Abstract
Autophagy can serve as a mechanism for survival of cells during nutrient deprivation by recycling cellular macromolecules and organelles transiently to provide essential metabolic substrates. However, autophagy itself causes metabolic stress to cells, and other cellular protective mechanisms likely cooperate with autophagy to promote cell survival during nutrient deprivation. In this study, we explored protective mechanisms in breast cancer cells in the setting of glucose deprivation. While breast cancer cells (MCF7 and T47D) survive in glucose-free medium for three days or more, autophagy is induced in this setting. Blocking autophagy pharmacologically with chloroquine or by knock-out of an essential autophagy gene, such as Beclin 1 or ATG7, markedly reduces the ability of cells to survive during glucose deprivation. Autophagy previously was shown to degrade p62, a protein that sequesters KEAP1, and KEAP1 in turn sequesters Nrf2, a master regulator of the antioxidant response. Hence, we investigated how the Nrf2 signaling pathway might be affected by glucose deprivation and autophagy. We found that while glucose deprivation does cause decreased cellular levels of p62, Nrf2 protein levels and activity unexpectedly increase in this setting. Moreover, this increase in Nrf2 activity provides important protection to breast cancer cells during glucose deprivation, since siRNA knockdown of Nrf2 markedly impairs survival during glucose deprivation. Antioxidants, N-acetyl cysteine and glutathione also protect these cells during glucose deprivation, leading us to conclude that Nrf2 signaling via its antioxidant activity has a critical and previously undescribed role of protecting cells during glucose deprivation-induced autophagy.
Keywords: KEAP1, Nrf2, Antioxidant response, Autophagy, P62, Beclin-1, ATG7
1. Text
Within irregularly vascularized tumors, cancer cells commonly are challenged by nutrient deprivation. Various cell survival mechanisms can help cells to withstand this metabolic stress, including autophagy, which can provide energy and essential nutrients by recycling intracellular macromolecules and organelles transiently to provide essential metabolic substrates. Accordingly, autophagy has been proposed to have an essential role in supporting the growth of cancerous tumors [1]. However, dissecting the role of autophagy in nutrient insufficiency has not been straightforward, in part because the contribution of autophagy to cell survival during experimental settings of glucose deprivation has been an area of dispute [2]. In multiple experiments, autophagy has been shown to protect cancer cells from the glycolytic inhibitor 2-deoxyglucose (2-DG) [3–5], but other experiments using a variety of cultured cancer cell lines did not find a protective role for autophagy in the setting of glucose deprivation [6].
Remarkably, investigations on how autophagy might protect cancer cells during glucose deprivation have given little attention to other cellular adaptations that could have complementary roles in cell survival – or competing roles in driving cell death - during metabolic stress. One cellular adaptation that warrants consideration for possible effects on cells during glucose deprivation is that of the Keap1/ Nrf2 pathway. Previous studies have found that autophagy leads to reduced levels of p62 [7], which in turn would be expected to allow Keap1 bind the Nrf2 transcription factor and thus lead to increased degradation of Nrf2 protein. Considering the importance of Nrf2 signaling in affecting how cells adapt to various types of stress and the expectation that autophagy might affect cellular Nrf2 levels, we investigated the roles of both autophagy and Nrf2 in the setting of nutrient deprivation.
2. Results
2.1. Survival of breast cancer cells during glucose deprivation is dependent on autophagy
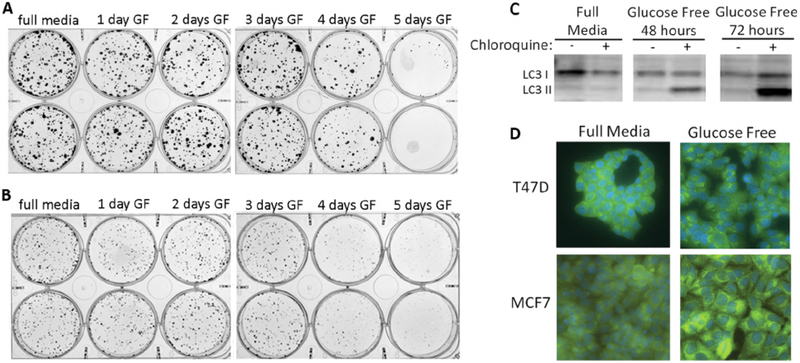
To investigate mechanisms of cell survival during glucose deprivation in breast cancer cells, we measured effects of glucose deprivation on clonogenic cell survival in MCF7 and T47D cells by culturing cells in glucose-free medium for variable periods of time, followed by culturing in full medium to allow growth of cell colonies. As seen in Fig. 1 (panels A and B), cells generally tolerate up to approximately three days of glucose-free media, with decreased rate of replication during the glucose deprivation but retained ability to proliferate when full media is restored. To determine whether autophagy is induced by this glucose deprivation, we measured conversion of the microtubule-associated protein light chain 3 (LC3) protein from the LC3I form to the LC3II form, which is considered to be characteristic of autophagy [8]. Indeed, the ratio of LC3II: LC3I increases progressively with time of glucose deprivation, particularly after the addition of chloroquine, which enhances detection of increases in LC3II by raising lysosomal pH and preventing fusion and degradation of lysosomal proteins [8] (Fig. 1, panel C). Glucose deprivation for three days also results in the appearance of a punctate staining pattern of LC3, which indicates the presence autophagosomes typically seen in macroautophagy (Fig. 1, panel D).
Fig. 1. Glucose deprivation causes autophagy in breast cancer cells.
Breast cancer cells (MCF7, panel A; T47D, panel B) plated at clonal density were cultured in glucose-free medium for up to 5 days, and then allowed to form colonies in full medium for an additional 8–10 days. Panel C: glucose deprivation in MCF7 cells causes increases in LC3 II to LC3I ratio, particularly when completion of autophagy is blocked by chloroquine. Panel D: Glucose deprivation in T47D and MCF7 cells results in appearance of punctate foci of LC3, typical of autophagosomes in macroautophagy.
We then tested whether autophagy contributes to survival of breast cancer cells during glucose deprivation, first by examining how chloroquine treatment, which blocks completion of autophagy [8], affects survival. As seen in Fig. 2 (panel A), addition of chloroquine causes substantial loss of cell viability when cells are grown in glucose-free conditions for three days. By contrast, the same dose of chloroquine has modest effects on cell growth in full media conditions. As reported in other experimental systems, cell death in this setting is associated with changes typical for apoptosis, including cell fragmentation (Fig. 2, panel B) and increases in cleaved poly-ADP-ribose polymerase (Fig. 2, panel C).
Fig. 2. Autophagy is protective of cell survival during glucose deprivation.
Panel A shows effects of chloroquine, which blocks completion of autophagy, on survival of MCF7 cells cultured with full media or glucose-free medium for 3 days, followed by full medium for 10 days. In panel B, cultures grown in glucose-free medium are seen to fragment in the presence of chloroquine, suggesting that apoptosis is induced when autophagy is blocked. Panel C confirms that poly-ADP ribose polymerase is cleaved in glucose-deprived cells cultured with chloroquine. Panel D shows that, with CRISPR/ Cas9 knock-out of ATG7 or Beclin 1, MCF7 cells have markedly reduced survival after glucose deprivation for 3 days.
To confirm the role of autophagy in cell survival, we then used a CRISPR-Cas9 approach to knock out two genes that are critical for autophagy, ATG7 and Beclin-1. Fig. 2 (panel D) shows that targeted knockout of these genes in MCF7 cultures results in substantially decreased survival after glucose deprivation. Survival of cells during glucose deprivation is unaffected by transfection with a control vector, and knockout of either ATG7 or Beclin-1 does not affect survival of cells cultured with full media. Thus, glucose deprivation leads to induction of autophagy in breast cancer cells, and this autophagy contributes to survival of cells during glucose deprivation lasting for several days.
2.2. p62 levels decrease but Nrf2 levels increase during glucose deprivation
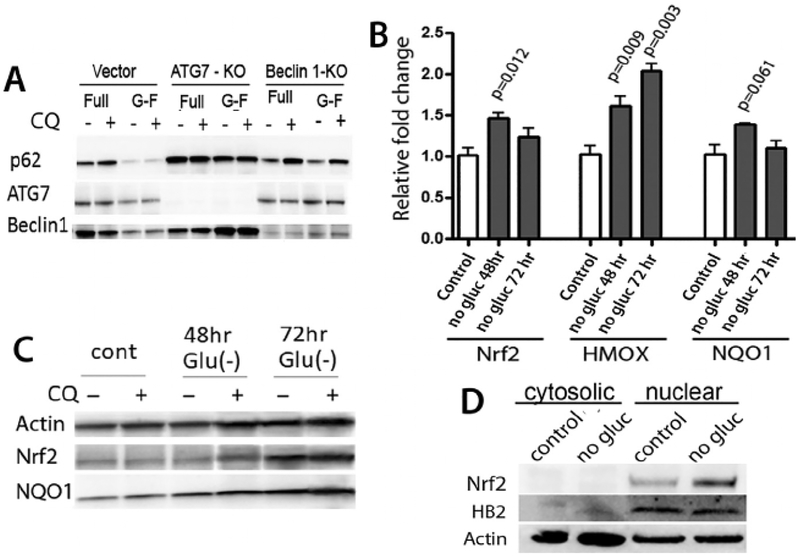
Previous studies demonstrated that autophagy can lead to degradation of autolysosomes containing aggregates of p62 and LC3 [7], resulting in decreased levels of p62. Consistent with these previously reported findings, we also observed decreased levels of p62 in cultures of MCF7 cells at 72 h of glucose deprivation, when compared to control cultures with full glucose media (Fig. 3, panel A). Decreases in levels of p62 were evidently dependent on autophagy, since p62 levels were not substantially changed by glucose deprivation in cells that had ATG7 or Beclin 1 knock-out.
Fig. 3. Glucose deprivation leads to decreased levels of p62 and increased levels of Nrf2 in MCF7 breast cancer cells.
Panel A shows decreased levels of p62 in MCF7 cells cultured for 72 h in glucose-free medium (G-F). These decreases in p62 are dependent on autophagy and do not occur in cells with knock-out of Belcin 1 or ATG7. Panel B: Time dependent changes in transcripts for Nrf2 and its target genes, heme oxygenase 1 (HMOX) and nitroquinone oxidase 1 (NQO1), during glucose deprivation. Fold changes were calculated relative to control cultures in complete media, and -actin was used for as endogenous control for each measurement. Measurements from glucose-deprived cultures were then compared to respective control cultures by t-test, with p values shown. Panel C: Immunoblots show that protein levels of both Nrf2 and NQO1 increase after 72 h glucose deprivation, and Panel D shows that the increased levels of Nrf2 protein are predominantly within the nuclear fraction of fractionated cells.
Previous investigations have described how p62 sequesters Keap1 into aggregates and how increased levels of p62 in cells with a deficiency of autophagy thus have sequestration of Keap1 [9–11]. By contrast, decreases in p62 during autophagy would be expected to allow Keap1 bind the Nrf2 transcription factor and thus lead to increased degradation of Nrf2 protein. To test how autophagy occurring during glucose-deprivation affects Nrf2 activity in breast cancer cells, we first determined how glucose deprivation affects levels of Nrf2 protein as measured by immunoblot. As shown in Fig. 3 (panels B and C), in spite of these decreases in p62 levels, transcript and protein levels of Nrf2 increase in MCF7 cells when cultured in the absence of glucose, with particularly prominent increases in Nrf2 protein levels in nuclear fractions of glucose-deprived cells (Fig. 3, panel D).
Glucose deprivation for 48 h or 72 h also resulted in increased transcript levels of the Nrf2-regulated transcript heme oxygenase-1 (Fig. 3, panel B). Although another Nrf2-regulated transcript, NADPH dehydrogenase quinone 1 (NQO1), did not show increases in levels of mRNA that meet criteria for statistical significance (p = 0.061) at the time points measured, cells grown in glucose deficient media for 72 h did show substantially increased levels of the NQO1 protein (Fig. 3, panel C), consistent with increased Nrf2 activity in these cultures. Remarkably, increases in Nrf2 protein levels occur even in glucose-deprived cells treated with chloroquine (Fig. 3, panel C), indicating that these increases in Nrf2 levels are a not direct consequence of autophagy.
2.3. Induction of the Nrf2 transcription factor is critical for survival of breast cancer cells during nutrient deprivation-induced autophagy
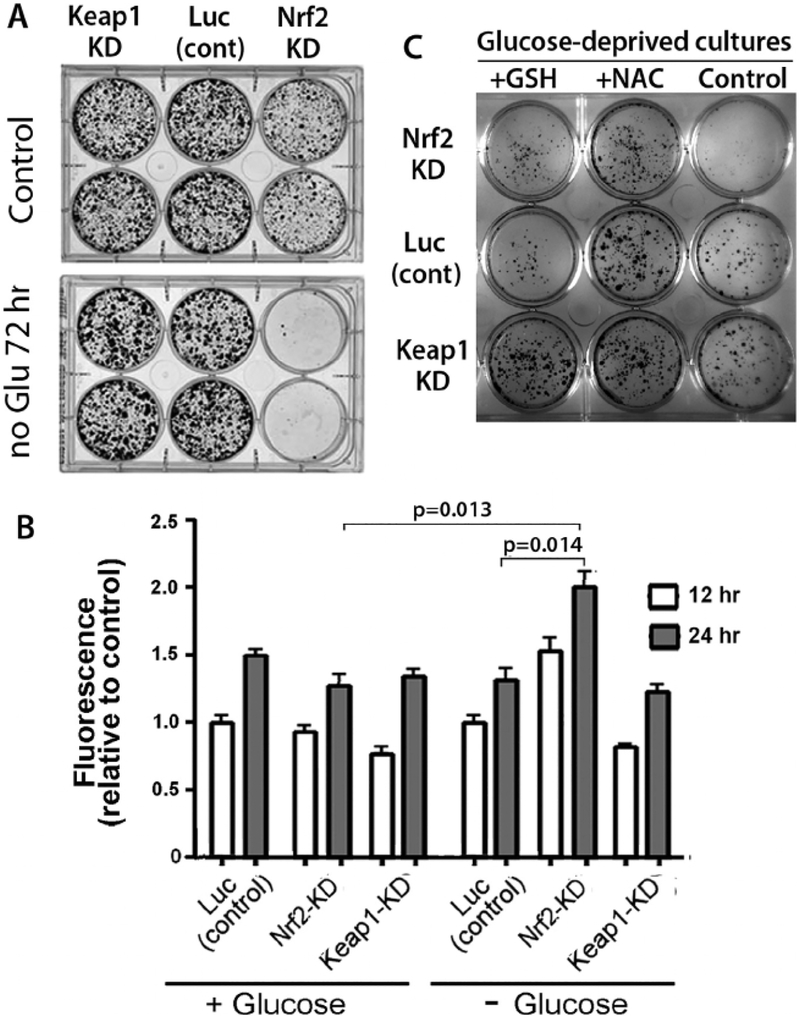
To test whether these unexpected increases in Nrf2 protein and activity have a role in survival of glucose-deprived breast cancer cells, we transfected shRNA designed to knock-down levels of Keap1 or Nrf2 transcripts. Significantly, neither Keap1 shRNA nor Nrf2 shRNA significantly affects growth of cells in full media, and with glucose deprivation, cells depleted of Keap1 show no effect or only a modestly improved survival compared to control cultures (Fig. 4, panel A). This result likely reflects the increased Nrf2 activity occurring with glucose deprivation alone, even without knockdown of Keap1. Remarkably, however, cells depleted of Nrf2 have substantially impaired survival after glucose deprivation compared to cells that were transfected with a control luciferase shRNA, indicating that Nrf2 has a critical role in survival of these cells during glucose deprivation (Fig. 4, panel A).
Fig. 4. Nrf2 provides antioxidant protection of breast cancer cells during glucose deprivation.
Panel A demonstrates that survival of MCF7 cells during glucose deprivation is dependent on Nrf2, since knock-down of Nrf2 leads to markedly decreased survival in cells cultured in glucose-deficient medium for 3 days. Panel B shows measurements of reactive oxygen species using the fluorescent marker, DCFDA. Note the significantly greater levels of ROS in cells that had knock-down of Nrf2 compared to cells with knock-down of KEAP1, consistent with Nrf2 activity controlling levels of ROS during glucose deprivation. In panel C, cultures of cells with KEAP1 knock-down, Nrf2 knock-down, or luciferase siRNA control transfection were cultured for 72 h in glucose free medium, followed by growth in full medium for 10 days. Addition of 2.0 mM N-acetyl cysteine (NAC) or 2.5 mM glutathione (GSH) to medium for the full 72 h resulted in improved survival of cultured cells.
2.4. Protective effects of Nrf2 are mediated by antioxidant activity
Since Nrf2-regulated transcripts contribute to cell survival by reducing oxidative stress [12,13], we next tested whether chemical antioxidants, N-acetyl cysteine (NAC) and glutathione (GSH), could protect cell survival during autophagy. Indeed, adding either of these two antioxidants (2 mM NAC or 2.5 mM GSH) improved survival of Nrf2-depleted cells from glucose deprivation for 72 h (Fig. 4, Panel B), suggesting that the protective role of increased Nrf2 during glucose deprivation involves coordinating an anti-oxidant response.
To directly examine how glucose deprivation and Nrf2 activity affect cellular levels of ROS, we used 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA), a cell-permeable form of fluorescein that is commonly used as an indicator for ROS in cells, to measure change in levels of ROS between 12 h and 24 h of glucose deprivation in MCF7 cells transfected with shRNA coding for Nrf2, Keap1, or luciferase (control), possibly reflecting increased Nrf2 activity in glucose-deficient cells. Consistent with a role of Nrf2 in decreasing levels of ROS during nutrient deprivation, ROS levels were significantly increased between 12 and 24 h by glucose deprivation in cells with knock-down of Nrf2 when compared to control cultures measured at the same times, consistent with Nrf2 having an antioxidant effect during glucose deprivation (Fig. 4, panel C). The modest decrease in ROS observed in cells with knock-down of Keap1 was not significant, likely reflecting the Nrf2 pathway already having been activated in those cells. Thus, Nrf2 activity is critical for decreasing levels of oxidants during glucose deprivation and this activity complements autophagy in protecting these breast cancer cells in this nutrient deprivation setting.
3. Discussion
Our studies found that cultured breast cancer cells (MCF7 and T47D cell lines) are capable of survival for at least three days when cultured in the absence of glucose, mimicking a situation that might be encountered in poorly vascularized areas of tumors. This glucose deprivation results in autophagy, as evidenced by an increased LC3 II: LC3 I ratio and punctate staining for LC3 II, and this autophagy response is critical for survival of these cells during glucose deprivation. Blocking autophagy with the pharmacological agent, chloroquine, or by knockdown of the essential autophagy genes ATG7 or Beclin1, results in rapid death by apoptosis when cells are cultured in glucose-free medium.
Previous experiments have not definitively defined the role of autophagy in glucose deprivation [2]. Several published experiments reported that knockdown of genes essential for autophagy in cells deprived of serum or amino acids leads to cell death by apoptosis [14,15], and autophagy has also been shown to protect cells from the glycolytic inhibitor 2-deoxyglucose (2-DG) [3–5]. While incubating cells with 2-deoxyglucose might be expected to mimic the situation of glucose deprivation, experiments testing glucose-free medium in a variety of cultured cell lines found no protective role for autophagy in the setting of glucose deprivation [6].
The apparent discrepancy between these results from previous studies on glucose deprivation and the experiments reported here cannot be explained readily. As detailed above, we did find that another survival pathway, activation of Nrf2, is also critical for survival of breast cancer cells deprived of glucose, and it is possible that previous experiments used cells or conditions that did not result in activation of Nrf2. Another possible explanation for discrepant results could be a result of some cancers being highly dependent on glutamine as a source of energy as well as biosynthetic substrates [16,17], and for these cells, glucose deprivation might have different consequences on cell survival.
The induction of autophagy and the protective effects of this process during glucose deprivation could be particularly important for survival of cancer cells, which commonly experience situations of metabolic stress. In fact, substantial data points to autophagy contributing to cancer development in a variety of experimental settings [18], and particularly in cancers with RAS mutations, experimental data indicates that autophagy is critical for growth and survival of cancer cells [19–22]. In most cancerous tumors, the disordered microenvironment is associated with metabolic stress due to variable hypoxia and nutrient insufficiency, and in this setting autophagy appears to have an essential role in recycling intracellular macromolecules and organelles to transiently provide essential metabolic substrates in such settings [1]. Thus, our observations are consistent with breast cancer cells inducing autophagy as a protective response to glucose deprivation.
Considering what protective molecular pathways might be triggered by autophagy in the setting of glucose deprivation, we focused on the Nrf2 pathway, since p62 can sequester Keap1 and thus affect levels of Nrf2. Autolysosomes containing p62- and LC3-positive bodies are degraded by autophagy [7], resulting in decreased levels of p62, an LC3-interacting, ubiquitin-associated protein [9–11], which in turn would be expected to allow Keap1 to bind the Nrf2 transcription factor and lead to increased degradation of Nrf2 protein. While we did find that p62 levels decrease in breast cancer cells during glucose deprivation, we surprisingly found that Nrf2 protein levels and Nrf2 activity are actually increased in the setting of glucose deprivation. These increases in Nrf2 protein levels and activity were not dependent on autophagy, since Nrf2 increased even in cells with knock-out of the ATG7 or Beclin1 autophagy genes.
Although the Nrf2 pathway can be activated by oxidative stress[23], we note that previous experiments found that detachment of mammary epithelial cells from an extracellular matrix results in a loss of glucose transport and corresponding ATP deficiency, leading to increased fatty acid oxidation [24]. Thus, we suggest that Nrf2 levels increase in breast cancer cells during glucose deprivation might be initiated by increased fatty acid oxidation, although we found increased Nrf2 activity in glucose-deficient cells even when cultures that were treated with the antioxidants, N-acetyl cysteine or glutathione. Thus, our experimental data show that Nrf2 levels are increased in response to glucose deprivation, in spite of decreases in p62, and although the protective activity of Nrf2 is likely mediated by antioxidant activity, increases in ROS might not be directly responsible for the increases in Nrf2.
While an important issue to address in future studies will be to determine how Nrf2 is activated by glucose deprivation, we focused our studies on to investigating what role the Nrf2 pathway might play in cell survival during glucose deprivation. Comparing cell survival in MCF7 cells that had been transfected with shRNA targeting KEAP1 (to increase Nrf2) or targeting Nrf2 (to decrease levels of this transcript), we found that loss of Nrf2 led to markedly decreased survival in glucose-deprived cultures. Thus, in addition to autophagy, induction of Nrf2 is critical for cell survival in the setting of glucose deprivation.
Nrf2 is a transcription factor known to activate multiple enzymes with antioxidant properties, including NADPH dehydrogenase quinone 1 (NQO1). We found that increased Nrf2 levels correlated with increased levels of NQO1, indicating that Nrf2 activity, as well as the level of Nrf2 protein, is increased during glucose deprivation. To explore how glucose deprivation and Nrf2 activity affect cellular levels of ROS, we used 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA), a cell-permeable, chemically reduced form of fluorescein, as an indicator for ROS in cells and found that cells with knockdown of Nrf2 showed time-dependent increases in ROS when cultured in glucose-free medium. By contrast, cells with knockdown of Keap1 showed significantly less ROS when cultured in glucose-free medium. These results show that cellular levels of ROS are increased during glucose deprivation, and that Nrf2 is important for attenuating this oxidative stress.
Since these data point to Nrf2 activity playing a critical role for cell survival during nutrient deprivation-induced autophagy, we tested whether the antioxidants, N-acetyl cysteine and glutathione, could also protect cell survival during autophagy. Indeed, these two antioxidants protect survival of Nrf2-depleted cells experiencing glucose deprivation, suggesting that the role of increased Nrf2 for protecting cells during glucose deprivation involves coordinating an anti-oxidant response.
In summary, glucose deprivation encountered by cancer cells is not necessarily lethal to these cells because of protective mechanisms, which include autophagy and induction of Nrf2. Interestingly, these two survival mechanisms appear to be activated independently of one another, and in fact, the activation of the Nrf2 pathway that we observed is contrary to expectations, based on previous studies that have investigated connections between autophagy and Nrf2. Understanding to what extent these mechanisms contribute to survival of cancerous cells in tumors could provide opportunities for therapeutic intervention, including the potential use of pharmacologic agents that target the Nrf2 pathway [25].
4. Materials and methods
4.1. Cell culture
MCF7 and T47D human breast cancer cells were purchased from ATCC (Manassas, VA), verified by DNA fingerprinting, and tested routinely for mycoplasma contamination. DMEM medium, RPMI1640 medium(including glucose-free RPMI1640), and fetal bovine serum (FBS) were purchased from GIBCO, and DMEM glucose-free medium was made from DME base (Sigma Aldrich) dissolved in ultrapure water and supplemented with sodium bicarbonate, sodium pyruvate, L-glutamine and 0.1%FBS. MCF7 cells were routinely cultured in DMEM media and T47D cells were routinely cultured in RPMI, both supplemented with 10% FBS. For experiments that tested the effects of glucose deprivation, all cultures were grown in media (with or without glucose) supplemented with 0.1%FBS.
4.2. Reagents
Primary antibodies to ATG7, Beclin1, cleaved PARP (Asp214), and PARP were purchased from Cell Signaling Technology, antibody to LC3B was purchased from Novus Biologicals, and antibody to p62 was purchased from Santa Cruz. Antibody to actin antibody was purchased from Sigma-Aldrich. Anti-rabbit and anti-mouse secondary antibodies were purchased from Cell Signaling Technology. Chloroquine, a pharmacological inhibitor of autophagy, was dissolved in water to make a 25 mM stock concentration. Puromycin was purchased from Sigma and dissolved in ultrapure water to make a 10 mg/ml stock solution. Alexa Fluor 488 Donkey anti-rabbit IGg and Alexa Fluor 594 Goat anti-mouse IGg were purchased from Invitrogen. Prolong Gold antifade reageant with DAPI by Molecular Probes Life Technologies was used to stain the nucleus in immunofluorescence experiments.
4.3. Clonogenic assays
For clonogenic assays, MCF7 and T47D were plated at equal densities of 500–3000 cells per well. After 24 h incubation, cell media was changed to complete or glucose-free medium, as specified in experiments. After culturing cells for specified times, media was changed to complete media and cells were allowed to grow for a total time in complete medium of 10 days before being fixed with 100% methanol and stained with 0.5% crystal violet (Sigma).
4.4. Immuno uorescence
MCF7 cells were plated at a density of 1500 per chamber slide well. After incubation with complete or glucose free media for 48 h, slides were washed with PBS and fixed for 10 min in zinc-formalin. Cells were then permeablized with 0.1% Triton-PBS for 10 min and then blocked with 5% BSA in PBS for 1 h before adding primary antibody (1:200 dilution) and incubating overnight at 4 °C. The slides were then washed 3 times for 10 min each and then incubated with secondary antibody (1:200 dilution) (Alexa Fluor 488 Donkey anti-rabbit IGg and Alexa Fluor 594 goat anti-mouse IGg) for 1 h in the dark. Slides were washed again and DAPI was added to the slide and covered with a coverslip. Images were captured using a Nikon eclipse 50i microscope.
4.5. Immunoblots
After experimental treatments, cells were harvested in TNE lysis buffer plus protease inhibitor and protein concentration was determined using Pierce BCA assay (Thermo-Fisher Scientific). Equal quantities of protein were loaded for each sample, separated by SDS-PAGE on a 10% or 10–20% gel Tris-HCl gel (Bio-Rad), and incubated overnight with primary antibody at recommended concentrations in 4 °C. After washing and a 1.5 h incubation with anti-rabbit or anti-mouse secondary antibody at room temperature, membranes were developed with SuperSignal West Femto Max Sensitivity Substrate (Thermo Fisher Scientific). For experiments where nuclear and cytoplasmic protein fractions were evaluated individually, cell fractions were isolated using NE-PER Nuclear and Cytoplasmic Extraction Kit from Thermo Fisher Scientific according to manufacturer’s instructions. For probing immunoblot membranes with multiple antibodies against proteins of similar size, membranes were stripped with Stripping Buffer (Thermo-Fisher Scientific).
4.6. Flow Cytometry
T47D and MCF7 cells were grown to sub-confluency in 100 mm cell culture plates in complete medium or glucose-free medium for the final 72 h. Cells were detached with 0.25% trypsin-EDTA, collected by centrifugation, washed and re-suspended in PBS, and fixed with 5 ml of a 1:1 methanol: acetone mixture. Fixed cells were then centrifuged for 5 min at 2000 RPM and the cell pellet was re-suspended in 5 ml of PBS and centrifuged again for 5 min at 2000 RPM. After aspirating PBS, 0.25 ml of 5 g/ml RNase was added for 15 min at 37 °C. Next, 0.25 ml of 100 g/ml propidium iodide (Sigma) solution was added for an overnight incubation, and cells were analyzed by FacsCalibur flow cytometer (BD company, Flanklin Lakes, NJ, USA).
4.7. Silencing Keap1 and Nrf2 by shRNA
For silencing expression of Nrf2 and Keap1, pLKO plasmids expressing shRNA duplexes were co-transfected with helper plasmids into 293 cells to produce lentivirus, which was then purified by filtration and used to infect cells as previously described [26]. The sense sequence used for Nrf2 was 5′-GATCCGTAAGAAGCCAGATGTTAATTCA AGAGACATTCTTCGGTCTACAATTTTTTTTGAAA-3′, and the sense sequence used for Keap1 was 5′- CCGGGCCTTAATTCAGCTGAGTGTTCT CGAGAACACTCAGCTGAATTAAGGCTTTTTTG-3′. Control luciferase expression vector was purchased from Sigma.
4.8. CRISPR/Cas-9 Transfection
LentiCRISPRv2 plasmid (Plasmid #52961) was digested and dephosphorylated with BsmB1 for 30 min at 37 °C. The digested plasmid was purified using QIAquick gel extractraction kit and eluted in distilled water. Next, oligonucleotides for ATG7 and Beclin 1 were phosphorylated by T4 poly-nucleotide kinase and annealed at 37 °C for 30 min, followed by incubation at 95 °C for 5 min and ramping down to 25 °C at 5 °C/min. Digested lentivirus and annealed oligonucleotides were followed by a ligation reaction and incubated at room temperature for 10 min. The ligated DNA were transformed into Stbl3 bacteria and positive colonies were selected on ampicillin contained LB agar media and cultured in ampicillin contained LB media. Supernatant was harvested and DNA was then purified using Invitrogen PureLink HiPure Plasmid Filter Midiprep Kit. Correct insertion was verified using sequencing. lentiCRISPR with inserted sequences were co-transfected into HEK293T cells with packaging plasmids pVSVg and psPAX2. MCF7 cells were then transfected twice, allowed to grow for 3–4 days cells and selected for a week. Cells were diluted into a 96 well plate and single colonies were picked. Knockdown of ATG7 and Beclin1 were verified by immunoblot and Cas9 antibody. Knockdown cells were maintained in DMEM with 1 g/ml of puromycin. The lentiCRISPR v2, a gift from Dr. Feng Zhang (Addgene plasmid # 52961), was used with the following oligonucleotide sequences.
| GENE | REFERENCE SEQUENCE | OLIGONUCLEOTIDE SEQUENCE |
|---|---|---|
| ATG7 | NM_006395.2 | Forward 1: 5′ CACCGAATCAAGTATGATGAGAACA3′ Reverse 1: 5′ AAACTGTTCTCATCATACTTGATTC3′ Forward 2: 5′ AAACTGTTCTCATCATACTTGATTC3′ Reverse 2: 5′ AAACGGACGACTCACAGTGCACTGC3′ |
| Beclin1 | NM_003766.3 | Forward 1: 5′ AAACGGACGACTCACAGTGCACTGC3′ Reverse 1: 5′ AAACGCATGGTGCTGTTGTTGGACC3′ Forward 2: 5′ CACCGGCCAACAGCTTCACTCTGAT3′ Reverse 2: 5′ AAACATCAGAGTGAAGCTGTTGGCC3′ |
4.9. Quantitative reverse transcription-PCR measurements of transcript levels
For quantitative measurements of mRNA levels, cDNA synthesis was performed with ABI high-capacity cDNA reverse transcription kit according to manufacturer’s instructions using 1 g of mRNA from MCF7 Nrf2shRNA, MCF7 Keap1shRNA, and MCF7 LucshRNA cells. The cDNA was then diluted 5-fold in water and used for real time PCR using Taqman primer/probe mix and ABI gene expression mastermix. Primer probes used for measured genes were: Hs00168547_m1 (NQO1), Hs00975961_g1 (Nrf2), and Hs00202227_m1 (Keap1). Statistical significance was determined by unpaired t−test in GraphPad Prism4 (GraphPad Software, Inc., La Jolla, CA).
4.10. Measurements of Reactive Oxygen Species
MCF7 were plated at a density of 7000–10,000 cells per well in a 96 well plate and after 24 h incubation, media was changed to complete DMEM or DMEM glucose-free media. At 12 and 24 h time points, cells were aspirated and washed with PBS. Cell-permeant 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), dissolved in ethanol and diluted to a final concentration of 20uM in PBS, was added to the wells and incubated for 30 min. Cells were then aspirated, and 10%FBS in PBS was added and incubated for 30 min. Wells were aspirated and fluorescence was measured using Filter Max F5 (Multi-mode Microplate Reader, Molecular Devices) and Soft Max Pro 6.3 software at an excitation of 495 and emission of 520. Statistical significance was determined by unpaired t-test in GraphPad Prism4.
References
- [1].Rabinowitz JD, White E, Autophagy and metabolism, Science 330 (6009) (2010) 1344–1348, 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Moruno F, Perez-Jimenez E, Knecht E, Regulation of autophagy by glucose in Mammalian cells, Cells 1 (3) (2012) 372–395, 10.3390/cells1030372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Xi H, Kurtoglu M, Liu H, Wangpaichitr M, You M, Liu X, Savaraj N, Lampidis TJ, 2-Deoxy-D-glucose activates autophagy via endoplasmic reticulum stress rather than ATP depletion, Cancer Chemother. Pharmacol 67 (4) (2011) 899–910, 10.1007/s00280-010-1391-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Altman BJ, Jacobs SR, Mason EF, Michalek RD, MacIntyre AN, Coloff JL, Ilkayeva O, Jia W, He YW, Rathmell JC, Autophagy is essential to suppress cell stress and to allow BCR-Abl-mediated leukemogenesis, Oncogene 30 (16) (2011) 1855–1867, 10.1038/onc.2010.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jeon JY, Kim SW, Park KC, Yun M, The bifunctional autophagic flux by 2-deoxyglucose to control survival or growth of prostate cancer cells, BMC Cancer 15 (2015) 623, 10.1186/s12885-015-1640-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ramirez-Peinado S, Leon-Annicchiarico CL, Galindo-Moreno J, Iurlaro R, Caro-Maldonado A, Prehn JH, Ryan KM, Muñoz-Pinedo C, Glucose-starved cells do not engage in prosurvival autophagy, J. Biol. Chem 288 (42) (2013) 30387–30398, 10.1074/jbc.M113.490581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T, p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy, J. Biol. Chem 282 (33) (2007) 24131–24145. [DOI] [PubMed] [Google Scholar]
- [8].Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and Interpretation of Assays for Monitoring Autophagy (3rd edition). Autophagy, 2016, 12(1), pp. 1–222. doi: 10.1080/15548627.2015, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. , The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1, Nat. Cell Biol 12 (3) (2010) 213–223, 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- [10].Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD, A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62, Mol. Cell Biol 30 (13) (2010) 3275–3285, 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jiang T, Harder B, Rojo M de la Vega, P.K. Wong, E. Chapman, D.D. Zhang, P62 links autophagy and Nrf2 signaling, Free Radic. Biol. Med 88 (Pt B) (2015) 199–204, 10.1016/j.freeradbiomed.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ma Q, Role of nrf2 in oxidative stress and toxicity, Annu. Rev. Pharmacol. Toxicol 53 (53) (2013) 401–426, 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kensler TW, Wakabayashi N, Biswal S, Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway, Annu. Rev. Pharmacol. Toxicol 47 (2007) 89–116. [DOI] [PubMed] [Google Scholar]
- [14].Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T, et al. , Inhibition of macro-autophagy triggers apoptosis, Mol. Cell Biol 25 (2005) 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim SE, Park HJ, Jeong HK, Kim MJ, Kim M, Bae ON, Baek SH, Autophagy sustains the survival of human pancreatic cancer PANC-1 cells under extreme nutrient deprivation conditions, Biochem. Biophys. Res. Commun 463 (3) (2015) 205–210, 10.1016/j.bbrc.2015.05.022. [DOI] [PubMed] [Google Scholar]
- [16].Wise DR, Thompson CB, Glutamine addiction: a new therapeutic target in cancer, Trends Biochem. Sci 35 (8) (2010) 427–433, 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hensley CT, Wasti AT, DeBerardinis RJ, Glutamine and cancer: cell biology, physiology, and clinical opportunities, J. Clin. Investig 123 (9) (2013) 3678–3684, 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].White E, The role for autophagy in cancer, J. Clin. Investig 125 (1) (2015) 42–46, 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. , Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis, Genes Dev 25 (5) (2011) 460–470, 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J, Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation, Mol. Biol. Cell 22 (2) (2011) 165–178, 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, et al. , Pancreatic cancers require autophagy for tumor growth, Genes Dev 25 (7) (2011) 717–729, 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lock R, Kenific CM, Leidal AM, Salas E, Debnath J, Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion, Cancer Discov 4 (4) (2014) 466–479, 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nguyen T, Nioi P, Pickett CB, The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress, J. Biol. Chem 284 (20) (2009) 13291–13295, 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS, Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment, Nature 461 (7260) (2009) 109–113, 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Singh A, Venkannagari S, Oh KH, Zhang YQ, Rohde JM, Liu L, Nimmagadda S, Sudini K, Brimacombe KR, Gajghate S, et al. , Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors, ACS Chem. Biol 11 (11) (2016) 3214–3225, 10.1021/acschembio.6b00651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Singh A, Wu H, Zhang P, Happel C, Ma J, Biswal S, Expression of ABCG2 (BCRP) is regulated by Nrf2 in cancer cells that confers side population and chemoresistance phenotype, Mol. Cancer Ther 9 (8) (2010) 2365–2376, 10.1158/1535-7163.MCT-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]