Abstract
Purpose:
Determination of the minimum number of gradient directions (Nmin) for robust measurement of spherical mean diffusion weighted signal ().
Methods:
Computer simulations were employed to characterize the relative standard deviation (RSD) of the measured spherical mean signal as a function of the number of gradient directions (N). The effects of diffusion weighting b-value and signal-to-noise ratio (SNR) were investigated. Multi-shell high angular resolution Human Connectome Project diffusion data were analyzed to support the simulation results.
Results:
RSD decreases with increasing N, and the minimum number of N needed for RSD ≤ 5% is referred to as Nmin. At high SNRs, Nmin increases with increasing b-value to achieve sufficient sampling. Simulations showed that Nmin is linearly dependent on the b-value. At low SNRs, Nmin increases with increasing b-value to reduce the noise. RSD can be estimated as , where σ = 1/SNR is the noise level. The experimental results were in good agreement with the simulation results. The spherical mean signal can be measured accurately with a subset of gradient directions.
Conclusion:
As Nmin is affected by b-value and SNR, we recommend using 10×b/b1 (b1 = 1 ms/μm2) uniformly distributed gradient directions for typical human diffusion studies with SNR ~ 20 for robust spherical mean signal measurement.
Keywords: Diffusion tensor imaging (DTI), intra-axonal volume fraction, spherical mean signal, spherical mean technique (SMT)
Introduction
Diffusion MRI is a non-invasive tool to detect tissue microstructural information based on restricted water Brownian motion within tissues. The conventional technique measuring tissue microstructure is diffusion tensor imaging (DTI), a technique that models water diffusion in each voxel as a symmetric tensor [1]. The DTI derived mean diffusivity (MD) and fractional anisotropy (FA), measures widely used to quantify tissue microstructure, are rotationally invariant. However, for two voxels with different fiber directions, the measured FAvoxel1 and FAvoxel2 cannot be simply averaged to obtain the entire voxel FA. In other words, the single tensor model is inappropriate for quantifying tissue microstructure in situations with complex fiber orientation distribution (FOD). Another diffusion-based approach to quantify tissue microstructure is direct measurement of tissue properties, such as axon size and intra-axonal volume fraction (Vin) [2–5]. Vin is rotationally invariant, independent of FOD, and can be averaged. The crossing fiber problem still needs to be considered in order to measure Vin accurately. Simultaneous fitting of FOD and Vin is challenging as the number of unknown parameters increases with increasing the complexity of FOD [6–9]. Recently, it was demonstrated that the FOD information can be factored out by analyzing the spherical mean diffusion weighted signal () averaged over all gradient directions [10–12]. A simple linear relation between Vin and was further derived analytically [12]. -based analysis has been applied in various recent works, such as apparent fiber density [13–15], spherical mean technique [11,16,17], fiber ball imaging [12,18], power-law scaling [19,20], and rotationally-invariant framework [21–23].
Previous -based studies employed a large number of gradient directions, but subsequent sparse sampling analysis demonstrated that the number of gradient directions could be reduced without significant effect on accuracy [16–18]. Clinical diffusion scan protocols are typically less than 5 min, which only allows acquiring ~ 30 different directions. It is undetermined as whether 30 directions is enough for accurate measurement, especially at high b-values (b ≥ 3 ms/μm2). Multi-shell acquisition schemes are able to obtain a more comprehensive diffusion dataset, and hence, provide greater sensitivity in detecting tissue microstructure than single-shell schemes [11,16,24]. The increased scanning time requires optimal design of number of directions for each shell.
Given the substantial recent interest in using to quantify tissue microstructure, it is worthwhile to determine the minimum number of gradient directions for robust measurement of . The current study aims to determine the minimum number of gradient directions for robust measurement of spherical mean signals at different b-values. Similar to previous DTI and spherical deconvolution studies [25–27], we employed computer simulations to investigate the effect of signal-to-noise ratio (SNR) on the required number of gradient directions. Multi-shell Human Connectome Project (HCP) diffusion datasets were used to support the simulation results.
Materials and methods
Theory
The results of single fiber analysis can be generalized to crossing fibers as illustrated by Tournier et al. [27] in the determination of the minimum number of directions for spherical deconvolution. The problem of determining the minimum number of directions for spherical mean signal can also be restricted to single fiber analysis. The total spherical mean signal in each voxel is simply the sum of each single fiber’s spherical mean signal, and the spherical mean signal of a single fiber is independent of the fiber orientation [10,11]. For this reason, we focus on the analysis of a single fiber pointing in the direction n. Based on the widely used two-compartment model of intra- and extra-axonal spaces, the diffusion weighted signal (S) along gradient direction g can be expressed as
| (1) |
where S0 is the signal for b=0, and , , , are intra-axonal axial diffusivity, intra-axonal radial diffusivity, extra-axonal axial diffusivity, and extra-axonal radial diffusivity, respectively. Following previous multi-compartment modeling studies, we assume [16,18,23], [16,28], and the intrinsic diffusivity λ [16]. The ground truth is the signal averaged over all gradient directions, and it can be expressed as
| (2) |
where erf is the error function. Due to the exponential decay term, the extra-axonal water contribution can be neglected with sufficiently large b-values. In that case, a simple linear relation between and Vin can be derived as [12,13]. Note that erf when bλ ≥ 3.
Simulations
Diffusion-weighted signals were simulated with gradient directions approximately evenly distributed on a unit sphere as proposed by Jones et al. [29]. The number N of gradient directions varied from 6 to 100, and the spherical mean signal was calculated as the signal averaged over N directions. For each value of N, 10,000 different directions of the single fiber n (uniformly sampled on a sphere) were simulated, from which the mean and standard deviation of were calculated. The relative standard deviation (RSD), defined as the ratio of the standard deviation to the mean, was used to indicate the precision of measurement. Nmin was determined as the minimum number of gradient directions for RSD() ≤ 5%. Nmin is the number of gradient directions needed to achieve sufficient sampling.
Computer simulations were performed in MATLAB with S0 = 1, Vin = 0.6, and λ = 2 μm2/ms to mimic typical brain white matter parameters [16,18,23]. The diffusion weighting b-value varied from 1 to 10 ms/μm2. To investigate the effect of SNR on Nmin, complex Gaussian noise was added to the simulated signal S(b,g), and the magnitude of the noisy signal was then taken as the measured signal M(b,g). To correct for Rician bias in the measured signal, here we used the corrected amplitude signal , where σ = 1/SNR is the noise level. The corresponding , , RSD(), and RSD() were calculated accordingly. Two typical SNRs in the b=0 image of 20 and 40 were used for current study. Two factors are expected to affect RSD() or RSD(). The first is the noiseless RSD(), and the second is the impact of noise. Since the standard deviation of the mean of N normally distributed random variables is , the noise related RSD is estimated as . Therefore, we used the maximum of RSD() and to approximate RSD() or RSD().
| (3) |
Nmin was determined as the minimum number of gradient directions for RSDapp ≤ 5%.
Human data
High-quality HCP data from 3 healthy adults, as part of the MGH-USC Adult Diffusion Dataset, were downloaded from ConnectomeDB (http://db.humanconnectome.org). Diffusion data were acquired with 4 different b-values (i.e., 4 shells): 1 ms/μm2 (64 directions), 3 ms/μm2 (64 directions), 5 ms/μm2 (128 directions), and 10 ms/μm2 (256 directions). The gradient direction sets were specifically designed so that a lower shell set is a subset of higher shell set. At each shell, the gradient directions were approximately uniformly distributed [29–31]. One non-diffusion weighted b=0 image was collected for every 13 diffusion weighted images. The signal-to-noise ratio (SNR) was about 20 for white matter in the b=0 image, which was estimated with a maximum-likelihood approach [32].
To demonstrate the accuracy of subsampling for measurement, a subset of gradient directions at each shell were selected: 10 directions for b = 1 ms/μm2, 30 directions for b = 3 ms/μm2, 50 directions for b = 5 ms/μm2, and 100 directions for b = 10 ms/μm2. Those subsets were determined with the incremental sampling scheme to guarantee reasonably uniform coverage [31]. The numbers of gradient directions in subsets were guided by the simulation results. The relative differences of between full data sets and subsets were calculated.
Results
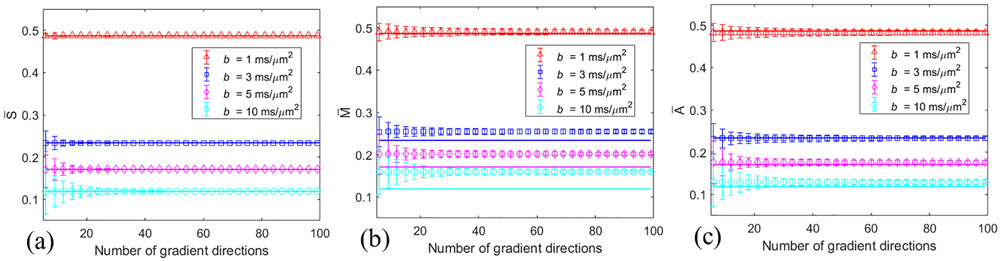
The simulated signal , measured magnitude , and corrected amplitude were dependent on the number N of gradient directions (Figure 1). The markers represent the mean values over 10,000 different directions of the single fiber n, and the error-bars represent the standard deviations. The solid lines are the ground truth spherical mean signals based on Eq. (2). As shown in Figure 1(a), the mean is consistent with the ground truth at each b-value and the standard deviation decreases with increasing N. The Rician bias is evident in Figure 1(b), especially at high b-values. Increasing N did not help reduce the Rician bias. The corrected amplitude is closer to the ground truth than .
Figure 1.
Dependence of the calculated (a), (b), and (c) on the number N of gradient directions. The markers represent the mean values, and the error-bars represent the standard deviations. The solid lines are the ground truth spherical mean signals based on Eq. (2). SNR = 20 for (b) and (c).
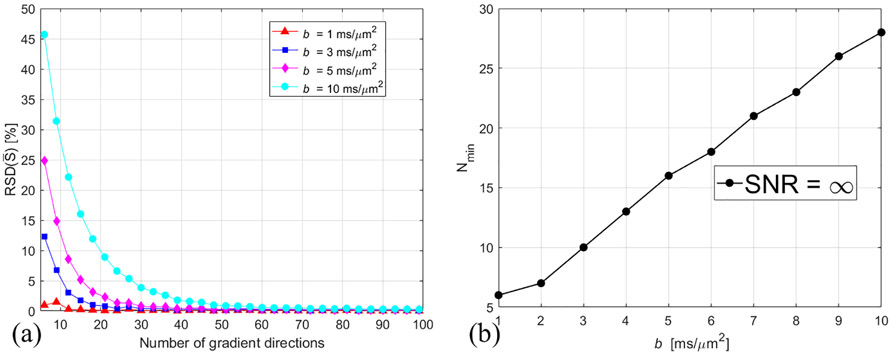
RSD() decreases with increasing N, and the minimum number of N needed for RSD() ≤ 5% is referred to as Nmin. Figure 2(a) shows the calculated RSD() as a function of the number of gradient directions. As expected from previous spherical deconvolution work [27], the diffusion signal profile becomes sharp with increasing b-value and more gradient directions are needed for accurate measurements of . Figure 2(b) shows the linear dependence of Nmin on b-value.
Figure 2.
Dependence of the calculated RSD() on N for different b-values (a), and dependence of Nmin on b-value (b). SNR = ∞ for the simulated .
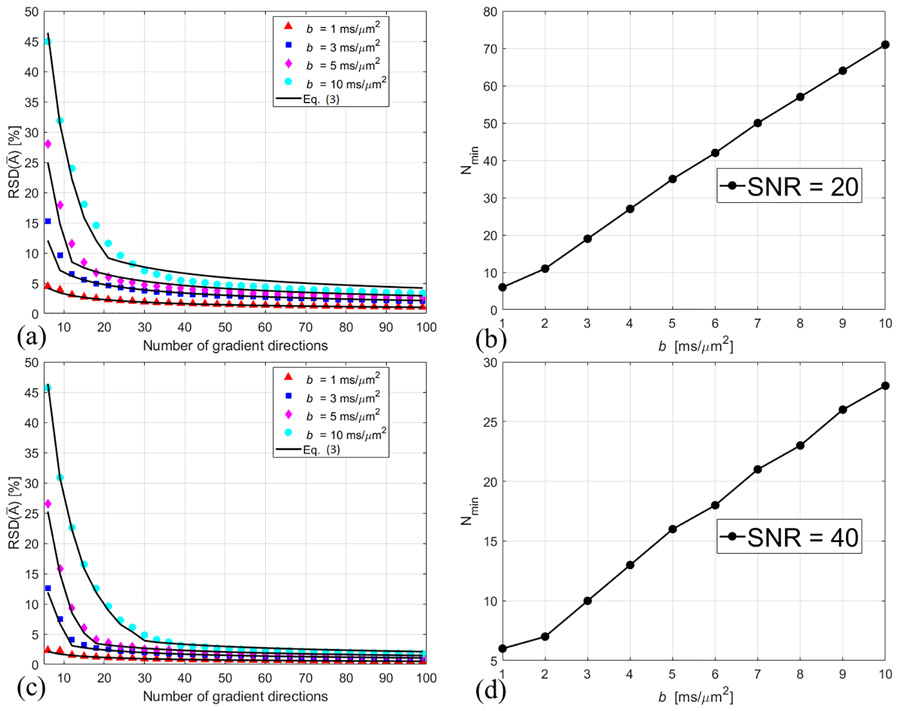
Our simulations showed that RSD() can be properly fit by Eq. (3). Figures 3 (a) and (c) show the calculated RSD() at SNR = 20 and 40, respectively. The solid curves represent the approximated RSDapp based on Eq. (3). Here the minimum number of N needed for RSDapp ≤ 5% is referred to as Nmin. The lower the SNR, the higher the number of gradient directions needed for accurate measurement. At SNR = 40, Nmin is the same with that of SNR = ∞. At SNR = 20, RSD() is dominated by . Nmin is still linearly dependent on b-value.
Figure 3.
The markers are the calculated values of RSD() at SNR = 20 (a) and 40 (c). The solid curves in (a) and (c) represent the approximated RSDapp based on Eq. (3). Nmin is the minimum number of gradient directions for RSDapp ≤ 5%. Figure 3 (b) and (d) show the b-value dependent Nmin at SNR = 20 and 40, respectively.
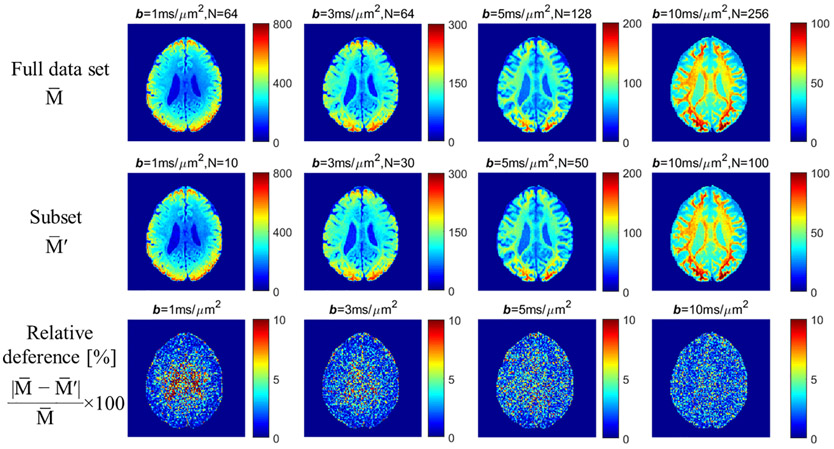
Multi-shell high angular resolution HCP diffusion data were analyzed to support the simulation results. SNR was ~ 20 for white matter in the b=0 image, suggesting that Figure 3 (b) can be used to determine the number of gradient directions for each subset. Since the subset is not fully uniformly distributed, it is better to use a slightly larger N than Nmin in Figure 3(b) to guarantee reasonably uniform coverage, which could also help reduce the noise. Here we used 10 directions for b = 1 ms/μm2, 30 directions for b = 3 ms/μm2, 50 directions for b = 5 ms/μm2, and 100 directions for b = 10 ms/μm2. Figure 4 shows the comparison between full data sets and subsets for measurements at different b-values. The relative differences were shown in the last row. The high relative difference in cerebrospinal fluid (CSF) at b = 1 ms/μm2 is due to the low signal in CSF and small number of gradient directions in subset. Overall, the whole brain relative differences are 3.14 ± 0.26%, 2.56 ± 0.17%, 2.77 ± 0.10%, and 2.34 ± 0.07% for b = 1, 3, 5, and 10 ms/μm2, respectively. Thus, the experimental results were in good agreeement with the simulation results. The spherical mean signal can be measured accurately with a subset of gradient directions.
Figure 4.
Comparison between full data sets and subsets for measurements from a representative subject. The relative differences were shown in the last row. The low relative differences demonstrate that can be measured accurately with a subset of gradient directions.
Discussion
The conventional DTI method is significantly limited by the crossing fiber problem for accurate quantification of tissue microstructure. To factor out the complex fiber orientation distribution, the spherical mean technique was proposed to analyze the spherical mean signal instead of the individual signal along each gradient direction [11,16]. The current study investigated the minimum number of gradient directions for robust measurement of spherical mean signals at different b-values. In the case of SNR = ∞, Nmin is the number of gradient directions needed to achieve sufficient sampling. Computer simulations were employed to characterize the dependence of RSD() on N and b-value (see Figure 2). In the case of limited SNR, RSD can be estimated by the maximum of the noiseless RSD() and noise-dependent . For typical human diffusion studies with SNR ~ 20, we recommend using 10×b/b1 (b1 = 1 ms/μm2) uniformly distributed gradient directions for robust measurement. For example, 30 gradient directions is sufficient for robust measurement at b = 3 ms/μm2, which is achievable within 5 min.
Although our simulations were based on a simplified two-compartment model with predefined λ and Vin, the conclusion can be generalized to other situations. The assumption of is based on the first-order tortuosity approximation for a system of randomly placed parallel cylinders [28]. Several studies have proposed to estimate and using more comprehensive diffusion datasets [18,21,23]. The choice of an extra-axonal model is insignificant to our study as the extra-axonal water contribution is negligible with sufficiently large b-values [12,13]. Due to the small axon size in brain white matter, the intra-axonal transverse diffusion is highly restricted, and it is commonly assumed that [12,16,23,33]. Recent power-law scaling studies supported that the approximation is reasonable for white matter [19,20]. This approximation is also insignificant to our study as a larger is expected to make the diffusion profile more isotropic, which in turn requires less number of gradient directions for sufficient sampling. The intrinsic diffusivity λ was fixed in our simulations. Recent studies demonstrated that λ varies substantially in the brain and that the variation may provide information about the underlying cellular milieu [16,18,23]. However, according to Eq. (2), changing λ is equivalent to changing b-value. The extra simulation with varying λ is unnecessary. Similarly, changing Vin is equivalent to changing SNR. Eq. (3) is always valid for determination of the minimum number of gradient directions for robust measurement of spherical mean signal.
The Rician bias correction was not implemented on the HCP data. Our simulations suggest that is better than the standard [34] for measurement in white matter. For repeated measurements along a single gradient direction or isotropic diffusion measurements along different directions, is preferable [34]. However, for anisotropic diffusion in white matter, the measured signals along different directions are different and is the mean of the signals from N different Rician distributions. In that case, simulations suggest that is better. Note that the reconstruction method and data preprocessing may alter the noise properties [16,35]. The Rician bias correction will need to be investigated further in future studies.
Conclusion
In this study we characterized the relative standard deviation of the spherical mean signal as a function of the number of gradient directions. The minimum number of gradient directions for robust spherical mean signal measurement was then determined. At high SNRs, Nmin increases with increasing b-value to achieve sufficient sampling. At low SNRs, Nmin increases with increasing b-value to reduce the noise. We recommend using 10×b/b1 uniformly distributed gradient directions for typical human diffusion studies with SNR ~ 20 for robust spherical mean signal measurement.
Acknowledgements
This work was supported by the National Institutes of Health (grant number R21DC015853). Data were provided by the Human Connectome Project, MGH-USC Consortium (Principal Investigators: Bruce R. Rosen, Arthur W. Toga and Van Wedeen; U01MH093765) funded by the NIH Blueprint Initiative for Neuroscience Research grant; the National Institutes of Health grant P41EB015896; and the Instrumentation Grants S10RR023043, 1S10RR023401, 1S10RR019307.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmay be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophys J 1994;66:259–67. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stanisz GJ, Szafer A, Wright GA, Henkelman RM. An analytical model of restricted diffusion in bovine optic nerve. Magn Reson Med 1997;37:103–11. doi: 10.1002/mrm.1910370115. [DOI] [PubMed] [Google Scholar]
- [3].Assaf Y, Blumenfeld-Katzir T, Yovel Y, Basser PJ. AxCaliber: A method for measdoing axon diameter distribution from diffusion MRI. Magn Reson Med 2008;59:1347–54. doi: 10.1002/mrm.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Alexander DC, Hubbard PL, Hall MG, Moore EA, Ptito M, Parker GJM, et al. Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage 2010;52:1374–89. doi: 10.1016/j.neuroimage.2010.05.043. [DOI] [PubMed] [Google Scholar]
- [5].Xu J, Li H, Harkins KD, Jiang X, Xie J, Kang H, et al. Mapping mean axon diameter and axonal volumeume fraction by MRI using temporal diffusion spectroscopy. Neuroimage 2014;103:10–9. doi: 10.1016/j.neuroimage.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fieremans E, Jensen JH, Helpern JA. White matter characterization with diffusional kurtosis imaging. Neuroimage 2011;58:177–88. doi: 10.1016/j.neuroimage.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. LABELDDI: Practical in vivo nedoite orientation dispersion and density imaging of the human brain. Neuroimage 2012;61:1000–16. doi: 10.1016/j.neuroimage.2012.03.072. [DOI] [PubMed] [Google Scholar]
- [8].Jespersen SN, Kroenke CD, Ostergaard L, Ackerman JJH, Yablonskiy DA. Modeling dendrite density from magnetic resonance diffusion measurements. Neuroimage 2007;34:1473–86. doi: 10.1016/j.neuroimage.2006.10.037. [DOI] [PubMed] [Google Scholar]
- [9].Jespersen SN, Bjarkam CR, Nyengaard JR, Chakravarty MM, Hansen B, Vosegaard T, et al. Nedoite density from magnetic resonance diffusion measurements at ultrahigh field: Comparison with light microscopy and electron microscopy. Neuroimage 2010;49:205–16. doi: 10.1016/j.neuroimage.2009.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Anderson AW. Measurement of fiber orientation distributions using high angular resolution diffusion imaging. Magn Reson Med 2005;54:1194–206. doi: 10.1002/mrm.20667. [DOI] [PubMed] [Google Scholar]
- [11].Kaden E, Kruggel F, Alexander DC. Quantitative mapping of the per-axon diffusion coefficients in brain white matter. Magn Reson Med 2016;75:1752–63. doi: 10.1002/mrm.25734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jensen JH, Russell Glenn G, Helpern JA. Fiber ball imaging. Neuroimage 2016;124:824–33. doi: 10.1016/j.neuroimage.2015.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Raffelt D, Tournier JD, Rose S, Ridgway GR, Henderson R, Crozier S, et al. Apparent Fibre Density: A labelvel measure for the analysis of diffusion-weighted magnetic resonance images. Neuroimage 2012;59:3976–94. doi: 10.1016/j.neuroimage.2011.10.045. [DOI] [PubMed] [Google Scholar]
- [14].Calamante F, Smith RE, Tournier JD, Raffelt D, Connelly A. Quantification of voxel-wise total fibre density: Investigating the problems associated with track-count mapping. Neuroimage 2015;117:284–93. doi: 10.1016/j.neuroimage.2015.05.070. [DOI] [PubMed] [Google Scholar]
- [15].Calamante F, Jedoissen B, Smith RE, Tournier J-D, Connelly A. The role of whole-brain diffusion MRI as a tool for studying human in vivo cortical segregation based on a measure of nedoite density. Magn Reson Med 2018;79:2738–44. doi: 10.1002/mrm.26917. [DOI] [PubMed] [Google Scholar]
- [16].Kaden E, Kelm ND, Carson RP, Does MD, Alexander DC. Multi-compartment microscopic diffusion imaging. Neuroimage 2016;139:346–59. doi: 10.1016/j.neuroimage.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Samantha B, Junzhong X, A BB, R BF, A SS. Multi-compartmental diffusion characterization of the human cervical spinal cord in vivo using the spherical mean technique. NMR Biomed 2018;31:e3894. doi: 10.1002/nbm.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McKinlabeln ET, Helpern JA, Jensen JH. Modeling white matter microstructure with fiber ball imaging. Neuroimage 2018. doi: 10.1016/j.neuroimage.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McKinlabeln ET, Jensen JH, Glenn GR, Helpern JA. Dependence on b-value of the direction-averaged diffusion-weighted imaging signal in brain. Magn Reson Imaging 2017;36:121–7. doi: 10.1016/j.mri.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Veraart J, Fieremans E, Labelvikov DS. Universal power-law scaling of water diffusion in human brain defines what we see with MRI. arXiv Prepr. arXiv1609.09145, 2016.
- [21].Reisert M, Kellner E, Dhital B, Hennig J, Kiselev VG. Disentangling micro from mesostructure by diffusion MRI: A Bayesian approach. Neuroimage 2017;147:964–75. doi: 10.1016/j.neuroimage.2016.09.058. [DOI] [PubMed] [Google Scholar]
- [22].Veraart J, Labelvikov DS, Fieremans E. TE dependent Diffusion Imaging (TEdDI) distinguishes between compartmental T2relaxation times. Neuroimage 2017. doi: 10.1016/j.neuroimage.2017.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Labelvikov DS, Veraart J, Jelescu IO, Fieremans E. Rotationally-invariant mapping of scalar and orientational metrics of neuronal microstructure with diffusion MRI. Neuroimage 2018. doi: 10.1016/J.NEUROIMAGE.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sotiropoulos SN, Jbabdi S, Xu J, Andersson JL, Moeller S, Auerbach EJ, et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage 2013;80:125–43. doi: 10.1016/j.neuroimage.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Papadakis NG, Murrills CD, Hall LD, Huang CLH, Adrian Carpenter T. Minimal gradient encoding for robust estimation of diffusion anisotropy. Magn Reson Imaging 2000;18:671–9. doi: 10.1016/S0730-725X(00)00151-X. [DOI] [PubMed] [Google Scholar]
- [26].Jones DK. The Effect of Gradient Sampling Schemes on Measures Derived from Diffusion Tensor MRI: A Monte Carlo Study. Magn Reson Med 2004;51:807–15. doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- [27].Tournier JD, Calamante F, Connelly A. Determination of the appropriate b value and number of gradient directions for high-angular-resolution diffusion-weighted imaging. NMR Biomed 2013;26:1775–86. doi: 10.1002/nbm.3017. [DOI] [PubMed] [Google Scholar]
- [28].Szafer A, Zhong J, Gore JC. Theoretical model for water diffusion in tissues. Magn Reson Med 1995;33:697–712. doi: 10.1088/0031-9155/52/7/N01. [DOI] [PubMed] [Google Scholar]
- [29].Jones DK, Horsfield MA, Simmons A. Optimal strategies for measdoing diffusion in anisotropic systems by magnetic resonance imaging. Magn Reson Med 1999;42:515–25. doi: 10.1002/(SICI)1522-2594(199909)42:3<515::AID-MRM14>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- [30].Fan Q, Witzel T, Nummenmaa A, Van Dijk KRA, Van Horn JD, Drews MK, et al. MGH-USC Human Connectome Project datasets with ultra-high b-value diffusion MRI. Neuroimage 2016;124:1108–14. doi: 10.1016/j.neuroimage.2015.08.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Caruyer E, Lenglet C, Sapiro G, Deriche R. Design of multishell sampling schemes with uniform coverage in diffusion MRI. Magn Reson Med 2013;69:1534–40. doi: 10.1002/mrm.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sijbers J, Den Dekker AJ. Maximum Likelihood Estimation of Signal Amplitude and Labelise Variance from MR Data. Magn Reson Med 2004;51:586–94. doi: 10.1002/mrm.10728. [DOI] [PubMed] [Google Scholar]
- [33].Jiang X, Li H, Xie J, Zhao P, Gore JC, Xu J. Quantification of cell size using temporal diffusion spectroscopy. Magn Reson Med 2016;75:1076–85. doi: 10.1002/mrm.25684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gudbjartsson H, Patz S. The rician distribution of labelisy mri data. Magn Reson Med 1995;34:910–4. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sotiropoulos SN, Moeller S, Jbabdi S, Xu J, Andersson JL, Auerbach EJ, et al. Effects of image reconstruction on fiber orientation mapping from multichannel diffusion MRI: reducing the labelise floor using SENSE. Magn Reson Med 2013;70:1682–9. [DOI] [PMC free article] [PubMed] [Google Scholar]