Abstract
Background
The lily is a perennial flowering plant belonging to the genus Lilium in the family Liliaceae. Most cultivated lily plants are propagated by bulbs. Therefore, numerous lily bulbs are frequently infected by diverse viruses causing viral diseases. To date, no study has examined the viromes of plants of one type with identical genetic backgrounds collected from different geographical regions.
Results
Here, we examined different viromes of the lily cultivar “Sorbonne” using 172 gigabytes of transcriptome data composed of 23 libraries from four different projects for the cultivar “Sorbonne.” We identified 396 virus-associated contigs from all but one library. We identified six different viruses, including Plantago asiatica mosaic virus (PlAMV), Cucumber mosaic virus (CMV), Lily symptomless virus (LSV), Tulip virus X (TVX), Lily mottle virus (LMoV), and Tobacco rattle virus (TRV). Of them, PlAMV was the most common virus infecting the lily. Scale and flower samples possessed a high number of virus-associated reads. We assembled 32 nearly complete genomes for the six identified viruses possessing the polyadenylate tails. Genomes of all six viruses were highly conserved in the lily cultivar “Sorbonne” based on mutation analysis. We identified defective RNAs from LSV, TVX, and PlAMV localized in the triple gene block region. Phylogenetic analyses showed that virus genomes are highly correlated with geographical regions and host plants.
Conclusions
We conducted comprehensive virome analyses of a single lily cultivar, “Sorbonne,” using transcriptome data. Our results shed light on an array of lily virome-associated topics, including virus identification, the dominant virus, virus accumulation in different plant tissues, virus genome assembly, virus mutation, identification of defective RNAs, and phylogenetic relationships of identified viruses. Taken together, we provide very useful methods and valuable results that can be applied in other virome-associated studies.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5138-3) contains supplementary material, which is available to authorized users.
Keywords: Genome, Lily, “Sorbonne”, Transcriptome, Virus, Virome
Background
Lilies are tall perennial herbaceous flowering plants, which are members of the genus Lilium in the family Liliaceae [1]. The flowers of lilies are large, colorful, and very often fragrant. Lilies are cultivated as cut flowers or grown in the garden. Numerous lily cultivars and interspecific hybrids have been developed [2]. To date, there are about 100 known lily species, which can be grouped into seven types [3].
In order to keep the genetic characteristics of lily flowers, lilies are usually clonally propagated using bulbs and scaling, which involves detaching scales from the bulb and planting individual scales to make new bulbs [4]. Due to the clonal propagation of lilies, most lily cultivars are frequently infected by diverse pathogenic microorganisms [5]. Of them, viruses infecting lily species cause serious damage to the quality and quantity of lily production. Since the first report of the three major viruses infecting the lily in 1944 (i.e., Lily symptomless virus (LSV) in the genus Carlavirus, Lily mottle virus (LMoV) in the genus Potyvirus, and Cucumber mosaic virus (CMV) in the genus Cucumovirus [6]), more than 20 viruses infecting the lily have been reported [7].
It is known that LMoV causes vein clearing, leaf mottle, mosaic, and chlorotic symptoms in infected lily plants, whereas LSV-infected plants are generally symptomless when they are grown under normal conditions [8]. In many cases, two or three different viruses infect lilies, resulting in severe economic losses [9]. Viruses infecting lilies are transmitted by mechanical transfer and aphids. For example, many viruses infecting lilies, such as LSV, LMoV, CMV, and Tulip breaking virus (TBV) in the genus Potyvirus, can be rapidly spread by aphids [8, 10, 11].
In order to diagnose viruses infecting lilies, many studies have developed diverse techniques. For example, a multiplex reverse-transcription (RT) polymerase chain reaction (PCR) [12], a multiplex Luminex bead array [13], a triplex IC-RT-PCR [9], polyclonal antisera [14], RT loop-mediated isothermal amplification [15, 16], and an immunochromatographic strip [17, 18] have been developed. Most studies have focused on the identification of three major viruses: LSV, LMoV, and CMV.
Due to the rapid advances in next-generation sequencing (NGS), co-infected known and novel viruses can be easily identified with full genomes [19–21]. For example, NGS of a small RNA library revealed co-infection of CMV, LSV, and a novel virus referred to as Lily yellow mosaic virus (LYMV) in the genus Potyvirus [22]. However, the NGS technique is not widely applied in the study of viruses infecting lilies.
Our previous studies have shown that plant transcriptomes are very useful for plant virome studies involving the identification of known and novel viruses, the assembly of virus genomes, mutation analyses, and the comparison of different viromes [23–26]. In this study, we carried out an in silico study of different viromes for the lily cultivar “Sorbonne.” Taking advantage of transcriptome data followed by bioinformatics analyses, we addressed diverse questions associated with lily viromes in this study.
Results
Identification of viruses infecting lilies
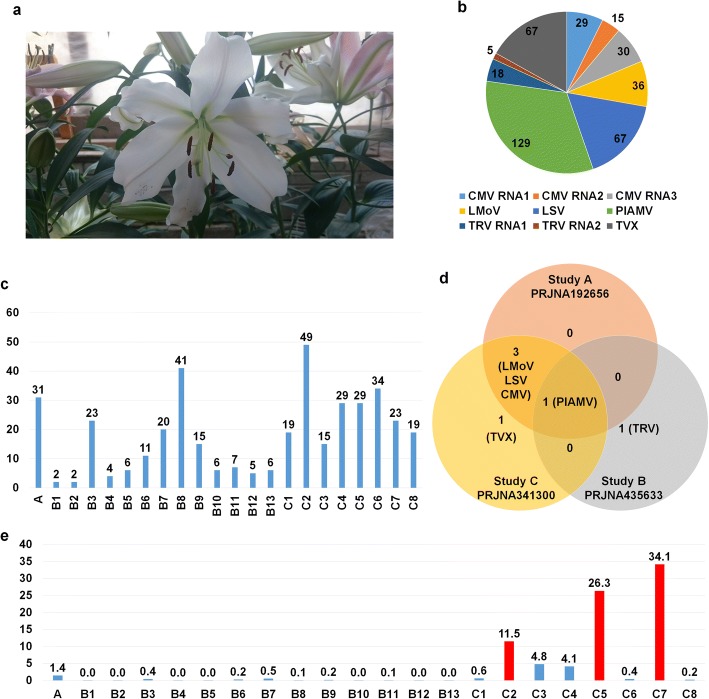
A total of 23 RNA-Sequencing (RNA-Seq) datasets derived from four different projects were associated with the lily cultivar “Sorbonne” (Fig. 1a). The 23 Sequence Read Archive (SRA) datasets were downloaded and subjected to de novo transcriptome assembly using the Trinity program (Additional file 1: Table S1). Assembled contigs for each library were used for a BLASTN search against the virus reference genome sequences derived from the viral genome database (https://www.ncbi.nlm.nih.gov/genome/viruses/). All but one library (SRR1390677 from PRJNA252055) contained virus-associated contigs. To simplify the names of the three different projects, we created the following names: Study A (PRJNA192656) containing a single library, Study B (PRJNA341300) containing 13 libraries, and Study C (PRJNA435633) containing eight libraries (Table 1). We identified 396 virus-associated contigs representing six viruses from 22 libraries (Fig. 1b). The identified viruses infecting “Sorbonne” were Plantago asiatica mosaic virus (PlAMV) and Tulip virus X (TVX) in the genus Potexvirus, CMV in the genus Cucumovirus, LSV in the genus Carlavirus, LMoV in the genus Potyvirus, and Tobacco rattle virus (TRV) in the genus Tobravirus. Based on the number of virus-associated contigs, PlAMV (129 contigs) was the dominant virus followed by CMV (74 contigs), LSV (67 contigs), TVX (67 contigs), LMoV (36 contigs), and TRV (23 contigs) (Fig. 1b). The number of virus-associated contigs in each library ranged from two contigs (B1 and B2) to 49 contigs (C2) (Fig. 1c). There were 14 libraries containing more than 10 virus-associated contigs. We next compared the identified viruses among the three different projects (Fig. 1d). Study A contained four viruses: LMoV, LSV, CMV, and PlAMV. Study B contained LMoV, LSV, CMV, PlAMV, and TVX, while Study C contained only two viruses, including PlAMV and TRV. In the case of Study B, we identified PlAMV-associated sequence reads but not assembled contigs. PlAMV was commonly identified in all three studies, while LMoV, LSV, and CMV were commonly identified in Study A and Study B. TVX and TRV were specifically identified in Study B and C, respectively.
Fig. 1.
Study of viromes for lily cultivar “Sorbonne”. a Flower of lily cultivar “Sorbonne” grown in greenhouse. Image was taken by WKC. b Pie chart showing number of virus-associated contigs from all examined transcriptome data. c Number of virus-associated contigs in each lily transcriptome. d Comparison of identified viruses in three different studies: A, B, and C. e Proportion (percentage) of virus-associated reads in each lily transcriptome. Virus-associated reads were obtained by BLASTN search against identified virus reference genome. Red bars indicate lily transcriptomes containing high proportion of virus-associated reads
Table 1.
Information of RNA-Seq libraries used for the virome study of lily cultivar “Sorbonne”
| Index | SRA accession No. | Tissues |
|---|---|---|
| A | SRR787322 | Pooled with different tissues such as basal roots, scales, leaves, epidermis, tepals, and stigmas |
| B1 | SRR6837843 | Upper stem |
| B2 | SRR6837844 | Upper stem |
| B3 | SRR6837845 | Lower stem |
| B4 | SRR6837846 | Apex |
| B5 | SRR6837847 | Upper stem |
| B6 | SRR6837848 | Lower stem |
| B7 | SRR6837850 | Lower stem |
| B8 | SRR6837851 | Root |
| B9 | SRR6837852 | Bulb |
| B10 | SRR6837853 | Lower stem |
| B11 | SRR6837854 | Root |
| B12 | SRR6837855 | Leaf |
| B13 | SRR6837856 | Upper stem |
| C1 | SRR4125791 | Leaf |
| C2 | SRR4128962 | Scale |
| C3 | SRR4128968 | Scale |
| C4 | SRR4131341 | Scale |
| C5 | SRR4131406 | Flower |
| C6 | SRR4142853 | Leaf |
| C7 | SRR4143329 | Flower |
| C8 | SRR4143339 | Flower |
Name of each library with respective SRA accession number and lily tissues used for library preparation were indicated
Proportion of virus-associated reads in the transcriptome
Viral abundance can vary depending on sample conditions and library preparation methods. All libraries in the three studies were prepared by mRNA isolation for cDNA library preparation. However, the sample conditions differed. For example, Study A used a mixture of six different tissues, including basal roots, scales, leaves, epidermal tissues, tepals, and stigmas (Table 1). Study B only used aerial bulbils, while Study C used three different tissues, including flowers, leaves, and bulb scales (Table 1). We analyzed the proportion of viral RNA in each library based on read numbers by a BLASTN search (Additional file 1: Table S2). The proportion of viral reads in Study A possessing a single library was 1.4%, whereas the proportion of viral reads in Study B containing 13 libraries ranged from 0 to 0.5% (Fig. 1e). Interestingly, the proportion of viral reads in Study C containing nine libraries was very high, ranging from 0.2 to 34.1%. In particular, samples for C2 (Scale 65) (11.5%), C5 (Flower 62) (26.3%), and C7 (Flower 64) (34.1%) were relatively high. Although there were three biological replicates for each condition, there were strong deviations among the three replicates for viral proportion.
Viral populations in samples co-infected by different viruses
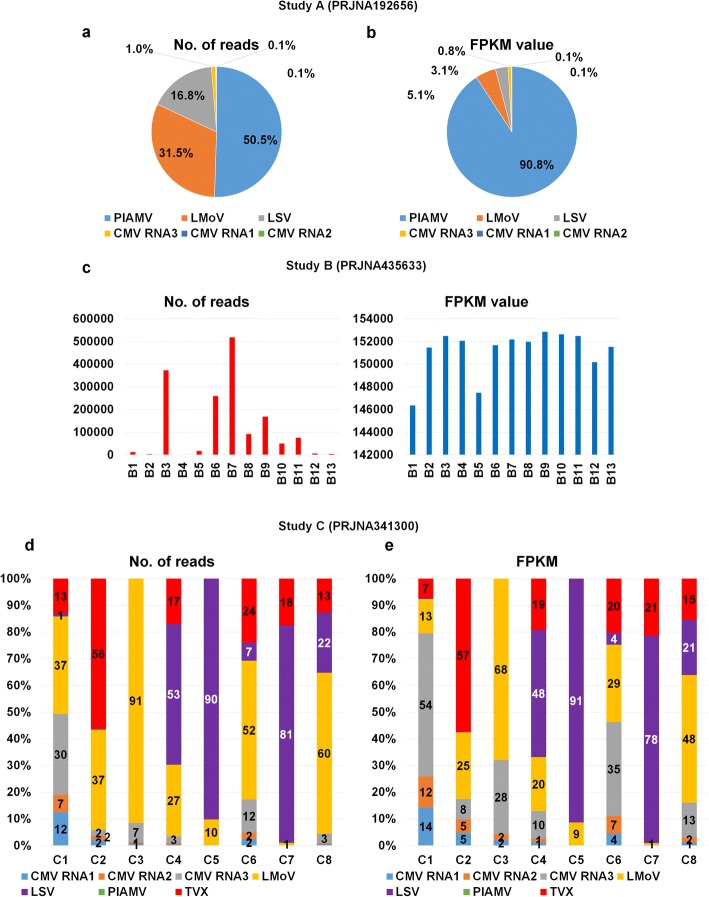
As we have shown, most libraries were co-infected by different viruses, except the 12 libraries containing only PlAMV in Study B. To reveal the viral populations in each library, we focused on two different numbers: number of virus-associated reads and number of Fragments Per Kilobase of transcript per Million (FPKM) mapped reads (Additional file 1: Table S2). Virus-associated reads are the total reads associated with the given virus, whereas FPKM values are normalized values using sequencing depth and genome size. In Study A, PlAMV (50.5%) was the dominant virus followed by LMoV (31.5%) and LSV (16.8%) based on the number of reads (Fig. 2a); however, the proportion of PlAMV (90.8%) was dramatically increased based on the FPKM value (Fig. 2b). We examined the proportion of PlAMV in the 13 libraries of Study B (Fig. 2c). FPKM values (blue bar) showed some deviation among libraries; however, the differences were not very significant. On the other hand, the number of reads associated with PlAMV (red bar) in each library changed significantly. In Study C using three different tissues, the virus population in each library changed dramatically. For instance, LMoV was the dominant virus in four libraries, whereas LSV was the dominant virus in three libraries based on the number of reads (Fig. 2d). In contrast, CMV composed of three RNAs was the dominant virus in two libraries (i.e., C1 and C6). In addition, there were strong deviations among the three replicates. For example, three libraries (C2, C3, and C4) were derived from scale samples; however, the viral populations of the three libraries were very different from each other. In RNA-Seq, the number of virus-associated reads is strongly correlated with the virus genome size. Therefore, it is important to use FPKM values that consider the size of the individual virus genome. The proportions of identified viruses in each library based on FPKM values differed from those based on the number of reads. For example, the proportion of CMV RNA3 with a relatively small genome size was dramatically increased in most libraries, such as C1, C3, and C6 (Fig. 2e). In contrast, the proportion of LMoV with a relatively large genome size was strongly decreased in most libraries. For instance, the proportion of LMoV was 91% based on the number of reads; however, the proportion of LMoV was reduced to 68% based on FPKM values in C3.
Fig. 2.
Proportion of identified viruses based on number of virus-associated reads and FPKM values. Pie charts showing proportion of identified viruses based on virus-associated reads (a) and FPKM values (b) in Study A. All virus-associated reads were identified by BLASTN search against identified virus reference genome. c Bar graphs showing change in PlAMV-associated reads (red bar) and FPKM values (blue bar) in different lily transcriptomes for Study B. Bar charts showing proportion of identified viruses based on virus-associated reads (d) and FPKM values (e) in Study C
Mapping of raw sequence reads and assembly of virus genomes
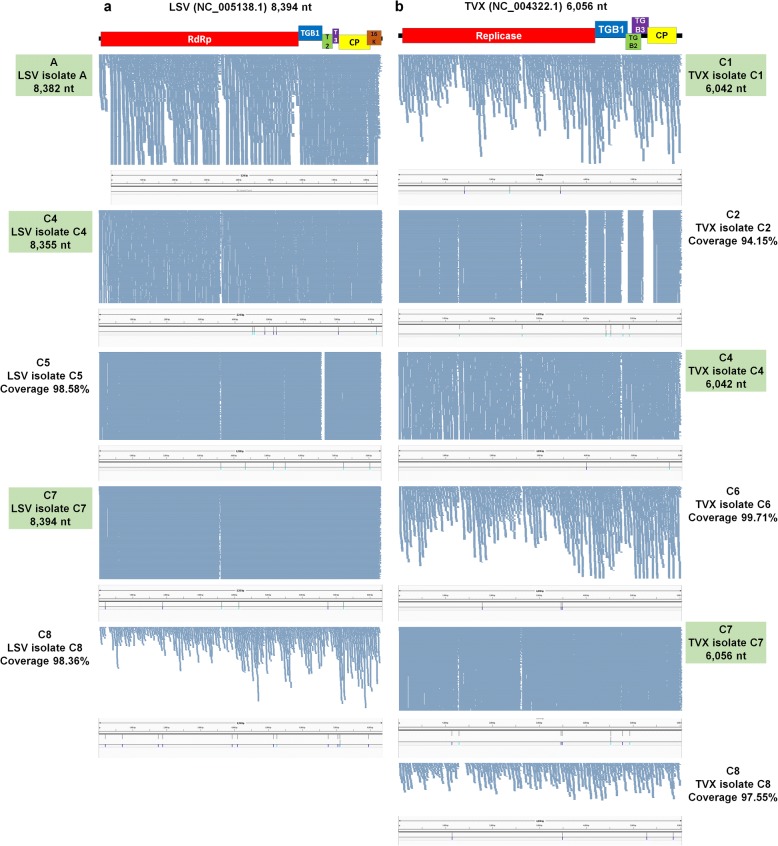
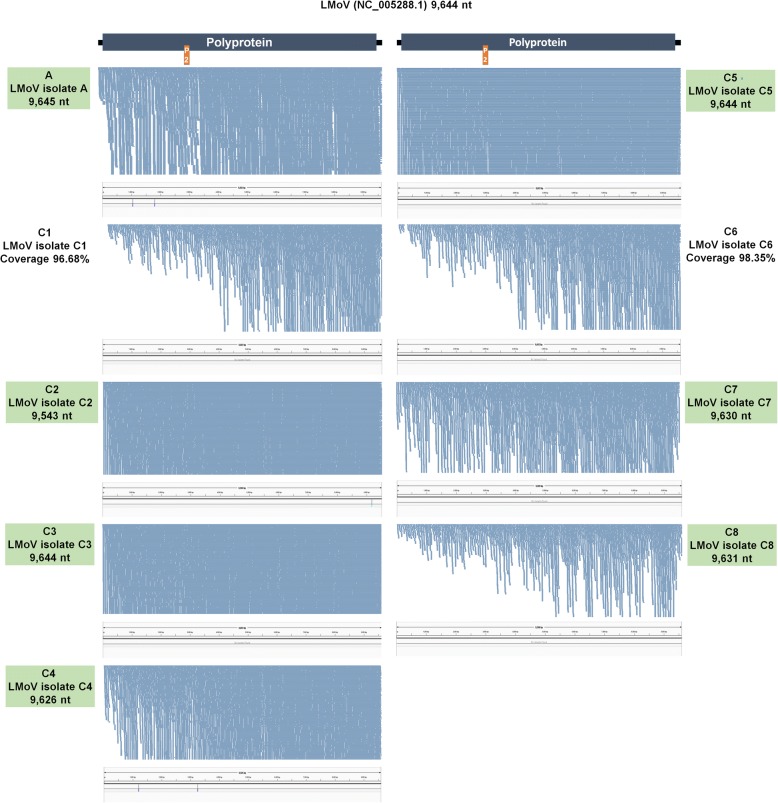
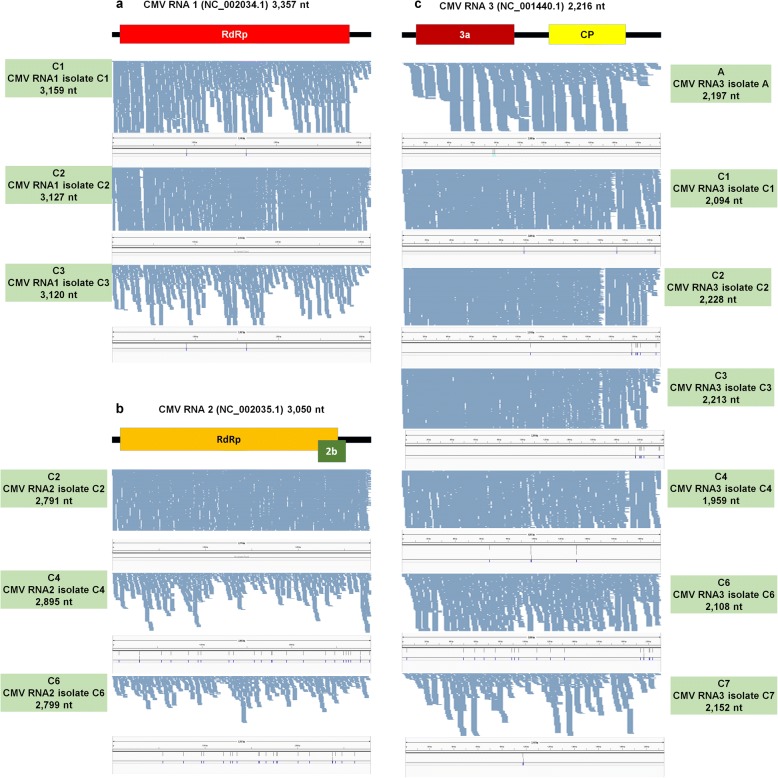
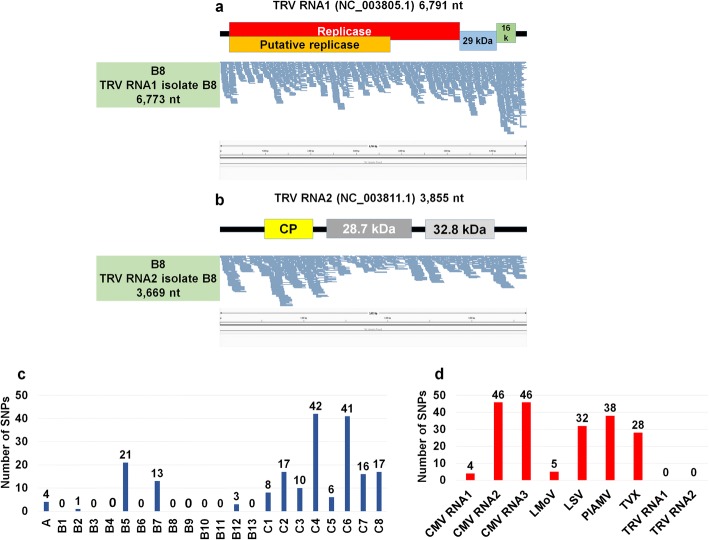
The sizes of virus-associated contigs ranged from 201 bp to 9661 bp (Table 2). Of the identified viruses, many contigs for LMoV were larger than 9000 bp, meaning they could nearly cover the complete genome of LMoV. In order to assemble the genomes of the identified viruses, we mapped raw sequence reads on each virus reference genome (Figs. 3, 4, 5, 6, 7). By mapping, sequence coverage of several identified virus isolates (e.g., five isolates for LSV, five isolates for TVX, nine isolates for LMoV, 11 isolates for PlAMV, three isolates for CMV RNA1, three isolates for CMV RNA2, seven isolates for CMV RNA3, one isolate for TRV RNA1, and one isolate for TRV RNA2) was more than 90%. In many cases, most regions of the target virus genome were fully mapped by virus-associated reads; however, some specific regions in some viruses were not mapped. Moreover, the number of mapped reads on LMoV was increased from the 5′ region to the 3′ region (Fig. 4). In the case of LSV isolate C5 with 98.58% coverage, a partial triple gene block (TGB) region was not mapped (Fig. 3a). Again, in TVX isolate C2 with 94.15% coverage, many TGB regions were not mapped (Fig. 3b). Moreover, three PlAMV isolates (i.e., B1, B10, and B11) contained unmapped regions (Fig. 5). Based on mapping and assembled virus-associated contigs, we obtained 32 nearly complete genomes: three genomes for CMV RNA1, three genomes for CMV RNA2, seven genomes for CMV RNA3, seven genomes for LMoV, three genomes for LSV, four genomes for PlAMV, one genome for TRV RNA1, one genome for TRV RNA1, and three genomes for TVX (Table 3). Genomes of some virus isolates lacking specific regions were not included.
Table 2.
Summary of identified virus-associated contigs
| Study | Project No. | SRA No. | Virus | No. of contigs | Minimum contig length (bp) | Maximum contig length (bp) |
|---|---|---|---|---|---|---|
| Study A | PRJNA192656 | SRR787322 | PlAMV | 4 | 6140 | 6149 |
| Study A | PRJNA192656 | SRR787322 | LMoV | 2 | 9656 | 9661 |
| Study A | PRJNA192656 | SRR787322 | LSV | 4 | 3611 | 4775 |
| Study A | PRJNA192656 | SRR787322 | CMV RNA3 | 3 | 289 | 2196 |
| Study A | PRJNA192656 | SRR787322 | CMV RNA1 | 12 | 262 | 731 |
| Study A | PRJNA192656 | SRR787322 | CMV RNA2 | 6 | 315 | 1013 |
| Study B | PRJNA435633 | SRR6837843 | PlAMV | 2 | 1562 | 4501 |
| Study B | PRJNA435633 | SRR6837844 | PlAMV | 2 | 788 | 5112 |
| Study B | PRJNA435633 | SRR6837845 | PlAMV | 23 | 204 | 5227 |
| Study B | PRJNA435633 | SRR6837846 | PlAMV | 4 | 382 | 1412 |
| Study B | PRJNA435633 | SRR6837847 | PlAMV | 6 | 232 | 3790 |
| Study B | PRJNA435633 | SRR6837848 | PlAMV | 11 | 275 | 4485 |
| Study B | PRJNA435633 | SRR6837850 | PlAMV | 20 | 201 | 5434 |
| Study B | PRJNA435633 | SRR6837851 | PlAMV | 18 | 277 | 2715 |
| Study B | PRJNA435633 | SRR6837851 | TRV RNA2 | 5 | 1224 | 3669 |
| Study B | PRJNA435633 | SRR6837851 | TRV RNA1 | 18 | 218 | 6773 |
| Study B | PRJNA435633 | SRR6837852 | PlAMV | 15 | 214 | 4188 |
| Study B | PRJNA435633 | SRR6837853 | PlAMV | 6 | 232 | 3861 |
| Study B | PRJNA435633 | SRR6837854 | PlAMV | 7 | 237 | 3729 |
| Study B | PRJNA435633 | SRR6837855 | PlAMV | 5 | 818 | 6145 |
| Study B | PRJNA435633 | SRR6837856 | PlAMV | 6 | 779 | 4446 |
| Study C | PRJNA341300 | SRR4125791 | CMV RNA3 | 2 | 917 | 1237 |
| Study C | PRJNA341300 | SRR4125791 | CMV RNA1 | 2 | 411 | 2774 |
| Study C | PRJNA341300 | SRR4125791 | CMV RNA2 | 1 | 2688 | |
| Study C | PRJNA341300 | SRR4125791 | TVX | 6 | 312 | 2162 |
| Study C | PRJNA341300 | SRR4125791 | LSV | 6 | 201 | 855 |
| Study C | PRJNA341300 | SRR4125791 | LMoV | 2 | 392 | 8952 |
| Study C | PRJNA341300 | SRR4128962 | TVX | 22 | 205 | 1442 |
| Study C | PRJNA341300 | SRR4128962 | LMoV | 12 | 332 | 5408 |
| Study C | PRJNA341300 | SRR4128962 | CMV RNA3 | 3 | 201 | 1762 |
| Study C | PRJNA341300 | SRR4128962 | CMV RNA1 | 2 | 378 | 2768 |
| Study C | PRJNA341300 | SRR4128962 | CMV RNA2 | 1 | 2791 | |
| Study C | PRJNA341300 | SRR4128962 | LSV | 9 | 231 | 2183 |
| Study C | PRJNA341300 | SRR4128962 | PlAMV | 0 | ||
| Study C | PRJNA341300 | SRR4128968 | LMoV | 7 | 282 | 5360 |
| Study C | PRJNA341300 | SRR4128968 | CMV RNA3 | 4 | 514 | 985 |
| Study C | PRJNA341300 | SRR4128968 | CMV RNA1 | 3 | 297 | 2751 |
| Study C | PRJNA341300 | SRR4128968 | CMV RNA2 | 1 | 2686 | |
| Study C | PRJNA341300 | SRR4131341 | LSV | 8 | 205 | 3626 |
| Study C | PRJNA341300 | SRR4131341 | LMoV | 2 | 4302 | 5349 |
| Study C | PRJNA341300 | SRR4131341 | TVX | 9 | 276 | 1914 |
| Study C | PRJNA341300 | SRR4131341 | CMV RNA3 | 5 | 246 | 873 |
| Study C | PRJNA341300 | SRR4131341 | CMV RNA1 | 3 | 376 | 1708 |
| Study C | PRJNA341300 | SRR4131341 | CMV RNA2 | 2 | 913 | 2004 |
| Study C | PRJNA341300 | SRR4131406 | LSV | 10 | 202 | 2593 |
| Study C | PRJNA341300 | SRR4131406 | LMoV | 4 | 1993 | 7253 |
| Study C | PRJNA341300 | SRR4131406 | TVX | 10 | 239 | 962 |
| Study C | PRJNA341300 | SRR4131406 | CMV RNA3 | 5 | 201 | 352 |
| Study C | PRJNA341300 | SRR4142853 | LMoV | 3 | 207 | 8811 |
| Study C | PRJNA341300 | SRR4142853 | TVX | 5 | 350 | 2141 |
| Study C | PRJNA341300 | SRR4142853 | CMV RNA3 | 2 | 898 | 1251 |
| Study C | PRJNA341300 | SRR4142853 | LSV | 15 | 210 | 1019 |
| Study C | PRJNA341300 | SRR4142853 | CMV RNA2 | 3 | 829 | 1054 |
| Study C | PRJNA341300 | SRR4142853 | CMV RNA1 | 6 | 205 | 795 |
| Study C | PRJNA341300 | SRR4143329 | LSV | 9 | 211 | 2604 |
| Study C | PRJNA341300 | SRR4143329 | TVX | 9 | 312 | 2018 |
| Study C | PRJNA341300 | SRR4143329 | LMoV | 1 | 9631 | |
| Study C | PRJNA341300 | SRR4143329 | CMV RNA3 | 3 | 432 | 924 |
| Study C | PRJNA341300 | SRR4143329 | CMV RNA1 | 1 | 217 | |
| Study C | PRJNA341300 | SRR4143329 | CMV RNA2 | 0 | ||
| Study C | PRJNA341300 | SRR4143339 | LMoV | 3 | 271 | 9003 |
| Study C | PRJNA341300 | SRR4143339 | LSV | 6 | 201 | 2547 |
| Study C | PRJNA341300 | SRR4143339 | TVX | 6 | 304 | 2137 |
| Study C | PRJNA341300 | SRR4143339 | CMV RNA3 | 3 | 254 | 1001 |
| Study C | PRJNA341300 | SRR4143339 | CMV RNA2 | 1 | 332 | |
| Study C | PRJNA341300 | SRR4143339 | CMV RNA1 | 0 |
Name of identified virus, number of virus-associated contigs, and lengths of minimum and maximum contigs, respectively, are indicated
Fig. 3.
Genome assembly, mapping of reads on virus genome, and mutation analyses for LSV and TVX. Genome organization of LSV (a) and TVX (b) based on corresponding reference genome. Green boxes indicate information of assembled virus genome, such as SRA number, name of isolate, and genome size. The positions of identified SNPs in each virus genome were visualized by the Tablet program
Fig. 4.
Genome assembly, mapping of reads on virus genome, and mutation analyses for LMoV. Genome organization of LMoV based on corresponding reference genome. Green boxes indicate information of assembled virus genome, such as SRA number, name of isolate, and genome size. The positions of identified SNPs in each virus genome were visualized by the Tablet program
Fig. 5.
Genome assembly, mapping of reads on virus genome, and mutation analyses for PlAMV. Genome organization of PlAMV based on corresponding reference genome. Green boxes indicate information of assembled virus genome, such as SRA number, name of isolate, and genome size. The positions of identified SNPs in each virus genome were visualized by the Tablet program
Fig. 6.
Genome assembly, mapping of reads on virus genome, and mutation analyses for CMV. Genome organization of three RNA fragments, RNA 1 (a), RNA 2 (b), and RNA 3 (c), of CMV genome. Green boxes indicate information of assembled virus genome, such as SRA number, name of isolate, and genome size. The positions of identified SNPs in each virus genome were visualized by the Tablet program
Fig. 7.
Genome assembly, mapping of reads on virus genome, and mutation analyses for TRV and number of identified SNPs. Genome organization of two RNA fragments, RNA1 (a) and RNA2 (b), of TRV genome. Green boxes indicate information of assembled virus genome, such as SRA number, name of isolate, and genome size. The positions of identified SNPs in each virus genome were visualized by the Tablet program. Number of identified SNPs associated with identified viruses in each transcriptome (c) and total number of identified SNPs for each virus (d)
Table 3.
List of assembled virus genomes
| Index | Virus genome | Study | Size (nt) | Accession No. |
|---|---|---|---|---|
| 1 | CMV-RNA1 | C1 | 3159 | MH360227 |
| 2 | CMV-RNA1 | C2 | 3127 | MH360228 |
| 3 | CMV-RNA1 | C3 | 3120 | MH360229 |
| 4 | CMV-RNA2 | C2 | 2791 | MH360230 |
| 5 | CMV-RNA2 | C4 | 2895 | MH360231 |
| 6 | CMV-RNA2 | C6 | 2799 | MH360232 |
| 7 | CMV-RNA3 | A | 2197 | MH360233 |
| 8 | CMV-RNA3 | C1 | 2094 | MH360234 |
| 9 | CMV-RNA3 | C3 | 2213 | MH360235 |
| 10 | CMV-RNA3 | C4 | 1959 | MH360236 |
| 11 | CMV-RNA3 | C6 | 2108 | MH360237 |
| 12 | CMV-RNA3 | C7 | 2152 | MH360238 |
| 13 | LMoV | A | 9645 | MH360239 |
| 14 | LMoV | C2 | 9543 | MH360240 |
| 15 | LMoV | C3 | 9644 | MH360241 |
| 16 | LMoV | C4 | 9626 | MH360242 |
| 17 | LMoV | C5 | 9644 | MH360243 |
| 18 | LMoV | C7 | 9630 | MH360244 |
| 19 | LMoV | C8 | 9631 | MH360245 |
| 20 | LSV | A | 8382 | MH360246 |
| 21 | LSV | C4 | 8355 | MH360247 |
| 22 | LSV | C7 | 8394 | MH360248 |
| 23 | PlAMV | A | 6119 | MH360249 |
| 24 | PlAMV | B5 | 6102 | MH360250 |
| 25 | PlAMV | B12 | 6109 | MH360251 |
| 26 | PlAMV | B13 | 6102 | MH360252 |
| 27 | TRV-RNA1 | B8 | 6773 | MH360253 |
| 28 | TRV-RNA2 | B8 | 3669 | MH360254 |
| 29 | TVX | C1 | 6042 | MH360255 |
| 30 | TVX | C4 | 6042 | MH360256 |
| 31 | TVX | C7 | 6056 | MH360257 |
| 32 | CMV-RNA3 | C2 | 2208 | MH360258 |
De novo-assembled virus genome in each library, genome size, and respective accession numbers are provided
Mutation rates of identified viruses
Most RNA viruses exhibit strong genetic variation within the infected host. In order to examine mutation positions and rates for identified viruses, we analyzed single nucleotide polymorphisms (SNPs) using assembled virus genome sequences as references. For example, the assembled viral genome of LSV isolate A was used for SNP calling of LSV genome in the library A while the assembled genome of LSV isolate C4 was used for SNP calling of LSV genome in the library C4. In the case of LSV, we identified no SNP in isolate A, seven SNPs in isolate C4, six SNPs in isolate C5, six SNPs in isolate C7, and 12 SNPs in isolate C8 (Fig. 3a). In TVX, we identified three SNPs in C1, six SNPs in C2, two SNPs in C4, three SNPs in C6, seven SNPs in C7, and four SNPs in C8 (Fig. 3b). In the case of LMoV, six isolates (C1, C3, C5, C6, C7, and C8) did not contain any SNP, while isolate A (two SNPs), isolate C2 (one SNP), and isolate C4 (two SNPs) possessed a small number of SNPs (Fig. 4). In the case of PlAMV, seven out of 11 isolates did not contain any SNP, while two, isolate B5 (21 SNPs) and isolate B7 (13 SNPs), contained several SNPs (Fig. 5). We also identified several SNPs in CMV RNAs (Fig. 6). For example, three were two SNPs for CMV RNA1 of isolate C1 and C3 (Fig. 6a). In the case of CMV RNA2, we identified 23 SNPs in isolate C4 and 20 SNPs in C5 (Fig. 6b). In CMV RNA3, we identified a relatively large number of SNPs as compared to the other two RNAs (Fig. 6c). In particular, isolates C2 and C3 contained many SNPs in the 3′ region. Of the seven CMV RNA genomes examined, all isolates contained at least one SNP (Fig. 6). There were no SNPs in TRV RNA1 (Fig. 7a) or RNA2 (Fig. 7b). Of the 22 libraries examined, nine libraries did not contain any SNPs, while some libraries, such as B5 (21 SNPs), C4 (42 SNPs), and C6 (41 SNPs), contained many SNPs (Fig. 7c). Based on each virus genome, CMV RNA2 and CMV RNA3 contained the largest number of SNPs (46 SNPs), while CMV RNA1 (four SNPs), LMoV (five SNPs), and two TRV RNA genomes (0 SNPs) did not contain any SNPs (Fig. 7d).
Virus genome assembly and phylogenetic analyses
We obtained 32 nearly complete genomes for all six identified viruses using the obtained virus-associated contigs and mapping of reads on the corresponding reference genome sequences (Table 3 and Additional file 1: Table S3). All obtained virus genome sequences were deposited in GenBank with respective accession numbers. In the case of CMV composed of three different RNAs, three CMV RNA1, three RNA2, and seven RNA3 genome sequences were obtained (Table 3). Seven genomes for LMoV, three genomes for LSV, four genomes for PlAMV, and three genomes for TVX were identified. In addition, we obtained a nearly complete genome for TRV containing two RNA fragments.
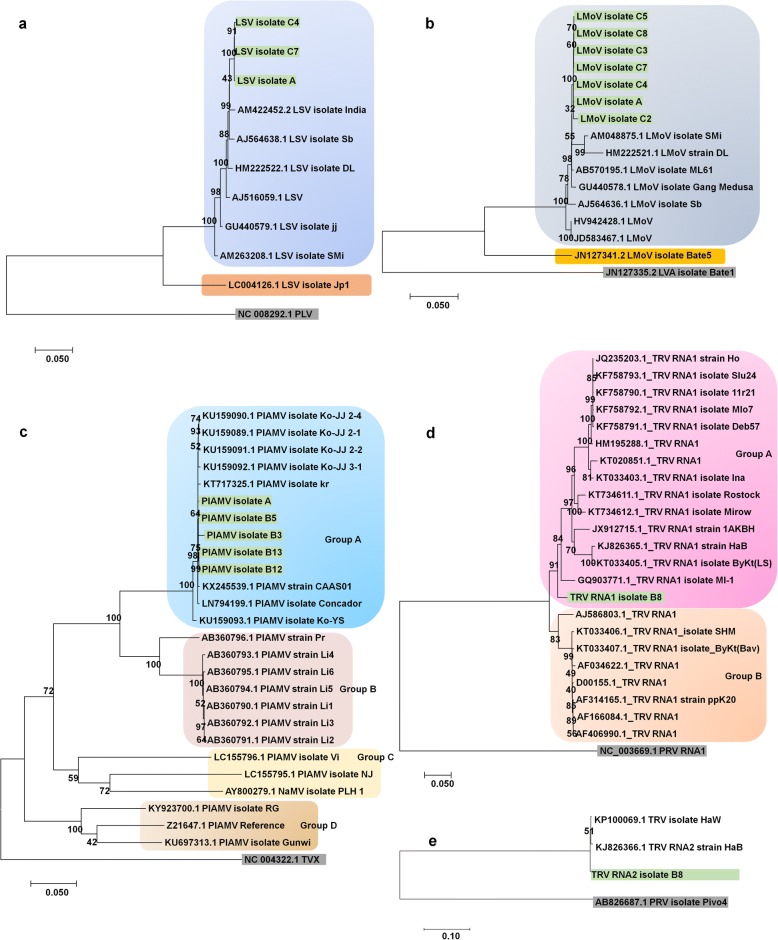
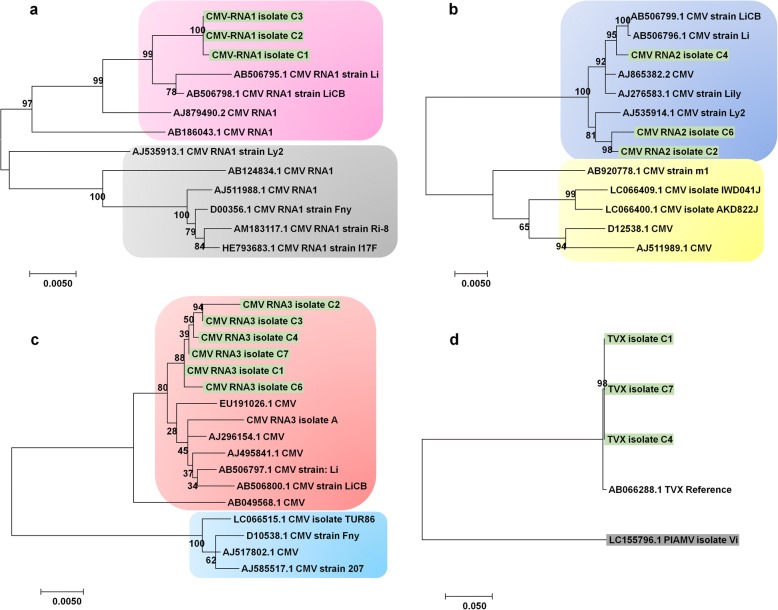
We next generated phylogenetic trees for the identified viruses using assembled genome sequences as well as publicly available virus genome sequences. A phylogenetic tree using 10 LSV genomes, nine LSV genomes including three isolates in this study is closely related except LSV isolate Jp1 (Fig. 8a). All nine LSV genomes were derived from Korea and China, while LSV isolate Jp1 originated from Japan. Similarly, the phylogenetic tree for 15 LMoV isolates showed two different phylogenetic clades (Fig. 8b). The first clade contained seven LMoV isolates from this study and seven LMoV isolates from previous studies. All 14 LMoV isolates have been identified from lilies in China, Korea, and Japan except LMoV isolate Bate5 derived from lilies in Australia [27]. The phylogenetic tree for PlAMV displayed four different groups of PlAMV isolates (Fig. 8c). Group A contained 13 PlAMV isolates, including five isolates in this study mostly derived from China and Korea, except isolate Concador from Hungary. Group B possessed seven isolates, all from Japan. PlAMV isolates in Groups C and D were highly divergent as compared to those in Groups A and B, which were derived from lilies. PlAMV isolates in Group C were derived from Viola grypoceras and Nandina domestica, while PlAMV isolates in Group D were derived from Rehmannia glutinosa, plantain, and Plantago asiatica. In the case of TRV possessing two RNA fragments, we generated two different phylogenetic trees (Fig. 8d and e). The phylogenetic tree using TRV RNA1 sequences showed two clades, Group A and Group B. Group A contained 15 TRV isolates along with isolate B8 from this study, whereas Group B included eight TRV isolates. In the case of TRV RNA2, there were only three TRV RNA2 sequences derived from tobacco and isolate B8 from lilies (Fig. 8e). In the case of CMV composed of three RNA fragments, there were numerous genome sequences. Therefore, only the best-matching genome sequences were used for phylogenetic tree construction (Fig. 9a, b, and c). Interestingly, all three and six isolates for CMV RNA1 and RNA3, respectively, were grouped together (Fig. 9a and c). In the case of CMV RNA2, all three isolates in this study were grouped together; other CMV isolates were from lilies. At the time of this study, there was only one TVX genome sequence available (Fig. 9d). The TVX genome sequence from this study showed strong genetic similarity to the TVX reference sequence from a tulip in Japan [28].
Fig. 8.
Phylogenetic relationships of LSV, LMoV, PlAMV, and TRV isolates with known isolates. a Phylogenetic tree of complete genomes for three LSV isolates, A, C4, and C7. Potato latent virus (PLV) was used as an outgroup. b Phylogenetic tree of complete genomes for seven LMoV isolates, A, C2, C3, C4, C5, C7, and C8. Lily virus A (LVA) was used as an outgroup. c Phylogenetic tree of complete genomes for five PlAMV isolates, A, B3, B5, B12, and B13. TVX was used as an outgroup. Phylogenetic tree of complete RNA1 (d) and RNA2 (e) for TRV isolate B8. Pepper ringspot virus (PRV) was used as an outgroup. Available genome sequences for LSV, LMoV, PlAMV, and TRV were also used for phylogenetic construction. Accession numbers and names of virus isolates or strains were provided. The isolates from this study were indicated by the color green
Fig. 9.
Phylogenetic relationships of CMV and TVX with known isolates. Phylogenetic trees of RNA1 (a), RNA2 (b), and RNA2 (c) for identified CMV isolates. Due to the presence of a large number of CMV genome sequences, only highly matched CMV isolates were used for BLASTN search. d Phylogenetic tree of TVX isolates, C1, C4, and C7. PlAMV was used as an outgroup. Available genome sequences for CMV and TVX were also used for phylogenetic construction. Accession numbers and names of virus isolates or strains were provided. The isolates from this study were indicated by the color green
Discussion
A virome is defined as a collection of nucleic acids that constitute viruses in a particular organism or environment [29]. Several previous studies on plant viromes have examined viruses using samples collected from diverse hosts or restricted regions [30, 31]. Moreover, some plant virome studies have examined viruses infecting single cultivars of diverse horticulture plants [23, 25, 26]. However, to date, no study has examined the viromes of plants of one type with identical genetic backgrounds collected from different geographical regions. Thus, we examined the different viromes of the lily cultivar “Sorbonne,” which is clonally propagated by bulbs and widely grown throughout the world.
Our first question was focused on whether the same plant cultivar has identical viromes among different plants grown in diverse regions. Our study clearly answered that question, in that the viromes of the lily cultivar “Sorbonne” were different, although the viruses, such as PlAMV, were commonly identified in three studies. This result demonstrated that some viruses might be transmitted by clonal propagation; however, other viruses could be newly infected under different environmental conditions (e.g., through co-cultivated plants or insect vectors). Thus, plants, viruses, and environments are important factors affecting the viromes of specific plants, as suggested previously [32].
Our next question was focused on how many viruses can infect a single lily plant. Our study showed that up to five different viruses (LMoV, LSV, CMV, TVX, and PlAMV) can infect a single lily plant. To date, LMoV, LSV, and CMV are the most common viruses infecting lilies, while PlAMV resulting in serious damage to lily production is considered one of the rapidly spreading viruses in Lilium species [33, 34]. Although cases of TVX and TRV infecting lilies have been reported [35–37], they have not been as well reported as the other four viruses. In addition, we did not find any novel viruses in our study.
NGS or high-throughput sequencing (HTS) is now the most important technique for plant virus diagnostics [38] and virus discovery [39]. In fact, virome studies using NGS not only reveal the presence of viruses but also provide nucleotide sequences associated with the identified viruses. The information of virus-associated sequences is a key factor for virome studies, illustrating virus genome assembly, number of virus-associated sequence reads, and virus mutation, as shown previously [23, 25, 26]. For instance, the choice of plant tissues and developmental stages is important for plant virus diagnostics using molecular methods. Interestingly, our study demonstrated that transcriptomes from scale and flower samples possessed high numbers of virus-associated reads, suggesting that those tissues are much more useful than normal leaves for virus diagnostics, at least in lilies.
Co-infection of diverse viruses in a single plant is now common in many horticulture plants. Furthermore, it is difficult to determine which virus is the main virus causing disease symptoms. Nevertheless, it is now possible to identify a dominant virus with virus-associated reads using transcriptome data. As indicated in our previous study [24, 26], this is done by using normalized values, such as FPKM, to calculate the proportions of individual viruses. With the co-infection of LMoV, LSV, CMV, and PlAMV in Study A, PlAMV was the dominant virus, suggesting a possible main virus causing disease symptoms. However, co-infection of five viruses in different tissues showed variable proportions of diverse viruses. Based on this result, we carefully hypothesized that different tissues and developmental stages affect virus populations.
Using virus-associated contigs and reads, we assembled nearly complete genomes of several viruses. Interestingly, all six viruses possessed the polyadenylate tails [40]. Therefore, we suggested that the possession of poly(A) tails in all six viruses might facilitate the assembly of virus genomes. The assembled genomes were used as reference genomes for virus mutation analysis and used for phylogenetic analyses. As compared to our previous studies [25, 26], genomes of identified viruses were highly conserved. Some viruses showed mutations; however, the mutation rates were very low, and the mutation positions in the genome were mostly random. In addition, a small number of SNPs might be associated with sequencing errors. Thus, we carefully hypothesized that all six viruses were highly conserved, at least in the lily cultivar “Sorbonne.” It is likely that the mutation rates of identified viruses might be different in other lily cultivars or under other environmental conditions.
Mapping of raw sequence reads on the viral genome revealed that three viruses (LSV, TVX, and PlAMV) produced defective RNAs. Interestingly, genomes of all three viruses had three partially overlapping open reading frames (ORFs) referred to as TGB, which is common in the order of Tymovirales. Moreover, some isolates produced defective RNAs that were preferentially localized in the TGB region. A previous study demonstrated that the defective RNA produced from Potato mop-top virus (PMTV) in the genus Pomovirus interferes with virus infection [41]. A recent study also revealed that defective RNA from Tomato black ring virus (TBRV) in the genus Nepovirus interfered with TBRV replication [42]. Thus, we suggested that the identified defective RNAs in our study could be defective interfering (DI) RNAs, which might be produced by the error-prone viral replicase [43]. However, the possible role of defective RNAs in our study as interfering RNAs should be further experimentally characterized.
With the help of the NGS technique, numerous virus genomes are now being assembled [44]. Similarly, we obtained 32 nearly complete virus genomes, which were further used for phylogenetic analyses. Our results showed that virus genomes are highly correlated with geographical regions. For instance, LSV isolates from Korea and China were closely related with one another but not with an isolate from Japan. Similarly, PlAMV isolates from China and Korea were grouped together, while PlAMV isolates from Japan were clustered together. However, LMoV isolates from China, Korea, and Japan were clustered together. Furthermore, PlAMV isolates from lilies were highly divergent as compared to those from other host plants, suggesting that plant host type is an important factor in distinguishing different virus isolates.
Conclusions
We conducted comprehensive virome analyses of a single lily cultivar, “Sorbonne,” using transcriptome data. Our results shed light on an array of lily virome-associated topics, including virus identification, the dominant virus, virus accumulation in different plant tissues, virus genome assembly, virus mutation, identification of defective RNAs, and phylogenetic relationships of identified viruses. Taken together, we provide very useful methods and valuable results that can be applied in other virome-associated studies.
Methods
Collection of RNA-Seq data for lily cultivar “Sorbonne” from SRA database
We searched RNA-Seq data for the lily cultivar “Sorbonne” in the SRA database of the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/sra) using “Lily” and “Sorbonne” as queries. There were 23 RNA-Seq datasets from four different projects. Detailed information on the RNA-Seq datasets can be found in Additional file 1: Table S1. Moreover, all detailed information on plant materials, library preparation, and RNA-Seq in each project can be found in the previous studies. Each library was newly indexed for simplicity based on the project. PRJNA192656, indexed as A, contained a single SRA dataset that was prepared from a mixture of six different tissues, including basal roots, scales, leaves, epidermal tissues, tepals, and stigmas for total RNA extraction [45]. PRJNA435633, indexed as B1 to B13, contained 13 SRA datasets that were prepared from different tissues. PRJNA341300, indexed as C1 to C8, included eight SRA datasets that were prepared from three different tissues (leaves, flowers, and scales) at different developmental stages [46]. The samples in Study A were derived from the gardens of the plants that were grown in the gardens of the institute of landscape architecture, Zhejiang University, Hangzhou, China. The samples in Study B were derived from the Institute of Botany, Chinese Academy of Sciences in Beijing, China. The samples in Study C were derived from Shanxi Agricultural University, Shanxi, China. We used a workstation with two 20-core CPUs and 256 GB of RAM installed with Ubuntu 16.04.4 LTS for all data analyses. All 23 SRA datasets were downloaded from the SRA database with corresponding accession numbers using the SRA toolkit (https://www.ncbi.nlm.nih.gov/sra/docs/toolkitsoft/). All downloaded SRA datasets were converted to fastq files using fastq-dump implemented in the SRA toolkit.
De novo transcriptome assembly and identification of virus-associated contigs
In order to identify virus-associated contigs in each RNA-Seq library, bioinformatics analyses were conducted as described previously [26]. In brief, single-end or paired-end sequenced fastq files in each library were subjected to de novo transcriptome assembly using the Trinity program (v2.6.5 Release) with default parameters [47]. All assembled contigs (transcriptomes) in each library were used for MEGABLAST [48] with a cut-off E-value of 1e− 6 to search against NCBI’s viral reference database downloaded from https://www.ncbi.nlm.nih.gov/genome/viruses/. Based on the BLAST results, we extracted only virus-associated contigs in FASTA format using the blastdbcmd program (https://www.ncbi.nlm.nih.gov/books/NBK279689/). The obtained virus-associated contigs were again subjected to BLASTN against NCBI’s nucleotide (NT) database using default parameters to delete lily endogenous virus-like sequences. Only pure virus-associated contigs were used for further analyses, such as virus genome assembly.
Virus genome assembly
In order to assemble complete or nearly complete genomes for identified viruses, we aligned virus-associated contigs in each library on the reference virus genome using the ClustalW program implemented in the MEGA7 program [49]. The missing gaps of the virus genome were filled by mapping raw sequence reads on the virus genome using a Burrows–Wheeler Aligner (BWA) program with default parameters [50]. ORFs and 5′ and 3′ untranslated regions (UTRs) were also manually checked by comparing the corresponding reference virus genome. The assembled virus genome sequences were deposited in NCBI’s GenBank database with respective accession numbers (Table 3).
Construction of phylogenetic trees for six identified viruses
For the construction of phylogenetic trees, virus genome sequences covering all ORFs were used. In the case of the four viruses composed of a single RNA, three LSV genomes, seven LMoV genomes, five PlAMV genomes, and three TVX genomes in this study were used. In the case of TRV composed of two RNA fragments and CMV composed of three RNA fragments, two and three different phylogenetic trees, respectively, were used. Assembled genome sequences in this study and the complete genomes of each virus available in GenBank were aligned together using the ClustalW program. The aligned nucleotide sequences were subjected to phylogenetic tree construction using the MEGA7 program with the maximum likelihood method based on the Tamura–Nei model and 1000 bootstrap replicates [49].
Identification of SNPs for identified viruses using transcriptome data
SNPs for six viruses in each transcriptome were analyzed, as described previously [26]. In our study, we used the assembled virus genome sequence in each library as a reference genome sequence to increase SNP specificity. We aligned the raw sequence reads in each library on the identified individual viral genome using the BWA program with default parameters. We converted the Sequence Alignment Map (SAM) files into Binary Alignment Map (BAM) files using SAMtools [51]. The sorted BAM files were used to generate the Variant Call Format (VCF) file format using the mpileup function of SAMtools for SNP calling. Finally, BCFtools implemented in SAMtools was used to call SNPs. The positions of identified SNPs on each viral genome were displayed by the Tablet program [52].
Additional file
Table S1. Information of RNA-Seq libraries for lily cultivar “Sorbonne.” Index indicates name of individual library from three different projects, which can be further divided into A, B, and C. Raw data can be downloaded from SRA database using respective accession numbers. Table S2. Summary of virus-associated reads, contigs, and FPKM values. Raw sequence reads in SRA format were converted to FASTA format using the SRA Toolkit. MEGABLAST was conducted using raw sequence reads and assembled contigs against virus reference genomes using e-value 1e-6 as a cutoff. To calculate FPKM values, raw sequence reads were mapped on the reference virus genome using the BWA program followed by the pileup program implemented in the BBMap package. Table S3. Assembled virus genome sequences. Orange shells indicate assembled virus genomes covering ORFs, while light yellow shells indicate partial virus genome sequences. (XLSX 116 kb)
Acknowledgements
We would like to express our deepest gratitude to Hyang Sook Kim, Jin Kyong Cho, and Mi Kyong Kim for all their support and commitment associated with lily cultivation. This work is dedicated to the memory of my father, Tae Jin Cho (1946–2015).
Funding
This work was partially supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through the Agri-Bio industry Technology Development Program, funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA) (117120–01) and a National Research Foundation of Korea (NRF) grant funded by the Korea government Ministry of Education (No. NRF-2018R1D1A1B07043597).
Availability of data and materials
The raw datasets in this study are freely available in the Sequence Read Archive (SRA) repository with project numbers PRJNA192656, PRJNA252055, PRJNA435633, and PRJNA341300. The 32 virus genome sequences obtained from this study were also deposited in GenBank, NCBI, with respective accession numbers (MH360227 to MH360258).
Abbreviations
- BWA
Burrows–-Wheeler Aligner
- CMV
Cucumber mosaic virus
- DI
Defective interfering
- HTS
High-throughput sequencing
- LMoV
Lily mottle virus
- LSV
Lily symptomless virus
- NGS
Next-generation sequencing
- ORF
Open reading frame
- PCR
Polymerase chain reaction
- PlAMV
Plantago asiatica mosaic virus
- PMTV
Potato mop-top virus
- RNA-Seq
RNA-Sequencing
- RT
Reverse-transcription
- SNP
Single nucleotide polymorphism
- SNP
Single nucleotide polymorphism
- TBRV
Tomato black ring virus
- TGB
Triple gene block
- TRV
Tobacco rattle virus
- TVX
Tulip virus X
- UTR
Untranslated region
- VCF
Variant Call Format
Authors’ contributions
YJ was the main author of this paper. YJ performed most of the bioinformatics analyses. YJ and WKC interpreted the data and discussed the results. This project was conceived and designed by YJ and WKC. YJ and WKC wrote the manuscript. All the authors have read, revised, and approved the manuscript.
Ethics approval and consent to participate
This study did not include the use of any animals, human or otherwise, so did not require ethical approval.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yeonhwa Jo, Email: yeonhwajo@gmail.com.
Won Kyong Cho, Phone: +82-2-880-4687, Email: wonkyong@gmail.com.
References
- 1.Du Y-p, Bi Y, Zhang M-f, Yang F-p, Jia G-x, Zhang X-h. Genome size diversity in Lilium (Liliaceae) is correlated with karyotype and environmental traits. Front Plant Sci. 2017;8:1303. doi: 10.3389/fpls.2017.01303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim K-B, Barba-Gonzalez R, Zhou S, Ramanna M, Van Tuyl JM. Interspecific hybridization in lily (Lilium): taxonomic and commercial aspects of using species hybrids in breeding. Floriculture Ornamental Plant biotechnol. 2008;5:138–145. [Google Scholar]
- 3.Marasek-Ciolakowska A, Nishikawa T, Shea DJ, Okazaki K. Breeding of lilies and tulips—interspecific hybridization and genetic background—. Breed Sci. 2018;68(1):35–52. doi: 10.1270/jsbbs.17097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmonds J, Cumming B. Propagation of Lilium hybrids. II. Production of plantlets from bulb-scale callus cultures for increased propagation rates. Sci Hortic. 1976;5(2):161–170. doi: 10.1016/0304-4238(76)90078-9. [DOI] [Google Scholar]
- 5.Chastagner Gary A., van Tuyl Jaap M., Verbeek Martin, Miller William B., Westerdahl Becky B. Handbook of Plant Disease Management. Cham: Springer International Publishing; 2017. Diseases of Lily; pp. 1–61. [Google Scholar]
- 6.Brierley P, Smith FF. Studies on Lily Virus Diseases: the neerotic-fleck Complex in Lilium longiflorum. Phytopathology. 1944;34(6):529–55.
- 7.Rivas EB, Bôdi ECA, Harakava R, Gregori F, Gonçalves MC. Occurrence and molecular analysis of quarantine virus in lily cultivation areas in Brazil. Pesq Agrop Brasileira. 2016;51(5):615–622. doi: 10.1590/S0100-204X2016000500023. [DOI] [Google Scholar]
- 8.Asjes CJ. Control of aphid-borne lily symptomless virus and lily mottle virus in Lilium in the Netherlands. Virus Res. 2000;71(1–2):23–32. doi: 10.1016/S0168-1702(00)00185-4. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang Y, Xie Z, Yang G, Guo Z, Wang L. Simultaneous detection of three lily viruses using triplex IC-RT-PCR. J Virol Methods. 2017;249:69–75. doi: 10.1016/j.jviromet.2017.08.018. [DOI] [PubMed] [Google Scholar]
- 10.Loebenstein G, Raccah B. Control of non-persistently transmitted aphid-borne viruses. Phytoparasitica. 1980;8(3):221–235. doi: 10.1007/BF03158322. [DOI] [Google Scholar]
- 11.Mowat W, ŠTEFANAC Z. Aphid-transmitted viruses from lilies in Britain. Ann Appl Biol. 1974;76(3):281–287. doi: 10.1111/j.1744-7348.1974.tb01368.x. [DOI] [Google Scholar]
- 12.Kwon JY, Ryu KH, Choi SH. Reverse transcription polymerase chain reaction-based system for simultaneous detection of multiple lily-infecting viruses. Plant Path J. 2013;29(3):338. doi: 10.5423/PPJ.NT.04.2013.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim MS, Kim SM, Choi SH. Simultaneous detection of three lily-infecting viruses using a multiplex Luminex bead array. J Virol Methods. 2016;231:34–37. doi: 10.1016/j.jviromet.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Kim JH, Yoo HN, Bae EH, Jung YT. Development of an indirect ELISA and immunocapture RT-PCR for lily virus detection. J Microbiol Biotechnol. 2012;22(12):1776–1781. doi: 10.4014/jmb.1208.08042. [DOI] [PubMed] [Google Scholar]
- 15.He X, Xue F, Xu S, Wang W. Rapid and sensitive detection of lily symptomless virus by reverse transcription loop-mediated isothermal amplification. J Virol Methods. 2016;238:38–41. doi: 10.1016/j.jviromet.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Zhao B, Yang D, Zhang Y, Xu Y, Zhao X, Liang J, Fan X, Du Y, Zhu Z, Shi B. Rapid visual detection of lily mottle virus using a loop-mediated isothermal amplification method. Arch Virol. 2018;163(2):545–548. doi: 10.1007/s00705-017-3618-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Wang Y, Meng J, Xie Z, Wang R, Kutcher HR, Guo Z. Development of an immunochromatographic strip test for rapid detection of lily symptomless virus. J Virol Methods. 2015;220:13–17. doi: 10.1016/j.jviromet.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Wang Y, Yang W, Xie Z, Wang R, Kutcher HR, Guo Z. A rapid immunochromatographic test to detect the lily mottle virus. J Virol Methods. 2015;220:43–48. doi: 10.1016/j.jviromet.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Pecman A, Kutnjak D, Gutiérrez-Aguirre I, Adams I, Fox A, Boonham N, Ravnikar M. Next generation sequencing for detection and discovery of plant viruses and Viroids: comparison of two approaches. Front Microbiol. 2017;8:1998. doi: 10.3389/fmicb.2017.01998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrero RA, Napier KR, Cunnington J, Liefting L, Keenan S, Frampton RA, Szabo T, Bulman S, Hunter A, Ward L. An internet-based bioinformatics toolkit for plant biosecurity diagnosis and surveillance of viruses and viroids. BMC Bioinformatics. 2017;18(1):26. doi: 10.1186/s12859-016-1428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Q, Ding S-W, Zhang Y, Zhu S. Identification of viruses and viroids by next-generation sequencing and homology-dependent and homology-independent algorithms. Annu Rev Phytopathol. 2015;53:425–444. doi: 10.1146/annurev-phyto-080614-120030. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Jia A, Qiao Y, Xiang J, Zhang Y, Wang W. Virome analysis of lily plants reveals a new potyvirus. Arch Virol. 2018;163(4):1079–1082. doi: 10.1007/s00705-017-3690-9. [DOI] [PubMed] [Google Scholar]
- 23.Jo Y, Choi H, Cho JK, Yoon J-Y, Choi S-K, Cho WK. In silico approach to reveal viral populations in grapevine cultivar Tannat using transcriptome data. Sci Rep. 2015;5:15841. doi: 10.1038/srep15841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jo Y, Choi H, Kim S-M, Kim S-L, Lee BC, Cho WK. Integrated analyses using RNA-Seq data reveal viral genomes, single nucleotide variations, the phylogenetic relationship, and recombination for apple stem grooving virus. BMC Genomics. 2016;17(1):579. doi: 10.1186/s12864-016-2994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jo Y, Choi H, Kim S-M, Kim S-L, Lee BC, Cho WK. The pepper virome: natural co-infection of diverse viruses and their quasispecies. BMC Genomics. 2017;18(1):453. doi: 10.1186/s12864-017-3838-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jo Y, Lian S, Chu H, Cho JK, Yoo S-H, Choi H, Yoon J-Y, Choi S-K, Lee BC, Cho WK. Peach RNA viromes in six different peach cultivars. Sci Rep. 2018;8(1):1844. doi: 10.1038/s41598-018-20256-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wylie SJ, Luo H, Li H, Jones MG. Multiple polyadenylated RNA viruses detected in pooled cultivated and wild plant samples. Arch Virol. 2012;157(2):271–284. doi: 10.1007/s00705-011-1166-x. [DOI] [PubMed] [Google Scholar]
- 28.Yamaji Y, Kagiwada S, Nakabayashi H, Ugaki M, Namba S. Complete nucleotide sequence of tulip virus X (TVX-J): the border between species and strains within the genus Potexvirus. Arch Virol. 2001;146(12):2309–2320. doi: 10.1007/s007050170004. [DOI] [PubMed] [Google Scholar]
- 29.Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, Azam F, Rohwer F. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci. 2002;99(22):14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coetzee B, Freeborough M-J, Maree HJ, Celton J-M, Rees DJG, Burger JT. Deep sequencing analysis of viruses infecting grapevines: virome of a vineyard. Virology. 2010;400(2):157–163. doi: 10.1016/j.virol.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 31.Gu Y-H, Tao X, Lai X-J, Wang H-Y, Zhang Y-Z. Exploring the polyadenylated RNA virome of sweet potato through high-throughput sequencing. PLoS One. 2014;9(6):e98884. doi: 10.1371/journal.pone.0098884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roossinck MJ. Plants, viruses and the environment: ecology and mutualism. Virology. 2015;479:271–277. doi: 10.1016/j.virol.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 33.Parrella G, Greco B, Pasqualini A, Nappo AG. Plantago asiatica mosaic virus found in protected crops of lily hybrids in Southern Italy. Plant Dis. 2015;99(9):1289. doi: 10.1094/PDIS-03-15-0281-PDN. [DOI] [Google Scholar]
- 34.Hammond J, Bampi D, Reinsel M. First report of Plantago asiatica mosaic virus in imported Asiatic and oriental lilies (Lilium hybrids) in the United States. Plant Dis. 2015;99(2):292. doi: 10.1094/PDIS-08-14-0792-PDN. [DOI] [PubMed] [Google Scholar]
- 35.Derks A. Tobacco rattle virus in lilies. Neth J Plant Pathol. 1975;81(1–2):78–80. doi: 10.1007/BF02650334. [DOI] [Google Scholar]
- 36.Tomassoli L, Benetti M. Tobacco rattle virus in triple infection in lily. Adv Hortic Sci. 1988;2(3):117–19.
- 37.Ward L, Tang J, Quinn B, Martin E, Clover G. First report of tulip virus X in New Zealand. Plant Pathol. 2008;57(6):1172. [Google Scholar]
- 38.Massart S, Olmos A, Jijakli H, Candresse T. Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 2014;188:90–96. doi: 10.1016/j.virusres.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 39.Roossinck MJ, Martin DP, Roumagnac P. Plant virus metagenomics: advances in virus discovery. Phytopathology. 2015;105(6):716–727. doi: 10.1094/PHYTO-12-14-0356-RVW. [DOI] [PubMed] [Google Scholar]
- 40.Li W, Zhang Y, Zhang C, Pei X, Wang Z, Jia S. Presence of poly (A) and poly (A)-rich tails in a positive-strand RNA virus known to lack 3′ poly (A) tails. Virology. 2014;454:1–10. doi: 10.1016/j.virol.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Lukhovitskaya NI, Thaduri S, Garushyants SK, Torrance L, Savenkov EI. Deciphering the mechanism of defective interfering RNA (DI RNA) biogenesis reveals that a viral protein and the DI RNA act antagonistically in virus infection. J Virol. 2013;JVI:03322. doi: 10.1128/JVI.03322-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasiów-Jaroszewska B, Minicka J, Zarzyńska-Nowak A, Budzyńska D, Elena SF. Defective RNA particles derived from tomato black ring virus genome interfere with the replication of parental virus. Virus Res. 2018;250:87–94. doi: 10.1016/j.virusres.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 43.Pathak KB, Nagy PD. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses. 2009;1(3):895–919. doi: 10.3390/v1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blawid R, Silva J, Nagata T. Discovering and sequencing new plant viral genomes by next-generation sequencing: description of a practical pipeline. Ann Appl Biol. 2017;170(3):301–314. doi: 10.1111/aab.12345. [DOI] [Google Scholar]
- 45.Du F, Wu Y, Zhang L, Li X-W, Zhao X-Y, Wang W-H, Gao Z-S, Xia Y-P. De novo assembled transcriptome analysis and SSR marker development of a mixture of six tissues from Lilium oriental hybrid ‘Sorbonne’. Plant Mol Biol Report. 2015;33(2):281–293. doi: 10.1007/s11105-014-0746-9. [DOI] [Google Scholar]
- 46.Du F, Fan J, Wang T, Wu Y, Grierson D, Gao Z, Xia Y. Identification of differentially expressed genes in flower, leaf and bulb scale of Lilium oriental hybrid ‘Sorbonne’and putative control network for scent genes. BMC Genomics. 2017;18(1):899. doi: 10.1186/s12864-017-4303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24(16):1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Durbin R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milne I, Stephen G, Bayer M, Cock PJ, Pritchard L, Cardle L, Shaw PD, Marshall D. Using tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2012;14(2):193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Information of RNA-Seq libraries for lily cultivar “Sorbonne.” Index indicates name of individual library from three different projects, which can be further divided into A, B, and C. Raw data can be downloaded from SRA database using respective accession numbers. Table S2. Summary of virus-associated reads, contigs, and FPKM values. Raw sequence reads in SRA format were converted to FASTA format using the SRA Toolkit. MEGABLAST was conducted using raw sequence reads and assembled contigs against virus reference genomes using e-value 1e-6 as a cutoff. To calculate FPKM values, raw sequence reads were mapped on the reference virus genome using the BWA program followed by the pileup program implemented in the BBMap package. Table S3. Assembled virus genome sequences. Orange shells indicate assembled virus genomes covering ORFs, while light yellow shells indicate partial virus genome sequences. (XLSX 116 kb)
Data Availability Statement
The raw datasets in this study are freely available in the Sequence Read Archive (SRA) repository with project numbers PRJNA192656, PRJNA252055, PRJNA435633, and PRJNA341300. The 32 virus genome sequences obtained from this study were also deposited in GenBank, NCBI, with respective accession numbers (MH360227 to MH360258).