Abstract
While the pathogenesis of schizophrenia (SCZ) is proposed to involve alterations of neural circuits via synaptic dysfunction, the underlying molecular mechanisms remain poorly understood. Recent exome sequencing studies of SCZ have uncovered numerous single nucleotide variants (SNVs); however, the majority of these SNVs have unknown functional consequences, leaving their disease relevance uncertain. Filling this knowledge gap requires systematic application of quantitative and scalable assays to assess known and novel biological functions of genes. Here we demonstrate loss-of-function effects of multiple rare coding SNVs found in SCZ subjects in the GIT1 (G protein-coupled receptor kinase interacting ArfGAP 1) gene using functional cell-based assays involving co-expression of GIT1 and PAK3 (p21 protein (Cdc42/Rac)-activated kinase 3). Most notably, a GIT1-R283W variant reported in four independent SCZ cases was defective in activating PAK3 as well as MAPK (Mitogen-activated protein kinase). Similar functional deficits were found for a de novo SCZ variant GIT1-S601N. Additional assays revealed deficits in GIT1-R283W’s capacity to stimulate PAK phosphorylation in cultured hippocampal neurons. Also, GIT1-R283W showed deficits in the induction of GAD1 (Glutamate decarboxylase 1) protein expression. Extending these functional assays to ten additional rare GIT1 variants revealed the existence of an allelic series with the majority of the SCZ case variants exhibiting loss-of-function towards MAPK activation in a manner correlated with loss of PAK3 activation. Taken together, we propose that rare variants in GIT1, along with other genetic and environmental factors, cause dysregulation of PAK3 leading to synaptic deficits in SCZ.
Keywords: schizophrenia, synapse, exome, GIT1, PAK3, GAD1, NMDA receptor, GRIN2A
Introduction
Schizophrenia (SCZ) (MIM 181500) is a severe neuropsychiatric disorder with a complex, polygenic pattern of inheritance that affects ~1% of the general population. Characteristics of this disease include positive symptoms (delusions, hallucinations, disorganized speech, or catatonic behavior), negative symptoms (flattened affect, avolition), and cognitive symptoms (poor executive functioning, working memory deficits, and trouble paying attention). Twin studies have long indicated a strong genetic component to the cause of SCZ (1), and recent large-scale human genetic studies have made begun to make progress in identifying both common (2–4) and rare (5–10) genetic variation associated with elevated risk for SCZ (reviewed in (11)). Analyses of functional commonalities amongst genes affected by SCZ-associated genetic variation suggest that several key pathways may be affected, including components of the ARC (Activity-regulated cytoskeleton-associated protein) and NMDA (N-methyl-D-aspartate) receptor complexes found in the postsynaptic region of glutamatergic neurons, those mRNAs that are targets of FMRP (Fragile X mental retardation protein) (12), and proteins involved in regulating actin cytoskeleton dynamics (6, 8, 10). However, much remains to be understood regarding precisely how this genetic variation impacts the underlying molecular, cellular and circuit-level mechanisms that may influence risk for SCZ. Given the polygenic nature of SCZ, validating the functional effects of specific gene variants that converge on key cellular pathways is likely to be essential for delineating the complex etiopathogenesis of the disorder.
One gene connected to SCZ by genetic studies and convergent pathway analysis is GIT1, where a non-synonymous, de novo coding SNV (GIT1-S601N) was reported in a large set of Bulgarian trios that implicated pathways involved in actin filament dynamics (8). GIT1 is highly expressed in the human and rodent central nervous system (CNS) (13) (Supplemental Figure 1) where it can be found both presynaptically (14–17) and postsynaptically (15, 16) in both excitatory (16) and inhibitory (16, 18) synapses. At synapses, GIT1 is reported to regulate pre-synaptic vesicle recycling and release probability (17, 19), to promote dendritic spine growth and synapse formation (20), and to regulate AMPA and GABAA receptor synaptic localization (15, 18). Consistent with the importance of these multiple roles of GIT1 at synapses, whole body GIT1 knockout (GIT1−/−) mice have deficits in behavioral models of learning and memory, including fear conditioning, novel object recognition, operant conditioning, and spatial learning (21–23). However, it remains uncertain whether these phenotypes are due to loss of GIT1 function in neurons, in other cells in the brain, or possibly in cells outside of the brain.
While its complete three dimensional structure is not known, analysis of the primary amino acid sequence of GIT1 and functional studies have mapped several well defined structural domains (Figure 1A), including an N-terminal GTPase-activating domain for the ADP-ribosylation factor (Arf) family of small GTPases (ArfGAP; (24)), an Ankyrin repeat domain (3XANK; (25)), a Spa2-homology domain, named after the yeast protein Spa2 that functions in actin cytoskeletal organization (SHD; (26)), a coiled-coil domain that mediates dimerization (14), a synaptic localization domain (SLD; (16)), and a C-terminal focal adhesion targeting (FAT) domain that binds paxillin (27, 28). GIT1 acts as a subunit of a larger complex by binding constitutively to the Rac/Cdc42 Guanine Nucleotide Exchange Factors (GEFs) ARHGEF6 or ARHGEF7 (also known as α-PIX and β-PIX, respectively). Because GIT1 dimerizes and ARHGEF6/7 trimerize, the GIT/PIX complex is thought to be an oligomeric pentamer (29). Through the SHD (26), GIT1 is known to bind the pre-synaptic protein Piccolo, and to the post-synaptic proteins FAK1 (Focal Adhesion Kinase 1) and Liprin-α. Through scaffolding functions, GIT1 and ARHGEF6/7 bound together interact with multiple other proteins, including members of the p21 protein (Cdc42/Rac)-activated kinase (PAK) family that bind to the ARHGEF6/7 SH3 domain (30). In the case of the GIT1-ARHGEF6/7 complex interacting protein PAK3, its role in cognitive function is highlighted by the fact that disruptive mutation of PAK3 causes nonsyndromic X-linked intellectual disability (31), as does mutation of ARHGEF6 (32, 33). Knockdown of PAK3 or overexpression of functionally impaired PAK3 mutants suppresses spine maturation and excitatory synapse formation in hippocampal neurons (34, 35). PAK3 may also play a role in regulating GABAergic inhibitory interneuron morphology (36), and forebrain-specific expression of a dominant negative PAK has been shown to alter cortical synaptic morphology and block memory consolidation (37). GIT1 has also been reported to be a scaffold for mitogen-activated protein kinase (MAPK) signaling, by binding to a complex of MAP2K1 (MEK1; Mitogen-activated protein kinase kinase 1) and MAPK1/3 (Mitogen-activated protein kinase 1 and 3; also known as ERK2/1 or p42/p44, respectively) (38).
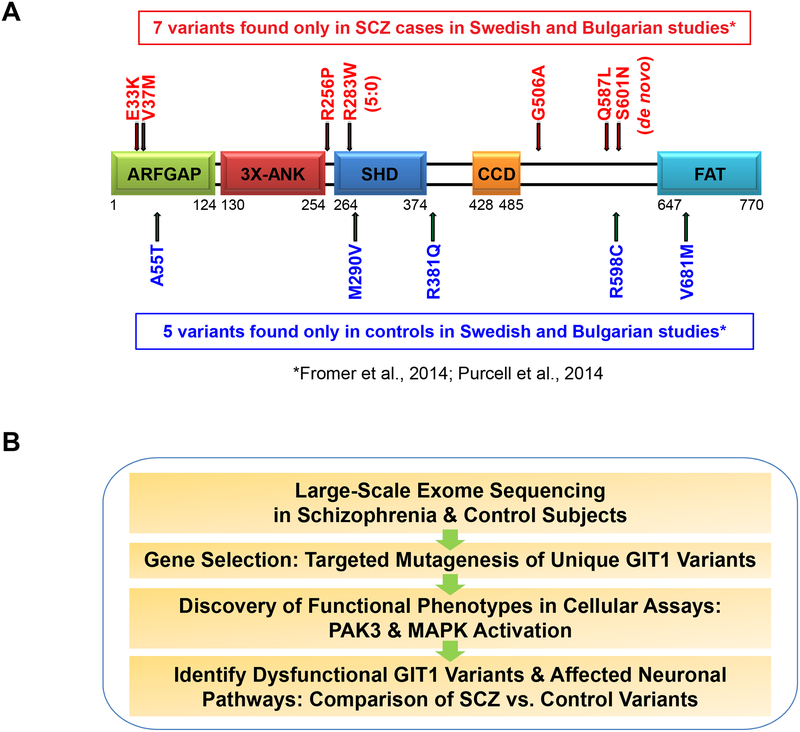
Figure 1. GIT1 domain structure, variants, and study design.
(A) GIT1 structural and functional domains (see Introduction for description). Locations of unique GIT1 coding variants are shown above (SCZ cases; in red) or below (controls; in blue) the domain structure diagram. The SHD domain (Spa2 Homology Domain) is a key protein-protein interaction region known to mediate interactions with ARHGEF6/7-PAK, MAP2K1 (MEK1), FAK, PYK2, PLCO, and PPFIA2. (B) Summary of study design. Coding variants unique to SCZ cases or controls identified in large-scale human exome sequencing studies were tested in a range of biological assays in cell lines and primary cultured neurons to characterize and compare the effect of different variant effects and reveal potentially relevant functional changes. See also Figures S4, Table S1, and Table S2.
Here we apply a functional genomic strategy to systematically characterize a total of 12 rare GIT1 coding variants identified by SCZ-exome sequencing using cellular assays in HEK293 cells and primary cultured rat neurons. Our data indicate that GIT1 variants found uniquely in SCZ subjects have deficits in the coupling between GIT1 and PAK3, as well as MAPK. We also identify changes in the expression of a critical regulator of inhibitory neurotransmission, GAD1. Taken together, our data support the notion that genetic variation in GIT1 and its network of interacting proteins may, along with other genetic and environmental factors, impact risk for SCZ.
Materials and Methods
Antibodies and western blotting
The following antibodies were used for western blotting and immunoprecipitation experiments.
Tag antibodies:
anti-FLAG (Cell Signaling, #2044), anti-Myc-Tag (Cell Signaling, #2278)
Total protein antibodies:
anti-ARHGEF6 (Cell Signaling, #4573), anti-DLG2 (Neuro Mab, #73–284), anti-DLG4 (Neuro Mab, #73–028), anti-GABRG2 (Synaptic Systems, #224 003), anti-GAD1 (Chemicon, #MAB5406), anti-GAPDH (Cell Signaling, #2118), anti-GAPDH (Cell Signaling, #8884), anti-GIT1 (Cell Signaling, #2919), anti-GRIA1 (Millipore, #ABN241), anti-GRIA2 (Millipore, #AB1768), anti-GRIK2/3 (GLUR6/7) (Millipore, #04–921), anti-GRM5 (Millipore, #AB5675), anti-MEK1 (MAP2K1) (Cell Signaling, #9146), anti-NLGN1 (Neuro Mab, #73–158), anti-PAK1 (Cell Signaling, #2602), anti-PAK3 (Cell Signaling, #2609), anti-β-Tubulin (Sigma, #T8660), anti-SLC17A7 (VGLUT1) (Synaptic Systems, #135303).
Phosphospecific antibodies:
anti-phospho-AKT (Ser473) (Cell Signaling, #4058), anti-phosphop44/42 MAPK (ERK1/2) (Thr202/Tyr204) (Cell Signaling, #9101), anti-phospho-GRIA1 (Ser845) (Millipore, #04–1073), anti-phospho-MEK1 (MAPK2K1) (Ser298) (Cell Signaling, #9128), anti-phospho-PAK1 (Ser144)/PAK2 (Ser141)/PAK3 (Ser139) (Cell Signaling, #2606), anti-phospho-PAK1 (Ser199/204)/PAK2 (Ser192/197)/PAK3 (Ser200/205) (Cell Signaling, #2605), anti-phospho-PAK1 (Thr423)/PAK2 (Thr402)/PAK3 (Thr421) (Cell Signaling, #2601), anti-phospho-SRC Family (Tyr416) (Cell Signaling, #2101).
All primary antibodies were diluted 1:1000 in an antibody dilution buffer (25 mM Tris, 0.15 M NaCl, 0.05% Tween-20, 5% BSA, 0.05% sodium azide). Primary antibodies were detected using the appropriate horseradish peroxidase conjugated secondary antibody (anti-mouse (GE Healthcare, #NA931), anti-rabbit (GE Healthcare, #NA9340)) and SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific, # 34096). Blots were imaged on the ChemiDoc MP System (Bio-Rad). Signal accumulation mode was used to acquire images at progressively longer exposure times. This allowed for acquisition of western blot images with band intensities within the linear range of the system. Densitometric analysis of the resulting 16-bit images was performed with ImageJ (http://imagej.nih.gov/ij/index.html). Images were rotated until the bands were horizontal. Then, a line region of interest was placed across the bands with the width of the line set thick enough to cover the bands. The “Plot Profile” function was used to plot the band intensities versus line length. Non-specific signal was removed using the Subtract Background function with rolling ball radius set to 50 pixels. Finally, band intensity was defined as the area under the curve for each plot.
DNA constructs and lentiviruses
Full-length, human GIT1 cDNA was obtained from Addgene (#15225) as initially characterized and deposition by Dr. Alan Rick Horwitz (59). This construct is the 770 amino acid +9 form and contains a C-terminal FLAG-tag, and was originally cloned from a cDNA library generated from human fetal brain. Full sequence information for this construct is available from Addgene. All human SNVs observed in SCZ subjects and controls were generated in this GIT1-FLAG construct with the Stratagene Quick-Change mutagenesis kit and confirmed by standard Sanger sequencing. Oligonucleotides used for mutagenesis were as follows:
| GIT1-E33K_sense | 5’-tgctggtgtgtgacaagtgctgcagcgtg-3’ |
| GIT1-E33K _antisense | 5’-cacgctgcagcacttgtcacacaccagca-3’ |
| GIT1-V37M_sense | 5’-gagtgctgcagcatgcaccggagcc-3’ |
| GIT1-V37M_antisense | 5’-ggctccggtgcatgctgcagcactc-3’ |
| GIT1-A55T_sense | 5’-cttcgccacagcacctggcctccca-3’ |
| GIT1-A55T_antisense | 5’-tgggaggccaggtgctgtggcgaag-3’ |
| GIT1-R381Q_sense | 5’-cgagctgtctctgcagagccagagtgacc-3’ |
| GIT1-R381Q_antisense | 5’-ggtcactctggctctgcagagacagctcg-3’ |
| GIT1-R256P_sense | 5’-gatggctgacagatctccgcaaaagtgcatgtctc-3’ |
| GIT1-R256P _antisense | 5’-gagacatgcacttttgcggagatctgtcagccatc-3’ |
| GIT1-R274W_sense | 5’-caggcgctcagcaactggctttttgaggaac-3’ |
| GIT1-R274W_antisense | 5’-gttcctcaaaaagccagttgctgagcgcctg-3’ |
| GIT1-M290V_sense | 5’-tttttgaggaactcgccgtggacgtgtatgacgag-3’ |
| GIT1-M290V_antisense | 5’-ctcgtcatacacgtccacggcgagttcctcaaaaa-3’ |
| GIT1-G506A_sense | 5’-ggcgccaggcgcgagcacacacc-3’ |
| GIT1-G506A _antisense | 5’-ggtgtgtgctcgcgcctggcgcc-3’ |
| GIT1-Q587L_sense | 5’-tgtcctgctccctggagggaagccg-3’ |
| GIT1-Q587L _antisense | 5’-cggcttccctccagggagcaggaca-3’ |
| GIT1-R598C_sense | 5’-gagcaaccacggcagtggagtgg-3’ |
| GIT1-R598C_antisense | 5’-ccactgccgtggcaggaaagcttgctc-3’ |
| GIT1-S601N_sense | 5’-tcccgccacggcaatggagccgac-3’ |
| GIT1-S601N_antisense | 5’-gtcggctccattgccgtggcggga-3’ |
| GIT1-V681M_sense | 5’-caagcatgacagcttcatgccctgctcagagaa-3’ |
| GIT1-V681M_antisense | 5’-ttctctgagcagggcatgaagctgtcatgcttg-3’ |
Selected GIT1 and GIT1 SNV constructs were then subcloned into the lentivirus expression vector pCDH-EF1-MCS-IRES-puro (System Biosciences #CD532A-2) and packaged into lentiviruses.
PAK3/MAPK activation assay in HEK293 cells
Human embryonic kidney (HEK) cell line 293FT (Life Technologies) was maintained in Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10% FBS (Life Technologies), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2 incubation. 293FT cells were seeded onto 6-well plates or 12-well plates 24 h before transfection. Cells were transfected using Lipofectamine 2000 (Life Technologies) at 80–90% confluency, according to manufacturer’s recommended protocol. Data were normalized to GAPDH and expressed relative to control levels as indicated. Phospho-PAK3 signals were divided by the expressed total PAK3 levels to normalize differences in expression that were observed in the presence of specific GIT1 SNV constructs.
GIT1-PAK3 co-immunoprecipitation assay
HEK293-FT cells were seeded into 6-well plates at moderate density. By the following day, the cells had reached ~80–90% confluency, and were transfected with the indicated GIT1, PAK3, and control (empty vector) expression constructs using Lipofectamine 2000 (Life Technologies). 24hrs later, transfected HEK293-FT cells were washed once with ice-cold PBS, and lysed with 600 μL of ice-cold lysis buffer (150 mM NaCl, 20 mM Tris-HCl pH 7.4, 1 mM EDTA, 1 mM EGTA, 1% Triton X-1000, protease inhibitor cocktail (Roche), phosphatase inhibitors cocktail (Roche)). Cells were lysed with a 20 min incubation on ice with occasional vortexing. Cell lysates were cleared by centrifugation at 20,000xg for 20min at 4oC. 500 μL of the resulting cleared cell lysate supernatants were incubated with 15 μL of FLAG-M2 antibody-coupled agarose beads (Sigma) overnight. Immune-complexes were washed four times with ice-cold lysis buffer. Washed immune-complexes were boiled in 50 μL of 2X SDS sample buffer, and subjected to immunoblot analysis using the indicated antibodies anti-FLAG (GIT1-FLAG) or anti-Myc (Myc-PAK3) antibodies. Results shown are the means calculated by combining data from 3 independent experiments.
Primary hippocampal neuron culture
Dissociated hippocampal neurons were prepared from Sprague Dawley rat embryos on E18-E19 (Charles River Labs). Hippocampal tissue was dissected in ice-cold 1× HBSS. Hippocampal tissue was washed 3× with 20 mL of ice-cold HBSS and then digested at 37°C for 10min in 5 mL of HBSS with trypsin (Life Technologies). Hippocampal tissue were then washed 3 times with HBSS and gently triturated in 2 mL of HBSS and plated at 100,000 cells per well in poly-D-lysine-coated 12-well plates (BD Biosciences). Neurons were maintained in Neurobasal media (Life Technologies), supplemented with 1× B27 (Life Technologies), and 1% penicillin/streptomycin at 37°C with 5% CO2 incubation.
RNA interference
The lentiviral pLKO.1 shRNA construct for targeting rat GIT1 (hairpin sequence: 5’-CCGGGCTAGTTGAGTGCCAGTATGACTCGAGTCATACTGGCACTCAACTAGCTTTTTG-3’) was obtained from Open Biosystems (TRCN0000106122). As a control, pLKO.1 with a non-targeting shRNA insert (5’-CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG-3’) was used.
Lentivirus preparation and infection
For the production of lentiviral vectors, a 15 cm tissue culture dish containing 70% confluent HEK293FT cells was transfected with 10 μg of transfer vector (GIT1-RNAi in pLKO.1 or GIT1 constructs in pCDH-EF-MCS-IRES-Puro), 9 μg Δ8.2, and 1 μg VSVG using LF2000 according to manufacturer’s protocol for 6–8 h. The medium was then replaced into DMEM containing 10% fetal bovine serum and 1% PenStrep (Invitrogen). Virus was collected on 2d post-transfection. The collected supernatant containing the virus was briefly spun at 1000 rpm and filtered through a 0.45 μm filter. Resulting virus mixture was concentrated 100X with LentiX-concentrator reagent (Clontech), according to manufacturer’s protocol. The pellet containing the virus was resuspended into Neurobasal medium (Invitrogen) and frozen for future use. For infecting primary hippocampal cultures in 12-well plates, 15 μL of viral resuspension was added to each well.
Structural modeling
A homology model was generated for the GIT1 ArfGAP domain using SwissModel program (http://swissmodel.expasy.org; (60)) and the atomic coordinates of the ArfGAP domain from human ACAP1 (ArfGAP with coiled-coil, ANK repeat and PH domain-containing protein 1; pdbid: 3JUE, (61)) as the template. Human GIT1 and human ACAP1 share 36% overall sequence identity in the central 80 amino acid stretch of the ArfGAP domain. To assess which regions of the homology model fall into highly conserved regions among several GAP domains and are thus the most reliable, we also generated a structure-based sequence alignment to compare 12 ArfGAPs with the GIT1 ArfGAP domain using PROMALS3D (http://prodata.swmed.edu/promals3d/promals3d.php; (62)) (Supplementary Figure S4B).
Results
Summary of rare GIT1 human genetic variants identified by exome sequencing
Whole exome sequencing data from a total of 3,159 SCZ cases and 3,789 healthy controls from the combination of a Swedish case-control study (10) and a Bulgarian trios study (8) available at the onset of our project collectively revealed a total of 37 coding SNVs in GIT1 (Supplementary Table S1), including a de novo variant (GIT1-S601N) found in the affected proband of a SCZ trio. Of particular interest for our functional analysis were coding SNVs that occur in multiple unrelated SCZ patients, but not in healthy controls (i.e. SCZ case-unique); or for comparison, coding SNVs that occur in controls, but not in patients (control-unique) in the study cohort. The locations of each SCZ case-unique (red) and control-unique (blue) coding SNVs are summarized in Figure 1A, constituting a total of 12 unique variants in GIT1 within the cohort. Prediction of the possible impact of each of these non-synonymous amino acid substitutions on GIT1 function by PolyPhen-2 suggested the SNVs spanned the full range from benign to likely damaging for both the disease and control groups (Supplemental Table S2), indicating that experimental assays are required to assess the impact of each mutation. With the release of the ExAC database (http://exac.broadinstitute.org/) containing exome sequencing data from 60,706 unrelated individuals, we were able to confirm that each of these 12 GIT1 SNVs are extremely rare (Supplemental Table S2). Additional instances of some of the 12 GIT1 SNVs considered here, as well as other novel GIT1 variants have been identified in other studies. However, none of these studies included the detailed phenotyping used in the Swedish and Bulgarian trio studies to classify SCZ cases and controls, hampering their interpretation in terms of potential relevance to SCZ. The current study focuses on the above noted 12 variants.
GIT1-R283W fails to activate PAK3 in human HEK293 cells
To advance a functional genomic strategy aiming at systematically characterizing the rare GIT1 variants identified by SCZ-exome sequencing in the whole exome sequencing studies of SCZ trios from Bulgaria and the Swedish case/control study, we set out to test the hypothesis that there would be a difference in functional consequences caused by coding SNVs found uniquely in SCZ cases compared to control SNVs (Figure 1B). Among the GIT1 variants that were unique to SCZ cases, the GIT1-R283W variant (arginine (R) at 283 changed to tryptophan (W)) was of particular interest. The GIT1-R283W variant was found in four independent cases of SCZ in a heterozygous state, and not in any of the matched healthy controls. Within the ExAC database, this variant was found in only one additional individual of unknown disease status, and further exome sequencing has identified another SCZ case with this variant bringing the total to five SCZ cases (S. Purcell and S. McCarroll, unpublished data). Mapping of the R283 residue in the GIT1 protein sequence revealed that it is located in within the first of the two repeated Spa2 subsequences in the SHD domain (Spa2 Homology Domain; PFAM:PF08518:GIT_SHD), which functions as a major signaling scaffold domain in GIT1 (Figure 1A). In addition, the R283W substitution replaces a positively charged residue with an uncharged hydrophobic residue, increasing the possibility this substitution could impact GIT1 function. Consistent with this notion, PolyPhen-2 predicts that the R283W variant is probably “damaging”. Thus, to test this prediction we began our functional studies of SCZ-associated GIT1 coding SNVs with analysis of GIT1-R283W.
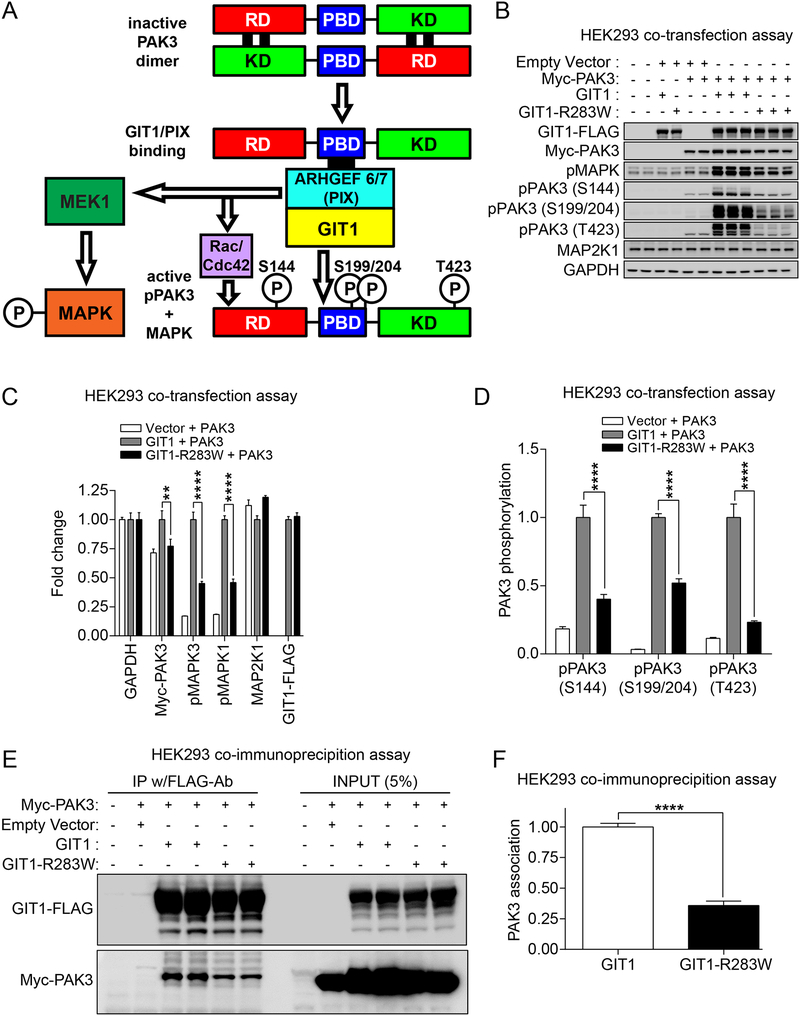
While developing functional assays to test GIT1 SNVs, we noted that a previous study on GIT1 from heterologous cells (38) suggested that the SHD domain (where the R283W variant in GIT1 is located) scaffolds MAP2K1 (MEK1) leading to activation of MAPK1/3 by phosphorylation (MAPK1 (T185/T187) and MAPK3 (T202/Y204)). Additional studies with GIT1−/− whole body knockout mice (23) indicated that GIT1 associates with PAK3 in the brain (Figure 2A). Accordingly, we co-transfected human HEK293 cells plus either a control empty vector or Myc-tagged PAK3 and either wild-type GIT1 or GIT1-R283W. As shown in Figure 2B,C, cotransfection of PAK3 and wild-type GIT1 in HEK293 cells resulted in substantial MAPK activation as measured by increased phosphorylation. GIT1 overexpression alone did not activate MAPK, and PAK3 overexpression alone produced only slight MAPK activation (lower band corresponding to MAPK3), as determined by measurement of phospho-MAPK levels, compared to mock transfected cells or vector transfected cells. Interestingly, in contrast to co transfection of wild-type GIT1 plus PAK3, co-transfection of GIT1-R283W plus PAK3 induced a lower level of phospho-MAPK in cells, suggesting that GIT1-R283W has a functional defect in MAPK activation. We further examined PAK3 activation itself in transfected cells by measuring the phosphorylation status of PAK3. As shown in Figure 2B,D, phosphorylation levels at the S144, S199/S204, and T423 residues of expressed PAK3 were significantly increased by co-expression of wild-type GIT1, but the expression of GIT1-R283W only weakly activated co-expressed PAK3 as judged by its phosphorylation state. Remarkably, when we performed similar co-transfection assays with PAK1, which shares 90% identity with PAK3 in amino acid sequence, we could not detect any significant activation of MAPK or PAK1 (Supplementary Figure 2D–F). Taken together, these results suggest that overexpressed GIT1 preferentially activates PAK3 in cells, and the alteration in the SHD domain by the GIT1-R283W variant found in multiple SCZ cases appears to have impaired PAK3 activation.
Figure 2. Functional assays of GIT1 sensitive to genetic variation.
(A) Schematic diagram depicting the biochemical basis of the GIT1-PAK3 co-transfection assay. Rac/Cdc42 Guanine Nucleotide Exchange Factor (ARHGEF6/7), also known as the PIX (p21-activated kinase interacting exchange factor) family, bind to the PIX binding domain (PBD) on PAK family members in the presence of GIT1, leading to conformational changes and activation of PAKs. Deactivated PAK dimers are arranged in a head-to-tail manner in an auto-inhibited state due to interaction of one N-terminal Regulatory Domain (RD) composed of an auto-inhibitory kinase domain with the opposing C-terminal Kinase Domain (KD). As reviewed in (63), upon GTP-bound Rac/Cdc42 family GTPase binding to the GTPase Binding Domain (GBD), PAKs undergo a conformational change enabling autophosphorylation in trans that switches PAKs to an active state. Whereas auto-phosphorylation of S144 reduces the interaction of the kinase auto-inhibitory domain and kinase domain, auto-phosphorylation of S199/S204 reduces PAK--ARHGEF6/7 (PIX) interaction. Activation-loop phosphorylation at T423, which is required for full catalytic activity, occurs through autophosphorylation in trans or from the activity of other kinases. (B-D) Effect of wild-type GIT1 and GIT1-R283W on PAK3 and MAPK activation in HEK293 cells. Cells were co-transfected with the indicated expression vectors and 24hrs later cells were harvested in 2X SDS sample buffer. (B) Protein samples were subjected to immunoblot analysis using the indicated antibodies. For pMAPK, the top band corresponds to phospho-MAPK3 (ERK1; p44) T202/Y204 and the bottom band corresponds to phospho-MAPK1 (ERK2; p42) T185/T187. (C) Elevation of phospho-MAPK3, phospho-MAPK1 and Myc-PAK3 levels with wild-type GIT1 but not GIT1-R283W. For immunoblots other than those for the phospho-PAKs, band intensities were normalized to GAPDH band intensities, and then to values observed in wild-type GIT1-transfected cells in order to obtain fold changes (data represent mean + SEM; N = 3; **** and ** = p < 0.0001 and p < 0.01, respectively; two-way ANOVA with post hoc across row comparisons using a Tukey correction). (D) Elevation of phospho-PAK3 levels with wild-type GIT1 but not GIT1-R283W. For phospho-PAK immunoblots, band intensities were normalized to total Myc-PAK3 signals, and then to values for wild-type GIT1-transfected cells, in order to obtain fold changes (data represent mean + SEM; N = 3; **** = p < 0.0001; two-way ANOVA with post hoc across row comparisons using a Tukey correction). (E) GIT1-R283W shows reduced complex formation with PAK3, compared to wild-type GIT1. HEK293 cells were transfected with the indicated expression vectors. One day after transfection, cells were lysed and immunoprecipitated with FLAG-M2 antibody-conjugated agarose. The resulting FLAG-M2-immunoprecipitates were analyzed by immunoblot analysis with anti-FLAG-M2 or anti-Myc antibodies. (F) Quantitation of the decreased association between Myc-PAK3 and wild-type GIT1 or GIT1-R283W. Co-immunoprecipitated PAK3 immunoblot band intensities were normalized to immunoprecipitated GIT1-FLAG values. Data represent mean + SEM; N = 4; **** = p < 0.0001; two-tailed t-test; group variances were not significantly different. See also Figure S2.
Since the SHD domain of GIT1 where the R283W variant is located is known to mediate direct interaction with ARHGEF6/7 proteins that then bind to PAKs (Figure 2A), we therefore wondered whether the R283W variant affects the association of GIT1 and PAK3. To test this, we co-transfected FLAG-tagged wild-type GIT1 or GIT1-R283W and Myc-PAK3 and immunoprecipitated with FLAG antibody, and compared the co-precipitated levels of Myc-PAK3. Indeed, GIT1-R283W showed reduced association with co-transfected PAK3, compared to wild-type GIT1 (Figure 2E,F). Thus, we concluded that the reduced association between GIT1-R283W and PAK3 provides at least one mechanism for the impaired PAK3 activation levels observed in our functional human HEK293 cell assays.
GIT1-R283W fails to increase PAK activation and GAD1 protein levels in neurons
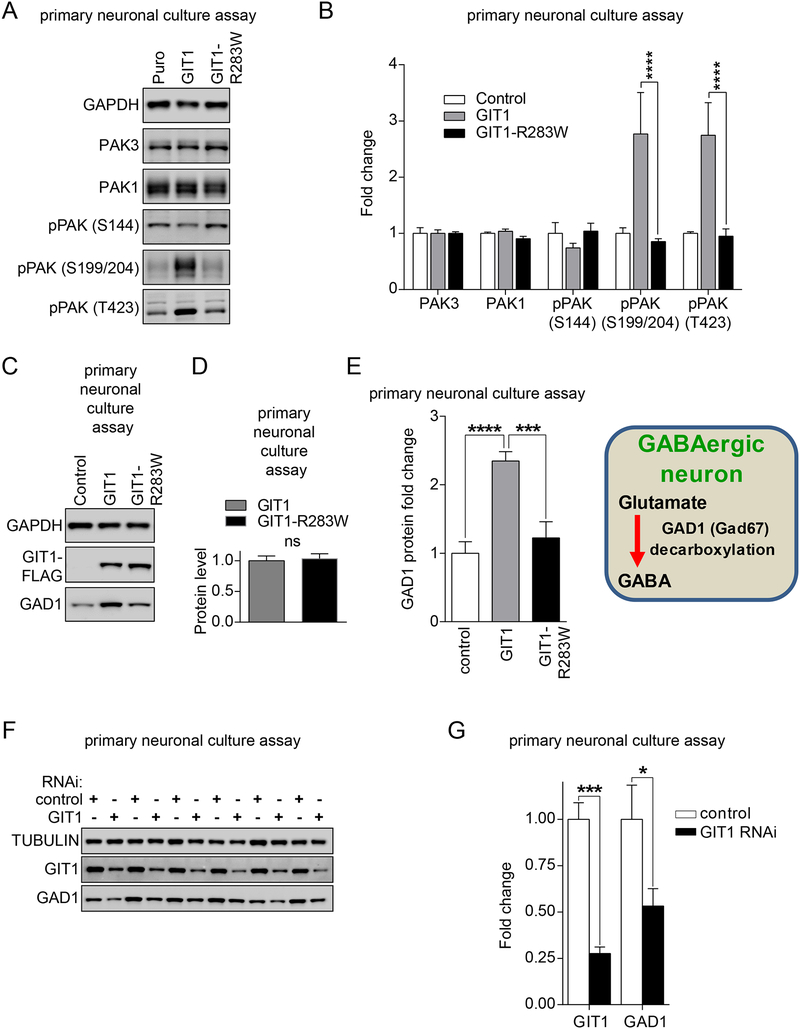
Having demonstrated functional deficits of GIT1-R283W in the human HEK293 assays, next we extended these findings to more context-relevant primary cultured rat hippocampal neurons. For these studies, we generated recombinant lentiviruses to enable the overexpression of either wild-type GIT1 or the GIT1-R283W variant in primary neurons. We infected primary rat hippocampal neurons after 16 days of in vitro (DIV) culture, a time point in which abundant synaptogenesis has occurred, and expressed the transgene for a total of seven days. Consistent with the result from overexpression in human HEK293 cells, lentiviral GIT1 overexpression in mature hippocampal neurons increased neuronal PAK activation, compared to neurons infected with the control lentivirus. However, lentiviral overexpression of the SCZ-unique GIT1-R283W variant failed to increase PAK activation in hippocampal neurons (Figure 3A,B), consistent with our findings in human HEK293 cells.
Figure 3. Effects of wild-type GIT1 and GIT1-R283W on neuronal PAK activation and GAD1 expression.
(A-E) Primary cultured hippocampal neurons at DIV 14–16 were infected with lentiviruses expressing the indicated wild-type GIT1, GIT1-R283W, or empty vector control (Puro) constructs. One week later, neurons were harvested in 2X SDS sample buffer and subjected to immunoblot analysis using the indicated antibodies (A). (B) Immunoblot intensities for PAK3, PAK1, and phospho-PAK (S144, S199/S204, or T423) were normalized to GAPDH values, and then compared to empty vector control lentivirus values to obtain fold changes. Significant elevation of pPAK (S199/S204) and pPAK (T423) levels occurred with expression of wild-type GIT1 but not GIT1-R283W. Data represent mean + SEM; N = 4; **** = p < 0.0001; two-way ANOVA with post hoc across row comparisons using a Tukey correction. (C) Immunoblot analysis for GAD1 upon expression of wild-type GIT1 compared to GIT1-R283W. (D) Relative levels of GIT1 and GIT1-R283W indicating no difference in expression. Data represent mean + SEM; N = 4; ns = not significant; two-tailed t-test. (E) Immunoblot intensities were normalized to GAPDH values, and then compared to empty vector control lentivirus values to obtain fold changes. Significant elevation of GAD1 protein levels occurred with expression of wild-type GIT1 but not GIT1-R283W. Data represent mean + SEM; N = 4; *** or **** = p < 0.001 or 0.0001, respectively; one-way ANOVA with post hoc comparisons of all means to each other using a Tukey correction. (F-G) Effect of GIT1 knockdown on GAD1 levels in hippocampal neurons. Cultured hippocampal neurons were infected with lentiviruses expressing either GIT1-RNAi or scrambled RNAi control. Five-seven days later, infected hippocampal neurons were harvested with 2X SDS sample buffers and analyzed with GIT1, GAD1, or tubulin antibodies. Band intensities were normalized to tubulin intensities, and then to scrambled RNAi controls to obtain fold changes. Data represent mean + SEM; N = 6; * or *** = p < 0.05 or 0.001, respectively; two-way ANOVA with post hoc across row comparisons using a Sidek correction. See also Figure S3.
Dysregulation of inhibitory GABAergic neurotransmission has been proposed as a critical aspect of SCZ pathophysiology (39). In particular, post-mortem human brain studies have repeatedly reported reduced levels of GAD1 (Glutamate decarboxylase 1), the rate-limiting enzyme in the biosynthesis of the major inhibitory neurotransmitter gamma-aminobutyric acid (GABA), in SCZ subjects compared to matched controls. In addition, GIT1 is expressed along with GAD1 in GABAergic inhibitory neurons (40). Thus, we tested the effect of GIT1 overexpression on GAD1 levels in the primary hippocampal neurons. As shown in Figure 3C,D,E, GIT1 and GIT1-R283W were expressed equally, but only wild-type GIT1 increased GAD1 protein levels. In contrast, a number of other markers of glutamatergic synaptic proteins were either not affected by wild-type GIT1 or GIT1-R283W, or only very modestly, as determined by immunoblot analyses (Supplemental Figure S3).
To complement these results from the lentiviral GIT1 overexpression study in neurons, we next asked whether knockdown of endogenous GIT1 in cultured neurons could lead to reduction of GAD1 protein levels. We infected cultured hippocampal neurons at DIV14 with lentiviruses either expressing GIT1-RNAi or scrambled-RNAi. Lentiviral overexpression of GIT1 RNAi for five-seven days strongly reduced endogenous levels of GIT1 in cultured neurons. RNAi-knockdown of endogenous GIT1 protein in mature hippocampal neurons also significantly reduced GAD1 protein levels, compared to neurons infected with lentivirus expressing scrambled RNAi (Figure 3F,G). Taken together with the overexpression studies where GIT1-R283W also failed to increase GAD1 protein levels, these results suggest that disruption of GIT1 activity may lead to impairment of GABAergic neuron function.
GIT1-S601N fails to increase PAK activation and GAD1 protein levels in neurons
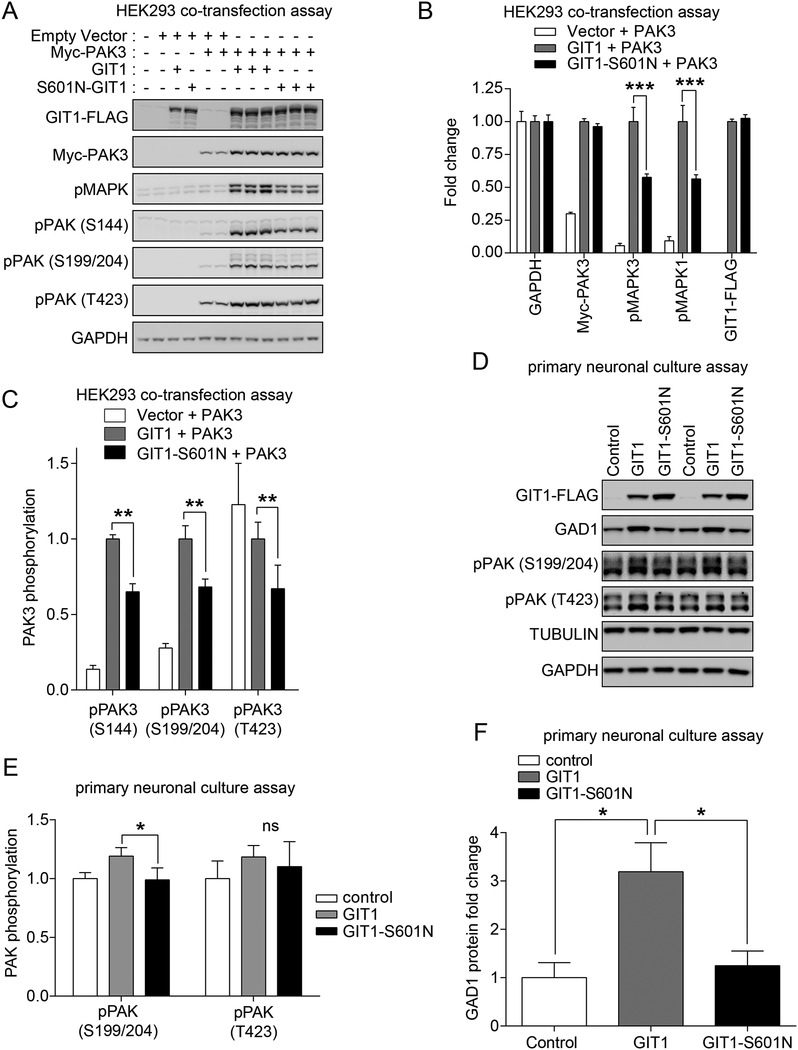
Having identified functional deficits with the GIT1-R283W, we turned our attention to other rare variants identified by exome sequencing from the Bulgarian trios and Swedish cohort that were unique to either SCZ cases or controls. We next selected the de novo GIT1-S601N variant (serine (S) at 601 changed to asparagine (N)) found in a proband with SCZ but not their unaffected mother or father, as the next variant to functionally characterize in order to test the generalizability of the functional deficits observed with GIT1-R283W. Accordingly, we co-expressed GIT1-S601N with PAK3 in the human HEK293 cell assay, and examined its effect on PAK3 and MAPK activation. As shown in Figure 4A–C, similar to GIT1-R283W, expression of PAK3 with the GIT1-S601N variant resulted in reduced phospho-PAK3 and phospho-MAPK levels compared to co-transfection of wild-type GIT1 plus PAK3. Furthermore, lentiviral-mediated expression of GIT1-S601N in cultured primary rat hippocampal neurons showed that this variant, like GIT1-R283W, was deficient in increasing phospho-PAK (S199/S204) levels (Figure 4D–E), although the deficiency was not as great as observed with GIT1-R283W. In further concordance with the results from GIT1-R283W, lentiviral overexpression of GIT1-S601N in cultured hippocampal neurons also failed to increase GAD1 protein levels, as compared to wild-type GIT1 (Figure 4D,F). Taken together, these results provide further evidence that genetic variation found in GIT1 in SCZ cases is associated with altered PAK3 signaling and pathways critical to the balance of inhibitory/excitatory neurotransmission.
Figure 4. Deficits of the SCZ de novo GIT1 variant S601N in PAK and MAPK activation and regulation of GAD1 levels.
(A-C) Effect of GIT1-S601N variant on PAK3 activation and MAPK activation in HEK293 cells. HEK293 cells were transfected with the indicated expression vectors. 24hrs later, cells were harvested in 2X SDS sample buffer. (A) Protein samples were subjected to immunoblot analysis using the indicated antibodies. (B) Band intensities for pMAPK, Myc-PAK3, or GIT1 were normalized to GAPDH. Normalized band intensities were compared to wild-type GIT1. Data represent mean + SEM; N = 3; ** = p < 0.01; two-way ANOVA with post hoc across row comparisons using a Sidek correction. (C) Phospho-PAK3 immunoblot band intensities were normalized to total Myc-PAK3 band intensities, and then to levels in wild-type GIT1-transfected cells in order to obtain fold changes. Data represent mean + SEM; N = 3; * and ** = p < 0.05 and 0.01, respectively; two-way ANOVA with post hoc across row comparisons using a Sidek correction. (D-G) Effect of the GIT1-S601N variant on PAK3 activation in hippocampal neurons. Cultured hippocampal neurons at DIV 14–16 were infected with lentiviruses expressing the indicated wild-type GIT1, GIT1-S601N, or empty vector control (Puro) constructs. One week later, hippocampal neurons were harvested in 2X SDS sample buffer, and subjected to immunoblot analysis (D) using the indicated antibodies. Immunoblot intensities were normalized with Tubulin intensities, and compared to empty vector control. (E) Relative expression levels of GIT1 and GIT1-S601N after lentiviral infection of cultured hippocampal neurons indicating increased levels of GIT1-S601N. Data represent mean + SEM; N = 3; * = p < 0.05; two-tailed t-test. (F) Deficit in pPAK (S199/S204) activation comparing GIT1 to GIT1-S601N with a trend but not a significant difference in pPAK (T423). Data represent mean + SEM; N = 3; * = p < 0.05; ns = not significant; two-tailed t-test. (G) GIT1 but not GIT1-S601N expression increases GAD1 protein levels in cultured hippocampal neurons. GAD1 band intensities were normalized with tubulin intensities, and compared to the empty vector control. Data represent mean + SEM; N = 3; * = p < 0.05 (two-tailed t-test).
Summary of functional characterization of additional GIT1 variants
We next completed the systematic functional characterization of additional GIT1 coding variants from the exome studies. While many studies identifying genetic variants by sequencing focus only on validating alleles found in cases, we reasoned that testing control unique variants would be equally critical to provide biological context for our findings of dysfunction GIT1 variants in SCZ subjects. Thus, we set out to systematically test a total of 10 additional GIT1 SNVs: five unique to SCZ cases (E33K, V37M, R256P, G506A, Q587L) and five unique to controls (A55T, M290V, R381Q, R598C, V681M) (Figure 1A).
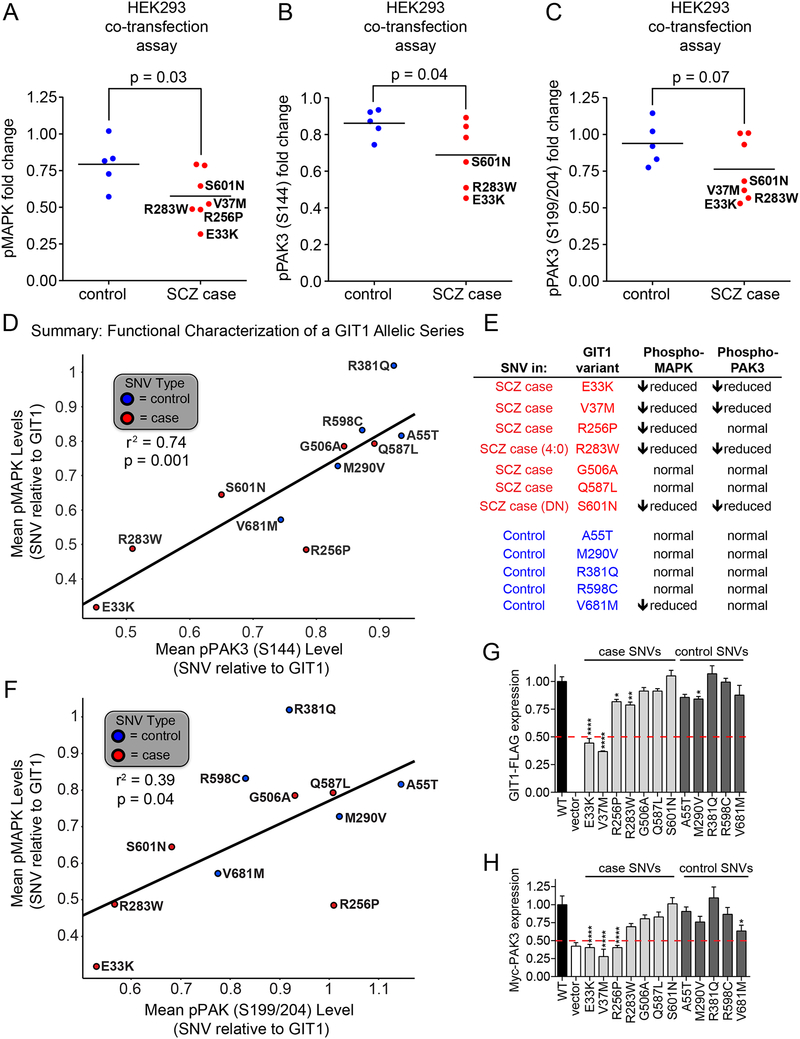
For functional evaluation of these 10 additional GIT1 variants, we again made mammalian over-expression vectors, and after confirming mutagenesis using DNA sequencing we co-expressed individual GIT1 variants with Myc-tagged PAK3 into human HEK293 cells and examined effects on MAPK activation, co-expressed PAK3 activation, as well as expression levels of GIT1 variants and PAK3. As summarized in Figure 5A, an aggregated analysis of the 10 additional GIT1 SNVs and both the GIT1-R283W and GIT1-S601N SNVs, revealed a significant (p=0.03) decrease in the phospho-MAPK levels after co-transfection of PAK3 and GIT1 SNVs from SCZ cases as compared to SNVs from controls. Here, in addition to the decreased MAPK activation with the two SCZ unique SNVs GIT1-R283W and GIT1-S601N, three other SCZ unique SNVs (E33K, V37M, R256P) showed reduced levels of phospho-MAPK accounting for a total of five out of seven of the unique GIT1 variants in SCZ subjects. In contrast, only one control unique SNV, GIT1-V681M, showed reduced phospho-MAPK levels.
Figure 5. Functional analysis of an allelic series of GIT1 coding variants in SCZ patients and controls.
(A-C) Plots of phospho-MAPK, phospho-PAK (S144), and phospho-PAK3 (S199/S204) as regulated by wild-type GIT1 and coding variants found in SCZ patients (red) or controls (blue) from co-transfection experiments with PAK3 in HEK293 cells. 24hrs after transfection, cells were harvested in 2X SDS sample buffer. Protein samples were subjected to immunoblot analysis using the indicated antibodies. Phospho-MAPK immunoblot band intensities were normalized to GAPDH levels, and then to levels in wild-type GIT1-transfected cells in order to obtain fold changes. Phospho-PAK3 immunoblot band intensities were normalized to total Myc-PAK3 band intensities, and then to levels in wild-type GIT1-transfected cells in order to obtain fold changes. Analysis includes all variants except GIT1-V37M was not run with pPAK3 (S144). Data points represent means; N = 4; * = p < 0.05 (one-tailed t-test); group variances were not significantly different. (D) Correlation between GIT1 coding variants’ degrees of activation of MAPK and PAK (S144) phosphorylation. A straight line fit (r2 = 0.74) is shown superimposed over the data points. Linear regression analysis indicated that this line fit had a statistically significant deviation from horizontal (p = 0.001). A summary table (E) of these results is shown to the right of the graph. (F) Relationship between abilities of coding variants from SCZ patients (red) and controls (blue) to activate MAPK and PAK3 (S199/204) when co-expressed with PAK3 in HEK293 cells. Plotted values were derived from immunoblot analyses as shown in Figure 2. A straight line fit is superimposed over the data points, suggesting a modest correlation between variant effects on MAPK and PAK3. In particular, the lower left region, highlighted by the red dashed lines, contains variants with little to no ability to activate MAPK and PAK3; three out of four of these variants were found uniquely in SCZ patients. (G) Expression of allelic series of GIT1 coding variants when transfected in HEK293 cells. GIT1 levels were measured by immunoblotting with anti-FLAG antibody, and normalized to GAPDH levels, and then to the level of wild-type GIT1 expression in order to obtain fold changes. Data represent mean + SEM; N = 3–6 for each variant; * = p < 0.05, ** = p < 0.01, **** = p < 0.0001; one-way ANOVA with post hoc across comparisons to wild-type with Dunnett correction. (H) Expression levels of Myc-PAK3 when co-expressed with wild-type GIT and coding variants in HEK293 cells. Myc-PAK3 levels were measured by immunoblotting with anti-Myc antibody, and normalized to GAPDH levels and then levels observed in cells transfected with wild-type GIT1 in order to obtain fold changes. Data represent mean + SEM; N = 3–6 for each variant; * = p < 0.05, **** = p < 0.0001; one-way ANOVA with post hoc across comparisons to wild-type GIT1 with Dunnett correction. In panels G and H, 50% decreases are indicated by red dashed lines; only GIT1 coding variants found in SCZ patients produce decreases below the 50% level.
Repeating this aggregate analysis for phospho-PAK3 (S144) levels also revealed a significant decrease (p=0.04) after co-transfection of PAK3 and GIT1 SNVs from SCZ cases as compared to controls (Figure 5B). A similar comparison for phospho-PAK3 (S199/S204) revealed a trend (p=0.07) of decreased phosphorylation levels amongst the SCZ unique SNVs compared to control unique SNVs (e.g. E33K, V37M), but this was not statistically significant (Figure 5C). Overall, a combined analysis of the relationship between phospho-PAK3 and phospho-MAPK levels showed that lower levels of pMAPK were significantly correlated with lower levels of pPAK3 (S144) (r2 =0.74; p = 0.001) (Figure 5D), with also a significant trend towards correlation with lower levels of pPAK3 (S199/S204) (r2=0.39; p = 0.04) (see Figure 5F).
While analyzing the aggregated data across all 12 GIT1 SNVs, we noted that the GIT1-E33K and GIT1-V37M SCZ-case variants showed dramatically reduced expression levels compared to wild-type GIT1 and the other variants, some of which had slightly reduced expression levels (Figure 5G). Additionally, PAK3 protein levels co-expressed with GIT1-E33K and GIT1-V37M were significantly lower than PAK3 levels co-expressed with GIT1 or control variants (Figure 5H). Thus, the various GIT1 variants found in SCZ subjects may affect GIT1 function through different mechanisms, including effects on the GIT1-PAK3 complex stability.
To gain insight into why the SCZ-variants E33K and V37M located in the ArfGAP domain affected GIT1 expression levels, whereas for example A55T did not, we constructed a homology model that allowed inspection of the local environment of each amino acid. As summarized in Supplementary Figure S4, this modeling revealed that both the E33 and V37 residues are located in a highly conserved α-helix that is central to the ArfGAP fold and flanked by regions of high sequence similarity. This α-helix with its adjacent residues provides two out of four cysteines that bind to the Zn2+ ion in the characteristic ArfGAP Zn2+-finger motif (CX2CX16CX2CX4R), a region important for the overall architecture of ArfGAP proteins (44). This is also quite near the arginine finger residue responsible for ArfGAP activity, R39 (45, 46). Thus, we speculate that the SCZ case-unique GIT1-V37M variant disrupts the stability of GIT1 by altering the Zn2+-finger motif and/or the arginine finger, while GIT1-E33K, which is located at a site that is more tolerable of such an amino acid change, may have a more complex effect leading to decreased protein levels (Supplementary Figure S4D–E). In contrast, the A55 residue was found to reside on a loop on the surface of GIT1 protruding out into the solvent in a poorly conserved region (Supplementary Figure S4C), which is consistent with the lack of functional effect of this control-unique variant on PAK3 or MAPK activation. Testing these predictions and gaining a more complete interpretation of structure-function relationships of GIT1 genetic variants will be facilitated by structure determination of GIT1 and/or its individual domains.
Discussion
The overall hypothesis of this study was that the systematic characterization of GIT1 variants found uniquely in SCZ subjects would reveal shared perturbations of biological function that may provide plausible molecular substrates mediating the dysfunction of neural circuits underlying the etiopathogenesis of SCZ. By systematically testing a total of 12 GIT1 SNVs, including a set of SNVs found uniquely in control subjects, our study showed that multiple rare GIT1 variants uniquely found in SCZ subjects had an impaired ability to induce PAK3 and MAPK phosphorylation, as well as increase or stabilize GAD1 protein levels in the case of two variants tested in primary neurons. Similar effects were observed with RNAi-mediated knockdown of GIT1 in primary cultured rat hippocampal neurons, suggesting that at least a subset of SCZ SNVs in GIT1 cause loss of function. Our finding of similar phenotypic deficits between different GIT1 variants is important as it suggests that the functional assays we utilized captured a sufficiently rich swath of biology. Overall, the correlation between observing functional deficits in any variant and having a SCZ case diagnosis is not expected to be 100% given the polygenic nature of SCZ etiology—i.e., it is expected that for a given functional assay, variants found in SCZ cases will have no functional deficits, while some coding variants found in controls will. Indeed, that certain SCZ-unique SNVs studied here (e.g. GIT1-G506A, GIT1-Q587L) had no detectable phenotypic difference in the biochemical assays utilized, and evidence for functional deficits in at least one control-unique SNV (e.g. GIT1-V681M) underscore the importance of utilizing functional measures when considering the impact of specific genetic variants identified by exome sequencing. As additional data from sequencing studies in SCZ emerges, it will also be important to re-evaluate our designations of ‘SCZ- or control-unique’ with the ultimate goal of comprehensively and quantitatively characterizing the case/control prevalence of all variants, and functionally testing the entire case/control allelic series. Overall, the identification of rare coding variants in GIT1 that are dysfunctional now sets the stage for dissecting the synaptic function of GIT1 through ‘rescue’ experiments following RNAi-mediated gene silencing, in neurons from GIT1 knockout mice, and ideally in GIT1 knock-in mice with specific variants, to better understand the contribution of different biochemical functions of GIT1 to neurodevelopment and synaptic plasticity. Using these models, in future studies it will be critical to directly measure the effects of biochemically dysfunctional GIT1 variants on excitatory and inhibitory synaptic structure and synaptic transmission. Moreover, since the studies described here only assayed a limited set of GIT1’s full functionality, which were guided by our initial observation of deficits in the GIT1-R283W variant and our overarching hypothesis that pathways implicated in non-syndromic X-linked intellectual disability may also be of relevance to cognitive deficits in schizophrenia (e.g. ARHGEF-PAK3 −31–33, 51)), it will also be important to assay both the case-unique and control-unique variants in additional functional assays not considered here.
SCZ-associated R283W and S601N GIT1 coding SNVs
The GIT1-R283W and GIT1-S601N coding SNVs from SCZ cases were both impaired in stimulating PAK3 and MAPK phosphorylation, with the GIT1-R283W variant having the most significant deficits of all the SNVs tested. In future studies, it will be interesting to further investigate the molecular mechanisms by which these SNVs cause impairment in GIT1 function. We show that the R283W SNV, located in the SHD domain of GIT1, has a reduced ability to associate with PAK3. Since ARHGEF6/7 are known to bridge between GIT1 and PAKs, this reduced association may be due to alteration of the GIT1-ARHGEF6/7-PAK complex. Given the location of R283W, this variant may also affect the association of GIT1 with other key synaptic proteins associated with psychiatric disease genetic risk, such as the presynaptic cytomatrix protein PCLO (Piccolo) and PPFIA2 (Protein Tyrosine Phosphatase, Receptor Type, F Polypeptide (PTPRF), Interacting Protein (Liprin), Alpha 2) (14, 47). Another interesting question is why GIT1-S601N has similar defects to GIT1-R283W, given that S601 is located well outside of the SHD domain of GIT1 in a region with no defined structural domain. GIT1-S601 is conserved between GIT1 and its related family member GIT2, is phosphorylated in HEK293 cells and in the mouse brain, and was suggested to be a potential PKA, PKC or GSK3 site (48, 49); our preliminary data suggests its phosphorylation is sensitive to inhibitors of the serine/threonine kinase GSK3 (Glycogen synthase kinase 3) (S.J.H., manuscript in preparation). It will be interesting to test whether S601 phosphorylation affects the tertiary protein structure of GIT1 to favor functional coupling between ARHGEF6/7 and PAK3, and whether the effects of GSK3 inhibitors on synaptic plasticity and behavior are mediated through alteration of GIT1 function.
Whereas HEK293 cells required the co-transfection of GIT1 and PAK3 to cause significant PAK3 phospho-activation, GIT1 expression alone in rat hippocampal neurons was sufficient to cause phospho-activation of PAK without the need for co-infection with PAK3 (Figure 3A, B). Based upon the fact that we observed a basal level of phospho-PAK in the primary hippocampal neurons, whereas the basal level of phospho-PAK was not detectable in HEK293 cells (Figure 2B), the differences between the two cell systems are likely due to differences in the basal level of PAK signaling, which is higher in hippocampal neurons than in HEK293 cells. We note, however, that PAK1 and GIT1 in HEK293 cells were unable to couple to produce the same phospho-activation of MAPK and PAK as seen with the combination of PAK3 and GIT1 (Supplemental Figure S2), which points to the specificity of the coupling observed between GIT1-PAK3 rather than simply of being a consequence of increasing PAK levels alone in HEK293 cells”.
Notably, dysregulation of MAPK activity due to loss-of-function alleles of GIT1 potentially links the GIT1 signaling network to other pathways implicated in intellectual disability and to potential SCZ GWAS genes that are known to cause alterations in MAPK activity; for example, SYNGAP1 (Synaptic GTPase-activating protein 1). SYNGAP1, an excitatory synapse-enriched Ras-GTPase activating protein, normally antagonizes MAPK activation by decreasing levels of Ras-GTP (50). The opposed directionality of regulation of MAPK due to loss of GIT1 or SYNGAP1 suggests that a fine balance of MAPK pathway activation is a requirement for proper synaptic plasticity. It will thus be of great interest to determine if the common genetic variation within the SYNGAP1 locus alters GIT1-PAK3 signaling. Extending this principle of GIT1 connectivity, we speculate that similar biochemical studies of allelic series of other GIT1-interacting proteins implicated by the emerging genetics of SCZ (e.g., CNKSR2, PTPRF) will provide a means to prioritize genes within loci implicated by GWAS and ultimately generate new insight into aspects of the shared dysfunction and the biochemical basis of SCZ pathogenesis.
Our identification of multiple rare GIT1 coding SNVs found in SCZ subjects that have impaired ability to induce PAK3 phosphorylation poses the question of whether PAK3 function and its downstream substrates could be dysregulated in SCZ patients. Indeed, PAK3 and ARHGEF6 loss-of-function mutations are sufficient to cause a form of X-linked intellectual disability (31, 32, 51), and are thought to work together in a complex (52). Additionally, in one large family pedigree, psychiatric symptoms occur in individuals with a PAK3 coding mutation (A365E) located in the kinase domain that abolishes PAK3 kinase activity (35, 51, 53). PAK1–3 mRNA levels have also recently shown to be differentially expressed in post-mortem studies of schizophrenic brains (54). PAK3 knockout mice have been generated, and initial testing indicates some cognitive impairment (55), although PAK3-PAK1 double knockouts have a more severe phenotype (56) that more closely resembles the knockout of ARHGEF6 (33) and the human disease state. Finally, PAK3 has recently been shown to control the surface expression of the AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid) receptor subunit GLUA1, pointing to another mechanism through which dysfunction of PAK3 signaling may lead to deficits in synaptic plasticity and cognition (57). Further behavioral, neurophysiological, and biochemical testing will be necessary to understand the deficits caused by loss of PAK3 in comparison to loss of GIT1.
In mice, GIT1 and PAKs have been shown to be required for GABAA receptor synaptic stability and inhibitory neurotransmission (18). Indeed, whole-body GIT1−/− knockout mice have reduced inhibitory synaptic transmission in the hippocampus as determined by postsynaptic potential (IPSP) frequency recordings in hippocampal CA1 pyramidal neurons (23). Given that SCZ patients have both cognitive impairment and reduced GAD1 expression (58), loss of function of GIT1 may contribute to the deficits in GAD1 expression in SCZ. Thus, an intriguing question is whether GIT1 genetic variants found in SCZ cases would show impaired inhibitory synaptic transmission in vivo; a question that can be addressed by generating GIT1-R283W knock-in mice.
In conclusion, while the pace of genetic variant discovery will undoubtedly continue to outpace the speed with which experimental neurobiological studies of these variants can be performed, the principles of network medicine with a focus on modules of human disease genes and convergent pathways in combination with rigorous biochemical and physiological studies holds promise to provide critically needed insight into the pathophysiology of SCZ. Such efforts, nucleated, for example around GIT1 and its interactors, may in turn advance efforts to develop next generation, targeted pharmacological agents to treat, and ideally prevent, SCZ and other neuropsychiatric disorders.
Supplementary Material
Acknowledgments
We thank members of the Stanley Center for Psychiatric Research and the Chemical Neurobiology Laboratory for helpful discussions and critical feedback. Dr. Guoping Feng (MIT) is thanked for critical feedback. This work was supported by funding from the Stanley Medical Research Institute and the National Institute of Mental Health (S.J.H, R01MH095088).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of general psychiatry. 2003;60(12):1187–92. [DOI] [PubMed] [Google Scholar]
- 2.Genome-wide association study identifies five new schizophrenia loci. Nature genetics. 2011;43(10):969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet (London, England). 2013;381(9875):1371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Molecular psychiatry. 2012;17(2):142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Need AC, McEvoy JP, Gennarelli M, Heinzen EL, Ge D, Maia JM, et al. Exome sequencing followed by large-scale genotyping suggests a limited role for moderately rare risk factors of strong effect in schizophrenia. American journal of human genetics. 2012;91(2):303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506(7487):179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timms AE, Dorschner MO, Wechsler J, Choi KY, Kirkwood R, Girirajan S, et al. Support for the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA psychiatry. 2013;70(6):582–90. [DOI] [PubMed] [Google Scholar]
- 10.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506(7487):185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature reviews Genetics. 2012;13(8):537–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146(2):247–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmalzigaug R, Phee H, Davidson CE, Weiss A, Premont RT. Differential expression of the ARF GAP genes GIT1 and GIT2 in mouse tissues. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 2007;55(10):1039–48. [DOI] [PubMed] [Google Scholar]
- 14.Kim S, Ko J, Shin H, Lee JR, Lim C, Han JH, et al. The GIT family of proteins forms multimers and associates with the presynaptic cytomatrix protein Piccolo. The Journal of biological chemistry. 2003;278(8):6291–300. [DOI] [PubMed] [Google Scholar]
- 15.Ko J, Kim S, Valtschanoff JG, Shin H, Lee JR, Sheng M, et al. Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23(5):1667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Webb DJ, Asmussen H, Horwitz AF. Synapse formation is regulated by the signaling adaptor GIT1. The Journal of cell biology. 2003;161(1):131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podufall J, Tian R, Knoche E, Puchkov D, Walter AM, Rosa S, et al. A presynaptic role for the cytomatrix protein GIT in synaptic vesicle recycling. Cell reports. 2014;7(5):1417–25. [DOI] [PubMed] [Google Scholar]
- 18.Smith KR, Davenport EC, Wei J, Li X, Pathania M, Vaccaro V, et al. GIT1 and betaPIX are essential for GABA(A) receptor synaptic stability and inhibitory neurotransmission. Cell reports. 2014;9(1):298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montesinos MS, Dong W, Goff K, Das B, Guerrero-Given D, Schmalzigaug R, et al. Presynaptic Deletion of GIT Proteins Results in Increased Synaptic Strength at a Mammalian Central Synapse. Neuron. 2015;88(5):918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25(13):3379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmalzigaug R, Rodriguiz RM, Bonner PE, Davidson CE, Wetsel WC, Premont RT. Impaired fear response in mice lacking GIT1. Neuroscience letters. 2009;458(2):79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menon P, Deane R, Sagare A, Lane SM, Zarcone TJ, O’Dell MR, et al. Impaired spine formation and learning in GPCR kinase 2 interacting protein-1 (GIT1) knockout mice. Brain research. 2010;1317:218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Won H, Mah W, Kim E, Kim JW, Hahm EK, Kim MH, et al. GIT1 is associated with ADHD in humans and ADHD-like behaviors in mice. Nature medicine. 2011;17(5):566–72. [DOI] [PubMed] [Google Scholar]
- 24.Premont RT, Claing A, Vitale N, Freeman JL, Pitcher JA, Patton WA, et al. beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(24):14082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagrodia S, Bailey D, Lenard Z, Hart M, Guan JL, Premont RT, et al. A tyrosinephosphorylated protein that binds to an important regulatory region on the cool family of p21-activated kinase-binding proteins. The Journal of biological chemistry. 1999;274(32):22393–400. [DOI] [PubMed] [Google Scholar]
- 26.Zhao ZS, Manser E, Loo TH, Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Molecular and cellular biology. 2000;20(17):6354–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang ZM, Simmerman JA, Guibao CD, Zheng JJ. GIT1 paxillin-binding domain is a four-helix bundle, and it binds to both paxillin LD2 and LD4 motifs. The Journal of biological chemistry. 2008;283(27):18685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmalzigaug R, Garron ML, Roseman JT, Xing Y, Davidson CE, Arold ST, et al. GIT1 utilizes a focal adhesion targeting-homology domain to bind paxillin. Cellular signalling. 2007;19(8):1733–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlenker O, Rittinger K. Structures of dimeric GIT1 and trimeric beta-PIX and implications for GIT-PIX complex assembly. Journal of molecular biology. 2009;386(2):280–9. [DOI] [PubMed] [Google Scholar]
- 30.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, et al. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Molecular cell. 1998;1(2):183–92. [DOI] [PubMed] [Google Scholar]
- 31.Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, et al. PAK3 mutation in nonsyndromic X-linked mental retardation. Nature genetics. 1998;20(1):25–30. [DOI] [PubMed] [Google Scholar]
- 32.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, et al. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nature genetics. 2000;26(2):247–50. [DOI] [PubMed] [Google Scholar]
- 33.Ramakers GJ, Wolfer D, Rosenberger G, Kuchenbecker K, Kreienkamp HJ, Prange-Kiel J, et al. Dysregulation of Rho GTPases in the alphaPix/Arhgef6 mouse model of X-linked intellectual disability is paralleled by impaired structural and synaptic plasticity and cognitive deficits. Human molecular genetics. 2012;21(2):268–86. [DOI] [PubMed] [Google Scholar]
- 34.Boda B, Alberi S, Nikonenko I, Node-Langlois R, Jourdain P, Moosmayer M, et al. The mental retardation protein PAK3 contributes to synapse formation and plasticity in hippocampus. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24(48):10816–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreis P, Thevenot E, Rousseau V, Boda B, Muller D, Barnier JV. The p21-activated kinase 3 implicated in mental retardation regulates spine morphogenesis through a Cdc42-dependent pathway. The Journal of biological chemistry. 2007;282(29):21497–506. [DOI] [PubMed] [Google Scholar]
- 36.Dai X, Iwasaki H, Watanabe M, Okabe S. Dlx1 transcription factor regulates dendritic growth and postsynaptic differentiation through inhibition of neuropilin-2 and PAK3 expression. The European journal of neuroscience. 2014;39(4):531–47. [DOI] [PubMed] [Google Scholar]
- 37.Hayashi ML, Choi SY, Rao BS, Jung HY, Lee HK, Zhang D, et al. Altered cortical synaptic morphology and impaired memory consolidation in forebrain- specific dominant-negative PAK transgenic mice. Neuron. 2004;42(5):773–87. [DOI] [PubMed] [Google Scholar]
- 38.Yin G, Haendeler J, Yan C, Berk BC. GIT1 functions as a scaffold for MEK1-extracellular signal-regulated kinase 1 and 2 activation by angiotensin II and epidermal growth factor. Molecular and cellular biology. 2004;24(2):875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis DA. Inhibitory neurons in human cortical circuits: substrate for cognitive dysfunction in schizophrenia. Current opinion in neurobiology. 2014;26:22–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86(6):1369–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balu DT, Coyle JT. The NMDA receptor ‘glycine modulatory site’ in schizophrenia: D-serine, glycine, and beyond. Current opinion in pharmacology. 2015;20:109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu W, MacDonald ML, Elswick DE, Sweet RA. The glutamate hypothesis of schizophrenia: evidence from human brain tissue studies. Annals of the New York Academy of Sciences. 2015;1338:38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taniguchi S, Nakazawa T, Tanimura A, Kiyama Y, Tezuka T, Watabe AM, et al. Involvement of NMDAR2A tyrosine phosphorylation in depression-related behaviour. The EMBO journal. 2009;28(23):3717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kahn RA, Bruford E, Inoue H, Logsdon JM Jr., Nie Z, Premont RT, et al. Consensus nomenclature for the human ArfGAP domain-containing proteins. The Journal of cell biology. 2008;182(6):1039–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ismail SA, Vetter IR, Sot B, Wittinghofer A. The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism. Cell. 2010;141(5):812–21. [DOI] [PubMed] [Google Scholar]
- 46.Meyer MZ, Deliot N, Chasserot-Golaz S, Premont RT, Bader MF, Vitale N. Regulation of neuroendocrine exocytosis by the ARF6 GTPase-activating protein GIT1. The Journal of biological chemistry. 2006;281(12):7919–26. [DOI] [PubMed] [Google Scholar]
- 47.Choi KH, Higgs BW, Wendland JR, Song J, McMahon FJ, Webster MJ. Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biological psychiatry. 2011;69(4):353–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb DJ, Mayhew MW, Kovalenko M, Schroeder MJ, Jeffery ED, Whitmore L, et al. Identification of phosphorylation sites in GIT1. Journal of cell science. 2006;119(Pt 14):2847–50. [DOI] [PubMed] [Google Scholar]
- 49.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rumbaugh G, Adams JP, Kim JH, Huganir RL. SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(12):4344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rejeb I, Saillour Y, Castelnau L, Julien C, Bienvenu T, Taga P, et al. A novel splice mutation in PAK3 gene underlying mental retardation with neuropsychiatric features. European journal of human genetics: EJHG. 2008;16(11):1358–63. [DOI] [PubMed] [Google Scholar]
- 52.Node-Langlois R, Muller D, Boda B. Sequential implication of the mental retardation proteins ARHGEF6 and PAK3 in spine morphogenesis. Journal of cell science. 2006;119(Pt 23):4986–93. [DOI] [PubMed] [Google Scholar]
- 53.Gedeon AK, Nelson J, Gecz J, Mulley JC. X-linked mild non-syndromic mental retardation with neuropsychiatric problems and the missense mutation A365E in PAK3. American journal of medical genetics Part A. 2003;120a(4):509–17. [DOI] [PubMed] [Google Scholar]
- 54.Datta D, Arion D, Corradi JP, Lewis DA. Altered Expression of CDC42 Signaling Pathway Components in Cortical Layer 3 Pyramidal Cells in Schizophrenia. Biological psychiatry. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng J, Meng Y, Hanna A, Janus C, Jia Z. Abnormal long-lasting synaptic plasticity and cognition in mice lacking the mental retardation gene Pak3. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25(28):6641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang W, Zhou Z, Asrar S, Henkelman M, Xie W, Jia Z. p21-Activated kinases 1 and 3 control brain size through coordinating neuronal complexity and synaptic properties. Molecular and cellular biology. 2011;31(3):388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hussain NK, Thomas GM, Luo J, Huganir RL. Regulation of AMPA receptor subunit GluA1 surface expression by PAK3 phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(43):E5883–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzalez-Burgos G, Cho RY, Lewis DA. Alterations in cortical network oscillations and parvalbumin neurons in schizophrenia. Biological psychiatry. 2015;77(12):1031–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manabe R, Kovalenko M, Webb DJ, Horwitz AR. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. Journal of cell science. 2002;115(Pt 7):1497–510. [DOI] [PubMed] [Google Scholar]
- 60.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42(Web Server issue):W252–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai M, Pang X, Lou J, Zhou Q, Zhang K, Ma J, et al. Mechanistic insights into regulated cargo binding by ACAP1 protein. The Journal of biological chemistry. 2012;287(34):28675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pei J, Grishin NV. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods in molecular biology (Clifton, NJ). 2014;1079:263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao ZS, Manser E. PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation. The Biochemical journal. 2005;386(Pt 2):201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.