Abstract
Objective
To identify modifiers of age at diagnosis of Parkinson disease (PD).
Methods
Genome-wide association study (GWAS) included 1,950 individuals with PD from the NeuroGenetics Research Consortium (NGRC) study. Replication was conducted in the Parkinson's, Genes and Environment study, including 209 prevalent (PAGEP) and 517 incident (PAGEI) PD cases. Cox regression was used to test association with age at diagnosis. Individuals without neurologic disease were used to rule out confounding. Gene-level analysis and functional annotation were conducted using Functional Mapping and Annotation of GWAS platform (FUMA).
Results
The GWAS revealed 2 linked but seemingly independent association signals that mapped to LPPR1 on chromosome 9. LPPR1 was significant in gene-based analysis (p = 1E-8). The top signal (rs17763929, hazard ratio [HR] = 1.88, p = 5E-8) replicated in PAGEP (HR = 1.87, p = 0.01) but not in PAGEI. The second signal (rs73656147) was robust with no evidence of heterogeneity (HR = 1.95, p = 3E-6 in NGRC; HR = 2.14, p = 1E-3 in PAGEP + PAGEI, and HR = 2.00, p = 9E-9 in meta-analysis of NGRC + PAGEP + PAGEI). The associations were with age at diagnosis, not confounded by age in patients or in the general population. The PD-associated regions included variants with Combined Annotation Dependent Depletion (CADD) scores = 10–19 (top 1%–10% most deleterious mutations in the genome), a missense with predicted destabilizing effect on LPPR1, an expression quantitative trait locus (eQTL) for GRIN3A (false discovery rate [FDR] = 4E-4), and variants that overlap with enhancers in LPPR1 and interact with promoters of LPPR1 and 9 other brain-expressed genes (Hi-C FDR < 1E-6).
Conclusions
Through association with age at diagnosis, we uncovered LPPR1 as a modifier gene for PD. LPPR1 expression promotes neuronal regeneration after injury in animal models. Present data provide a strong foundation for mechanistic studies to test LPPR1 as a driver of response to damage and a therapeutic target for enhancing neuroregeneration and slowing disease progression.
The underlying neurodegenerative process that causes Parkinson disease (PD) begins decades before the disease is diagnosed.1 The current view is that following an initial insult (e.g., toxicity, trauma, or genetic), the disease starts with an asymptomatic phase of unknown duration, followed by development of prodromal nonmotor symptoms such as constipation, anosmia, and sleep disorders. Years later, cardinal motor signs appear, at which point a diagnosis of PD is made. Age at onset of motor signs, and therefore the age at diagnosis of PD, is highly variable, ranging from teen ages to the 10th decade of life. The reason for this variation is unknown, and understanding it will likely shed light on factors that affect the rate of disease progression.
There is substantial evidence that genetic factors play a major role in age at onset of motor signs and age at diagnosis of PD.2–6 Genome-wide studies have identified numerous loci that associate with the risk of developing PD,7 but the risk factors do not explain the variation in age at onset.8–10 Three loci have been nominated as modifiers of age at onset in familial PD.11,12 The present study was aimed at identifying genetic modifiers for common idiopathic PD. We hypothesized that identification of the genetic basis to interindividual variability in age at diagnosis will provide insights into the intrinsic mechanisms that determine the rate of deterioration during preclinical disease.
Methods
This study was a case-control GWAS, followed by replication and functional annotation.
Standard protocol approvals, registrations, and patient consents
The study was approved by the institutional review boards at all participating institutions. Written informed consent was obtained from all patients and controls for participation in the study.
Participants
The study included 2 data sets. The NeuroGenetics Research Consortium (NGRC) data set13 was used for the discovery GWAS, gene-based test, and functional annotations. The Parkinson's, Genes and Environment (PAGE) study14 was used for replication. Participants' characteristics are shown in table 1 and figure e-1 (links.lww.com/NXG/A66).
Table 1.
Data sets and participants' characteristics
NGRC is a case-control study of genetically unrelated participants, including 2000 PD cases and 1986 controls.13 Patients were enrolled sequentially from movement disorder clinics in Portland (OR), Seattle (WA), Albany (NY), and Atlanta (GA). Controls were spouses of patients or community volunteers, self-reported as being free of neurologic disease. The eligibility criterion for cases was diagnosis of PD by a movement disorder specialist according to the UK Brain Bank criteria.15 The eligibility criteria for controls were no neurologic disease and genetically unrelated to patients. Age was defined as age at study entry. Age at diagnosis was extracted from medical records or ascertained by self-report. Age at onset of the first motor sign was obtained using a self-administered questionnaire. Age at onset and age at diagnosis were highly correlated in the NGRC (r2 = 0.91, p < 2E-16). All participants were whites of European descent.13
PAGE is a cross-sectional study nested in the longitudinal NIH-American Association of Retired Persons Diet and Health Study.14 Participants were enrolled in 1995–1997 (irrespective of PD) via a food frequency questionnaire mailing16 and in the 2004–2006 follow-up visit were asked if they had been diagnosed with a major chronic disease including PD. Participants who had been diagnosed with PD before enrollment (before 1998) were designated as prevalent PD (PAGEP, N = 209), participants who were diagnosed during follow-up (1998–2006) were designated as incident PD (PAGEI, N = 517), and participants who did not have PD were designated as controls (N = 1,549). All participants in this study were non-Hispanic whites.
Genotyping
NGRC participants were genotyped on Illumina HumanOmni1-Quad v1-0 B array and Immunochip array. Genotypes and samples were filtered by call rate, minor allele frequency (MAF) < 0.01, Hardy-Weinberg, and cryptic relatedness, as described before.13 Imputation was performed using IMPUTE v2.3.0,17 with the 1000G Phase3 integrated variant set (October 2014) as reference. Imputed single nucleotide polymorphisms (SNPs) with info score < 0.9 or MAF < 0.01 were excluded. A total of 8.5 million SNPs (900,000 genotyped and 7.6 million imputed) were used in the analysis.
PAGE participants were genotyped for rs73656147 (block 1) and rs17763929 (block 2). SNPs were chosen based on statistical significance and availability of predesigned validated TaqMan assay from Thermo Fisher (rs73656147 assay number = C__97534229_10; rs17763929 assay number = C__34297681_10).
Population structure
Principal component (PC) analysis18 is used to infer population-specific genetic differences, which arise from ancestry differences in allele frequencies and can obscure genetic association studies if not accounted for. NGRC PC analysis was conducted using a pruned subset of 100K SNPs from the GWAS as previously described.13 The top 3 PCs (effect sizes PC1 = 0.2%, PC2 = 0.06%, and PC3 = 0.06%) were included in the GWAS and adjusted for in all downstream analyses involving the NGRC. The PAGE data sets used for replication did not have ancestry informative markers (AIMs); however, a subset of the participants (396 of 726 PD cases) was previously genotyped with the Immunochip array. We conducted PC analysis using a pruned set of 20K SNPs from the Immunochip array, using PLINK. Tests were conducted once using the full PAGE data set, with no PC adjustment, and again with a PAGE subset, adjusting for PC1-3 (effect sizes PC1 = 0.48%, PC2 = 0.20%, and PC3 = 0.17%). NGRC and PAGE cluster with Europeans in the 1000G_Phase_3 global data set (figure e-2, links.lww.com/NXG/A67).
Statistics
Discovery
GWAS was conducted using PD cases only (1,950 NGRC participants with known age at diagnosis). Association between 8.5M SNPs and age at diagnosis was tested using Cox regression in ProbABEL v0.5.0.,19 specifying an additive genetic model, treating age at diagnosis as a quantitative trait, and adjusting for PC1-3. The statistical outcome of Cox regression was hazard ratios (HRs) and corresponding p values. Statistical significance was set at p < 5E-8. Manhattan plots and quantile-quantile (QQ) plots were generated using FUMA v1.3.0.20 Genomic inflation factor (λ) was calculated using the estlambda function in GenABEL v1.8 in R.21 LocusZoom22 was used to visualize the chr9:103,865,000–104,055,000 region (GWAS peak). Haploview v4.223 was used to generate linkage disequilibrium (LD) plots of D′ and r2 for SNPs in the chr9:103,865,000–104,055,000 region with GWAS p < 1E-4. LD between 2 SNPs was calculated using 1000G Phase3 v5 in LDlink.24 Linear regression was used to estimate and test differences in mean age at diagnosis (β). Conditional analysis was performed using coxph function in the survival v2.41 R package. Moving average plots (MAPs) were generated using the freqMAP v0.2 R package.25
Gene-based analysis was conducted using summary statistics from the GWAS and LD from the 1000G Phase3 EUR to map the GWAS SNPs to 18,985 protein-coding genes (hg19 build) and to calculate gene-based p values, using MAGMA v1.06,26 as implemented in FUMA v1.3.0.20 Statistical significance was set at Bonferroni-corrected p < 2.6E-6 (0.05/18,985).
Replication
Cox regression (coxph function in the survival v2.41 R package) was used to replicate the association of 2 SNPs with age at diagnosis. We used the same model as the NGRC (additive genetic model, treating age at diagnosis as a quantitative trait). Because of the availability of PCs only in a subset of PAGE, analyses were conducted twice: using the full PAGE data set without PC adjustment and using the subset that had AIMs and adjusting for PC1-3. PAGEI and PAGEP were treated separately and were combined using meta-analysis after testing for heterogeneity. If p of heterogeneity was <0.1, the fixed-effect model was used. Meta-analysis was performed using the metagen function in the meta v4.8 R package.
Functional annotation
Functional annotation was conducted in FUMA v1.3.0,20 using SNPs with GWAS p < 1E-6 and all variants in r2 ≥ 0.6 with them, and included CADD analysis,27 eQTL mapping,28 3D chromatin interaction mapping (Hi-C),29 annotation of enhancers,30 tissue-specific expression of genes identified via Hi-C and eQTL mapping,28 and their age-specific expression in the brain (BrainSpan.org). The false discovery rate (FDR) was used to correct for multiple testing. STRUM was used to predict the effect of a missense on the structural stability of a protein.31
Data availability
NGRC genotype and phenotype data are available at dbGaP ncbi.nlm.nih.gov/gap accession number phs000196.v3.p1.
Results
GWAS
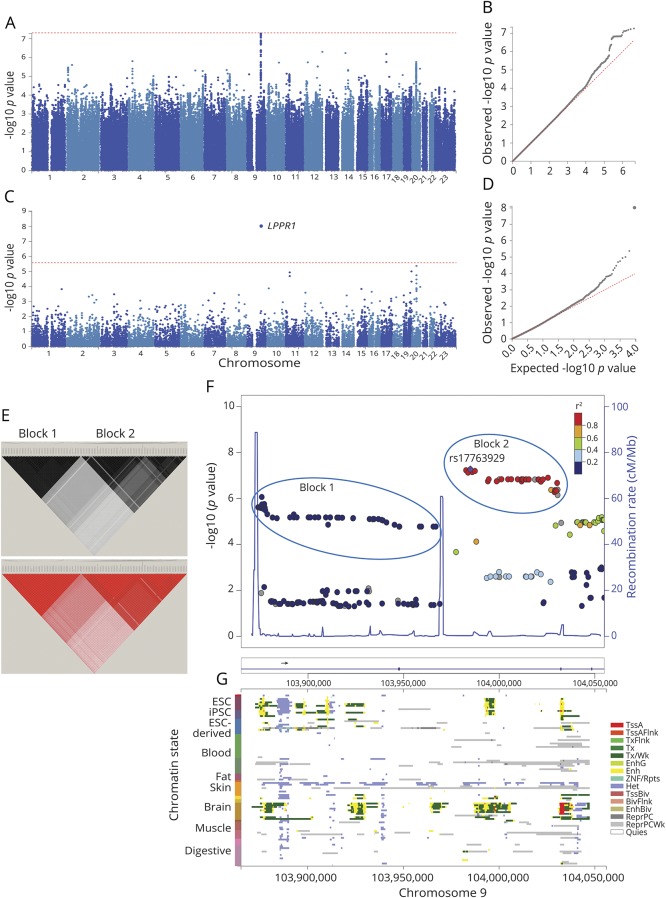
In SNP-based GWAS, the most significant signal for association, at p = 5E-8, mapped to LPPR1 on chromosome 9q31.1 (figure 1, A and B). In the gene-based test, LPPR1 achieved p = 1E-8, surpassing the genome-wide statistical significance threshold of p < 2.6E-6 (figure 1, C and D). The p values were not inflated (λ = 1.007 SNP based, λ = 1.04 gene based). Analysis of LD in the region revealed 2 haplotype blocks with seemingly independent signals for association (figure 1, E and F). There was strong LD among SNPs in each block, but weak LD between the blocks (r2 ≤ 0.2) because of a recombination hot spot between them (figure 1F). The 2 blocks were in a ∼200 Kb region inside LPPR1. Block 1 consisted of 51 SNPs with MAF∼0.01, which yielded HR = 2.02–1.88, with p = 9E-7 to 2E-5 for association with age at diagnosis. Block 2 consisted of 39 SNPs with MAF∼0.02, which yielded HR = 1.88–1.85, with p = 5E-8 to 7E-7. We chose 1 SNP to represent each block for replication: rs73656147 for block 1 (MAF = 0.01, HR = 1.95, p = 3E-6) and rs17763929 for block 2 (MAF = 0.02, HR = 1.88, p = 5E-8), both in Hardy-Weinberg (p > 0.3), with little correlation between them (r2 = 0.2). Conditional analysis conducted to determine whether the 2 blocks were tagging the same or different disease-associated variants was inconclusive because although the signals were weakened when adjusted for each other, neither was abolished when conditioned on the other (table e-1, links.lww.com/NXG/A69).
Figure 1. Results of genome-wide association study for age at diagnosis of PD.
Genome-wide association was tested between 8.5 million SNPs and age at diagnosis in 1,950 PD cases from the NGRC, using the Cox hazard ratio regression method and adjusting for principal components (PC1-3). (A) Manhattan plot of SNP-based GWAS. Tallest peak, at p = 5E-8, was on chromosome 9q31.1. (B) QQ plot of SNP-based GWAS. The observed p values were not inflated (λ = 1.007). (C) Manhattan plot of gene-based GWAS. LPPR1 was at p = 1E-8. Statistical significance threshold was p < 2.6E-6, which is Bonferroni corrected for the 18,985 protein-coding genes tested. (D) QQ plot of gene-based GWAS. The observed p values were not inflated (λ = 1.04). (E) r2 (top panel) and D' (bottom panel). Linkage disequilibrium (LD) across the SNPs that gave p < 1E-4 for association with age at diagnosis reveals 2 blocks represented by rs73656147 (left triangle) and rs17763929 (right triangle). (F) Magnified map of the associated region (chr9:103,865,000–104,055,000), showing that PD-associated SNPs map to LPPR1 and form 2 haplotype blocks separated by recombination hot spots (blue spikes). (G) Chromatin state of LPPR1 (Roadmap 111 Epigenomes), showing that active enhancers (yellow), transcription start site (red), and transcripts (green) of LPPR1 are seen only in stem cells and the brain and that the GWAS SNPs align with regulatory elements. ESC = embryonic stem cell; iPSC = induced pluripotent stem cell; TssA = active transcription start site (TSS); TssAFlnk = flanking active TSS; TxFlnk = transcription at gene 5′ and 3'; Tx = strong transcription; TxWk = weak transcription; EnhG = genic enhancers; Enh = enhancers; ZNF/Rpts = zinc-finger genes and repeats; Het = heterochromatin; TssBiv = bivalent/poised TSS; BivFlnk = flanking bivalent TSS/enhancer; EnhBiv = bivalent enhancer; ReprPC = repressed polycomb; ReprPCWk = weak repressed polyComb; Quies = quiescent.
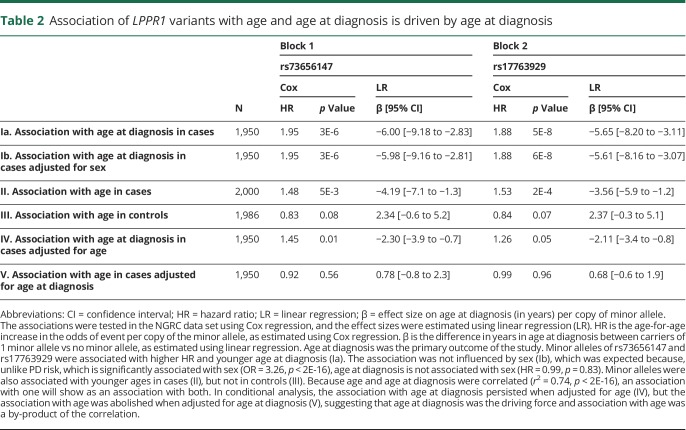
There are 2 caveats in interpreting statistical evidence for association with age at diagnosis. First, age at diagnosis is correlated with age (r2 = 0.74, p < 2E-16), which can result in spurious conclusions if the driving force responsible for the association is not identified. Second, tests of age at diagnosis are conducted using patients only without the benefit of controls. For example, an SNP that appears to be associated with earlier PD diagnosis may in fact be associated with an age-related event unrelated to PD. To interpret the statistical evidence for association with age at diagnosis, we examined whether and how allele frequencies vary by age in cases or in controls. Allele frequencies were plotted in a moving average window as a function of age (figure e-3, links.lww.com/NXG/A68). Starting at age 45 years, allele frequencies were the same in cases and controls. In controls, allele frequencies remained the same across the age spectrum, whereas in cases, they decreased sharply and significantly by age and by age at diagnosis. The effect was therefore in cases and not in controls. Next, conditional analysis was conducted to tease age from age at diagnosis (table 2). The minor alleles of rs73656147 and rs17763929 were associated with age, as was expected, given their association with age at diagnosis. However, the association with age at diagnosis persisted when adjusted for age, but the association with age was abolished when adjusted for age at diagnosis. Hence, age at diagnosis was the driving force, and association with age was a by-product of the correlation.
Table 2.
Association of LPPR1 variants with age and age at diagnosis is driven by age at diagnosis
To gauge robustness of the association signals with age at diagnosis and to test for heterogeneity, we stratified the data by 8 PD-relevant variables, tested the association of each SNP with age at diagnosis within each stratum, and compared the results across strata for evidence of heterogeneity (table e-2, links.lww.com/NXG/A70). The 8 categories of stratification were family history, sex, cigarette smoking, caffeine intake, nonsteroidal anti-inflammatory drugs use, recruitment site, Jewish heritage, and the European country of ancestral origin. The association signal for rs73656147 (block 1) was robust across all strata. rs17763929 (block 2) showed evidence of heterogeneity as a function of recruitment site and the European country of ancestral origin. Given these results, we tested the association of the 2 SNPs with PCs. rs17763929 was associated with PC1 (p = 7E-6) and PC3 (p = 8E-3), and rs73656147 was not (p > 0.05 for PC1-3), indicating the presence of population structure in block 2 but not in block 1.
Replication
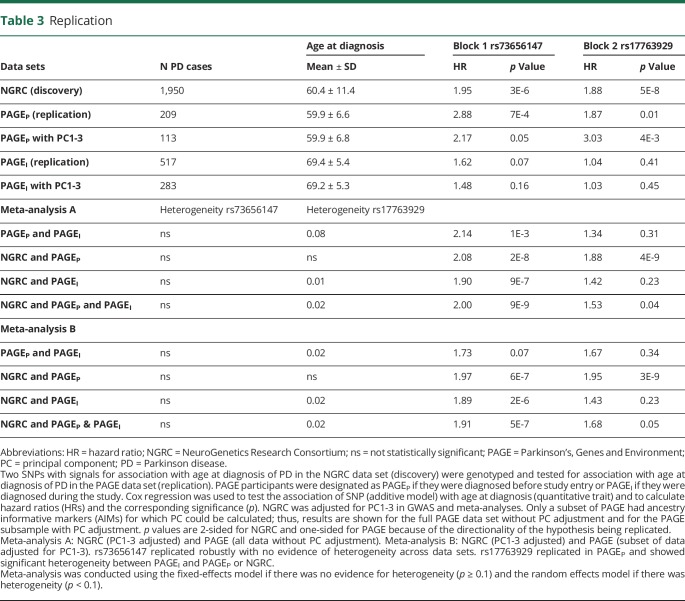
In comparison to NGRC, which had a 65-year range for age at diagnosis, the PAGE data sets had a narrower range of less than 30 years. Because PAGE participants were of similar age at study entry, the method of classifying the participants into prevalent PD (diagnosis before entry) vs incident PD (diagnosis after entry) inevitably assigned earlier ages at diagnosis to the prevalent group (PAGEP) and later diagnoses to the incident group (PAGEI). Mean age at diagnosis in PAGEP was 59.9 ± 6.6 years, which was similar to the NGRC (60.4 ± 11.4). PAGEI participants were on average 10 years older at diagnosis (69.4 ± 5.4, range 53–81 years). Given the disparity in the range and mean ages at diagnosis, we analyzed PAGEP and PAGEI separately.
Association of rs73656147 (block 1) with age at diagnosis replicated robustly (table 3). There was no evidence of heterogeneity between PAGEI and PAGEP in the association of rs73656147 with age at diagnosis, although the signal was stronger in PAGEP than in PAGEI, which is not surprising, given that the former is enriched in cases with earlier age at diagnosis. Nor was there evidence of heterogeneity between PAGE and NGRC for the association of rs73656147 with age at diagnosis. Meta-analysis yielded HR = 2.14, p = 1E-3 for replication and HR = 2.00, p = 9E-9 for replication and discovery. Mean difference in age at diagnosis per copy of rs73656147 minor allele was −6.0 (95% confidence interval: −9.18 to −2.83) years in the NGRC, −5.53 (−9.72 to −1.34) in PAGEP, −0.84 (−4.22 to 2.55) in PAGEI, and −4.08 (−7.45 to −0.70) in the meta-analysis of the 3 data sets.
Table 3.
Replication
Association of rs17763929 (block 2) with age at diagnosis showed significant heterogeneity between PAGEI and PAGEP (table 3), as it had within the NGRC (table e-2, links.lww.com/NXG/A70). The association with rs17763929 replicated in PAGEP but not in PAGEI. There was significant heterogeneity between PAGEI and NGRC, but not between PAGEP and NGRC. Meta-analysis of PAGEP and NGRC yielded HR = 1.88, p = 4E-9 for full PAGE data and HR = 1.95, p = 3E-9 for the PAGE subsample adjusted for PC1-3. Including PAGEI with PAGEP and NGRC in a random-effects meta-analysis diluted the effect size to HR = 1.53, p = 0.04. Mean difference in age at diagnosis per copy of rs17763929 minor allele was −5.65 (−8.20 to −3.11) years in the NGRC, −3.62 (−7.23 to −0.02) in PAGEP, and 0.62 (−1.34 to 2.58) in PAGEI.
Functional annotation
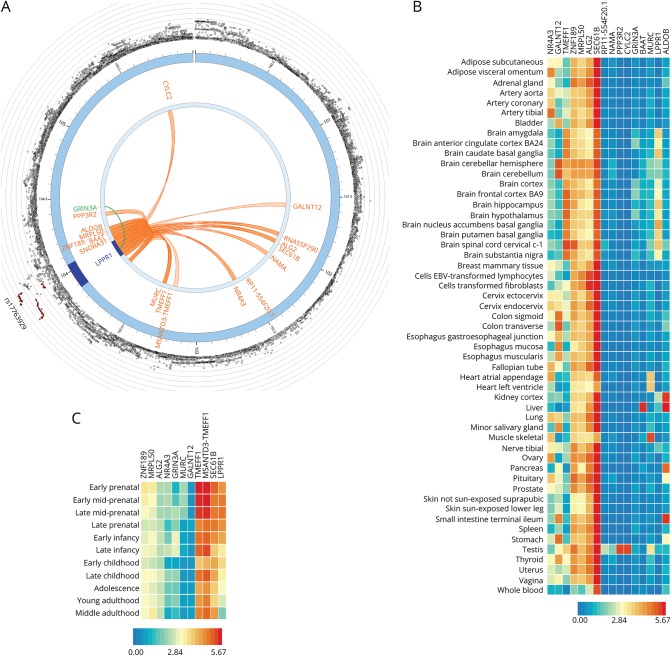
Hi-C analysis showed significant (FDR < 1E-6) chromatin interaction between the PD-associated LPPR1 SNPs and promoters of LPPR1 and several genes on chromosome 9 (figure 2, A). Some of the SNPs that were significant in Hi-C mapped to enhancers in the brain (table 4 and figure 1, G). Eleven of the genes identified through Hi-C are expressed in the brain: LPPR1, SEC61B, MSANTD3-TMEFF1, TMEFF1, GALNT12, MURC, GRIN3A, NR4A3, ALG2, MRPL50, and ZNF189 (figure 2, B and C). The expression of LPPR1 in the brain is the strongest in early prenatal stage and decreases with developmental stage and increasing age (figure 2, C).
Figure 2. Functionally significant genes.
(A) 3D chromatin interaction (Hi-C) and eQTL analysis. Hi-C revealed significant interaction between GWAS variants in LPPR1 and 17 other genes on chromosome 9 (FDR < 1E-6, shown in orange). An SNP in LPPR1 was associated with the expression of GRIN3A (FDR = 4E-4, shown in green). (B) Tissue-specific expression of LPPR1, GRIN3A, and genes in Hi-C with LPPR1. Colors reflect average expression (log2 transformed) from highest (red) to lowest/absent (blue). (C) Age-specific expression of the genes in the brain. LPPR1 expression decreases with age.
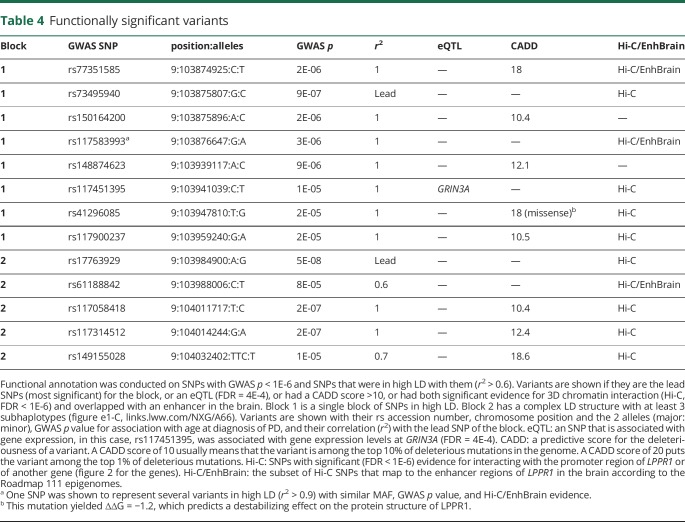
Table 4.
Functionally significant variants
CADD analysis, a scoring system for deleteriousness of genetic variants, identified 5 SNPs in block 1 and 3 in block 2, with CADD = 10–19 (table 4), which places them among the top 10% (CADD > 10) to 1% (CADD > 20) of most deleterious mutations in the genome.27 rs41296085 (CADD = 18, in block 1) is a missense (p.Ser12Ala) in exon 2, predicted to structurally destabilize the LPPR1 protein (∆∆G = −1.2). The remainder of the variants with high CADD scores are in introns. eQTL analysis revealed an association between rs117451395 (block 1) with expression levels of GRIN3A (FDR = 4E-4).
Discussion
There has been intense research on PD risk factors, which so far has resulted in identification of numerous causative genes, 40 susceptibility loci, several environmental factors, and a few genes that interact with the environmental factors to increase or reduce the risk of developing PD. In contrast, we know little about factors that affect the rate of disease progression. In this study, we attempted to identify genetic modifiers of age at diagnosis, a reflection of rate of progression, using an unbiased genome-wide approach, followed by independent replication, and functional annotation.
We uncovered evidence for association of genetic variants in neuronal plasticity-related gene 3 (LPPR1) with age at diagnosis of PD. Two signals of association were detected, each representing a haplotype block of SNPs. The variants that were associated with earlier age at diagnosis had low allele frequencies (MAF = 0.01–0.02), as were the variants that were previously found for age at onset of familial PD.11 The low allele frequencies may be one reason why modifier genes have been more difficult to detect than common variants that associate with risk.
The association with block 1 replicated robustly in both PAGEP and PAGEI. Block 2 signal replicated in PAGEP but not in PAGEI. Block 2 has a complex LD structure, with evidence of population substructure, which limits generalizability of results. Failure to capture a signal for block 2 in PAGEI may be because we had genotype on only 1 SNP in block 2 for PAGE, which did not fully capture the complexity of block 2. PAGEI participants being significantly older than NGRC and PAGEP participants may also be a factor. LPPR1 promotes neuroregeneration,32–34 but its expression diminishes with age to nearly undetectable level by age 40 years (figure 2C). One can speculate that some detrimental variants may not have an effect after a certain age when the gene is no longer expressed.
Functional annotation of the PD-associated variants in LPPR1 revealed the presence of several variants with predicted deleterious effects, including a missense that destabilizes the structure of LPPR1, a regulatory element that associates with expression levels of GRIN3A, and enhancers that interact with promoters of LPPR1 and several other genes in the brain. Some of the candidate genes that were identified via interaction with LPPR1 play key roles in pathways that are implicated in PD, including GRIN3A (which encodes a subunit of NMDA receptor involved in the glutamate-regulated ion channels in the brain), SEC61B (protein transport apparatus of the endoplasmic reticulum membrane), MURC (Rho kinase signaling), and MRPL50 (mitochondrial ribosomal protein).
LPPR1 is one of the 5 members of a brain-specific gene family that modulates neuronal plasticity during development, aging, and after brain injury.32–34 LPPR1 is the strongest driver of axonal outgrowth in the gene family. Studies in mice have shown that after neuronal injury, overexpression of LPPR1 enhances axonal growth, improves motor behavior, and promotes functional recovery.33,34 Extrapolating to our findings, we propose that LPPR1 is involved, not necessarily in the cause of PD, rather in response to damage, and influences the efficacy of regeneration and the subsequent rate of deterioration in preclinical PD. The actual cause of injury and neuronal death is not stipulated in this hypothesis; it could be head trauma, environmental toxins or genetic, but once the initial damage is incurred, it is the efficacy of intrinsic mechanisms of repair that determine the rate of disease progression. Present findings provide a strong foundation for mechanistic studies to investigate the role of LPPR1 in PD and determine its potential as a therapeutic target to impede disease progression.
Glossary
- AIM
ancestry informative marker
- FDR
false discovery rate
- HR
hazard ratio
- LD
linkage disequilibrium
- MAF
minor allele frequency
- MAP
moving average plot
- NGRC
NeuroGenetics Research Consortium
- PAGE
Parkinson's, Genes and Environment
- PC
principal component
- PD
Parkinson disease
Author contributions
Z.D. Wallen: statistical analysis, review, and critique of the manuscript. H. Chen: creation of the PAGE data set, acquisition of data, review, and critique of the manuscript. E.M. Hill-Burns: assembly and QC of phenotype and genotype data, imputation, review, and critique of the manuscript. S.A. Factor and C.P. Zabetian: creation of the NGRC data set, review, and critique of the manuscript. H. Payami: study concept, design and execution, creation of the NGRC data set, wrote the manuscript, and obtained funding.
Study funding
This work was supported by the National Institute of Neurological Disorders and Stroke grant R01NS036960. Additional support was provided by a Merit Review Award from the Department of Veterans Affairs grant 1I01BX000531; Office of Research & Development, Clinical Sciences Research & Development Service, Department of Veteran Affairs; the Close to the Cure Foundation; and the Sartain Lanier Family Foundation. Genome-wide array genotyping was conducted by the Center for Inherited Disease Research, which is funded by the NIH grant HHSN268200782096C. Collection of samples and data for replication (Parkinson, Genes, and Environment (PAGE) study) was supported by the intramural research program of NIH National Institute of Environmental Health Sciences grant Z01 ES101986. Funding agencies did not have a role in the design or execution of the study.
Disclosure
Z.D. Wallen, H. Chen, C.P. Zabetian, and E.M. Hill-Burns report no disclosures. S.A. Factor has received honoraria from Neurocrine, Lundbeck, Teva, Avanir, Sunovion Pharmaceuticals, Adamas, and UCB; has received research support from Ipsen, Medtronic, Teva, US WorldMeds, Sunovion Pharmaceuticals, Solstice, Vaccinex, Voyager, the CHDI Foundation, the Michael J. Fox Foundation, and the NIH; and receives publishing royalties from Demos, Blackwell Futura, and UpToDate. H. Payami has received research support from the NIH and the University of Alabama at Birmingham. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Obeso JA, Stamelou M, Goetz CG, et al. Past, present, and future of Parkinson's disease: a special essay on the 200th Anniversary of the Shaking Palsy. Mov Disord 2017;32:1264–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zareparsi S, Taylor TD, Harris EL, Payami H. Segregation analysis of Parkinson disease. Am J Med Genet 1998;80:410–417. [DOI] [PubMed] [Google Scholar]
- 3.Maher NE, Currie LJ, Lazzarini AM, et al. Segregation analysis of Parkinson disease revealing evidence for a major causative gene. Am J Med Genet 2002;109:191–197. [DOI] [PubMed] [Google Scholar]
- 4.McDonnell SK, Schaid DJ, Elbaz A, et al. Complex segregation analysis of Parkinson's disease: the Mayo clinic family study. Ann Neurol 2006;59:788–795. [DOI] [PubMed] [Google Scholar]
- 5.Hamza TH, Payami H. The heritability of risk and age at onset of Parkinson's disease after accounting for known genetic risk factors J Hum Genet 2010;55:241–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escott-Price V, Consortium IPsDG, Nalls MA, et al. Polygenic risk of Parkinson disease is correlated with disease age at onset. Ann Neurol 2015;77:582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang D, Nalls MA, Hallgrimsdottir IB, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat Genet 2017;49:1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nalls MA, Escott-Price V, Williams NM, et al. Genetic risk and age in Parkinson's disease: continuum not stratum. Mov Disord 2015;30:850–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lill CM, Hansen J, Olsen JH, Binder H, Ritz B, Bertram L. Impact of Parkinson's disease risk loci on age at onset. Mov Disord 2015;30:847–850. [DOI] [PubMed] [Google Scholar]
- 10.Pihlstrom L, Toft M. Cumulative genetic risk and age at onset in Parkinson's disease. Mov Disord 2015;30:1712–1713. [DOI] [PubMed] [Google Scholar]
- 11.Hill-Burns EM, Ross OA, Wissemann WT, et al. Identification of genetic modifiers of age-at-onset for familial Parkinson's disease. Hum Mol Genet 2016;25:3849–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinh J, Gustavsson EK, Vilarino-Guell C, et al. DNM3 and genetic modifiers of age of onset in LRRK2 Gly2019Ser parkinsonism: a genome-wide linkage and association study. Lancet Neurol 2016;15:1248–1256. [DOI] [PubMed] [Google Scholar]
- 13.Hamza TH, Zabetian CP, Tenesa A, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet 2010;42:781–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Huang X, Guo X, et al. Smoking duration, intensity, and risk of Parkinson disease. Neurology 2010;74:878–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry 1992;55:181–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schatzkin A, Subar AF, Thompson FE, et al. Design and serendipity in establishing a large cohort with wide dietary intake distributions: the National Institutes of Health-American Association of retired persons diet and Health study. Am J Epidemiol 2001;154:1119–1125. [DOI] [PubMed] [Google Scholar]
- 17.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 19.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 2010;11:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics 2007;23:1294–1296. [DOI] [PubMed] [Google Scholar]
- 22.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010;26:2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 24.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015;31:3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payami H, Kay DM, Zabetian CP, Schellenberg GD, Factor SA, McCulloch CC. Visualizing disease associations: graphic analysis of frequency distributions as a function of age using moving average plots (MAP) with application to Alzheimer's and Parkinson's disease. Genet Epidemiol 2010;34:92–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium G. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Berkum NL, Lieberman-Aiden E, Williams L, et al. Hi-C: a method to study the three-dimensional architecture of genomes. J Vis Exp 2010:e-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Consortium RE, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quan L, Lv Q, Zhang Y. STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 2016;32:2936–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savaskan NE, Brauer AU, Nitsch R. Molecular cloning and expression regulation of PRG-3, a new member of the plasticity-related gene family. Eur J Neurosci 2004;19:212–220. [DOI] [PubMed] [Google Scholar]
- 33.Broggini T, Schnell L, Ghoochani A, et al. Plasticity related gene 3 (PRG3) overcomes myelin-associated growth inhibition and promotes functional recovery after spinal cord injury. Aging (Albany NY) 2016;8:2463–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fink KL, Lopez-Giraldez F, Kim IJ, Strittmatter SM, Cafferty WBJ. Identification of intrinsic axon growth modulators for intact CNS neurons after injury. Cell Rep 2017;18:2687–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
NGRC genotype and phenotype data are available at dbGaP ncbi.nlm.nih.gov/gap accession number phs000196.v3.p1.