Abstract
Background
RUNXl plays a key regulatory role in the process of hematopoiesis and is a common target for multiple chromosomal translocations in human acute leukemia. Mutations of RUNX1 gene can lead to acute leukemia and affect the prognosis of AML patients. We aimed to identify pivotal genes and pathways involved in RUNX1-mutated patients of with acute myeloid leukemia (AML) and to explore possible molecular markers for novel therapeutic targets of the disease.
Material/Methods
The RNA sequencing datasets of 151 cases of AML were obtained from the Cancer Genome Atlas database. Differentially expressed genes (DEGs) were identified using edgeR of the R platform. PPI (protein–protein interaction) network clustering modules were analyzed with ClusterONE, and the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analyses for modules were performed.
Results
A total of 379 genes were identified as DEGs. The KEGG enrichment analysis of DEGs showed significantly enriched pathways in cancer, extracellular matrix (ECM)-receptor interaction pathway, and cyclic adenosine monophosphate (cAMP) signaling pathway. The top 10 genes ranked by degree were PRKACG, ANKRD7, RNFL7, ROPN11, TEX14, PRMT8, OTOA, CFAP99, NRXN1, and DMRT1, which were identified as hub genes from the protein–protein interaction network (PPI). Statistical analysis revealed that RUNX1-mutated patients with AML had a shorter median survival time (MST) with poor clinical outcome and an increased risk of death when compared with those without RUNX1 mutations.
Conclusions
DEGs and pathways identified in the present study will help understand the molecular mechanisms underlying RUNX1 mutations in AML and develop effective therapeutic strategies for RUNX1-mutation AML.
MeSH Keywords: Chemistry, Bioinorganic; Leukemia, Myeloid, Acute; Suppression, Genetic
Background
Acute myeloid leukemia (AML) is the most common type of adult leukemia and its incidence rate is increasing every year [1]. The treatment has advanced greatly in recent years, resulting in an increased remission rate of 50% to 80%. However, there are still many patients who do not respond well to multiple-induction chemotherapy and the early recurrence rate is still high. The 5-year overall survival (OS) is still at a low level of 30% to 40%, while recurrent and refractory patients are less than 15% [1,2], and 60% to 80% of patients with complete remission (CR) will eventually experience recurrence. Such recurrence is more likely to cause resistance to chemotherapy and failed remission of leukemia stem cells (LSCs) [3,4].
A variety of gene mutations have been found in AML patients. Runt-related transcription factor 1 (RUNXl) is a member of the RUNX transcription factor family and plays an important role in the determination of cell lineage differentiation, normal hematopoietic cell formation, and stem cell proliferation [5,6]. Many studies have shown that RUNX1 is involved in the differentiation of myeloid cells, B cells, and T cells. Recent studies have found that RUNXl also promotes leukemia cell proliferation, suggesting that RUNXl plays a different role in different hematological malignancies [7–9]. As a transcription factor, RUNXl regulates the expression of many hematopoietic-related genes that control hematopoietic cell differentiation, apoptosis, and self-renewal. The mutations of RUNXl often lead to acute leukemia [6] and seriously affect disease outcome [10].
The aim of the present study was to find the key genes and pathways involved in RUNX1 mutations of AML.
Material and Methods
RNA sequencing data
Identification of differentially expressed genes (DEGs)
EdgeR was used according to the user’s guide for screening differential expression of genes at gene levels [14,15]. We identified differentially expressed genes (DEGs) with fold change (FC) ≥2, and defined P value and false discovery rate (FDR) cut-offs of <0.05 to be statistically significant. A heat map and volcano plot of the DEGs were drawn using the ggplot package in the R platform. In order to annotate input genes, classify gene functions, identify gene conversions, and carry out gene ontology (GO) term analysis, we use integrated Discovery (DAVID) v. 6.8 (https://david.ncifcrf.gov/tools.jsp; accessed as in August 16, 2017) [16]. To analyze the DEGs at the functional level, GO enrichment and KEGG pathway analysis were performed using the DAVID online tool. P value <0.05 was considered statistically significant.
Integration of protein–protein interaction (PPI)
The Search Tool for the Retrieval of Interacting Genes (STRING) database (http://string.embl.de/; accessed August 16, 2017) is an online tool designed to evaluate the protein–protein interaction (PPI) information [17]. To evaluate the interactive relationships among DEGs, we mapped the DEGs to STRING in order to evaluate the interactive relationships among DEGs. Experimentally validated interactions with a combined score >0.4 were selected as significant. Using the PPI networks, module screening was performed using Molecular Complex Detection (MCODE) (scores >3 and nodes >4) in Cytoscape, an integrated bioinformatics platform [18].
Statistical analysis
We conducted the statistical analyses with SPSS version 20.0 and R 3.3.0.
Using the Cox proportional hazards regression model, hazard ratios (HRs) and their 95% confidence intervals (CIs) were calculated. FDR in edgeR was adjusted for multiple testing with the Benjamini-Hochberg procedure to control FDR [19–21]. A value of P<0.05 was considered as statistically significant.
Results
Gene expression dataset
Information for 151 patients with adult AML and a corresponding bone marrow RNA-Seq dataset were obtained from the TCGA database. These data were obtained from the cBioPortal for Cancer Genomics website.
GO and KEGG
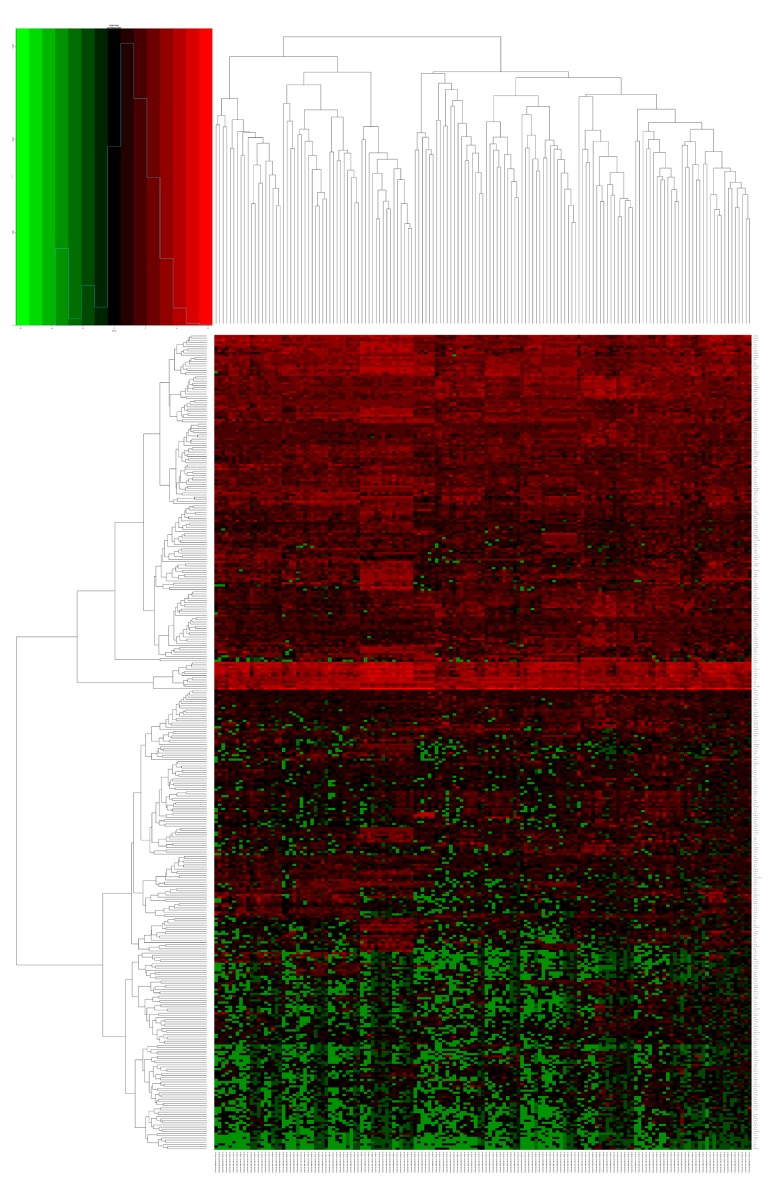
Altogether, we presented 379 DEGs (Figure 1) for further GO and KEGG pathway analyses using DAVID (Tables 1, 2). Heat maps of potential RUNX1-mutation-related DEGs are shown in Figure 2. The KEGG enrichment analysis of DEGs showed that these DEGs were significantly enriched in cancer pathways, as well as in the extracellular matrix-receptor interaction pathway and the cAMP signaling pathway.
Figure 1.
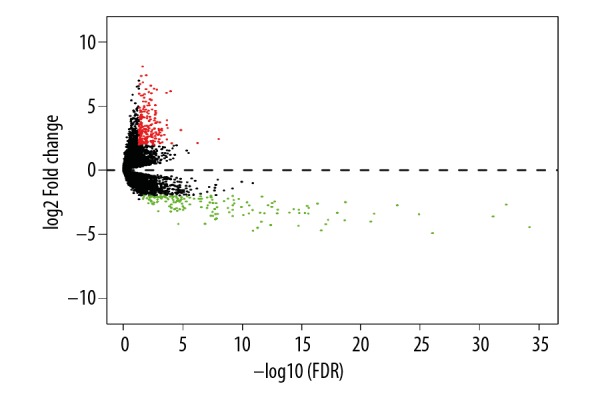
Volcano plot of the differentially expressed genes. Red: upregulation; green: downregulation; black: non-differentially expressed genes. P-adj, adjusted P value.
Table 1.
GO term analysis of RUNX1 mutation-DEGs associated with AML.
| Category | GO ID | Term | Count | % | P-value |
|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | GO: 0007155 | Cell adhesion | 20 | 3.86 | 0.001790 |
| GOTERM_BP_DIRECT | GO: 0001525 | Angiogenesis | 12 | 2.32 | 0.004496 |
| GOTERM_BP_DIRECT | GO: 0051897 | Positive regulation of protein kinase B signaling | 7 | 1.35 | 0.006020 |
| GOTERM_BP_DIRECT | GO: 0035360 | Positive regulation of peroxisome proliferator activated receptor signaling pathway | 3 | 0.58 | 0.007486 |
| GOTERM_BP_DIRECT | GO: 0043066 | Negative regulation of apoptotic process | 17 | 3.28 | 0.017451 |
| GOTERM_BP_DIRECT | GO: 0071356 | Cellular response to tumor necrosis factor | 7 | 1.35 | 0.020903 |
| GOTERM_BP_DIRECT | GO: 0050679 | Positive regulation of epithelial cell proliferation | 5 | 0.97 | 0.029629 |
| GOTERM_BP_DIRECT | GO: 2000147 | Positive regulation of cell motility | 3 | 0.58 | 0.029648 |
| GOTERM_BP_DIRECT | GO: 0050930 | Induction of positive chemotaxis | 3 | 0.58 | 0.033775 |
| GOTERM_BP_DIRECT | GO: 0072604 | Interleukin-6 secretion | 2 | 0.39 | 0.038175 |
| GOTERM_BP_DIRECT | GO: 0007568 | Aging | 8 | 1.55 | 0.043500 |
| GOTERM_BP_DIRECT | GO: 0071300 | Cellular response to retinoic acid | 5 | 0.97 | 0.047991 |
| GOTERM_BP_DIRECT | GO: 0005912 | Adherens junction | 6 | 1.16 | 0.002782 |
| GOTERM_BP_DIRECT | GO: 0008083 | Growth factor activity | 8 | 1.55 | 0.031109 |
| GOTERM_BP_DIRECT | GO: 0005112 | Notch binding | 3 | 0.58 | 0.042876 |
GO – gene ontology; DEGs – differentially expressed genes; AML – acute myeloid leukemia.
Table 2.
KEGG pathway analysis of RUNX1mutation-DEGs associated with AML.
| Pathway ID | Name | Count | % | P-value | Genes |
|---|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 20 | 3.87 | 5.14E-05 | PTGER1, PTGER3, LPAR4, RUNX1T1, FGF10, GNG11, MECOM, GLI2, GLI3, MMP2, CTNNA3, PRKACG, LAMB4, WNT7B, PTK2, LAMA3, LAMC3, NTRK1, NOS2, WNT8A |
| hsa04080 | Neuroactive ligand-receptor interaction | 12 | 2.32 | 0.009536961 | LEP, CRHR2, HCRTR1, PTGER1, GABRR3, GABRE, GLRB, PTGER3, PRSS2, LPAR4, CHRNA6, CTSG |
| hsa05146 | Amoebiasis | 7 | 1.35 | 0.01088093 | PRKACG, LAMB4, PTK2, LAMA3, LAMC3, NOS2, CTSG |
| hsa04512 | ECM-receptor interaction | 6 | 1.16 | 0.018487893 | LAMB4, LAMA3, LAMC3, COL6A5, SV2B, TNN |
| hsa05032 | Morphine addiction | 6 | 1.15 | 0.022018528 | PRKACG, GABRR3, GABRE, GNG11, PDE4C, KCNJ3 |
| hsa04024 | cAMP signaling pathway | 9 | 1.74 | 0.023512773 | PRKACG, PTGER3, NPY, ATP1B4, RYR2, PDE4C, CREB3L3, TNNI3, GLI3 |
| hsa05205 | Proteoglycans in cancer | 9 | 1.73 | 0.024791345 | PRKACG, WNT7B, PTK2, MRAS, IGF2, FLNC, MMP2, WNT8A, TWIST1 |
| hsa04726 | Serotonergic synapse | 6 | 1.16 | 0.046110307 | PRKACG, ALOX15B, MAOA, GNG11, KCNJ3, PLA2G4D |
KEGG – Kyoto Encyclopedia of Genes and Genomes; DEGs – differentially expressed genes; cAMP – cyclic adenosine monophosphate.
Figure 2.
Heat map of the RUNX1 mutation -related differentially expressed genes. Red: upregulation; green: downregulation.
Networks and module analysis
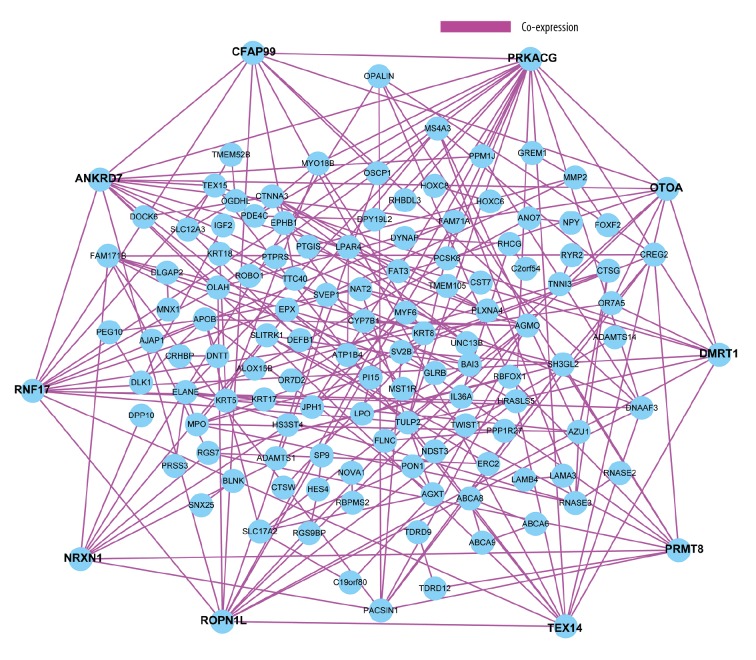
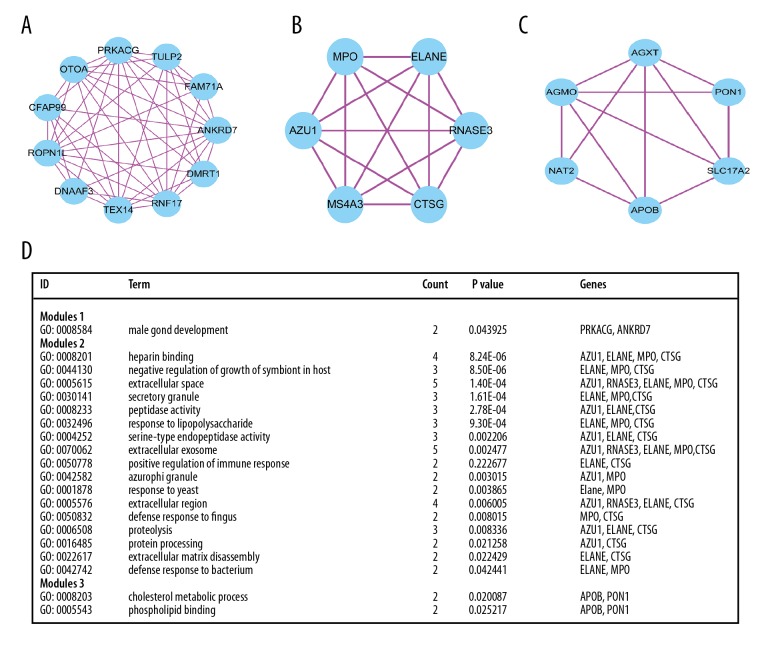
We constructed the protein–protein interactome networks and identified some RUNX1 mutation-associated hub genes (Figure 3). The top 10 genes ranked by degree were identified as hub genes, including PRKACG, ANKRD7, RNFL7, ROPN11, TEX14, PRMT8, OTOA, CFAP99, NRXN1, and DMRT1. PRKACG had the highest degree of nodes. Modules of genes in PPI networks were identified by the MCODE plugin in Cytoscape. The top 3 notable modules were chosen for bioinformatics analysis (Figure 4).
Figure 3.
The protein–protein interactome networks and hub genes.
Figure 4.
Top 3 modules from the PPI interaction networks. (A) module 1; (B) module 2; (C) module 3; (D) the enriched GO term of module 1 module 2 and module 3.
Survival analysis
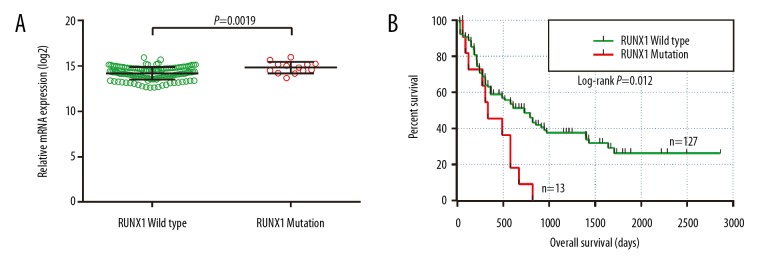
In the present study, we observed that RUNX1 mRNA expression was different between RUNX1 mutation and wild-type patients’ bone marrow tissues (Figure 5A). OS analysis indicated that RUNX1 mutant AML patients had a shorter MST than those without RUNX1 mutations (335 days vs. 608 days, log-rank P=0.012). Univariate Cox proportional hazards regression analysis showed that RUNX1 mutations were significantly associated with a poor clinical outcome and an increased risk of death (log-rank P=0.012, HR=2.145, 95%CI=1.156–3.982, Figure 5).
Figure 5.
The comparison of mRNA expression and survival between AML patients with RUNX1 mutation and wild type. (A) The mRNA expression of the RUNX1 gene in AML patients’ bone marrow tissue between RUNX1 with mutations and the wild type; (B) Kaplan-Meier survival curves for AML patients stratified by RUNX1 mutation. AML – acute myeloid leukemia; OS – overall survival.
Discussion
It is well accepted that genetic variants can be used as independent prognostic biomarkers for AML due to their potential effect on efficacy of chemotherapy. RUNX1 mutations are the dangerous element in AML. RUNX1 might be a novel biomarker for early diagnosis and a therapeutic target for treatment of AML. Therefore, further investigation is essential for better understanding of the biological roles of RUNX1 mutations in AML. In this study, the KEGG enrichment analysis showed that DEGs were notably abundant in pathways of cancer, ECM-receptor interaction, and cAMP signal. While the cancer pathway was expected and is in line with a previous study [22], it is unclear how the ECM-receptor interaction pathway and the cAMP signaling pathway affect the pathogenesis and prognosis of AML. ECM-receptor interaction pathway enrichment was reported in esophageal squamous cell carcinoma [23]. Li et al. found multiple DEGs and miRNAs were potential biomarkers for the prognosis of epithelial ovarian cancer (EOC) metastasis through the ECM-receptor interaction pathway [24] and affected tumor prognosis [25]. XuS et al. found that prostaglandin E2 (PGE2) receptor EP4 signaling activates the cyclic (c)-AMP signaling pathway, which may be closely related to the occurrence of prostate cancer [26]. It is reported by Kumar N1 that the cAMP signaling pathway was associated with non-small cell lung cancer [27]. Thus, it is possible that RUNX1 mutations may affect development and prognosis of AML through the ECM-receptor interaction pathway and cAMP signaling pathway.
Furthermore, we constructed the protein–protein interactome networks and identified some hub genes associated with RUNX1 mutations. The PRKACG gene, which had the highest degree of nodes, is a member of the protein kinase super-family. PRKACG is known to promote tumor progression because it affects the prognosis of colorectal cancer and leads to poor clinical outcomes [28]. Matos et al. [29] showed that RNF17/TDRD4 (cancer antigen) is a known cancer-associated hub gene of the individual GRNs markers, which are associated with cancer-related diagnostic and prognostic features. RNF17 is a potential liver cancer CT antigen and is associated with unfavorable prognosis [30]. ROPN1L variants are significantly associated with high risk of breast cancer in women [31]. Moreover, Tian et al. reported that ROPN1L was enriched in lung squamous cell carcinoma [32]. Lowe et al. [33] showed that ROPN1L gene was overexpressed in pancreatic cancer. However, there is no correlation between this gene and AML susceptibility and prognosis. Again, our results inspire further research on this gene. Testis-expressed gene 14 (Tex14) is a gene encoding a preferentially expressed protein kinase and promotes the survival of breast cancer [34]. DMRT1 gene is a candidate regulator of sexual development in vertebrates and encodes conserved transcription factor essential for gonadal function. Moreover, DMRT1 promotes testicular germ cell tumors (TGCT) progression [35]. PRMT8 is a type I PRMT that catalyzes the MMA ω-NG-monomethyl arginine and aDMA (aDMA (ω-NG, NG-asymmetric, dimethylarginine) modifications. Yanhong et al. [36] reported that PRMT8 is involved in gliomagenesis molecular pathogenesis. Simandi et al. showed that PRMT8 might also be relevant in the development of human brain malignancy [37]. Otoancorin (OTOA) was reported to be associated with ovarian and pancreatic cancer [38]. NRXN1, also known as KIAA0578, belongs to the neurexin family. Lee et al. found that NRXN1 may result in ALL leukemic cells harboring a novel 3-way translocation t(2;19;11) (p12;p13.3;q23), thereby affecting development of the disease [39].
However, no study has reported the association of CFAP99 or DMRT1 with AML or other cancers. Thus, it is important to further explore the role of CFAP99 and DMRT1 in the pathogenesis and prognosis of AML.
KEGG pathway enrichment showed the top 3 module genes. Genes enrichment of module 1 was prkacg and ankrd7. The genes of module 2 were most abundant in azu1, elane, mpo, ctsg, and rnase3. Furthermore, 3 module genes were abundant in apob and pon1. Apolipoprotein B (ApoB) belongs to the lipoprotein family, which plays important roles in lipid metabolism. In diabetes and metabolic syndrome, serum APOB levels are abnormal [40,41], which can promote the development of cancer [42,43]. Moreover, recent research revealed that high APOB levels can lead to increased risk of lung cancer and colorectal cancer [44]. Serum paraoxonase-1 (PON1) is a 45 kDa glycoprotein [45]. Goncalves et al. [46] found that PON1 rs854560 (L55M) may be related to increased incidence of childhood leukemia, while Çebi et al. [47] reported that PON1 activity can affect the pathology of AML disease.
Statistical analysis indicated that mutated leukemia patients had a shorter MST and poor clinical outcome and increased mortality compared to those without RUNX1 mutation. Our results are consistent with previous reports that RUNX1 mutation adversely affects overall survival (OS) and prognosis of AML patients [48–50]. Germline et al. [51] found that RUNX1 mutations can lead to increased risk of developing AML. The mutations of RUNX1 often result in poor clinical outcomes in AML patients [52]. Gaidzik et al. [49] reported that for AML patients carrying RUNX1-mutation, the rates of event-free survival (EFS), relapse-free survival (RFS), and overall survival (OS) were lower after intensive chemotherapy when compared with wild-type patients. Other studies also showed RUNX1 mutations were related to chemoresistance and increased chance of becoming refractory [53–55]. Tang et al. analyzed RUNX1 mutant AML patients treated with intensive chemotherapy and found that the complete remission rate was lower [54]. The mutations of RUNX1 may lead to increased mortality rate of acute myeloid leukemia. Stengel et al. recently reported that RUNX1-mutant AML showed a distinct pattern of genetic abnormalities and an adverse prognosis [56].
Conclusions
Our results indicate that mutated RUNX1 promotes poor OS in AML patients, which is consistent with previous reports. Results of bioinformatic analysis in the study also reveal that genes such as PRKACG, ANKRD7, RNFL7, ROPN11, TEX14, PRMT8, OTOA, CFAP99, NRXN1, and DMRT1, as well as pathways such as cancer pathways, ECM-receptor interaction pathway, and cAMP signaling pathway, may play a crucial role underlying the effect of RUNX1 mutations in AML prognosis. Our findings provide a novel biomarker for early diagnosis and a therapeutic target for personized treatment of AML. This study had certain limitations: the sample size was small, survival data were incomplete, and survival analysis was only focused on a single factor. Therefore, further studies in molecular pathogenesis and large-scale clinical tumor specimen validation are still needed.
Acknowledgments
The authors wish to thank the Cancer Genome Atlas and the cBioPortal for Cancer Genomics for their contribution of the AML sequencing dataset on open access.
Footnotes
Source of support: This work was supported in part by the National Nature Science Foundation of China (Grant No. 81560024), and Program of Scientific and Technology Project (2016012706-2), Guilin Science Research and Technology Development
Conflict of interest
None.
References
- 1.Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88(4):318–27. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemla A. Cancer statistics, 2013. Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Saito Y, Yuki H, Kuratani M, et al. A pyrrolo-pyrimidine derivative targets human primary AML stem cells in vivo. Sci Transl Med. 2013;5(181):181r. doi: 10.1126/scitranslmed.3004387. a52. [DOI] [PubMed] [Google Scholar]
- 4.Pabst C, Krosl J, Fares I, et al. Identification of small molecules that support human leukemia stem cell activity ex vivo. Nat Methods. 2014;11(4):436–42. doi: 10.1038/nmeth.2847. [DOI] [PubMed] [Google Scholar]
- 5.Link KA, Chou FS, Mulloy JC. Core binding factor at the crossroads: Determining the fate of the HSC. J Cell Physiol. 2010;222(1):50–56. doi: 10.1002/jcp.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim W, Barron DA, San Martin R, et al. RUNXl is essential for mesenchymal stem cell proliferation and myofibroblast differentiation. Proc Natl Acad Sci USA. 2014;111(46):16389–94. doi: 10.1073/pnas.1407097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Ami O, Friedman D, Leshkowitz D, et al. Addiction of t(8 21)and inv(16)acute myeloid leukemia to native RUNXl. Cell Rep. 2013;4(6):1131–43. doi: 10.1016/j.celrep.2013.08.020. [DOI] [PubMed] [Google Scholar]
- 8.Goyama S, Schibler J, Cunningham L, et al. Transcription/actor RUNXl promotes survival of acute myeloid leukemia cells. J Clin Invest. 2013;123(9):3876–88. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilkinson AC, Ballabio E, Geng H, et al. RUNXl is a key target in t(4;11)leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep. 2013;3(1):116–27. doi: 10.1016/j.celrep.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen Y, Zhu Y-M, Fan X, et al. Gene mutation patterns and their prognostic impact in a cohort of 11185 patients with acute myeloid leukemia. Blood. 2011;118:5593–603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6 doi: 10.1126/scisignal.2004088. pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B – Statistical Methodology. 1995;57:289–300. [Google Scholar]
- 20.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 21.Benjamini Y, Drai D, Elmer G, et al. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–84. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Zhang C, Feng R, et al. Investigating the microRNA-mRNA regulatory network in acute myeloid leukemia. Oncol Lett. 2017;14(4):3981–88. doi: 10.3892/ol.2017.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Shi X, Yang W, et al. Transcriptome profiling of lncRNA and co-expression networks in esophageal squamous cell carcinoma by RNA sequencing. Tumour Biol. 2016;37(10):13091–100. doi: 10.1007/s13277-016-5227-3. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Li H, Xu Y, et al. Identification of candidate biomarkers for epithelial ovarian cancer metastasis using microarray data. Oncol Lett. 2017;14(4):3967–74. doi: 10.3892/ol.2017.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ning X, Deng Y. Identification of key pathways and genes influencing prognosis in bladder urothelial carcinoma. Onco Targets Ther. 2017;10:1673–86. doi: 10.2147/OTT.S131386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu S, Zhou W, Ge J, Zhang Z. Prostaglandin E2 receptor EP4 is involved in the cell growth and invasion of prostate cancer via the cAMP PKA/PI3K Akt signaling pathway. Mol Med Rep. 2018;17(3):4702–12. doi: 10.3892/mmr.2018.8415. [DOI] [PubMed] [Google Scholar]
- 27.Kumar N, Prasad P, Jash E, et al. Insights into exchange factor directly activated by cAMP(EPAC) as potential target for cancer treatment. Mol Cell Biochem. doi: 10.1007/s11010-018-3294-z. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Li L, Huang Z, et al. Application of genome-wide gene chip for screening and identifying genes related to CD133(+)CD200(+) colorectal cancer stem cells. Nan Fang Yi Ke Da Xue Xue Bao. 2013;33(12):1787–91. [PubMed] [Google Scholar]
- 29.de Matos Simoes R, Dalleau S, Williamson KE, et al. Urothelial cancer gene regulatory networks inferred from large-scale RNAseq, Bead and Oligogene expression data. BMC Syst Biol. 2015;9:21. doi: 10.1186/s12918-015-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon H, Lee H, Kim HJ, et al. Tudor domain-containing protein 4 as a potential cancer/testis antigen in liver cancer. Tohoku J Exp Med. 2011;224(1):41–46. doi: 10.1620/tjem.224.41. [DOI] [PubMed] [Google Scholar]
- 31.Sehrawat B, Sridharan M, Ghosh S, et al. Potential novel candidate polymorphisms identified in genome-wide association study for breast cancer susceptibility. Hum Genet. 2011;130(4):529–37. doi: 10.1007/s00439-011-0973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian F, Zhao J, Fan X, et al. Weighted gene co-expression network analysis in identification of metastasis-related genes of lung squamous cell carcinoma based on the Cancer Genome Atlas database. J Thorac Dis. 2017;9(1):42–53. doi: 10.21037/jtd.2017.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe AW, Olsen M, Hao Y, et al. Gene expression patterns in pancreatic tumors, cells and tissues. PLoS One. 2007;2(3):e323. doi: 10.1371/journal.pone.0000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlin KL, Mondal G, Hartman JK, et al. The oncogenic STP axis promotes triple-negative breast cancer via degradation of the REST tumor suppressor. Cell Rep. 2014;9(4):1318–32. doi: 10.1016/j.celrep.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elzinga-Tinke JE, Dohle GR, Looijenga LH, et al. Etiology and early pathogenesis of malignant testicular germ cell tumors: Towards possibilities for preinvasive diagnosis. Asian J Androl. 2015;17(3):381–93. doi: 10.4103/1008-682X.148079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y, Melin BS, Rajaraman P, et al. Insight in glioma susceptibility through an analysis of 6p22.3, 12p13.33-12.1, 17q22-23.2 and 18q23 SNP genotypes in familial and non-familial glioma. Hum Genet. 2012;131(9):1507–17. doi: 10.1007/s00439-012-1187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simandi Z, Czipa E, Horvath A, et al. PRMT1 and PRMT8 regulate retinoic acid-dependent neuronal differentiation with implications to neuropathology. Stem Cells. 2015;33(3):726–41. doi: 10.1002/stem.1894. [DOI] [PubMed] [Google Scholar]
- 38.Muminova ZE, Strong TV, Shaw DR. Characterization of human mesothelin transcripts in ovarian and pancreatic cancer. BMC Cancer. 2004;4(1):19. doi: 10.1186/1471-2407-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee SG1, Park TS, Won SC, et al. Three-way translocation involving MLL, MLLT1, and a novel third partner, NRXN1, in a patient with acute lymphoblastic leukemia and t(2;19;11) (p12;p13.3;q23) Cancer Genet Cytogenet. 2010;197(1):32–38. doi: 10.1016/j.cancergencyto.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 40.Ryoo JH, Park SK. Association of apolipoprotein B and incidence of metabolic syndrome in Korean men: A 5-years’ follow-up study. Atherosclerosis. 2013;226(2):496–501. doi: 10.1016/j.atherosclerosis.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 41.Seo MH, Bae JC, Park SE, et al. Association of lipid and lipoprotein profiles with future development of type 2 diabetes in nondiabetic Korean subjects: A 4-year retrospective, longitudinal study. J Clin Endocrinol Metab. 2011;96(12):E2050–54. doi: 10.1210/jc.2011-1857. [DOI] [PubMed] [Google Scholar]
- 42.Cowey S, Hardy RW. The metabolic syndrome: A high-risk state for cancer? Am J Pathol. 2006;169(5):1505–22. doi: 10.2353/ajpath.2006.051090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atchison EA, Gridley G, Carreon JD, et al. Risk of cancer in a large cohort of U.S. veterans with diabetes. Int J Cancer. 2011;128(3):635–43. doi: 10.1002/ijc.25362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borgquist S, Butt T, Almgren P, et al. Apolipoproteins, lipids and risk of cancer. Int J Cancer. 2016;138(11):2648–56. doi: 10.1002/ijc.30013. [DOI] [PubMed] [Google Scholar]
- 45.Blatter MC, James RW, Messmer S, et al. Identification of a distinct human high-density lipoprotein subspecies defined by a lipoprotein-associated protein, K-45: Identity Iof K-45 with paraoxonase. Eur J Biochem. 1993;211:871–79. doi: 10.1111/j.1432-1033.1993.tb17620.x. [DOI] [PubMed] [Google Scholar]
- 46.Aguiar Gonçalves BA, Vasconcelos GM, Thuler LCS, et al. Brazilian Collaborative Study Group of Infant Acute Leukemia. NQO1 rs1800566 (C609T), PON1 rs662 (Q192R), and PON1 rs854560 (L55M) polymorphisms segregate the risk of childhood acute leukemias according to age range distribution. Cancer Causes Control. 2012;23:1811–19. doi: 10.1007/s10552-012-0060-5. [DOI] [PubMed] [Google Scholar]
- 47.Çebi A, Akgun E, Esen R, et al. The activities of serum paraoxonase and arylesterase and lipid profile in acute myeloid leukemia: Preliminary results. Eur Rev Med Pharmacol Sci. 2015;19(23):4590–94. [PubMed] [Google Scholar]
- 48.Ley TJ, Miller C, Ding L, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaidzik VI, Teleanu V, Papaemmanuil E, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2160–68. doi: 10.1038/leu.2016.126. [DOI] [PubMed] [Google Scholar]
- 50.Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood. 2016;127:29–41. doi: 10.1182/blood-2015-07-604496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jongmans MC, Kuiper RP, Carmichael CL, et al. Novel RUNX1 mutations in familial platelet disorder with enhanced risk for acute myeloid leukemia: Clues for improved identification of the FPD/AML syndrome. Leukemia. 2010;24:242–46. doi: 10.1038/leu.2009.210. [DOI] [PubMed] [Google Scholar]
- 52.Khan M, Cortes J, Kadia T, et al. Clinical outcomes and co-occurring mutations in patients with RUNX1-mutated acute myeloid leukemia. Int J Mol Sci. 2017;18(8) doi: 10.3390/ijms18081618. pii: E1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi S. Current findings for recurring mutations in acute myeloid leukemia. J Hematol Oncol. 2011;4:36. doi: 10.1186/1756-8722-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang JL, Hou HA, Chen CY, et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: Prognostic implication and interaction with other gene alterations. Blood. 2009;114(26):5352–61. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- 55.Gaidzik VI, Bullinger L, Schlenk RF, et al. RUNX1 mutations in acute myeloid leukemia: Results from a comprehensive genetic and clinical analysis from the AML study group. J Clin Oncol. 2011;29(10):1364–72. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- 56.Stengel A, Kern W, Meggendorfer M, et al. Number of RUNX1 mutations, wild-type allele loss and additional mutations impact on prognosis in adult RUNX1-mutated AML. Leukemia. 2018;32(2):295–302. doi: 10.1038/leu.2017.239. [DOI] [PubMed] [Google Scholar]