed] [Google Scholar]
- 10.Pahnke J, Fröhlich C, Paarmann K, Krohn M, Bogdanovic N, Arsland D, et al. Cerebral ABC transporter-common mechanisms may modulate neurodegenerative diseases and depression in elderly subjects. Arch Med Res. 2014;45:738–43. [DOI] [PubMed] [Google Scholar]
- 11.Krohn M, Lange C, Hofrichter J, Scheffler K, Stenzel J, Steffen J, et al. Cerebral amyloid-beta proteostasis is regulated by the membrane transport protein ABCC1 in mice. J Clin Invest. 2011;121:3924–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yvan-Charvet L, Welch C, Pagler TA, Ranalletta M, Lamkanfi M, Han S, et al. Increased inflammatory gene expression in ABC transporter-deficient macrophages: free cholesterol accumulation, increased signaling via toll-like receptors, and neutrophil infiltration of atherosclerotic lesions. Circulation. 2008;118:1837–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole SP. Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J Biol Chem. 2014;289:30880–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mason BL, Pariante CM, Thomas SA. A revised role for P-glycoprotein in the brain distribution of dexamethasone, cortisol, and corticosterone in wild-type and ABCB1A/B-deficient mice. Endocrinology. 2008;149:5244–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki M, Neway WE, Ohe T, Chen I, Rowe JF, Hochman JH, et al. In vitro substrate identification studies for p-glycoprotein-mediated transport: species difference and predictability of in vivo results. J Pharmacol Exp Ther. 2001;296:723–35. [PubMed] [Google Scholar]
- 16.Baltes S, Gastens AM, Fedrowitz M, Potschka H, Kaever V, Löscher W. Differences in the transport of the antiepileptic drugs phenytoin, levetiracetam and carbamazepine by human and mouse P-glycoprotein. Neuropharmacology. 2007;52:333–46. [DOI] [PubMed] [Google Scholar]
- 17.Feng B, Mills JB, Davidson RE, Mireles RJ, Janiszewski JS, Troutman MD, et al. In vitro P-glycoprotein assays to predict the in vivo interactions of P-glycoprotein with drugs in the central nervous system. Drug Metab Dispos. 2008;36:268–75. [DOI] [PubMed] [Google Scholar]
- 18.Helms HC, Abbott NJ, Burek M, Cecchelli R, Couraud PO, Deli MA, et al. In vitro models of the blood–brain barrier: an overview of commonly used brain endothelial cell culture models and guidelines for their use. J Cereb Blood Flow Metab. 2016;36:862–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devoy A, Bunton-Stasyshyn RK, Tybulewicz VL, Smith AJ, Fisher EM. Genomically humanized mice: technologies and promises. Nat Rev Genet. 2011;13:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dallas S, Salphati L, Gomez-Zepeda D, Wanek T, Chen L, Chu X, et al. Generation and characterization of a breast cancer resistance protein humanized mouse model. Mol Pharmacol. 2016;89:492–504. [DOI] [PubMed] [Google Scholar]
- 21.Sadiq MW, Uchida Y, Hoshi Y, Tachikawa M, Terasaki T, Hammarlund-Udenaes M. Validation of a P-glycoprotein (P-gp) humanized mouse model by integrating selective absolute quantification of human MDR1, Mouse Mdr1a and Mdr1b protein expressions with in vivo functional analysis for blood–brain barrier transport. PLoS One. 2015;10:e0118638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunner M, Langer O, Sunder-Plassmann R, Dobrozemsky G, Müller U, Wadsak W, et al. Influence of functional haplotypes in the drug transporter gene ABCB1 on central nervous system drug distribution in humans. Clin Pharmacol Ther. 2005;78:182–90. [DOI] [PubMed] [Google Scholar]
- 23.Loening AM, Gambhir SS. AMIDE: a free software tool for multimodality medical image analysis. Mol Imaging. 2003;2:131–7. [DOI] [PubMed] [Google Scholar]
- 24.Ohtsuki S, Schaefer O, Kawakami H, Inoue T, Liehner S, Saito A, et al. Simultaneous absolute protein quantification of transporters, cytochromes P450, and UDP-glucuronosyltransferases as a novel approach for the characterization of individual human liver: comparison with mRNA levels and activities. Drug Metab Dispos. 2012;40:83–92. [DOI] [PubMed] [Google Scholar]
- 25.Hoshi Y, Uchida Y, Tachikawa M, Inoue T, Ohtsuki S, Terasaki T. Quantitative atlas of blood–brain barrier transporters, receptors, and tight junction proteins in rats and common marmoset. J Pharm Sci. 2013;102:3343–55. [DOI] [PubMed] [Google Scholar]
- 26.Uchida Y, Tachikawa M, Obuchi W, Hoshi Y, Tomioka Y, Ohtsuki S, et al. A study protocol for quantitative targeted absolute proteomics (QTAP) by LC–MS/MS: application for inter-strain differences in protein expression levels of transporters, receptors, claudin-5, and marker proteins at the blood–brain barrier in ddY, FVB, and C57BL/6J mice. Fluids Barriers CNS. 2013;10:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaves C, Gomez-Zepeda D, Auvity S, Menet MC, Crete D, Labat L, et al. Effect of subchronic intravenous morphine infusion and naloxone-precipitated morphine withdrawal on P-gp and Bcrp at the rat blood–brain barrier. J Pharm Sci. 2016;105:350–8. [DOI] [PubMed] [Google Scholar]
- 28.Kamiie J, Ohtsuki S, Iwase R, Ohmine K, Katsukura Y, Yanai K, et al. Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm Res. 2008; 25:1469–83. [DOI] [PubMed] [Google Scholar]
- 29.Gomez-Zepeda D, Chaves C, Taghi M, Sergent P, Liu WQ, Chhuon C, et al. Targeted unlabeled multiple reaction monitoring analysis of cell markers for the study of sample heterogeneity in isolated rat brain cortical microvessels. J Neurochem. 2017;142:597–609. [DOI] [PubMed] [Google Scholar]
- 30.Maclean B, Tomazela DM, Abbatiello SE, Zhang S, Whiteaker JR, Paulovich AG, et al. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal Chem. 2010;82:10116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopec AM, Rivera PD, Lacagnina MJ, Hanamsagar R, Bilbo SD. Optimized solubilization of TRIzol-precipitated protein permits Western blotting analysis to maximize data available from brain tissue. J Neurosci Methods. 2017;280:64–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swain MD, Orzechowski KL, Swaim HL, Jones YL, Robl MG, Tinaza CA, et al. P-gp substrate-induced neurotoxicity in an Abcb1a knock-in/Abcb1b knock-out mouse model with a mutated canine ABCB1 targeted insertion. Res Vet Sci. 2013;94:656–61. [DOI] [PubMed] [Google Scholar]
- 34.Orzechowski KL, Swain MD, Robl MG, Tinaza CA, Swaim HL, Jones YL, et al. Neurotoxic effects of ivermectin administration in genetically engineered mice with targeted insertion of the mutated canine ABCB1 gene. Am J Vet Res. 2012;73:1477–84. [DOI] [PubMed] [Google Scholar]
- 35.Römermann K, Wanek T, Bankstahl M, Bankstahl JP, Fedrowitz M, Müller M, et al. (R)-[11C]verapamil is selectively transported by murine and human P-glycoprotein at the blood–brain barrier, and not by MRP1 and BCRP. Nucl Med Biol. 2013;40:873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wanek T, Römermann K, Mairinger S, Stanek J, Sauberer M, Filip T, et al. Factors governing P-glycoprotein-mediated drug–drug interactions at the blood–brain barrier measured with positron emission tomography. Mol Pharmaceutics. 2015;12:3214–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44:D726–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mealey KL. Therapeutic implications of the MDR-1 gene. J Vet Pharmacol Ther. 2004;27:257–64. [DOI] [PubMed] [Google Scholar]
- 40.Mealey KL, Bentjen SA, Gay JM, Cantor GH. Ivermectin sensitivity in collies is associated with a deletion mutation of the mdr1 gene. Pharmacogenetics. 2001;11:727–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated during and analyzed during the current study are available on reasonable request. Correspondence and requests for materials should be addressed to M.K. (markus.krohn@medisin.uio.no), J.P. (jens. pahnke@medisin.uio.no), W.L. (wolfgang.loescher@tihohannover.de), or O.L. (oliver.langer@ait.ac.at). All ENCODE data sets are publicly available at www.encodeproject.org.


