Abstract
Introduction
Dynamin 3 (DNM3) is a large GTPase that possesses mechanochemical properties and has been shown to be involved in malignancies. However, most studies about DNM3 are observational, and knowledge of the precise molecular mechanism of DNM3 remains limited.
Materials and methods
We constructed a PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 recombinant eukaryotic expression vector, which was then transfected into SW620 and LoVo cells. One cell line was divided into three groups. DNM3 mRNA and protein expression was analyzed by quantitative real-time PCR and Western blot assay. To investigate DNM3 biological activity in colon cancer SW620 and LoVo cell line, we performed cell proliferation, transwell migration, and invasion assay. Matrix metalloproteinase (MMP)-2 and MMP-9 protein expressions were detected by Western blot.
Result
We successfully constructed a PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 recombinant eukaryotic expression vector, and stable DNM3 expression was observed in SW620 and LoVo cell lines. The vector overexpressing DNM3 inhibited the proliferation, weak invasion, and migration ability of colon cancer SW620 and LoVo cells relative to those in the control group (all P<0.001). DNM3 downregulated the protein expression of MMP-2 and MMP-9.
Conclusion
DNM3 may weaken the malignant behavior of colon cancer and may have promoted the invasion and migration of colon cancer by regulating the expression of MMP-2 and MMP-9.
Keywords: dynamin 3, colon cancer, proliferation, migration, invasion
Introduction
Dynamin is a large GTPase of the dynamin superfamily1 that includes classical dynamins (dynamin-1,2,3) encoded by DNM1, DNM2, and DNM3 genes in eukaryotic cells,2 dynamin-like proteins that are involved in apoptosis,3 OPA1, mitofusins, Mx proteins, and guanylate-binding proteins.2 The sequence of dynamin-1,2,3 exhibitŝ80% homology. Furthermore, dynamins share a primary structure consisting of an amino-terminal GTPase domain, pleckstrin homology domain, carboxy-terminal proline/arginine-rich domain, middle domain, and GTPase effector domain.4 However, dynamins are expressed in different tissues. Dynamin 1 is limited to neuronal cells, dynamin 2 is found ubiquitously, and dynamin 3 is expressed in the lungs, testis, and brain.5 Dynamin possesses mechanochemical properties involving membrane trafficking processes, which includes vesicle scission, phagocytosis, cytokinesis, podosome formation, and endocytosis.6–11 Recently, DNM3 has been shown to be involved in malignancies.12–17 Inokawa et al and Shen et al found that DNM3 is hypermethylated in hepatocellular cancer (HCC).13,14 Zhang et al have further studied the mechanism of DNM3 in HCC.18 DNM3 weakens the proliferation and induces apoptosis of HCC cells by upregulating P53. Lee et al found that low DNM2 expression is associated with tumor invasion and metastasis in cervix carcinoma and upregulates matrix metalloproteinase 2 (MMP-2) expression.19 Inokawa et al found that inhibited expression of DNM2 or DNM3 upregulates MMP-2 in HCC.13 However, studies about the relationship between DNM3 and malignancies are still insufficient, and most of them are observational. The precise molecular mechanism is unclear. In this study, we constructed a eukaryotic expression vector that comprises DNM3 gene to analyze the effect of DNM3 gene on the proliferation, invasion, and migration abilities of MMP-2 and MMP-9 expression in human colon cancer SW620 and LoVo cells.
Materials and methods
Materials
High Fidelity Prime Script™ RT-PCR Kit and DNA Ligation Kit Ver.2.1 were purchased from Takara (Shiga, Japan). Ampicillin was purchased from Generay (Shanghai, China). Universal DNA Purification Kit was purchased from Tiangen (Beijing, China). Endo-free Plasmid Mini Kit II was purchased from Omega (Norcross, GA, USA). Competent Escherichia coli DH5α was purchased from Transgen Biotech (Beijing, China). Luria Bertani broth, agarose, and agar were purchased from Generay. Colon cancer cell lines, SW620 and LoVo, were obtained from Geneseed (Guangzhou, China). FBS and Roswell Park Memorial Institute 1640 (RPMI-1640) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Lipofectamine™ 2000, Opti-MEM, and Trizol were purchased from Thermo Fisher Scientific. Puromycin was purchased from MP Biomedicals (Solon, OH, USA). PrimeScript™ RT reagent kit with gDNA Erase and SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) were purchased from Takara. BCA protein assay kit was purchased from Beyotime (Shanghai, China). Mini GEL tank, NUPAGE 10% BT GEL 1.0 MM 12W Western, NuPAGE™ LDS sample buffer (4×), and NuPAGE™ MES SDS running buffer (20×) were purchased from Thermo Fisher Scientific. DNM3 antibody was purchased from GeneTex (Irvine, CA, USA), and anti-MMP-2, anti-MMP-9, and GAPDH antibody were purchased from Abcam (Cambridge, MA, USA). Pierce™ ECL Western blot substrate was purchased from Thermo Fisher Scientific. Cell Counting Kit-8 (CCK-8) was purchased from Dojindo (Tokyo, Japan). Matrigel was purchased from BD Biosciences (San Jose, CA, USA).
Construction of the PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 vector
Obtaining and identification of DNM3
The DNM3 RNA was obtained from the gene bank of Gene-seed Biotech Co., Ltd (China). A pair of specific primers containing EcoRI and NotI restriction enzyme cutting sites was designed as follows: DNM3-EcoRI-F: 5′-CCGGAATTCATGGGGAACCGGGAGATGGAGGAGC-3′ and DNM3-NotI-R: 5′-ATTTGCGGCCGCCTAAATGTTCTCATCATCTTTATAC-3′. The primers were designed and synthesized by Sangon Biotech Co., Ltd (China). DNM3 complementary DNA (cDNA) was synthesized by High Fidelity Prime Script™ RT-PCR Kit according to the manufacturer’s instructions. The PCR products were analyzed by 1% agarose gel electrophoresis.
Purification of DNM3 cDNA and vector
The PCR products and PCDH-CMV-MCS-EF1a-GFP-Puro plasmids were digested by EcoRI and NotI restriction enzymes. The target fragments and the vector were then purified with a Universal DNA purification kit (according to the manufacturer’s instructions).
Construction and identification of the recombinant eukaryotic expression vector
For the formation of the recombinant vector PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3, the digested and purified PCR products and plasmids were ligated with DNA Ligation Kit Ver.2.1. The recombinant vector was transformed into competent E. coli DH5α. The recombinant vector was amplified in positive transformants, which were selected by ampicillin. The positive clones with DNM3 were identified by PCR and then sequenced. Approximately 5 µL of PCR products were analyzed by 2% agarose gel electrophoresis, and the positive vector was sequenced by Geneseed Biotech Co., Ltd.
Expression of the recombinant PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 eukaryotic expression vector in colon cancer SW620 and LoVo cells
Plasmid extraction
The recombinant vector PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 and the empty vector plasmid PCDH-CMV-MCS-EF1a-GFP-Puro were transformed to competent E. coli DH5α. The bacterial strains were amplified in 1× LB supplemented with ampicillin, and the vector and plasmid were extracted with an Endo-free Plasmid Mini Kit II.
Cell stable transfection
Colon cancer SW620 and LoVo cells were cultured in RPMI-1640 containing 10% FBS at 37°C in a humidified atmosphere of 5% CO2. Puromycin dose used for the selection of positive clones was determined by challenging SW620 and LoVo cells with gradient concentration of puro-mycin. The optimal dose (2 µg/mL) was determined by titration. Vector PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 and plasmid PCDH-CMV-MCS-EF1a-GFP-Puro were transfected into SW620 and LoVo cells by Opti-MEM and Lipofectamine™ 2000 reagent according to the manufacturer’s instructions. Approximately 48 hours after transfection, the cells were passaged (1:10) and cultured in RPMI-1640 medium containing 10% FBS and 2 µg/mL of puromycin. Puromycin-resistant clones were visible ~10 days after the selection. The puromycin-resistant clones were then selected and cultured. After 2 weeks of cloning, four cell lines with stable expression of PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 and PCDH-CMV-MCS-EF1a-GFP-Puro were obtained. The cells were divided into six groups, namely no treatment SW620, empty plasmid PCDH-CMV-MCS-EF1a-GFP-Puro SW620-P, PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 SW620-DNM3, no treatment LoVo, empty plasmid PCDH-CMV-MCS-EF1a-GFP-Puro LoVo-P, and PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 LoVo-DNM3.
Detection of DNM3 mRNA by quantitative real-time PCR
Total RNA was extracted from the transfected cells by Trizol, and cDNA was synthesized with a PrimeScript™ RT reagent kit with gDNA Erase according to the manufacturer’s instructions. Real-time PCR was performed with a SYBR® Premix Ex Taq™ II (Tli RNaseH Plus). The gene-specific primer pairs were as follows: β-actin-F: 5′-GCATGGGTCAGAAGGATTCCT-3′, β-actin-R: 5′-TCGTCCCAGTTGGTGACGAT-3′, and DNM3-F: 5′-GC TGGCAGAGAGGAAGTTTTTC-3′, DNM3-R: 5′-TTT GGTAGGGTATCCCGAATG-3′. The data were analyzed through the 2−ΔΔCT method.
Detection of DNM3, MMP-9, and MMP-2 by Western blot assay
The cells were collected and lysed, and the total proteins were extracted. The concentration of protein was determined with a BCA protein assay kit. Approximately 15 µg of protein samples mixed with the NuPAGE™ LDS sample buffer (4×) were loaded on a NUPAGE 10% BT GEL 1.0 MM 12W Western and processed in the Mini GEL Tank with NuPAGE™ MES SDS running buffer according to the manufacturer’s instructions. The gel was wet-transferred onto polyvinylidene fluoride membranes. After blocking the membranes with 5% milk (western blocking buffer) at room temperature for 2 hours, the membranes were incubated with the corresponding primary antibodies: DNM3, 1:300; MMP-2, 1:1,000; and GAPDH, 1:1,000 at 4°C overnight and secondary antibodies (1:2,000) at room temperature for 2 hours simultaneously. The bands were detected by Pierce™ ECL Western blot substrate. Finally, the blot was photographed with a VersaDoc 5000 Imager (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The image was scanned for the relative value of protein expression in gray scale by ImageJ software 1.45.
Cell proliferation assay
To analyze the effects of DNM3 on proliferation of colon cancer SW620 and LoVo cells, we performed CCK-8 assay according to the manufacturer’s instructions. The cells were plated onto 96-well plates at a density of 3,000 cells per well. Each group was seeded to 16 well cells, and every four succeeding wells were cultured for 12, 24, 48, and 72 hours. Approximately 10 µL of CCK-8 was added to the wells and incubated for 2 hours. The absorbance value (OD) of the cells was measured using Multiskan Mk3 (Thermo Fisher Scientific) with a 450 nm filter.
Transwell migration and invasion assay
Transwell chambers were covered with 50 µL of Matrigel diluted with serum-free RPMI-1640 (1:7) and incubated at 37°C for 2 hours (this step was omitted in the transwell migration assay). Approximately 5×105 cells were suspended in 100 µL of serum-free RPMI-1640 and 600 µL of RPMI-1640 with 10% FBS and were seeded in the upper and lower chambers, respectively. After incubation at 37°C in 5% CO2 for 48 hours (36 hours for the migartion assay), the cells were fixed and stained for 15 minutes in a crystal violet solution in PBS. The cells were viewed under the microscope.
Statistical analysis
All the experiments were carried out in triplicates, and the data were expressed as mean ± SD of six independent experiments. Data were analyzed with SPSS21.0 software. A P-value of <0.001 indicated significant difference. Statistical comparisons among three groups and between two groups were conducted with Virance and LSD test, respectively. The experimental charts were produced by GraphPad Prism5.
Results
mRNA expression levels of DNM3 in colon cancer SW620 and LoVo cells
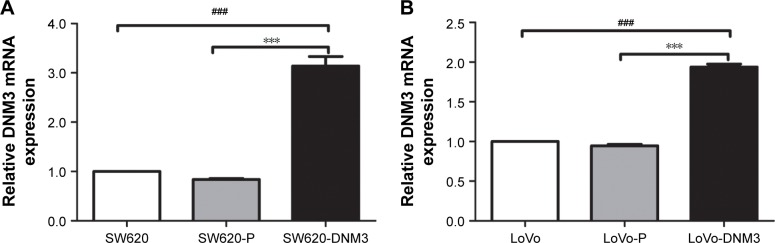
After transfection, the DNM3 expression was analyzed by quantitative real-time PCR, which showed that the DNM3 transfected group was significantly overexpressed compared to the control groups (all P<0.001, Figure 1).
Figure 1.
DNM3 mRNA was analyzed by quantitative real-time PCR.
Notes: (A) SW620-DNM3 groups significantly overexpressed DNM3 compared with the SW620 and SW620-P groups. (B) LoVo-DNM3 groups significantly overexpressed DNM3 relative to the LoVo and LoVo-P groups. SW620: no treatment, SW620-P: empty plasmid PCDH-CMV-MCS-EF1a-GFP-Puro, SW620-DNM3: PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3, LoVo: no treatment, LoVo-P: empty plasmid PCDH-CMV-MCS-EF1a-GFP-Puro, LoVo-DNM3: PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3, ***P<0.001. ###P<0.001.
DNM3 protein expression and its effect on MMP-9 and MMP-2 protein expression levels
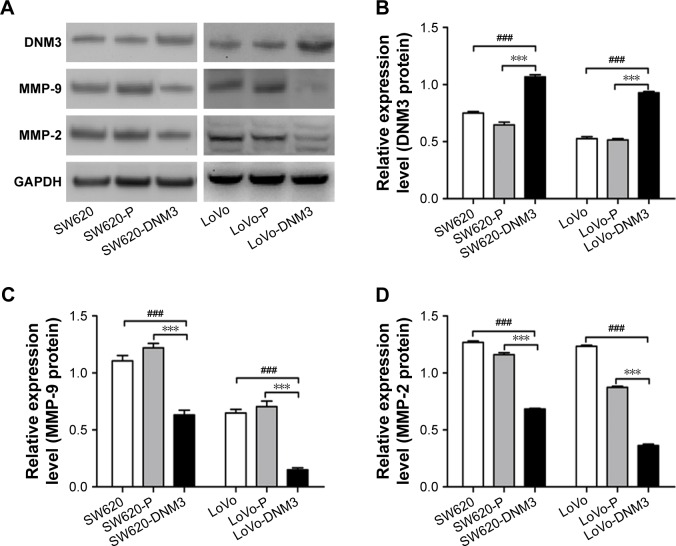
The expression levels of DNM3, MMP-9, and MMP-2 proteins were analyzed by Western blot. The result showed that the DNM3 transfected group was significantly overexpressed compared with each of the control group (all P<0.001), and DNM3 overexpression group downregulated the MMP-9 (all P<0.001) and MMP-2 (all P<0.001) protein expression levels compared with the control groups (Figure 2).
Figure 2.
DNM3, MMP-9, and MMP-2 proteins were analyzed by Western blot.
Notes: (A) Gray scale image of protein expression. (B) Cells transfected with DNM3 significantly overexpressed DNM3 protein compared with the control groups. (C) Cells transfected with DNM3 significantly downregulated MMP-9 protein compared with the control groups. (D) Cells transfected with DNM3 significantly downregulated MMP-2 protein compared with the control groups, ***P<0.001. ###P<0.001.
Abbreviation: MMP, Matrix metalloproteinase.
Effect of DNM3 on the proliferation of colon cancer SW620 and LoVo cells
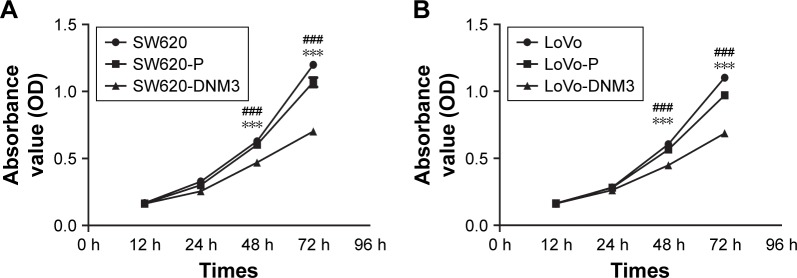
The OD values of the six groups were measured every 12 hours after transfection. The mean value was calculated. As shown in Figure 3, the survival number of either SW620 or SW620-P group was higher than that of SW620-DNM3 group (P<0.001, Figure 3A). The survival number of either the LoVo or LoVo-P group was higher than that the LoVo-DNM3 group (P<0.001, Figure 3B). DNM3 overexpression was associated with decreased proliferation in SW620 and LoVo cells.
Figure 3.
SW620 and LoVo cell growth curves. Compared with the control groups, DNM3 overexpression significantly inhibited cell proliferation.
Notes: (A) SW620 cell growth curves. Compared with the SW620 and SW620-P groups, SW620-DNM3 group significantly inhibited cell proliferation. (B) LoVo cell growth curves. Compared with the LoVo and LoVo-P groups, LoVo-DNM3 group significantly inhibited cell proliferation, ***P<0.001. ###P<0.001.
Effect of DNM3 on migration and invasion of colon cancer SW620 and LoVo cells
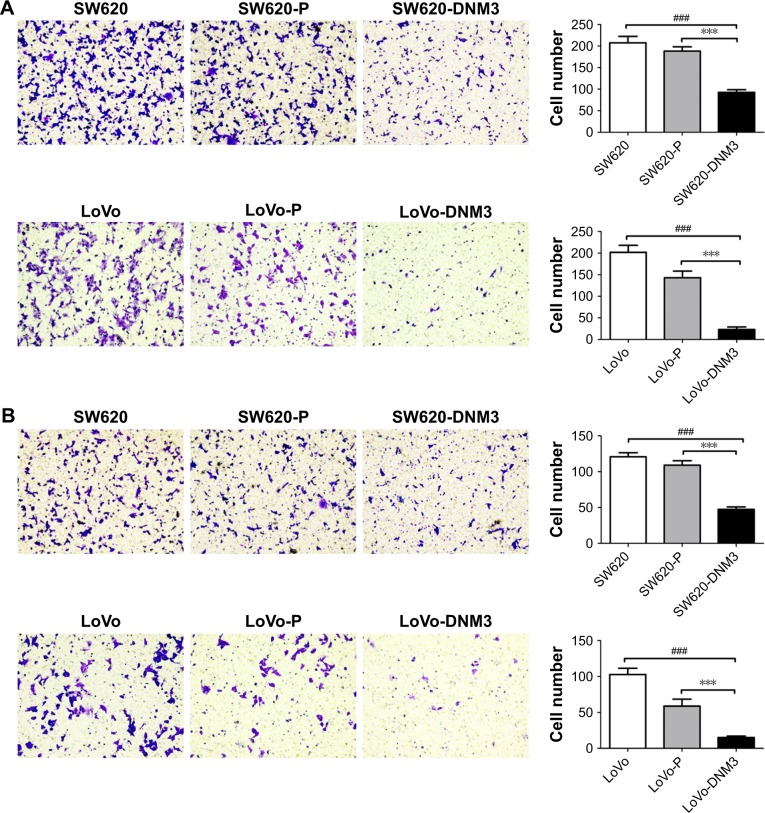
As shown in Figure 4, DNM3 overexpression weakened the ability of cell invasion and migration. SW620-DNM3 group migration ability was weaker than that of either the SW620 or SW620-P group (all P<0.001, Figure 4A). The migration ability of the LoVo-DNM3 group was weaker than that of either the LoVo or LoVo-P group (all P<0.001, Figure 4A). The invasion ability of the SW620-DNM3 group was weaker than that of either the SW620 or SW620-P group (all P<0.001, Figure 4B). The invasion ability of the LoVo-DNM3 group was weaker than that of either the LoVo or LoVo-P group (all P<0.001, Figure 4B).
Figure 4.
Microscopic pictures of transwell migration and invasion assay.
Notes: (A) Migration assay, DNM3 overexpression groups significantly decreased the ability of migration. (B) Invasion assay, DNM3 overexpression groups significantly decreased the ability of invasion. The results were expressed as fold change relative to the corresponding negative control. Data are presented as the mean ± SD, n=3. SW620: no treatment, SW620-P: empty plasmid PCDH-CMV-MCS-EF1a-GFP-Puro, SW620-DNM3: PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3, LoVo: no treatment, LoVo-P: empty plasmid PCDH-CMV-MCS-EF1a-GFP-Puro, LoVo-DNM3: PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3, ***P<0.001. ###P<0.001.
Discussion
DNM3 gene can be a predictor for prognosis and is a molecular marker for diagnosis and gene therapy of malignant disease.17 Booken et al16 indicated that DNM3 is overexpressed in Sezary syndrome (SS) in a screen for differential gene expression in peripheral blood mononu-clear cells of ten healthy controls and ten SS patients by Affymetrix genechip. Shen et al14 analyzed the genome-wide DNA methylation profiles in tumor and adjacent nontumor specimens of 62 HCC patients and demonstrated that DNM3 is hypermethylated in HCC. Inokawa et al13 showed that DNM3 is hypermethylated in HCC. Methylation downregulates DNM3 expression and is associated with poor prognosis, which was determined by analyzing the disease-specific survival of 48 HCC patients. Teicher et al15 reported that liposarcoma had 1q24.3 amplifications involving DNM3.
With an increased attention of DNM3, the precise molecular mechanism should be clarified. Marino et al12 performed the MTT and migration assays in breast cancer cells and found that the migration ability of breast cancer cells is weakened after stable silencing of DNM3. However, cell proliferation ability exhibits no difference. In our study, we successfully constructed a recombinant eukaryotic expression vector containing DNM3. The result showed that PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 could be overexpressed in colon cancer SW620 and LoVo cells after transfection. We detected the biological activities in colon cancer SW620 and LoVo cells. To determine whether DNM3 inhibits cell proliferation, we performed a CCK-8 assay. The OD values of the DNM3 groups were lower than those of the control groups, which indicated that DNM3 could inhibit cell proliferation. Moreover, cell invasion and migration assay results showed that DNM3 can weaken invasion and migration ability and these results were different from the results of a previous study.12 The main reason for this result may be related to the use of different cancer cells in the two studies, and our research methods were different. Marino et al12 silenced the DNM3 gene, but we used the overexpressed DNM3 gene. DNM3 is worthy of further study. To further explore the molecular mechanisms of motility inhibition induced by DNM3, we examined the expression of two major members of the MMP family, MMP-2 and MMP-9. We analyzed the relative expression level histogram and showed that overexpression of DNM3 can downregulate MMP-2 and MMP-9 protein expression.
Conclusion
In conclusion, we constructed a PCDH-CMV-MCS-EF1a-GFP-Puro-DNM3 recombinant eukaryotic expression vector, which indicated that DNM3 overexpression can inhibit proliferation, weak invasion, and migration ability in colon cancer SW620 and LoVo cells. DNM3 might be involved in the invasion and migration of colon cancer by regulating the expression of MMP-2 and MMP-9. DNM3 might weaken the malignant behavior of colon cancer.
Acknowledgments
This work was supported by a grant from the Technology Planning Project of Guangdong Province, China (No 2014A020212291) and Technology Planning Project of Zhanjiang, China (No 2017A01011).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Shpetner HS, Vallee RB. Dynamin is a GTPase stimulated to high levels of activity by microtubules. Nature. 1992;355(6362):733–735. doi: 10.1038/355733a0. [DOI] [PubMed] [Google Scholar]
- 2.Praefcke GJ, Mcmahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5(2):133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 3.Frank S, Gaume B, Bergmann-Leitner ES, et al. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1(4):515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 4.Orth JD, Mcniven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol. 2003;15(1):31–39. doi: 10.1016/s0955-0674(02)00010-8. [DOI] [PubMed] [Google Scholar]
- 5.Cao H, Garcia F, Mcniven MA. Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell. 1998;9(9):2595–2609. doi: 10.1091/mbc.9.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrais D, Merrifield CJ. Dynamics of endocytic vesicle creation. Dev Cell. 2005;9(5):581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Okamoto PM, Herskovits JS, Vallee RB. Role of the basic, proline-rich region of dynamin in Src homology 3 domain binding and endocytosis. J Biol Chem. 1997;272(17):11629–11635. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 8.Ochoa GC, Slepnev VI, Neff L, et al. A functional link between dynamin and the actin cytoskeleton at podosomes. J Cell Biol. 2000;150(2):377–390. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gold ES, Underhill DM, Morrissette NS, et al. Dynamin 2 is required for phagocytosis in macrophages. J Exp Med. 1999;190(12):1849–1856. doi: 10.1084/jem.190.12.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson HM, Skop AR, Euteneuer U, Meyer BJ, Mcniven MA. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr Biol. 2002;12(24):2111–2117. doi: 10.1016/s0960-9822(02)01390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruchten AE, Mcniven MA. Dynamin as a mover and pincher during cell migration and invasion. J Cell Sci. 2006;119(Pt 9):1683–1690. doi: 10.1242/jcs.02963. [DOI] [PubMed] [Google Scholar]
- 12.Marino N, Collins JW, Shen C, et al. Identification and validation of genes with expression patterns inverse to multiple metastasis suppressor genes in breast cancer cell lines. Clin Exp Metastasis. 2014;31(7):771–786. doi: 10.1007/s10585-014-9667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inokawa Y, Nomoto S, Hishida M, et al. Dynamin 3: a new candidate tumor suppressor gene in hepatocellular carcinoma detected by triple combination array analysis. Onco Targets Ther. 2013;6:1417–1424. doi: 10.2147/OTT.S51913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J, Wang S, Zhang YJ, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55(6):1799–1808. doi: 10.1002/hep.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teicher BA. Searching for molecular targets in sarcoma. Biochem Pharmacol. 2012;84(1):1–10. doi: 10.1016/j.bcp.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Booken N, Gratchev A, Utikal J, et al. Sézary syndrome is a unique cutaneous T-cell lymphoma as identified by an expanded gene signature including diagnostic marker molecules CDO1 and DNM3. Leukemia. 2008;22(2):393–399. doi: 10.1038/sj.leu.2405044. [DOI] [PubMed] [Google Scholar]
- 17.Zhang HJ, Yuan GL, Liang QL, et al. Progress of dynamin 3 in tumors. Int J Clin Exp Med. 2017;10(11):15060–15063. [Google Scholar]
- 18.Zhang Z, Chen C, Guo W, Zheng S, Sun Z, Geng X. DNM3 Attenuates Hepatocellular Carcinoma Growth by Activating P53. Med Sci Monit. 2016;22(1):197–205. doi: 10.12659/MSM.896545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YY, do IG, Park YA, et al. Low dynamin 2 expression is associated with tumor invasion and metastasis in invasive squamous cell carcinoma of cervix. Cancer Biol Ther. 2010;10(4):329–335. doi: 10.4161/cbt.10.4.12275. [DOI] [PubMed] [Google Scholar]