Abstract
Background
Circular RNAs (circRNAs) are a class of noncoding RNAs with closed loop structures. There has been growing evidence showing that circRNAs are involved in the pathogenesis of human diseases including various carcinomas. Our study is aimed to investigate the association between a new circRNA named circ-DLG1 (hsa_circ_0007203) and esophageal squamous cell carcinoma (ESCC) carcinogenesis.
Methods
The circ-DLG1 expression levels were detected by real-time quantitative reverse transcription-polymerase chain reaction in cells, tissues, and plasmas. The correlation between circ-DLG1 expression and clinicopathologic features was then analyzed. The effect of circ-DLG1 expression on cell proliferation was evaluated in vitro by CCK8 assay and clone formation experiment. Finally, a network of circ-DLG1 with its targeted miRNA interactions and corresponding mRNAs was constructed.
Results
circ-DLG1was found to be significantly upregulated in ESCC cells, tissues, and plasmas compared to normal cases. Furthermore, in vitro assays, TE10 and KYSE180, of the ESCC cell lines demonstrated that knockdown of circ-DLG1 reduced cell proliferation significantly. Prediction and annotation revealed that circ-DLG1 was able to sponge to 20 miRNAs and 60 corresponding target mRNAs.
Conclusion
Our study indicated that upregulation of circ-DLG1 promoted esophageal cell proliferation ability and might serve as a novel biomarker for ESCC.
Keywords: ESCC, spong, upregulation, biomarker, miRNA
Introduction
Esophageal cancer (EC) is one of the common causes of cancer-related death all over the world, with a huge difference of morbidity and mortality in different areas. China is one of the countries with a significantly high morbidity, with pathological type of esophageal squamous cell carcinoma (ESCC).1 Lack of early-stage diagnosis and metastasis always leads to poor prognosis of EC.2 Therefore, the key to reduce morbidity and mortality is to explain the molecular mechanisms underlying the development of EC and to improve early screening techniques.
Circular RNAs (circRNAs), different from the canonical splicing of linear RNAs, represent a novel class of conserved endogenous RNAs that are able to regulate gene expression in many species.3–5 To date, literature searches have shown that more than 100,000 circRNAs have been predicted in animals, plants, humans, mice, zebrafish, fruit flies, nematodes, yeast, rice, and fungal cells.6–8 Some reports have shown that circRNAs can preserve high cell and tissue specificity in different mammalian cells.9 As for functional studies, circRNAs are reported to be involved in complex RNA–RNA interaction networks and play a role in post-transcriptional gene regulation. In addition, circRNAs can bind directly to proteins or via RNA-mediated indirect interactions. More and more evidence shows that circRNAs can bind miRNAs, and is rich in functional miRNA-binding sites used primarily to regulate cancer progression; and these circRNA–miRNA axes are involved in a variety of disease pathways, such as cancer metastasis, apoptosis, and invasion.10–12
Our study is aimed to investigate the association between a new circRNA named circ-DLG1 (hsa_circ_0007203) and ESCC. The circ-DLG1 expression levels were detected by real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) in cells, tissues, and plasmas. The correlation between circ-DLG1 expression and clinicopathologic features was analyzed. The effect of circ-DLG1 expression on cell proliferation was evaluated in vitro by knocking down technique. Finally, a network of circ-DLG1 with its targeted miRNA interactions and corresponding mRNAs was constructed.
Materials and methods
Patients and clinical samples
A total of 63 plasmas, from 28 healthy individuals and 35 patients with ESCC, were obtained in Nanjing First Hospital. A total of 55 EC tissues and corresponding 55 adjacent nontumor tissue samples were collected from patients with ESCC undergoing surgery in Nanjing First Hospital. None of the patients received radiotherapy or chemotherapy prior to surgery, and all tumors were accurately staged according to the TNM staging system. Each patient signed a written informed consent form prior to participating in the study, and the experiment was approved by the Medical Ethics Committee of Nanjing Medical University.
RNase R and actinomycin treatment
Within RNase R treatment, 2 mg of total RNA was incubated for 20 minutes at 37°C with or without 3 U mg−1 of RNase R (Epicentre Technologies, Madison, WI, USA), and the resulting RNA was subsequently purified using an RNeasy MinElute Cleanup Kit (Qiagen). For actinomycin D treatment, 2 mg of total RNA was incubated at 37°C with or without 1 mg actinomycin (Actinomycin D, Sigma, Chengdu, China), and the resulting RNA was detected separately at 0, 6, 12, and 18 hours.
Cell line, cell culture, and transfection
Human ESCC cell lines, K140, K70, K150, K180, and T10, were established from samples extracted from patients with ESCC by Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. KYSE series cells and human normal esophageal cell line HET-1 were generous gifts from Dr Zhi-hua Liu at the State Key Laboratory of Molecular Oncology, Cancer Institute, Chinese Academy of Medical Sciences (Beijing, China). K180 and T10 cells were transfected with 50 nM siRNAs targeting circ-DLG1 (si-circ-DLG1) and an negative control (si-NC) using the Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) based on the orders provided by the manufacturer. The target sequences of circ-DLG1 siRNAs are listed below: siRNA-1: 5′-ACGAGAGATAAAGGGCTTCTT-3′; siRNA-2: 5′-GAGAGATAAAGGGCTTCTTCT-3′; siRNA-3: 5′-CGAGAGATAAAGGGCTTCTTC-3′. After 48 hours, knockdown of circ-DLG1 was tested via qRT-PCR.
RNA isolation, reverse transcription, and qRT-PCR
Total RNA was extracted from cancerous/noncancerous specimens or cell lines using the RNA Isolation Kit and was refrigerated at −40°C. RNA was reverse transcribed into cDNA using the Prime-Script TM one-step RT-PCR Kit. The expression level of circ-DLG1 was detected via qRT-PCR using the following primer sequences: forward: 5′-AAACGAGAGATAAAGGGCTTCT-3′; reverse: 5′-ACTGCTTTAGAAACTGGGGAGT-3′. Glyceralde-hyde 3-phosphate dehydrogenase (GAPDH) was applied as an internal control. Circ-DLG1 levels were normal-ized to GAPDH using the following primer sequences: forward: 5′-CTGGGCTACACTGAGCACC-3′; reverse: 5′-AAGTGGTCGTTGAGGGCAATG-3′. qRT-PCR reactions were performed by the ABI7500 System (Applied Biosystems, Foster City, CA, USA) and the SYBR Green PCR Master Mix.
CCK-8 assay
The relative cell growth rate was observed using the CCK-8 assay. K180 and T10 cells were plated at 3 × 103 cells/well in 96-well plates with four wells for each condition, and then transfected with si-circ-DLG1 or si-NC. The absorbance at 450 nm was measured to evaluate the cell viability every 24 hours with a microplate reader.
Clone formation experiment
K180 and T10 cells were transfected with 50 nM si-circ-DLG1 or si-NC. We selected cells within the logarithmic growth phase and digested them with 0.25% trypsin into single cell, respectively. The cells were cultured in 1640 medium with 10% FBS and incubated at 37°C in 5% CO2 and saturated humidity for 7–10 days. The culture was ended when a macroscopic clone appeared in the dish. Invert the plate and superimpose a gridded clear film and count the number of colonies that have more than 10 cells in the microscope (low magnification) to calculate the rate of clone formation at last.
Statistical analyses
Comparison of continuous data was analyzed using an independent t-test between the two groups. All statistical analyses were performed using SPSS for Windows v.17.0 and GraphPad software. For all results, P<0.05 was considered statistically significant.
Results
The biological structure of circ-DLG1
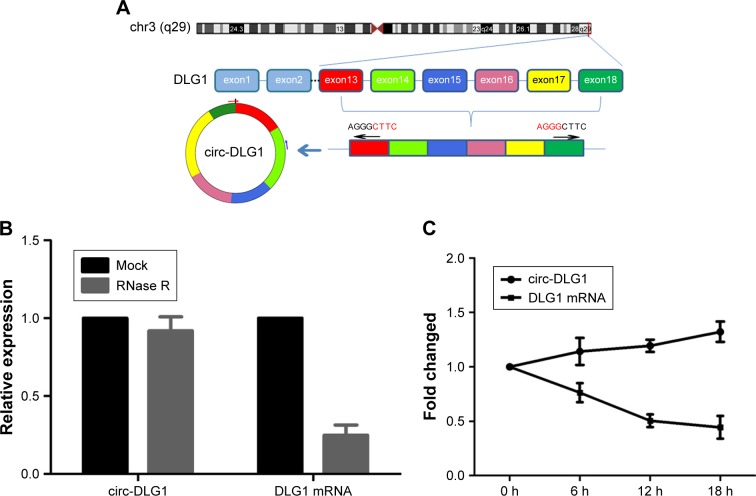
First, we explored the mechanisms of how circ-DLG1 was formed. circ-DLG1 was derived from the exon 13 to exon 18 of the gene DLG1 (Figure 1A). The RNase R exonuclease and actinomycin experiments were applied to verify the circular characteristic of circ-DLG1. The results showed that circ-DLG1 was resistant to digestion with the RNase R exonuclease or actinomycin (Figure 1B–C). In summary, we concluded that circ-DLG1 was a stable circRNA.
Figure 1.
(A) Schematics showed that circ-DLG1 was derived from exons 13 to 18 of DLG1. (B) qRT-PCR for the abundance of circ-DLG1 and DLG1 mRNA in ESCC cells treated with RNase R. (C) qRT-PCR for the abundance of circ-DLG1 and DLG1 mRNA in ESCC cells treated with actinomycin.
Abbreviations: ESCC, esophageal squamous cell carcinoma; qRT-PCR, quantitative reverse transcription-polymerase chain reaction.
Expression of circ-DLG1 in clinical samples of patients with ESCC
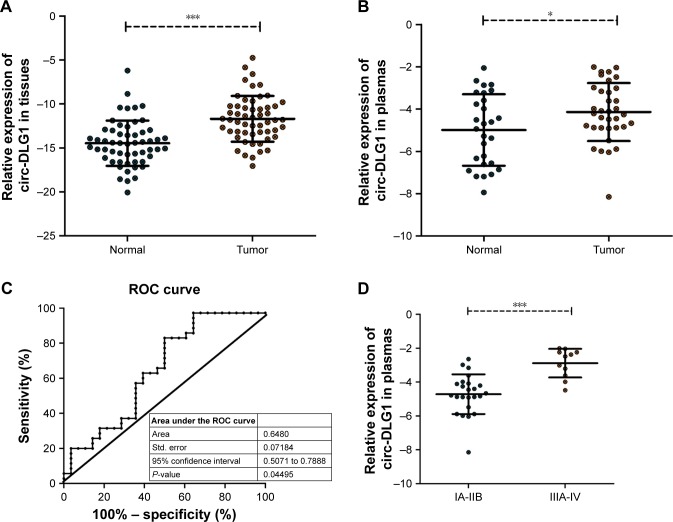
Through qRT-PCR, the circ-DLG1 expression levels were tested in a total of 63 plasmas, 28 from healthy individuals and 35 from patients with ESCC, and 55 paired primary ESCC tissues and the adjacent normal tissues. The results revealed that the expression of circ-DLG1 in ESCC plasmas was significantly higher than that in normal plasmas (Figure 2A, P<0.0001). Meanwhile, the expression of circ-DLG1 in ESCC tissues was significantly higher than that in normal tissues, coherent with the results of plasmas (Figure 2B, P<0.05). Moreover, we performed an ROC curve to assess the diagnostic value of circ-DLG1 in distinguishing ESCC plasmas from normal cases and the area under the ROC curve was 0.6480 with a cut off value of −4.924 (Figure 2C). In terms of the over-regulation of circ-DLG1 in ESCC plasmas, we next evaluated the association between circ-DLG1 and clinicopathologic parameters. As shown in Figure 2D, the expression level of circ-DLG1 was signifi-cantly associated with TNM stage (P<0.0001). However, circ-DLG1 expression was not associated with age, gender, nodal status, diameter, or differentiation (Table 1).
Figure 2.
Relative circ-DLG1 expression and its clinical significance in ESCC plasmas and tissues.
Notes: (A) The expression of circ-DLG1 levels was significantly higher in ESCC plasmas than that in normal plasmas. (B) The expression of circ-DLG1 expression levels was significantly higher in ESCC tissues than that in adjacent noncancerous tissues. (C) The AUC of distinguishing ESCC plasmas from healthy controls was 0.6480. (D) The expression level of circ-DLG1 was significantly associated with TNM stage. ***P<0.0001, *P<0.05.
Abbreviations: AUC, area under the ROC curve; ESCC, esophageal squamous cell carcinoma.
Table 1.
The relationship between circ-DLG1 expression levels (−ΔCt) in gastric cancer plasmas and clinicopathologic factors of patients
| Variables | No of patients (%) | Mean ± SE | P-value |
|---|---|---|---|
|
|
|
|
|
| Age (year) | 0.465 | ||
| ≥60 | 27 | −4.04±1.26 | |
| <60 | 8 | −4.45±1.75 | |
| Gender | 0.887 | ||
| Female | 15 | −4.18±1.57 | |
| Male | 20 | −4.10±1.24 | |
| Diameter (cm) | 0.208 | ||
| ≥3 | 24 | −3.93±1.26 | |
| <3 | 11 | −4.57±1.56 | |
| Differentiation | 0.630 | ||
| Middle–Low | 29 | −4.19±1.39 | |
| High | 6 | −3.88±1.36 | |
| Lymphatic metastasis | 0.440 | ||
| No | 7 | −3.84±1.00 | |
| Yes | 28 | −4.21±1.45 | |
| TNM stage | 0.000* | ||
| IA–IIB | 24 | −4.71±1.17 | |
| IIIA–IV | 11 | −2.88±0.84 |
Note:
P<0.05.
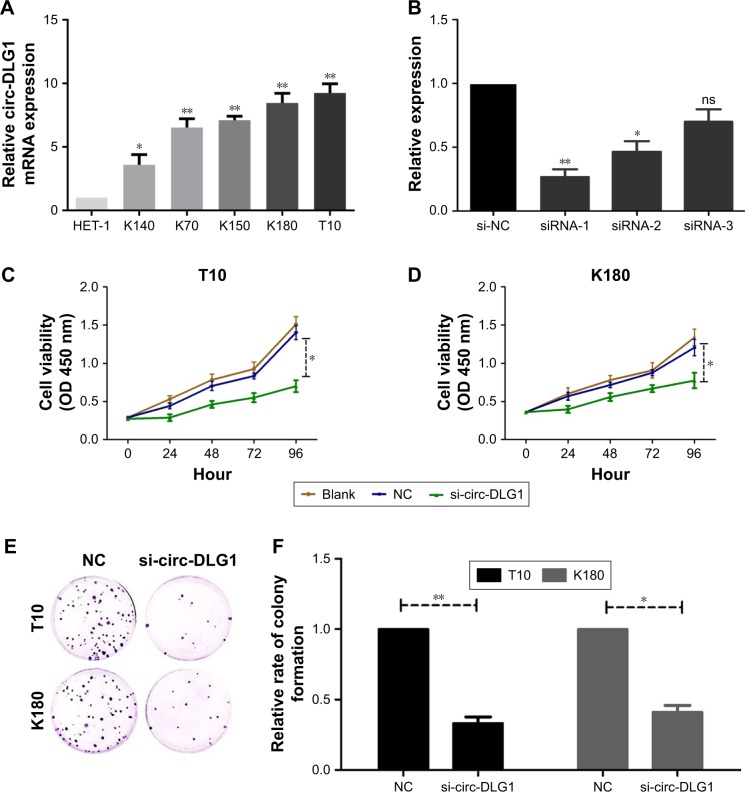
Impact proliferation of ESCC cells by knockdown of circ-DLG1
The expression of circ-DLG1 in five ESCC cell lines was examined by qRT-PCR, revealing that the K180 and T10 cells had the highest expression levels compared with HET-1A, the normal esophageal epithelia cell line (Figure 3A). The ESCC cells in 6-well plate were transfected with three different si-circ-DLG1s or si-NC using the Lipofectamine 2000 Transfection Reagent. At 48 hours after treatment, the expression of circ-DLG1 was effectively knocked down in both ESCC cells (Figure 3B). Then, we selected the most effective one (siRNA-1) to transfect ESCC cells by the same method. The clone formation experiment and CCK8 assay
Figure 3.
The expression of circ-DLG1 in ESCC cell lines.
Notes: (A) The expression of circ-DLG1 in K140, K70, K150, K180, and T10 cells was higher than that in HET-1 significantly. (B) Knockdown of circP-DLG1 was confirmed via qRT-PCR, demonstrating the effective knockdown in ESCC cells. (C) Knockdown of circ-DLG1 inhibited T10 cell proliferation significantly. (D) Knockdown of circ-DLG1 inhibited K180 cell proliferation significantly. (E) Cloning formation experiment showed that knockdown of circ-DLG1 inhibited K180 and T10 cell proliferation significantly. (F) Cell clone formation ratio after knockdown of circ-DLG1 was lower than that of the control group in K180 and T10 cells. **P<0.001, *P<0.05.
Abbreviations: ESCC, esophageal squamous cell carcinoma; ns, not significant; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; NC, normal control.
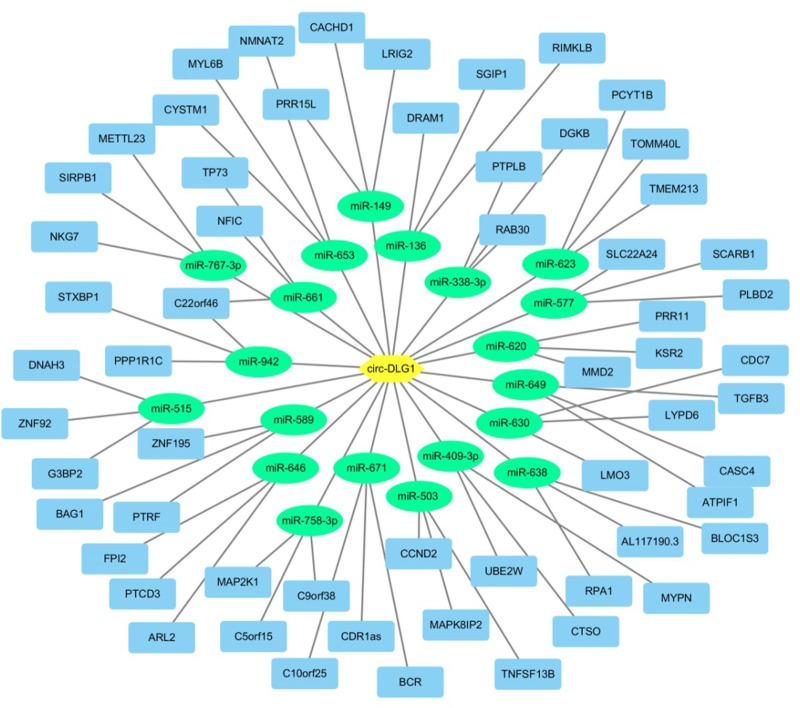
Circ-DLG1 could serve as a sponge for multiple miRNAs
To explore the ability of circ-DLG1 to bind with miRNAs, we performed a circRNA–miRNA interaction network prediction on Circular RNA Interactome database. A total of 20 miRNAs (ie, miR-515, miR-942, miR-589, miR-136, miR-620, and miR-630) and corresponding target mRNAs were predicted to have an interaction with circ-DLG1 via the Target Scan Human database. The relative interaction relationships between circ-DLG1 and its targets are displayed in Figure 4.
Figure 4.
Circ-DLG1 could serve as a sponge for multiple miRNAs.
Note: A schematic drawing showing the miRNAs and mRNAs associated with circ-DLG1.
Discussion
In this study, we found that circ-DLG1 presented a higher level expression in ESCC plasmas and tissues compared with that in normal controls. Furthermore, the verified results showed that it also presented higher level expression in ESCC cell lines than in HET-1A. Knocking down of circ-DLG1 expression inhibited cell proliferation significantly, indicating that circ-DLG1 might promote the procession of ESCC. This was not the first circRNA which has been confirmed in the diagnosis and prognosis of EC. Previous reports have shown that the expression of circITCH, which is a circRNA generated from several exons of itchy E3 ubiquitin protein ligase (ITCH) in ESCC was significantly lower than that in paracancerous tissues, not only acting as a sponge to adsorb tumor-associated miR-7, miR-17, and miR-214, but also increasing the expression of ITCH. Moreover, decreased expression of circ-ITCH activates the Wnt pathway and promotes tumor proliferation.13 In addition, Xia et al found that increased expression of hsa_circ_0067934 in ESCC was associated with the prognosis, staging, and grade of EC, and could promote the proliferation and migration of EC cells.14 These results make circRNAs a novel class of potential biomarkers for diagnosis and prognosis of ESCC in clinical blood samples and tissues. Yet the present methods to detect and characterize circRNAs are still immature and limited. Although more and more circRNAs with known functions have been studied and reported, there are still a large number of circRNAs in EC, whose functions remain unknown. A deeper research of circRNAs biogenesis is needed for further exploring the exact mechanism.
Increasing evidence indicates that circRNAs are not only the by-products of missplicing or splicing errors, but also play an important role in epithelial–mesenchymal transition (EMT). For instance, Zhong found that circRNA-MYLK might work as competing endogenous RNA for miR-29a, promoting EMT by activating VEGFA/VEGFR2 and downstream Ras/ERK signaling pathway in breast cancer. In this study, we found that inhibition of circ-DLG1 expression decreased the proliferation of ESCC cells significantly. However, whether circ-DLG1 participates in the EMT process require further experimental confirmation.
It has been reported currently that circRNAs are involved in regulating gene expression as a sponge of miRNAs. For example, in 2013, two different groups of researchers at the same time demonstrated for the first time that circRNA functions in the form of miRNA adsorption.15,16 Therefore, we assumed circ-DLG1 might also work through their miRNA-mediated effects in ESCC. A total of 20 miRNAs and corresponding target mRNAs were predicted to have an interaction with circ-DLG1 in this study. Among these miRNAs, miR-942 and miR-630 have been previously investigated and confirmed involved in suppressing or promoting the formation of ESCC. Ge et al reported that miR-942 expression is significantly upregulated in ESCC and associated with poor prognosis in patients with ESCC. Overexpression of miR-942 promotes, whereas inhibition of miR-942 decreases, the tumor sphere formation, the CD90+ subpopulation cells, and the expression of pluripotency-associated markers. In vivo assay shows that miR-942 overexpressing cells form larger tumors and display higher tumourigenesis. Furthermore, they demonstrated that miR-942 upregulates the Wnt/β-catenin signaling activity by directly targeting sFRP4, GSK3β, and TLE1, which are multiple level negative regulators of the Wnt/β-catenin signaling cascade.17 However, to the best of our knowledge, in the process of circRNAs sponging miRNAs, circRNA expressions are commonly negatively correlated with miRNA expressions. But in this study, circ-DLG1 and miR-942 were both upregulated in ESCC, suggesting that this hypothesis might not be established. Using miRNA expression profiling analysis, Jin found that miR-630 was markedly downregulated in three ESCC tissue samples compared with that in paired normal esophageal tissues. Decreased miR-630 expression was associated with poor overall survival in these patients. In addition, they also explored the biological function of miR-630 by targeting Slug and investigated the correlation between miR-630 expression and EMT progression in vivo and in vitro ectopic miR-630 expression could inhibit proliferation, invasion, and metastasis, whereas miR-630 knockdown induced proliferation, invasion, metastasis, and EMT traits.18 Contrary to the results of miR-942, miR-630 was significantly downregulated in ESCC, while circ-DLG1 was upregulated. These predictions revealed that circ-DLG1 might work through miR-630-mediated effects in ESCC, but the specific relationship still requires luciferase reporter gene assay and expression regulation experiment to confirm.
When illustrating the results, there are some problems that should not be ignored. At first, all clinical samples are from one hospital. It is recommended that more samples from different hospitals or even different races are detected. Moreover, experimental identification and characterization of the associated molecules, such as miRNAs, are suggested in the future. At last, previous study reported that some cir-cRNAs can save, classify, or localize RNA-binding protein (RBPs), and probably regulate the function of RBPs by acting as competing elements. Hence, the deeper researches of circ-DLG1 are necessary for this mechanism.
Conclusion
Our study demonstrated that circ-DLG1 had higher-level expression in ESCC cells, tissues, and plasmas. Upregulation of circ-DLG1 promoted ESCC cell proliferation significantly. Circ-DLG1 might work as a novel biomarker for ESCC, but how circ-DLG1 has an interaction with targeted miRNA and their corresponding mRNAs need to be studied further. In future, we believe that molecular mechanisms by which circRNAs function as miRNA sponges to regulate ESCC occurrence and development will become a profound research field.
Footnotes
Disclosure
This work was funded by Taikang Xianlin Drum Tower Hospital Youth Research Project (TKKY3918) for JR and Jiangsu Natural Science Foundation (BK20151292) for WS. The authors report no other conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97(2):142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 3.Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi: 10.2147/OTT.S123220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F, Nazarali AJ, Ji S. Circular RNAs as potential biomarkers for cancer diagnosis and therapy. Am J Cancer Res. 2016;6(6):1167–1176. [PMC free article] [PubMed] [Google Scholar]
- 5.Yao T, Chen Q, Fu L, Guo J. Circular RNAs: biogenesis, properties, roles, and their relationships with liver diseases. Hepatol Res. 2017;47(6):497–504. doi: 10.1111/hepr.12871. [DOI] [PubMed] [Google Scholar]
- 6.Nigro JM, Cho KR, Fearon ER, et al. Scrambled exons. Cell. 1991;64(3):607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 7.He J, Xie Q, Xu H, Li J, Li Y. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144. doi: 10.1016/j.canlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 8.Kumar L, Shamsuzzama HR, Haque R, Baghel T, Nazir A. Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol Neurobiol. 2016;53:1–11. doi: 10.1007/s12035-016-0213-8. [DOI] [PubMed] [Google Scholar]
- 9.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 11.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.du Z, Fei T, Verhaak RG, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20(7):908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 16.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Ge C, Wu S, Wang W, et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget. 2015;6(13):10964–10977. doi: 10.18632/oncotarget.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin L, Yi J, Gao Y, et al. MiR-630 inhibits invasion and metastasis in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin. 2016;48(9):810–819. doi: 10.1093/abbs/gmw073. [DOI] [PubMed] [Google Scholar]