Abstract
Tripartite motif (TRIM) proteins are a versatile family of ubiquitin E3 ligases involved in a multitude of cellular processes. Studies in recent years have demonstrated that many TRIM proteins play central roles in the host defense against viral infection. While some TRIM proteins directly antagonize distinct steps in the viral life cycle, others regulate signal transduction pathways induced by innate immune sensors, thereby modulating antiviral cytokine responses. Furthermore, TRIM proteins have been implicated in virus-induced autophagy and autophagy-mediated viral clearance. Given the important role of TRIM proteins in antiviral restriction, it is not surprising that several viruses have evolved effective maneuvers to neutralize the antiviral action of specific TRIM proteins. Here, we describe the major antiviral mechanisms of TRIM proteins as well as viral strategies to escape TRIM-mediated host immunity.
Keywords: TRIM protein, antiviral restriction, innate immunity, virus infection, interferon
INTRODUCTION
Tripartite motif (TRIM) proteins are a family of >80 distinct members in humans that have a conserved structural arrangement of three N-terminal domains: RING–B-box–coiled-coil (1). TRIM proteins can be found in most eukaryotes, with the total number of TRIM genes correlating with the evolutionary degree of the organism (e.g., ~64 members in mice, ~20 in worms, <10 in flies), indicating extensive evolution (2).
The really interesting new gene (RING) domain, which is present in most TRIM proteins, contains a zinc finger motif and is known to possess E3 ubiquitin ligase activity (3). TRIM proteins are able to conjugate a wide range of different polyubiquitin linkage types, which determine the fate of the modified target protein. For example, whereas K48-linked ubiquitination induces proteasomal degradation of the substrate, K63-linked polyubiquitin often modulates the substrate’s activity, its subcellular localization, or its ability to interact with other proteins (4). In addition to attaching ubiquitin to their targets, several TRIM proteins catalyze the conjugation of ubiquitin-like (UBL) proteins, such as ISG15 or SUMO, demonstrating enzymatic flexibility of the RING domain. Activity of the RING domain is crucial for the antiviral activity of most TRIM proteins because it allows them to modulate the function of a wide variety of substrates—both host and virus derived—via ubiquitin or UBL modifications. A few TRIM proteins, however, lack a RING domain, and in at least one TRIM protein, TRIM20, the N-terminal RING domain is replaced by a PYRIN domain (5). TRIM proteins generally contain one or two B-box domains that also contain zinc-binding motifs similar to the RING domain (6). Whereas the functional role of the B-box domains remains enigmatic, they have been shown to modulate higher-order self-assembly, E3 ligase activity, and/or protein interactions of certain TRIM proteins. The coiled-coil domain (CCD) directly mediates homomeric self-association of TRIM proteins and in some instances also heteromeric assemblies, which for several TRIM proteins are crucial for their antiviral activities (7).
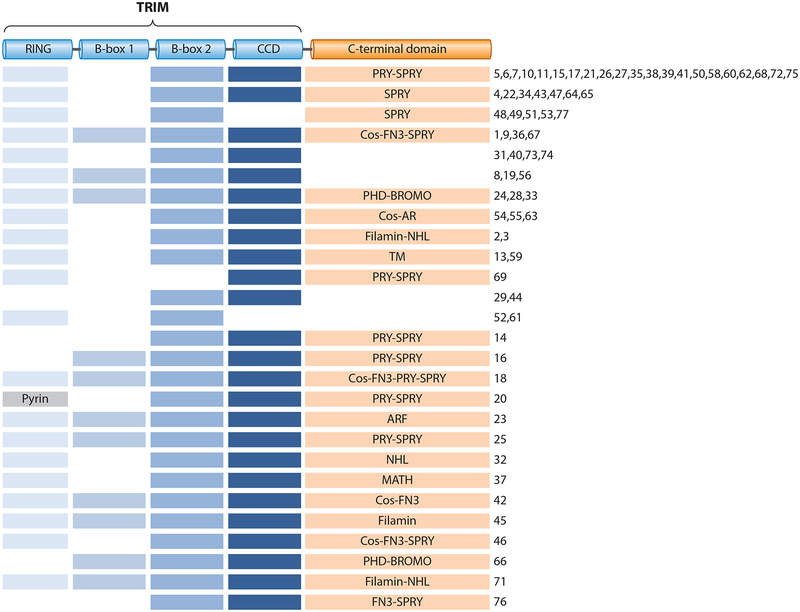
TRIM proteins can be classified into several subgroups based on their overall domain structure and distinct C-terminal domains (Figure 1). The C-terminal domain usually acts as a protein-protein interaction scaffold but can also have enzymatic activity or bind nucleic acids (2). Most commonly found are the ~61 amino acid long PRY domain and the ~140 amino acid long SPRY domain, either in combination (PRY-SPRY) or individually. SPRY domains are found in other human protein families as well and are evolutionarily conserved in mammals, plants, and fungi. In contrast, the PRY-SPRY fusion domain (also referred to as B30.2) is found only in vertebrates (8). The SPRY and PRY-SPRY domains usually serve as protein-protein interaction platforms and exhibit remarkable protein-binding specificities. In addition, the PRY-SPRY domains can have Fc receptor–like properties, as seen with TRIM21 (9), or can mediate binding to RNA molecules (10, 11). The C-terminal subgroup one signature (Cos) domain usually mediates binding to the cytoskeleton, in particular the microtubule network (12). Furthermore, several TRIM proteins possess NHL repeats, which are composed of a six-bladed β propeller and are generally thought to mediate protein-protein interactions (13). Other C-terminal domains found in TRIM family members are fibronectin type 3 (FN3) domains, which can contain binding sites for DNA and heparin; plant homeodomains (PHDs), which bind nuclear substrates; bromodomains, which recognize acetylated lysine residues; meprin and tumor necrosis factor receptor–associated factor homology (MATH) domains, which can mediate self-association; and filamin domains, which have actin cytoskeleton–binding capability (2). Unique among the TRIM proteins is TRIM23, which is the only known family member that encodes an ADP-ribosylation factor (ARF) domain, harboring GTPase activity (14).
Figure 1.
Domain structure of tripartite motif (TRIM) proteins. The TRIM, consisting of a really interesting new gene (RING) domain, a B-box 1 and/or B-box 2 domain, and a coiled-coil domain (CCD), is followed by distinct C-terminal domains: SPRY-associated domain (PRY), SPIa and the ryanodine receptor domain (SPRY), C-terminal subgroup one signature domain (Cos), fibronectin type 3 domain (FN3), plant homeodomain (PHD), bromodomain (BROMO), acid-rich region (AR), filamin domain, NHL repeats (NHL), transmembrane domain (TM), ADP-ribosylation factor domain (ARF), and meprin and tumor necrosis factor receptor–associated factor homology domain (MATH). Numbers indicate individual TRIM proteins.
TRIM proteins have been implicated in many cellular processes, including signal transduction, oncogenesis, gene regulation, regulation of cell death, and antimicrobial immunity. In this review, we discuss the mechanisms that are used by TRIM proteins to mediate an antiviral host response in mammalian cells. The antiviral strategies employed by TRIM proteins fall into three broad categories: modulation of innate immune sensing or signaling, direct restriction of viruses, and regulation of autophagy-mediated antiviral defenses. Finally, we detail how viral pathogens antagonize the antiviral function of specific TRIM proteins to evade host immunity.
TRIM PROTEINS MODULATING INNATE SIGNALING
The innate immune system plays a vital role in sensing incoming viruses and limiting their replication, as well as inducing a tailored adaptive immune response that ultimately controls or eradicates the infection. This process starts with the recognition of conserved pathogen-associated molecular patterns (PAMPs) by pattern-recognition receptors (PRRs) (15). Through a series of highly regulated phosphorylation and ubiquitination events of adaptor molecules and kinases, stimulation of PRRs results in the activation and nuclear translocation of the transcription factors IRF3/7 and NF-κB, which in turn induce transcriptional upregulation of type I interferons (IFNs; predominantly IFNα/β) and proinflammatory cytokines (15). Type I IFNs are of paramount importance in the establishment of an antiviral state by inducing the expression of a plethora of IFN-stimulated genes (ISGs) that can have direct antiviral activity or alter cellular processes to establish an environment hostile to viral replication (16). Over the past decade, TRIM proteins have emerged as important positive and negative regulators of PRR signaling pathways. In this section, we detail the role of TRIM proteins in modulating four major innate signaling pathways, namely, the ones initiated by RIG-I-like receptors (RLRs), the DNA sensor cGAS, Toll-like receptors (TLRs), and the IFNα/β receptor (IFNAR) (Figure 2).
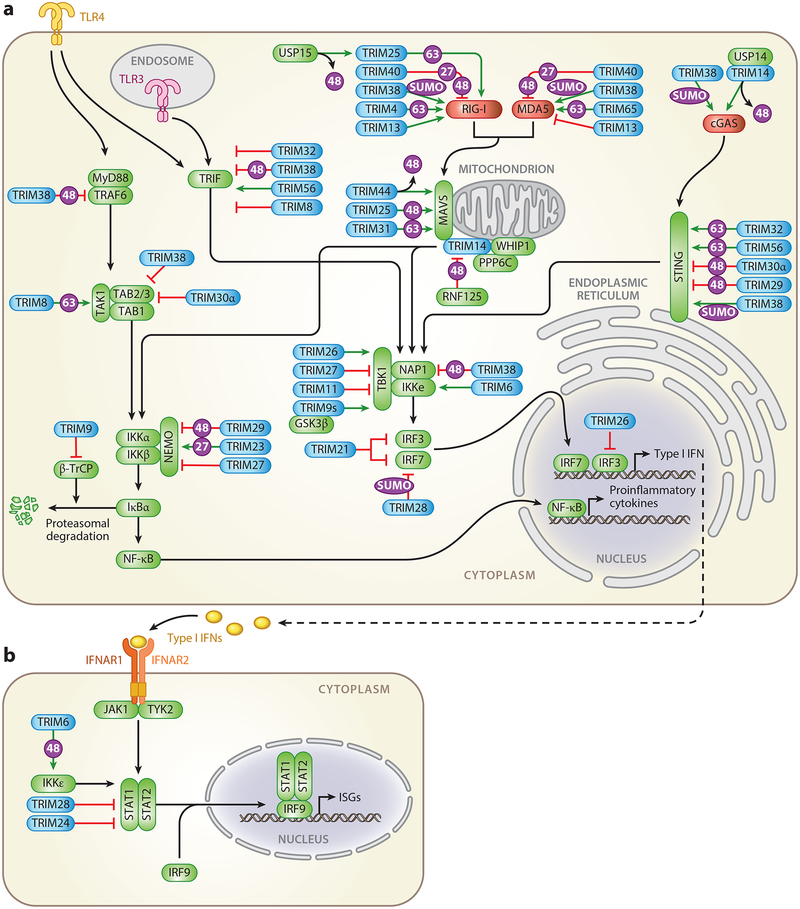
Figure 2.
TRIM-mediated regulation of innate immune signaling pathways. Overview of the TRIM proteins (blue) that positively (green arrows) or negatively (red lines) regulate (a) PRR-mediated induction of type I IFNs and proinflammatory cytokines or (b) IFNAR-induced expression of ISGs. Intracellular receptors are illustrated in red. Signaling intermediates or transcription factors are illustrated in green. Abbreviations: cGAS, cyclic GMP-AMP synthase; 27, K27-linked polyubiquitination; 48, K48-linked polyubiquitination; 63, K63-linked polyubiquitination; IFN, interferon; IFNAR, IFNα/β receptor; IRF, interferon regulatory factor; ISG, interferon-stimulated gene; MAVS, mitochondrial antiviral signaling protein; MDA5, melanoma differentiation-associated protein 5; PRR, pattern-recognition receptor; RIG-I, retinoic acid-inducible gene-I; STAT, signal transducer and activator of transcription; STING, stimulator of interferon genes; SUMO, SUMOylation; TLR, Toll-like receptor; TRIM, tripartite motif protein; USP, ubiquitin-specific peptidase, WHIP, Werner helicase interacting protein 1.
TRIM Proteins Regulating RLR Signaling
DExD/H-box RNA helicases of the RLR family are essential sensors of RNA virus infection that are expressed in most cell types and transcriptionally upregulated following viral infection (17). While the prototypic sensor retinoic acid-inducible gene-I (RIG-I) recognizes 5’− triphosphorylated viral RNA with short dsRNA stretches, the closely related RLR member melanoma differentiation-associated protein 5 (MDA5) detects longer dsRNA or viral RNA aggregates. Several studies have demonstrated the importance of RIG-I and MDA5 in innate sensing of a wide spectrum of RNA viruses as well as some DNA viruses (18, 19). RIG-I and MDA5 contain a central helicase domain and a C-terminal domain (CTD), which are both required for RNA binding, as well as two N-terminal caspase activation and recruitment domains (CARDs) essential for downstream signaling (15). Following activation, RIG-I and MDA5 undergo conformational changes and are recruited to their common adaptor mitochondrial antiviral signaling protein (MAVS). MAVS in turn recruits the IKK-related kinases IKKε and TBK1 to activate IRF3/7, as well as the canonical IKKα/β/γ complex, to promote degradation of the NF-κB inhibitor IκBα, leading to NF-κB activation (15). The activity of RIG-I and MDA5 is strictly regulated by posttranslational modifications to allow rapid RLR activation yet prevent excessive immune activation (20). Several TRIM proteins have emerged as key regulators of these modifications and are imperative in the tight control of the RLR-triggered antiviral response (Figure 2a).
TRIM25 was the first immunomodulatory TRIM protein identified; it controls RIG-I antiviral activity by decorating its CARDs with K63-linked polyubiquitin (21). This nondegradative type of ubiquitination induces RIG-I oligomerization and its recruitment to MAVS to trigger downstream antiviral gene expression. In addition, RIG-I can be activated by noncovalently attached K63-linked ubiquitin chains synthesized by TRIM25 (22). TRIM25 protein stability is regulated by the deubiquitinating enzyme USP15, which reverses K48-linked ubiquitination and subsequent degradation of TRIM25 (23). Besides TRIM25, TRIM4 (as well as the E3 ligases Riplet and MEX3C) reportedly modifies RIG-I with K63-polyubiquitin to facilitate its activation and antiviral response (24). Whether these E3 ligases play redundant roles in RIG-I regulation, or whether they have cell type–specific or tissue-specific roles, remains to be determined.
TRIM65 promotes K63-linked ubiquitination of the MDA5 helicase domain, thereby enhancing IRF3 activation and IFN production in response to MDA5 agonists or encephalomyocarditis virus (EMCV) infection (25). TRIM65−deficient mice were more susceptible to EMCV infection and showed compromised IFN production. Furthermore, TRIM13 was shown to suppress MDA5-mediated IFN production (26). TRIM13−/− mice and cells showed increased cytokine production upon EMCV infection compared to wild-type mice and cells (26). In contrast, TRIM13 appears to positively regulate RIG-I signaling through an unknown mechanism (26, 27). TRIM40 modifies both RIG-I and MDA5 with K27- and K48-linked polyubiquitin, leading to proteasomal degradation of the sensors and inhibition of antiviral signaling (28). TRIM40 deficiency enhanced RNA virus–induced IFNβ production in mice and improved survival following vesicular stomatitis virus challenge (28). TRIM38 was recently identified as an E3 SUMO ligase for RIG-I and MDA5. SUMOylation of the sensors prevents their K48-polyubiquitin-dependent degradation, improving RLR stability and thereby potentiating antiviral responses (29). Furthermore, SUMOylation of specific lysine residues in the CARDs of RIG-I and MDA5 was shown to be essential for their subsequent dephosphorylation by PP1 (29). Dephosphorylation by PP1 is required for activation of both RIG-I and MDA5 (30), which in the case of RIG-I licenses TRIM25-mediated ubiquitination. At the later stages of infection, deSUMOylation of RIG-I and MDA5 by SENP2 aids in terminating the antiviral response (29).
TRIM14, which lacks an E3 ubiquitin ligase domain, forms a complex with Werner helicase interacting protein 1 (WHIP) and the phosphatase PPP6C that is constitutively associated with MAVS at the outer mitochondrial membrane (31). Upon RIG-I activation, WHIP binds to and stabilizes dsRNA-bound, K63-ubiquitinated RIG-I, promoting its translocation to the mitochondria. Dephosphorylation of RIG-I by PPP6C further amplifies RIG-I-mediated antiviral signaling (31). TRIM14 has been reported to recruit the adaptor protein IKKγ/NEMO to the MAVS signalosome, bridging RIG-I to the downstream transcription factors IRF3 and NF-κB (32). The stability of TRIM14 is regulated by the ubiquitin ligase RNF125, which induces its proteasomal degradation (33). Recently, TRIM31 was reported to catalyze K63-linked polyubiquitination of MAVS at K10, K311, and K461, which promoted MAVS aggregate formation and type I IFN induction upon RNA virus infection (34). TRIM44 counteracts the K48-linked polyubiquitination and proteasomal degradation of MAVS, potentiating antiviral responses (35). TRIM44 contains an atypical ubiquitin-binding zinc finger UBP domain and may act as a deubiquitinase rather than an E3 ligase. Another study suggested that K48-linked polyubiquitination and destabilization of the larger MAVS isoform by TRIM25 led to the release of downstream signaling components, thereby enhancing IFN production (36).
Several TRIM proteins regulate signaling by RLRs (and also other sensors) by targeting downstream kinases or IRFs. For example, the short isoform of TRIM9, TRIM9s, promotes TBK1 phosphorylation and IRF3 activation by bridging TBK1 with GSK3β, a protein that aids in the oligomerization of TBK1 (37). At the same time, TRIM9s suppresses NF-κB activation and proinflammatory cytokine production upon vesicular stomatitis virus or herpes simplex virus 1 (HSV-1) infection, essentially biasing the innate response to type I IFN production (37). Interestingly, the longer isoform of TRIM9 (TRIM9l) was previously shown to inhibit NF-κB responses by interfering with β-TrCP-mediated IκBα degradation (38). TRIM11 interacts with TBK1 and inhibits IRF3-mediated IFNβ production (39). TRIM26 reportedly also interacts with TBK1 and restricts RNA virus infection by bridging TBK1 and NEMO to facilitate downstream signaling (40). However, TRIM26 was also shown to negatively regulate antiviral IFN production by promoting K48-linked polyubiquitination and proteasomal degradation of phosphorylated, nuclear IRF3 (41). TRIM28 specifically targets IRF7 for inhibitory SUMOylation (42), while TRIM23 has been suggested to modify NEMO with K27-linked polyubiquitin, thereby enhancing innate signaling by RIG-I, MDA5, and TLR3 (43). Finally, a comprehensive cDNA screen by Versteeg et al. (27) showed that roughly half of the 75 tested TRIM family proteins regulated RIG-I signaling. Another overexpression screen identified 16 TRIMs that induced NF-κB and/or AP1 activation (44), strengthening the concept that TRIM proteins are key regulators of innate and proinflammatory signal transduction.
TRIM Proteins Modulating cGAS-STING Signaling
In recent years, various cytosolic viral DNA sensors have been identified, most prominently cyclic GMP-AMP synthase (cGAS) (45). Many studies have established that cGAS is crucial for sensing DNA viruses, and more recently, cGAS has been shown to act antivirally against retroviruses and several positive-strand RNA viruses. Upon activation by DNA binding, cGAS produces the second messenger cGAMP, which activates the stimulator of interferon genes (STING) to induce transcriptional upregulation of type I IFNs (46). Although the characterization of posttranslational modifications that regulate the cGAS-STING pathway is still in its infancy (20), recent studies have demonstrated that several TRIM proteins modulate cGAS-STING-dependent antiviral signaling via regulatory modifications (Figure 2a).
TRIM14 was shown to be upregulated upon viral infection and to recruit the deubiquitinating enzyme USP14 to revert degradative ubiquitination of cGAS, improving its stability and enhancing the antiviral response against HSV-1 (47). Analogous to its role in RIG-I and MDA5 SUMOylation, TRIM38 was found to induce SUMOylation of cGAS and STING (48). TRIM38 catalyzed multiple SUMOylation events of cGAS to prevent its K48-linked polyubiquitination and proteasomal degradation. Similarly, SUMOylation of STING by TRIM38 prevented STING degradation by the chaperone-mediated autophagy pathway (48). Together, these modifications ensure robust IRF3 activation and production of antiviral effectors, and TRIM38 deficiency severely impaired the antiviral response to HSV-1 in vivo (48). TRIM56 and TRIM32 were found to induce K63-linked polyubiquitination of STING upon cytosolic DNA stimulation to facilitate the induction of an antiviral response (49, 50). However, more recent studies suggested that instead of directly ubiquitinating STING, TRIM56 and TRIM32 may synthesize unanchored polyubiquitin chains that activate NEMO and, together with IKKβ and TBK1, ultimately induce IRF3 and NF-κB activation (51, 52). TRIM29, which is specifically expressed in lung alveolar macrophages and airway epithelial cells, was reported to negatively regulate innate immune responses to DNA viruses by mediating degradative ubiquitination of STING (53). Similarly, mouse-specific TRIM30α promotes degradation of STING through K48-linked polyubiquitination (54).
TRIM Proteins Modulating TLR Signaling
TLRs sense pathogen-derived nucleic acid, lipid, or protein components at the cell surface or in endosomal compartments (55). Major TLRs involved in virus recognition are endosomally localized TLR3, TLR7, TLR8, and TLR9. Upon ligand binding, TLRs dimerize and activate signaling cascades that ultimately lead to NF-κB- and IRF-mediated induction of proinflammatory cytokines. All TLRs except TLR3 signal through the adaptor molecule MyD88, which recruits the kinases IRAK1/4 and E3 ubiquitin ligase TRAF6. Autoubiquitination of TRAF6 is recognized by TAB2, which activates the kinase TAK1, ultimately inducing IKKα/β/γ and NF-κB activation (55). Alternatively, TLR3 (and also TLR4) can activate TRIF-mediated signaling, which results in activation of IRF3 and IRF7 via TRAF3 and TBK1/IKKε (55). Several steps in TLR signaling pathways are regulated by TRIM proteins (Figure 2a).
TRIM38 negatively affects TLR signaling through destabilization of several signaling intermediates required for TBK1/IKKε and IRF3/7 activation, including TRAF6 (56), TRIF (57), TAB2/3 (58), and NAP1 (59). Similarly, TLR signaling is suppressed by murine TRIM30α, which is induced upon TLR stimulation and promotes lysosomal degradation of TAB2/3 (60). TRIM32 negatively regulates TLR3/4 responses by inducing autophagy-mediated degradation of TRIF (61). Conversely, TRIM56 was reported to interact with TRIF and play a positive role in TLR3-mediated IFN/ISG production (62), while TRIM62 was shown to promote activation of the TRIF branch of TLR4 signaling (44). TRIM21 functions as a negative feedback modulator by promoting ubiquitination and proteasomal degradation of IRF3/7 following viral TLR stimulation (63, 64). Upregulation of TRIM27 in response to viral infection was shown to inhibit antiviral responses by targeting the IKK complex and TBK1 (65–67). TRIM29 acts as a negative regulator of proinflammatory cytokine production in alveolar macrophages by inducing NEMO degradation (68). Finally, TNFα signaling and IL-1β signaling, which activate the same MyD88-dependent pathway as TLRs, are potentiated by TRIM8 through K63-linked ubiquitination of TAK1 (69), while TRIM8 suppresses TRIF-mediated responses by disrupting the TRIF-TBK1 interaction (70).
TRIM-Mediated Regulation of IFNAR Signaling
Secreted type I IFNs induce signal transduction pathways in an autocrine and paracrine fashion through binding to the IFNα/β receptor, a heterodimer of IFNAR1/2 that is expressed on virtually all nucleated cells (71). Receptor ligation activates the kinases JAK1 and TYK2 and induces phosphorylation of signal transducer and activator of transcription (STAT)1 and STAT2, which together with IRF9 form the interferon-stimulated gene factor 3 (ISGF3) complex, which translocates into the nucleus and induces transcription of an extensive set of antiviral ISGs (71). TRIM6, TRIM24, and TRIM28 have been implicated in regulation of IFNAR signal transduction (Figure 2b). Besides its role in the IFN induction pathway, IKKε is also required for optimal IFNAR signaling and ISG production (72). TRIM6 was found to interact with IKKε and synthesize atypical unanchored K48-linked ubiquitin chains that are vital for optimal IKKε activation and subsequent STAT1 phosphorylation (73). In contrast, TRIM24 and TRIM28 negatively regulate IFNAR signaling. Whereas TRIM24 suppresses transcription of STAT1 by binding to the STAT1 promoter (74), TRIM28 associates with STAT1 and inhibits STAT1-mediated IRF1 gene expression (75).
DIRECT ANTIVIRAL RESTRICTION BY TRIM PROTEINS
Retroviral Restriction by TRIM Proteins
Several TRIM proteins interfere with retroviral infection by directly inhibiting various stages of the infectious cycle (Figure 3a). TRIM5α, which is probably the most well-characterized antiviral restriction factor, interacts with the intact viral capsid lattice and forms a complementary lattice that induces premature virion disassembly, blocking viral infection (76). This process also releases viral PAMPs into the cytosol that may be detected by cytoplasmic innate sensors that in turn promote antiviral responses (77). Additionally, TRIM5α itself reportedly functions as an immune sensor that, upon retroviral capsid binding, catalyzes the synthesis of unanchored K63-linked ubiquitin chains that activate the TAK1-containing complex and AP1- and NF-κB-mediated signaling (78). TRIM5α-mediated retroviral restriction is known to be highly species specific (76, 79, 80), but a recent study nuanced the view of TRIM5α’s species specificity by showing that human TRIM5α is able to restrict human immunodeficiency virus type 1 (HIV-1) infection in Langerhans cells, a specific dendritic cell subset that represents a primary target of HIV-1 infection (81). Binding of HIV-1 to the surface C-type lectin receptor Langerin, which is uniquely expressed on Langerhans cells, routes HIV-1 into TRIM5α-dependent autophagic degradation and prevents these cells from becoming infected (81). In contrast, HIV-1 entry mediated by the DC-SIGN receptor expressed on conventional dendritic cell subsets was shown to bypass TRIM5α-mediated degradation (81).
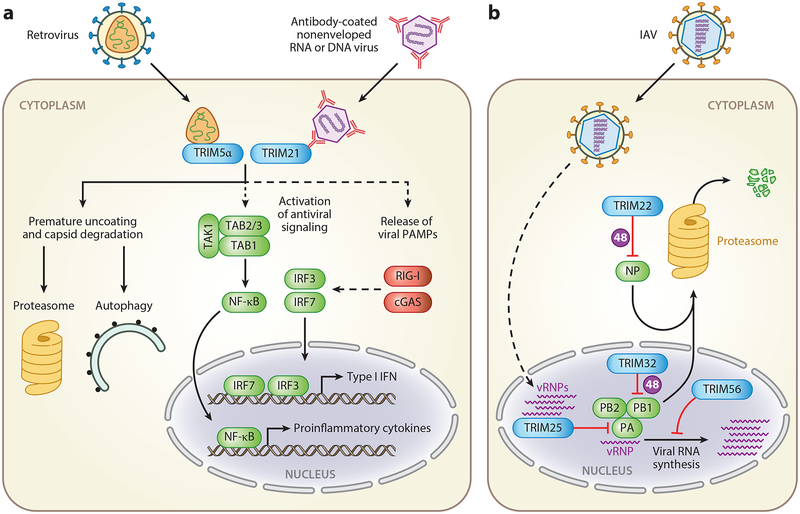
Figure 3.
Direct antiviral effects of TRIM proteins. (a) TRIM5α restricts retroviral infection by directly interacting with the viral capsid and inducing premature uncoating and capsid disassembly. This process may release viral PAMPs that can be sensed by cytosolic PRRs. Both proteasomal and autophagic degradative mechanisms have been implicated in TRIM5α-mediated virus restriction. In addition, TRIM5α catalyzes noncovalent K63-linked ubiquitin chains that activate the TAK1 signaling complex and thereby activate NF-κB and AP1. TRIM21 is a cytosolic sensor of antibody-opsonized, nonenveloped RNA and DNA viruses. Proposed antiviral mechanisms for TRIM21 include proteasomal degradation of incoming viral particles; activation of signaling pathways leading to NF-κB, AP1, and IRF activation; and release of viral PAMPs for detection by RIG-I or cGAS and potentially other intracellular PRRs. (b) Several TRIM proteins directly interfere with IAV replication. TRIM22 induces proteasomal degradation of the viral nucleoprotein NP. In the nucleus, TRIM32 induces proteasomal degradation of the viral polymerase subunit PB1, while TRIM25 interacts with vRNPs to prevent the viral polymerase from accessing the viral RNA template. TRIM56 blocks IAV RNA synthesis through an unknown mechanism. Solid lines indicate direct or well-established interactions. Dashed lines indicate indirect correlations. Abbreviations: 48, K48-linked polyubiquitination; cGAS, cyclic GMP-AMP synthase; IAV, influenza A virus; IFN, interferon; PAMP, pathogen-associated molecular pattern; PRR, pattern-recognition receptor; TRIM, tripartite motif protein; vRNP, viral ribonucleoprotein.
TRIM22, which is closely related to TRIM5, suppresses retroviral transcription by regulating binding of the transcription factor Sp1 to the long terminal repeats and by interfering with trafficking of the viral Gag protein to the plasma membrane (82–84). Enhanced TRIM22 expression in peripheral blood mononuclear cells of HIV-1-infected patients was associated with lower viral load (85). In addition to its role in retroviral restriction, several reports have described an antiviral activity of TRIM22 against influenza A virus (IAV) (86), hepatitis B virus (HBV) (87), hepatitis C virus (HCV) (88, 89), and EMCV (90). Several other TRIM proteins, including TRIM11 (91), TRIM28 (92), and TRIM37 (93), directly or indirectly interfere with retroviral infections. Moreover, a screening of 36 human and 19 mouse TRIM proteins for antiretroviral activity revealed ~20 TRIMs that interfered with either the early or the late stage of retroviral replication (94).
TRIM21, an Intracellular Antibody Receptor
TRIM21 is a high-affinity intracellular antibody receptor that binds the Fc portion of immunoglobulin (Ig) molecules through its PRY-SPRY domain, thereby contributing to the restriction of various nonenveloped viruses such as adenoviruses and rhinoviruses (95–98) (Figure 3a). The important role of TRIM21 in the antiviral response was illustrated by the fact that mice lacking TRIM21 succumbed to an otherwise nonlethal dose of mouse adenovirus 1(95). As for TRIM5α, multiple mechanisms have been proposed for how TRIM21 restricts viral infection. Following viral entry, TRIM21 neutralizes infection by targeting viral particles for degradation by the proteasome in conjunction with the ATPase p97 (99, 100). Additionally, TRIM21 acts as an innate immune sensor that, upon recognition of opsonized virions, activates NF-κB, AP1, and IRF signaling through the generation of unanchored K63-linked polyubiquitin (101, 102). Moreover, a delayed and more specific induction of immune pathways is achieved by TRIM21-mediated premature uncoating of the virus particle and release of viral PAMPs for exposure to RIG-I, cGAS, and potentially other intracellular sensors (102).
TRIM19/PML
TRIM19, or promyelocytic leukemia (PML) protein, is a key component of PML nuclear bodies, which are dynamic protein aggregates containing many proteins including PML, hDaxx, Sp100, and ATRX. They have been implicated in a multitude of cellular processes, such as cell cycle regulation, apoptosis, senescence, the DNA damage response, and intrinsic resistance to viral infection (103). PML nuclear bodies are known to restrict many DNA viruses, including herpesviruses, adenoviruses, papillomaviruses, and parvoviruses, as well as RNA viruses such as IAV, vesicular stomatitis virus, and HIV-1 (104–106). The mechanisms of PML nuclear body–mediated virus restriction have been most extensively characterized for herpesviral infections and include engulfment and epigenetic silencing of incoming viral genomes, as observed for HSV-1 and human cytomegalovirus, or entrapment of newly synthesized nucleocapsids, as in the case of varicella zoster virus (107, 108). Besides conferring these direct antiviral functions, PML is also emerging as a positive regulator of IFN induction during retroviral and herpesviral infections (reviewed in 109).
TRIM25 and ZAP
Recently, TRIM25 was identified as an important regulator of zinc finger antiviral protein (ZAP) (110, 111). Originally described to suppress retroviral infections, ZAP has now been shown to confer antiviral activity against several other viruses, including alphaviruses, filoviruses, and HBV (112). Although the exact antiviral mechanism employed by ZAP remains somewhat elusive, ZAP was shown to recruit the exosome to target viral RNA for degradation, inhibit translation of incoming viral genomes, and promote RIG-I-mediated IFNβ induction (reviewed in 112). Two recent studies have identified that TRIM25 acts as a key regulator of ZAP’s antiviral activity by mediating both K48- and K63-linked polyubiquitination of ZAP (110, 111). Furthermore, RNA binding by TRIM25 was suggested to be required for licensing TRIM25-mediated ZAP ubiquitination (11).
Other TRIM Proteins Directly Restricting Viral Infections
Several other TRIM proteins were shown to directly target viral components to restrict replication (Figure 3b). For example, TRIM32 targets the polymerase subunit PB1 of several IAV strains for ubiquitination and subsequent proteasomal degradation (113). TRIM25 was recently suggested to interact with viral ribonucleoproteins of IAV in the nucleus and to inhibit mRNA chain elongation by preventing access of the viral polymerase to the viral RNA template (114). Furthermore, IAV infection is restricted by TRIM56, which blocks IAV RNA synthesis (115), and TRIM22, which induces proteasomal degradation of the viral nucleoprotein NP (86).
Other viruses are also directly targeted by TRIM proteins. TRIM52 targets the NS2A protein of Japanese encephalitis virus for proteasomal degradation (116). TRIM56 was reported to restrict infection by the flaviviruses bovine viral diarrhea virus, yellow fever virus, and dengue virus, as well as human coronavirus OC43 and HIV-1 (117–119). TRIM14 suppresses HCV replication by targeting the NS5a protein for degradation (120). Moreover, a cDNA screen identified multiple TRIM proteins that interfered with HBV infection (121), although the molecular mechanisms of TRIM-mediated HBV restriction remain to be determined.
TRIM PROTEINS REGULATING VIRUS-INDUCED AUTOPHAGY
A major cellular homeostatic process mediating destruction of damaged organelles, surplus proteins, and even viral cargo is autophagy. During autophagy, cytoplasmic contents are engulfed by double-layered membranes called autophagosomes and degraded upon fusion with the lysosome. Initially, autophagy was described as a nonspecific cellular response toward nutrient depletion, but it has become evident that autophagy also mediates degradation of cargos via specific receptors such as p62 or NDP52 (122). Autophagy is implicated in diverse physiological functions, including inflammation, cancer, and antimicrobial activities (123). In regards to viral infection, autophagy can have either a proviral or antiviral role, dependent on the virus, cell type, and host organism. Whereas autophagy promotes the replication of several positive-strand RNA viruses, it restricts HSV-1, Sindbis virus, and HIV-1. Multiple mechanisms for autophagy-mediated antiviral restriction have been identified, including the direct degradation of viral components, leading to virus clearance, and the exposure of PAMPs to PRRs, promoting innate immune signaling.
Several recent studies demonstrated that TRIM proteins are important modulators of both nonviral and virus-induced autophagy. In a cDNA screen, 31 out of 62 tested TRIM proteins induced autophagy (124). Furthermore, individual depletion of 24 TRIM proteins interfered with IFNγ-induced autophagy (125), and silencing of 22 TRIM proteins diminished autophagy upon mTOR inhibition (126). The precise mode of action in the autophagy pathway, however, has been determined for only a few TRIM proteins. Whereas some TRIM proteins act as key regulators of the core autophagy machinery, others function as viral cargo receptors in a highly virus-specific manner.
TRIM23, a Key Regulator of Core Autophagic Components
While several TRIM proteins have been implicated in virus-induced autophagy, the precise mechanisms of their action in the autophagy pathway have been determined for very few, among them TRIM23 (Figure 4a). TRIM23 depletion or gene targeting abrogated autophagy induction by several viruses (e.g., HSV-1, Sindbis virus, adenovirus), which correlated with increased replication of these viruses (124). TRIM23 is unique among TRIM family proteins as it exhibits two enzymatic functions: E3 ubiquitin ligase function in the RING domain and GTPase function in the C-terminal ARF domain (14). Upon viral infection, TRIM23 modifies itself at the ARF domain with atypical K27-linked polyubiquitin, which stimulates the GTPase cycling activity of the ARF domain, a process necessary for TRIM23’s autophagy function and antiviral activity (124). Following virus-induced autophagy, TRIM23 interacts with TBK1, which is well known to regulate the IFN response and recently has been implicated in autophagy (127). Mechanistically, the GTP-GDP hydrolysis activity of TRIM23 facilitates TBK1 self-association and transautophosphorylation, thereby activating the kinase (124). TBK1 proceeds to phosphorylate p62 to enhance cargo recognition, ultimately inducing autophagic degradation of viral components (124).
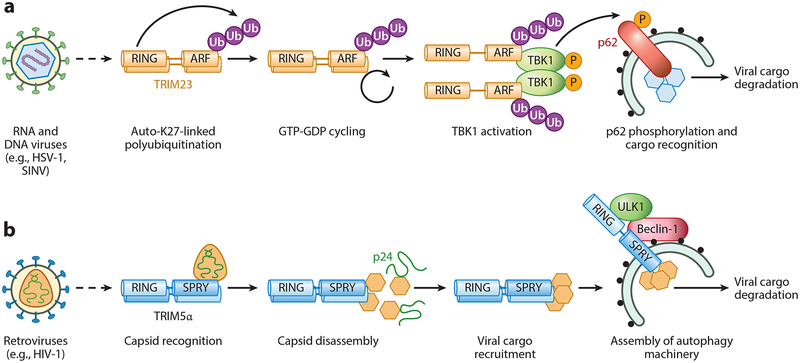
Figure 4.
TRIM proteins regulating virus-induced autophagy. TRIM proteins can promote viral autophagy in at least two different ways. For example, TRIM23 (panel a) activates key molecules of the autophagic machinery, whereas TRIM5α (panel b) recognizes and recruits viral cargo to the autophagic machinery for degradation. (a) The E3 enzymatic activity of TRIM23 is activated upon infection by diverse viral pathogens (e.g., HSV-1, SINV), which leads to nondegradative K27-linked autopolyubiquitination of its C-terminal ARF domain. K27-linked ubiquitination in turn stimulates ARF GTPase activity. Upon recruitment of TBK1 to the ARF domain of TRIM23, ubiquitin-induced GTP-GDP cycling of the ARF domain facilitates TBK1 dimerization and trans−autophosphorylation and thereby promotes activation of TBK1’s catalytic activity. TBK1-mediated phosphorylation of the autophagy receptor p62 enables viral cargo recognition by p62 and ultimately causes degradation of the cargo. (b) TRIM5α recognizes the capsid protein of HIV-1 (p24), causing premature virus uncoating, which blocks HIV-1 infection. At least two destructive mechanisms reportedly lead to p24 degradation: the proteasome (not illustrated) and autophagic degradation. Autophagy is induced by the p24-TRIM5α complex in a Beclin-1- and ULK1-dependent manner at budding autophagosomes, ultimately leading to p24 degradation. Abbreviations: ARF, ADP-ribosylation factor; HIV-1, human immunodeficiency virus type 1; HSV-1, herpes simplex virus type 1; RING, really interesting new gene; SINV, Sindbis virus; SPRY, SPIa and the ryanodine receptor; TRIM, tripartite motif protein; Ub, ubiquitin.
TRIM5α, a Viral Cargo Receptor Triggering Autophagic Degradation
Besides restricting retroviral infection through nondegradative and proteasomal mechanisms, TRIM5α was recently shown to act as a viral cargo receptor inducing autophagic degradation of viral components (Figure 4b). Recognition of the HIV-1 capsid (p24) by the SPRY domain of TRIM5α assembles two key molecules in autophagy, ULK1 and Beclin-1, to induce autophagic flux and thereby cause degradation of p24 (81, 126). However, HIV-1 restriction by TRIM5α still occurs in cells lacking key autophagic components, suggesting that autophagy is not solely responsible for antiviral restriction (128).
VIRAL MANIPULATION OF TRIM PROTEINS
Viruses have evolved strategic maneuvers to antagonize the antiviral function of TRIM proteins, including inhibition of their E3 ligase activity, sequestration of specific TRIM proteins, or induction of their proteasomal degradation. On the other hand, some viral pathogens divert the activity of TRIM proteins or induce their gene expression in a way that directly or indirectly promotes virus replication and/or spread. In this section, we discuss how diverse viral pathogens manipulate TRIM proteins and the impact of these interactions on viral infection and antiviral immunity.
Viral Antagonism of TRIM Proteins
Given the important role of TRIM25 in the RIG-I antiviral pathway, it is not surprising that viruses have evolved a diverse collection of strategies to neutralize TRIM25’s activity, thereby preventing RIG-I activation and IFN-mediated immunity. The NS1 protein of IAV forms a complex with TRIM25, which inhibits TRIM25 dimerization and thereby prevents its E3 ligase activity from modifying RIG-I with K63-polyubiquitin (129). Interestingly, the CCDs of different TRIM25 orthologs show high sequence variability, and accordingly, human-, avian-, and mouse-adapted IAV strains bind to TRIM25 in a species-specific fashion (130). The nucleocapsid (N) protein of severe acute respiratory syndrome virus binds to the RIG-I-recruiting module of TRIM25, the C-terminal SPRY domain, thereby interfering with RIG-I binding to TRIM25 (131). This strategy of TRIM25 and RIG-I antagonism is also conserved in the related Middle East respiratory syndrome virus (131), providing a molecular mechanism of how the N proteins of coronaviruses thwart the host immune response. Severe fever with thrombocytopenia syndrome virus (SFTSV) has evolved a completely different mechanism to inhibit TRIM25. Using its nonstructural protein NSs, SFTSV sequesters TRIM25 as well as RIG-I and TBK1 to cytoplasmic structures that colocalize with Rab5 (132). Redistribution of TRIM25, RIG-I, and TBK1 by NSs away from appropriate signaling platforms at mitochondria-associated membranes then dampens type I IFN induction. Intriguingly, at least one virus uses a viral protein–independent strategy to suppress TRIM25 activity and thereby attenuate RIG-I signaling. The subgenomic flavivirus RNA encoded by an epidemic strain of dengue virus that emerged in Puerto Rico in 1994 bound to TRIM25 and interfered with its deubiquitination and stabilization by USP15 (10, 23).
Many viruses encode effector proteins that disrupt TRIM19/PML-containing ND10 subnu clear structures and thereby interfere with intrinsic immunity, and some of these viral antagonists directly target TRIM19. For example, the immediate early protein IE1 of human cytomegalovirus directly binds to the CCD region of TRIM19 to prevent TRIM19 auto-SUMOylation, disrupting nuclear bodies (133, 134). Structural analysis of IE1 revealed that its globular core, which is the region that binds to TRIM19, exhibits features that are highly similar to those of the CCD regions of TRIM proteins, indicating structural mimicry (133). Interestingly, IE1 interacts not only with TRIM19 but also with TRIM5α and TRIM33 (133, 135), suggesting that the unique CCD-like structure of the IE1 core region allows it to interact with and manipulate multiple TRIM proteins, although this requires further investigation. The related murine gammaherpesvirus 68 uses a different strategy to incapacitate TRIM19. Upon interaction with TRIM19, murine gamma-herpesvirus 68 ORF75c induces the ubiquitination and subsequent proteasomal degradation of TRIM19 (136).
Other viruses that target specific TRIM proteins for proteasomal degradation are HSV-1 and Nipah virus. The ICP0 protein of HSV-1 utilizes its own RING E3 ligase activity to induce polyubiquitination and degradation of TRIM27 (137). The matrix protein of Nipah virus targets TRIM6 for degradation, thereby blocking the activation of IKKε and the ensuing type I IFN–mediated antiviral responses (138). Several other henipaviruses (e.g., Hendra, Cedar, and Ghana viruses) also effectively inhibited the IFN response mediated by TRIM6 and IKKε (138).
Viral Commandeering of TRIM Proteins
Viruses use a variety of strategies to commandeer TRIM proteins for promoting their own replication. Conceptually, hijacked TRIM proteins either directly alter the activity of viral proteins or modulate signaling cascades that establish a milieu in the infected host cell that benefits the virus. For example, Japanese encephalitis virus induces transcriptional expression of TRIM21, which then negatively regulates the IFNβ pathway (139). Similarly, Epstein-Barr virus induces the expression of TRIM29, which in turn modifies STING with K48-linked polyubiquitin, inducing its degradation (53). The NS5 protein of yellow fever virus utilizes TRIM23 to promote its own ubiquitination; ubiquitination of NS5 promotes its binding to STAT2 and thereby triggers NS5-mediated interference of the IFNAR signaling cascade (140). Recently, it has been shown that the Ebola virus VP35 protein, which is a major IFN antagonist of the virus and also regulates the activity of the viral polymerase, recruits TRIM6, which in turn ubiquitinates VP35 and promotes both its IFN-antagonistic and polymerase-promoting functions (141). Accordingly, the replication of infectious Ebola virus in TRIM6−deficient cells was reduced as compared to wild-type cells, supporting a proviral function of TRIM6 during Ebola virus infection. The UL144 protein of human cytomegalovirus is known to activate NF-κB via TRAF6 and TAK1, ultimately upregulating the expression of macrophage-derived chemokine (or CCL22), which may function to subvert the Th1 immune response (142). UL144 directly interacts with TRIM23, which leads to K63-linked autoubiquitination of TRAF6 and thereby causes NF-κB induction by UL144 (143).
CONCLUDING REMARKS
Over the past decade we have learned many amazing things about TRIM proteins and their roles during viral infection. While it has been well established that TRIM proteins can act as antiviral restriction factors and master immune regulators, completely new functions of TRIM proteins have recently been discovered, such as their ability to regulate autophagy. Interestingly, emerging evidence also suggests that several TRIM proteins influence RNA metabolism or microRNA processing, or even have RNA-binding capacities themselves. For example, TRIM25 has RNA-binding activity (11, 144), and TRIM32, TRIM65, and TRIM71 have been described as regulators of microRNA processing and RNA interference (145–148). It warrants further investigation whether processing of host microRNAs by these TRIMs influences infection biology, and whether some of these TRIMs could even process virus-encoded microRNAs.
Apart from their involvement in antiviral immunity, many TRIM proteins play powerful roles in cancer-related processes, such as modulation of the tumor suppressor protein p53 among many other tumor-related molecules (149, 150). Furthermore, several TRIM proteins (e.g., TRIM11, TRIM25, TRIM32, and TRIM66) are upregulated in certain cancers. As many oncogenic viruses are known to manipulate critical pathways involved in oncogenesis or cancer progression, it is tempting to speculate that some of these TRIM proteins are involved in virus-induced tumorigenesis as well. Lastly, it remains to be seen whether viruses target specific TRIM proteins to either block autophagy-mediated virus clearance or, on the other hand, promote autophagy for their efficient replication. Elucidating virus-TRIM interactions and their impact on pathogenesis may reveal new molecular targets for preventing and treating viral infectious diseases in humans.
ACKNOWLEDGMENTS
We apologize to all colleagues whose contributions were not discussed and cited owing to space constraints. Current research in the Gack laboratory is supported by National Institutes of Health grants (R01 AI087846, R01 AI127774, R21 AI118509, and R21 AI133361) and an award from the PML Consortium. K.M.J.S. was supported by a fellowship from the German Research Foundation (SP 1600/1–1).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Hatakeyama S 2017. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem. Sci 42:297–311 [DOI] [PubMed] [Google Scholar]
- 2.Ozato K, Shin DM, Chang TH, Morse HC. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol 8:849–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joazeiro CA, Weissman AM. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–52 [DOI] [PubMed] [Google Scholar]
- 4.Davis ME, Gack MU. 2015. Ubiquitination in the antiviral immune response. Virology 479–80:52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertin J, DiStefano PS. 2000. The PYRIN domain: a novel motif found in apoptosis and inflammation proteins. Cell Death Differ. 7:1273–74 [DOI] [PubMed] [Google Scholar]
- 6.Meroni G 2012. Genomics and evolution of the TRIM gene family. Adv. Exp. Med. Biol 770:1–9 [DOI] [PubMed] [Google Scholar]
- 7.Sanchez JG, Chiang JJ, Sparrer KMJ, Alam SL, Chi M, et al. 2016. Mechanism of TRIM25 catalytic activation in the antiviral RIG-I pathway. Cell Rep. 16:1315–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Cruz AA, Babon JJ, Norton RS, Nicola NA, Nicholson SE. 2013. Structure and function of the SPRY/B30.2 domain proteins involved in innate immunity. Protein Sci. 22:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foss S, Watkinson R, Sandlie I, James LC, Andersen JT. 2015. TRIM21: a cytosolic Fc receptor with broad antibody isotype specificity. Immunol. Rev 268:328–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, et al. 2015. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudhury NR, Heikel G, Trubitsyna M, Kubik P, Nowak JS, et al. 2017. RNA-binding activity of TRIM25 is mediated by its PRY/SPRY domain and is required for ubiquitination. BMC Biol. 15:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Short KM, Cox TC. 2006. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem 281:8970–80 [DOI] [PubMed] [Google Scholar]
- 13.Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. 2001. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105:281–89 [DOI] [PubMed] [Google Scholar]
- 14.Vitale N, Moss J, Vaughan M. 1996. ARDi, a 64-kDa bifunctional protein containing an 18-kDa GTP-binding ADP-ribosylation factor domain and a 46-kDa GTPase-activating domain. Biochemistry 93: 1941–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–20 [DOI] [PubMed] [Google Scholar]
- 16.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol 32:513–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goubau D, Deddouche S, Reis e Sousa C. 2013. Cytosolic sensing of viruses. Immunity 38:855–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang JJ, Sparrer KMJ, van Gent M, Lässig C, Huang T, et al. 2017. Viral unmasking of cellular 5S rRNA pseudogene transcripts induces RIG-I-mediated immunity. Nat. Immunol 19:53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlee M 2013. Master sensors of pathogenic RNA—RIG-I like receptors. Immunobiology 218:1322–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang C, Gack MU. 2017. Post-translational control of intracellular pathogen sensing pathways. Trends Immunol. 38:39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gack MU, Shin YC, Joo CH, Urano T, Liang C, et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–20 [DOI] [PubMed] [Google Scholar]
- 22.Zeng W, Sun L, Jiang X, Chen X, Hou F, et al. 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pauli EK, Chan YK, Davis ME, Gableske S, Wang MK, et al. 2014. The ubiquitin-specific protease USP15 promotes RIG-I–mediated antiviral signaling by deubiquitylating TRIM25. Sci. Signal 7:ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan J, Li Q, Mao AP, Hu MM, Shu HB. 2014. TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J. Mol. Cell Biol 6:154–63 [DOI] [PubMed] [Google Scholar]
- 25.Lang X, Tang T, Jin T, Ding C, Zhou R, Jiang W. 2017. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. J. Exp. Med 214:459–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narayan K, Waggoner L, Pham ST, Hendricks GL, Waggoner SN, et al. 2014. TRIM13 is a negative regulator of MDA5-mediated type I interferon production. J. Virol 88:10748–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Versteeg GA, Rajsbaum R, Sanchez-Apáricio MT, Maestre AM, Valdiviezo J, et al. 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38:384–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao C, Jia M, Song H, Yu Z, Wang W, et al. 2017. The E3 ubiquitin ligase TRIM40 attenuates antiviral immune responses by targeting MDA5 and RIG-I. Cell Rep. 21:1613–23 [DOI] [PubMed] [Google Scholar]
- 29.Hu MM, Liao CY, Yang Q, Xie XQ, Shu HB. 2017. Innate immunity to RNA virus is regulated by temporal and reversible sumoylation of RIG-I and MDA5. J. Exp. Med 214:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wies E, Wang MK, Maharaj NP, Chen K, Zhou S, et al. 2013. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 38:437–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan P, He L, Cui J, Qian C, Cao X, et al. 2017. Assembly of the WHIP-TRIM14-PPP6C mitochondrial complex promotes RIG-I-mediated antiviral signaling. Mol. Cell 68:293–307 [DOI] [PubMed] [Google Scholar]
- 32.Zhou Z, Jia X, Xue Q, Dou Z, Ma Y, et al. 2014. TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I–like receptor-mediated innate immune response. PNAS 111:E245–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia X, Zhou H, Wu C, Wu Q, Ma S, et al. 2017. The ubiquitin ligase RNF125 targets innate immune adaptor protein TRIM14 for ubiquitination and degradation. J. Immunol 198:4652–58 [DOI] [PubMed] [Google Scholar]
- 34.Liu B, Zhang M, Chu H, Zhang H, Wu H, et al. 2016. The ubiquitin E3 ligase TRIM31 promotes aggregation and activation of the signaling adaptor MAVS through Lys63-linked polyubiquitination. Nat. Immunol 18:214. [DOI] [PubMed] [Google Scholar]
- 35.Yang B, Wang J, Wang Y, Zhou H, Wu X, et al. 2013. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J. Immunol 190:3613–19 [DOI] [PubMed] [Google Scholar]
- 36.Castanier C, Zemirli N, Portier A, Garcin D, Bidère N, et al. 2012. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 10:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin Y, Liu Q, Tian S, Xie W, Cui J, Wang RF. 2016. TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3β to TBK1. Cell Res. 26:613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi M, Cho H, Inn KS, Yang A, Zhao Z, et al. 2014. Negative regulation of NF-κB activity by brain-specific TRIpartite Motif protein 9. Nat. Commun 5:4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y, Song B, Park C, Kwon KS. 2013. TRIM11 negatively regulates IFNβ production and antiviral activity by targeting TBK1. PLOS ONE 8:e63255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ran Y, Zhang J, Liu LL, Pan ZY, Nie Y, et al. 2016. Autoubiquitination of TRIM26 links TBK1 to NEMO in RLR-mediated innate antiviral immune response. J. Mol. Cell Biol. 8:31–43 [DOI] [PubMed] [Google Scholar]
- 41.Wang P, Zhao W, Zhao K, Zhang L, Gao C. 2015. TRIM26 negatively regulates interferon-β production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PLOS Pathog. 11:e1004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang Q, Deng H, Li X, Wu X, Tang Q, et al. 2011. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol 187:4754–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arimoto KI, Funami K, Saeki Y, Tanaka K, Okawa K, et al. 2010. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. PNAS 107:15856–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, et al. 2013. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol 87:257–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol 32:461–88 [DOI] [PubMed] [Google Scholar]
- 46.Chen Q, Sun L, Chen ZJ. 2016. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol 17:1142. [DOI] [PubMed] [Google Scholar]
- 47.Chen M, Meng Q, Qin Y, Liang P, Tan P, et al. 2016. TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol. Cell 64:105–19 [DOI] [PubMed] [Google Scholar]
- 48.Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, et al. 2016. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 45:555–69 [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Hu MM, Wang YY, Shu HB. 2012. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem 287:28646–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, et al. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33:765–76 [DOI] [PubMed] [Google Scholar]
- 51.Wang Q, Liu X, Cui Y, Tang Y, Chen W, et al. 2014. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 41:919–33 [DOI] [PubMed] [Google Scholar]
- 52.Fang R, Wang C, Jiang Q, Lv M, Gao P, et al. 2017. NEMO–IKKβ are essential for IRF3 and NF-κB activation in the cGAS–STING pathway. J. Immunol 199:3222–33 [DOI] [PubMed] [Google Scholar]
- 53.Xing J, Zhang A, Zhang H, Wang J, Li XC, et al. 2017. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat. Commun 8:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Lian Q, Yang B, Yan S, Zhou H, et al. 2015. TRIM30αis a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLOS Pathog. 11:e1005012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gay NJ, Symmons MF, Gangloff M, Bryant CE. 2014. Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol 14:546. [DOI] [PubMed] [Google Scholar]
- 56.Zhao W, Wang L, Zhang M, Yuan C, Gao C. 2012. E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J. Immunol 188:2567–74 [DOI] [PubMed] [Google Scholar]
- 57.Xue Q, Zhou Z, Lei X, Liu X, He B, et al. 2012. TRIM38 negatively regulates TLR3-mediated IFN-β signaling by targeting TRIF for degradation. PLOS ONE 7:e46825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu MM, Yang Q, Zhang J, Liu SM, Zhang Y, et al. 2014. TRIM38 inhibits TNFα- and IL-1β–triggered NF-κB activation by mediating lysosome-dependent degradation of TAB2/3. PNAS 111:1509–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao W, Wang L, Zhang M, Wang P, Yuan C, et al. 2012. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I–mediated IFN-β production and antiviral response by targeting NAP1. J. Immunol 188:5311–18 [DOI] [PubMed] [Google Scholar]
- 60.Shi M, Deng W, Bi E, Mao K, Ji Y, et al. 2008. TRIM30α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol 9:369. [DOI] [PubMed] [Google Scholar]
- 61.Yang Q, Liu TT, Lin H, Zhang M, Wei J, et al. 2017. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLOS Pathog. 13:e1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen Y, Li NL, Wang J, Liu B, Lester S, Li K. 2012. TRIM56 is an essential component of the TLR3 antiviral signaling pathway. J. Biol. Chem 287:36404–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higgs R, Ní Gabhann J, Larbi NB, Breen EP, Fitzgerald KA, Jefferies CA. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol 181:1780–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higgs R, Lazzari E, Wynne C, Ní Gabhann J, Espinosa A, et al. 2010. Self protection from anti-viral responses—Ro52 promotes degradation of the transcription factor IRF7 downstream of the viral Toll-like receptors. PLOS ONE 5:e11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng Q, Hou J, Zhou Y, Yang Y, Xie B, Cao X. 2015. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res. 25:1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng Q, Hou J, Zhou Y, Yang Y, Cao X. 2016. Type I IFN–inducible downregulation of microRNA-27a feedback inhibits antiviral innate response by upregulating Siglec1/TRIM27. J. Immunol 196:1317–26 [DOI] [PubMed] [Google Scholar]
- 67.Zha J, Han KJ, Xu LG, He W, Zhou Q, et al. 2006. The Ret finger protein inhibits signaling mediated by the noncanonical and canonical IκB kinase family members. J. Immunol 176:1072–80 [DOI] [PubMed] [Google Scholar]
- 68.Xing J, Weng L, Yuan B, Wang Z, Jia L, et al. 2016. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nat. Immunol 17:1373–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li Q, Yan J, Mao AP, Li C, Ran Y, et al. 2011. Tripartite motif 8 (TRIM8) modulates TNFα- and IL-1β–triggered NF-κB activation by targeting TAK1 for K63-linked polyubiquitination. PNAS 108:19341–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ye W, Hu MM, Lei CQ, Zhou Q, Lin H, et al. 2017. TRIM8 negatively regulates TLR3/4-mediated innate immune response by blocking TRIF–TBK1 interaction. J. Immunol 199:1856–64 [DOI] [PubMed] [Google Scholar]
- 71.Ivashkiv LB, Donlin LT. 2013. Regulation of type I interferon responses. Nat. Rev. Immunol 14:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.tenOever BR, Ng SL, Chua MA, McWhirter SM, García-Sastre A, Maniatis T. 2007. Multiple functions of the IKK-related kinase IKKε in interferon-mediated antiviral immunity. Science 315:1274–78 [DOI] [PubMed] [Google Scholar]
- 73.Rajsbaum R, Versteeg GA, Schmid S, Maestre AM, Belicha-Villanueva A, et al. 2014. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity 40:880–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tisserand J, Khetchoumian K, Thibault C, Dembeĺé D, Chambon P, Losson R. 2011. Tripartite motif 24 (Trim24/Tif1α) tumor suppressor protein is a novel negative regulator of interferon (IFN)/signal transducers and activators of transcription (STAT) signaling pathway acting through retinoic acid receptor α (Rarα) inhibition. J. Biol. Chem 286:33369–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kamitani S, Ohbayashi N, Ikeda O, Togi S, Muromoto R, et al. 2008. KAP1 regulates type I interferon/STAT1-mediated IRF-1 gene expression. Biochem. Biophys. Res. Commun 370:366–70 [DOI] [PubMed] [Google Scholar]
- 76.Grütter MG, Luban J. 2012. TRIM5 structure, HIV-1 capsid recognition, and innate immune signaling. Curr. Opin. Virol 2:142–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kutluay SB, Perez-Caballero D, Bieniasz PD. 2013. Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLOS Pathog. 9:e1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pertel T, Hausmann S, Morger D, Züger S, Guerra J, et al. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848. [DOI] [PubMed] [Google Scholar]
- 80.Yap MW, Nisole S, Lynch C, Stoye JP. 2004. Trim5αprotein restricts both HIV-1 and murine leukemia virus. PNAS 101:10786–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ribeiro CMS, Sarrami-Forooshani R, Setiawan LC, Zijlstra-Willems EM, van Hamme JL, et al. 2016. Receptor usage dictates HIV-1 restriction by human TRIM5α in dendritic cell subsets. Nature 540: 448–52 [DOI] [PubMed] [Google Scholar]
- 82.Kajaste-Rudnitski A, Marelli SS, Pultrone C, Pertel T, Uchil PD, et al. 2011. TRIM22 inhibits HIV-1 transcription independently of its E3 ubiquitin ligase activity, Tat, and NF-κB-responsive long terminal repeat elements. J. Virol 85:5183–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Turrini F, Marelli S, Kajaste-Rudnitski A, Lusic M, Van Lint C, et al. 2015. HIV-1 transcriptional silencing caused by TRIM22 inhibition of Sp1 binding to the viral promoter. Retrovirology 12:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barr SD, Smiley JR, Bushman FD. 2008. The interferon response inhibits HIV particle production by induction of TRIM22. PLOS Pathog. 4:e1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Singh R, Gaiha G, Werner L, McKim K, Mlisana K, et al. 2011. Association of TRIM22 with the type 1 interferon response and viral control during primary HIV-1 infection. J. Virol 85:208–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Di Pietro A, Kajaste-Rudnitski A, Oteiza A, Nicora L, Towers GJ, et al. 2013. TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation. J. Virol 87:4523–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao B, Duan Z, Xu W, Xiong S. 2009. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 50:424–33 [DOI] [PubMed] [Google Scholar]
- 88.Yang C, Zhao X, Sun D, Yang L, Chong C, et al. 2015. Interferon alpha (IFNα)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell. Mol. Immunol 13:94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Medrano LM, Rallón N, Berenguer J, Jiménez-Sousa MA, Soriano V, et al. 2016. Relationship of TRIM5 and TRIM22 polymorphisms with liver disease and HCV clearance after antiviral therapy in HIV/HCV coinfected patients. J. Transl. Med 14:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eldin P, Papon L, Oteiza A, Brocchi E, Lawson TG, Mechti N. 2009. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J. Gen. Virol 90: 536–45 [DOI] [PubMed] [Google Scholar]
- 91.Yuan T, Yao W, Tokunaga K, Yang R, Sun B. 2016. An HIV-1 capsid binding protein TRIM11 accelerates viral uncoating. Retrovirology 13:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, et al. 2011. The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9:484–95 [DOI] [PubMed] [Google Scholar]
- 93.Tabah AA, Tardif K, Mansky LM. 2014. Anti-HIV-1 activity of Trim 37. J. Gen. Virol 95:960–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. 2008. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLOS Pathog. 4:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vaysburd M, Watkinson RE, Cooper H, Reed M, O’Connell K, et al. 2013. Intracellular antibody receptor TRIM21 prevents fatal viral infection. PNAS 110:12397–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Keeble AH, Khan Z, Forster A, James LC. 2008. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. PNAS 105:6045–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rhodes DA, Trowsdale J. 2007. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol. Immunol 44:2406–14 [DOI] [PubMed] [Google Scholar]
- 98.Rhodes DA, Isenberg DA. 2017. TRIM21 and the function of antibodies inside cells. Trends Immunol. 38:916–26 [DOI] [PubMed] [Google Scholar]
- 99.Hauler F, Mallery DL, McEwan WA, Bidgood SR, James LC. 2012. AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization. PNAS 109:19733–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mallery DL, McEwan WA, Bidgood SR, Towers GJ, Johnson CM, James LC. 2010. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). PNAS 107:19985–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.McEwan WA, Tam JCH, Watkinson RE, Bidgood SR, Mallery DL, James LC. 2013. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nat. Immunol 14:327–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Watkinson RE, McEwan WA, Tam JCH, Vaysburd M, James LC. 2015. TRIM21 promotes cGAS and RIG-I sensing of viral genomes during infection by antibody-opsonized virus. PLOS Pathog. 11:e1005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bernardi R, Pandolfi PP. 2007 Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol 8: 1006–16 [DOI] [PubMed] [Google Scholar]
- 104.Chelbi-Alix MK, Quignon F, Pelicano L, Koken MHM, de Thé H. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol 72:1043–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Geoffroy MC, Chelbi-Alix MK. 2011. Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res 31:145–58 [DOI] [PubMed] [Google Scholar]
- 106.Dutrieux J, Maarifi G, Portilho DM, Arhel NJ, Chelbi-Alix MK, Nisole S. 2015. PML/TRIM19-dependent inhibition of retroviral reverse-transcription by Daxx. PLOS Pathog. 11:e1005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tavalai N, Stamminger T. 2009. Interplay between herpesvirus infection and host defense by PML nuclear bodies. Viruses 1:1240–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reichelt M, Wang L, Sommer M, Perrino J, Nour AM, et al. 2011. Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLOS Pathog. 7:e1001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scherer M, Stamminger T. 2016. Emerging role of PML nuclear bodies in innate immune signaling. J. Virol 90:5850–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng X, Wang X, Tu F, Wang Q, Fan Z, Gao G. 2017. TRIM25 is required for the antiviral activity of zinc finger antiviral protein. J. Virol 91:e00088–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li MMH, Lau Z, Cheung P, Aguilar EG, Schneider WM, et al. 2017. TRIM25 enhances the antiviral action of zinc-finger antiviral protein (ZAP). PLOS Pathog. 13:e1006145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li MMH, MacDonald MR, Rice CM. 2015. To translate, or not to translate: viral and host mRNA regulation by interferon-stimulated genes. Trends Cell Biol. 25:320–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fu B, Wang L, Ding H, Schwamborn JC, Li S, Dorf ME. 2015. TRIM32 senses and restricts influenza A virus by ubiquitination of PB1 polymerase. PLOS Pathog. 11:e1004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Meyerson NR, Zhou L, Guo YR, Zhao C, Tao YJ, et al. 2017. Nuclear TRIM25 specifically targets influenza virus ribonucleoproteins to block the onset of RNA chain elongation. Cell Host Microbe 22: 627–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu B, Li NL, Shen Y, Bao X, Fabrizio T, et al. 2016. The C-terminal tail of TRIM56 dictates antiviral restriction of influenza A and B viruses by impeding viral RNA synthesis. J. Virol 90:4369–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fan W, Wu M, Qian S, Zhou Y, Chen H, et al. 2016. TRIM52 inhibits Japanese encephalitis virus replication by degrading the viral NS2A. Sci. Rep 6:33698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J, Liu B, Wang N, Lee YM, Liu C, Li K. 2011. TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection. J. Virol 85:3733–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu B, Li NL, Wang J, Shi PY, Wang T, et al. 2014. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J. Virol 88:13821–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kane M, Zang TM, Rihn SJ, Zhang F, Kueck T, et al. 2016. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe 20:392–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang S, Chen Y, Li C, Wu Y, Guo L, et al. 2016. TRIM14 inhibits hepatitis C virus infection by SPRY domain-dependent targeted degradation of the viral NS5A protein. Sci. Rep 6:32336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang S, Guo JT, Wu JZ, Yang G. 2013. Identification and characterization of multiple TRIM proteins that inhibit hepatitis B virus transcription. PLOS ONE 8:e70001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deretic V, Saitoh T, Akira S. 2013. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol 13:722–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Levine B, Kroemer G. 2008. Autophagy in the pathogenesis of disease. Cell 132:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sparrer KMJ, Gableske S, Zurenski MA, Parker ZM, Full F, et al. 2017. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat. Microbiol 2:1543–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, et al. 2015. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol 210:973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, et al. 2014. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell 30:394–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, et al. 2012. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37:223–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Imam S, Talley S, Nelson RS, Dharan A, O’Connor C, et al. 2016. TRIM5α degradation via autophagy is not required for retroviral restriction. J. Virol 90:3400–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, et al. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLOS Pathog. 8:e1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hu Y, Li W, Gao T, Cui Y, Jin Y, et al. 2017. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol 91:e02143–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Santiago FW, Covaleda LM, Sanchez-Aparicio MT, Silvas JA, Diaz-Vizarreta AC, et al. 2014. Hijacking of RIG-I signaling proteins into virus-induced cytoplasmic structures correlates with the inhibition of type I interferon responses. J. Virol 88:4572–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Scherer M, Klingl S, Sevvana M, Otto V, Schilling EM, et al. 2014. Crystal structure of cytomegalovirus IE1 protein reveals targeting of TRIM family member PML via coiled-coil interactions. PLOS Pathog. 10:e1004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schilling EM, Scherer M, Reuter N, Schweininger J, Muller YA, Stamminger T. 2017. The human cytomegalovirus IE1 protein antagonizes PML nuclear body-mediated intrinsic immunity via the inhibition of PML de novo SUMOylation. J. Virol 91:e02049–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Martínez F, Tang Q. 2014. Identification of cellular proteins that interact with human cytomegalovirus immediate-early protein 1 by protein array assay. Viruses 6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sewatanon J, Ling PD. 2013. Murine gammaherpesvirus 68 ORF75c contains ubiquitin E3 ligase activity and requires PML SUMOylation but not other known cellular PML regulators, CK2 and E6AP, to mediate PML degradation. Virology 440:140–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Conwell SE, White AE, Harper JW, Knipe DM. 2015. Identification of TRIM27 as a novel degradation target of herpes simplex virus 1 ICP0. J. Virol 89:220–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bharaj P, Wang YE, Dawes BE, Yun TE, Park A, et al. 2016. The matrix protein of Nipah virus targets the E3-ubiquitin ligase TRIM6 to inhibit the IKKεkinase-mediated type-I IFN antiviral response. PLOS Pathog. 12:e1005880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Manocha GD, Mishra R, Sharma N, Kumawat KL, Basu A, Singh SK. 2014. Regulatory role of TRIM21 in the type-I interferon pathway in Japanese encephalitis virus-infected human microglial cells. J. Neuroinflamm 11:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Laurent-Rolle M, Morrison J, Rajsbaum R, Macleod JML, Pisanelli G, et al. 2014. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe 16:314–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bharaj P, Atkins C, Luthra P, Giraldo MI, Dawes BE, et al. 2017. The host E3-ubiquitin ligase TRIM6 ubiquitinates the Ebola virus VP35 protein and promotes virus replication. J. Virol 91:e00833–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Poole E, King CA, Sinclair JH, Alcami A. 2006. The UL144 gene product of human cytomegalovirus activates NFκB via a TRAF6-dependent mechanism. EMBO J. 25:4390–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Poole E, Groves I, Macdonald A, Pang Y, Alcami A, Sinclair J. 2009. Identification of TRIM23 as a cofactor involved in the regulation of NF-κB by human cytomegalovirus. J. Virol 83:3581–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, et al. 2013. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol 20:1122. [DOI] [PubMed] [Google Scholar]
- 145.Schwamborn JC, Berezikov E, Knoblich JA. 2009. The TRIM-NHL protein TRIM32 activates micro-RNAs and prevents self-renewal in mouse neural progenitors. Cell 136:913–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li S, Wang L, Fu B, Berman MA, Diallo A, Dorf ME. 2014. TRIM65 regulates microRNA activity by ubiquitination of TNRC6. PNAS 111:6970–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, et al. 2009. The let-7 target gene mouse lin−41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell Biol 11:1411–20 [DOI] [PubMed] [Google Scholar]
- 148.Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. 2012. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat. Commun 3:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Elabd S, Meroni G, Blattner C. 2016. TRIMming p53’s anticancer activity. Oncogene 35:5577–84 [DOI] [PubMed] [Google Scholar]
- 150.Crawford LJ, Johnston CK, Irvine AE. 2018. TRIM proteins in blood cancers. J. Cell Commun. Signal 12:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]