Abstract
Inorganic polyphosphate (polyP) constitutes one of the most conserved and ubiquitous molecules in biology. Recent work in bacteria demonstrated that polyP increases oxidative stress resistance by preventing stress-induced protein aggregation, and promotes biofilm formation by stimulating functional amyloid formation. To gain insights into these two seemingly contradictory functions of polyP, we investigated the effects of polyP on the folding model lactate dehydrogenase (LDH). We discovered that presence of polyP during the thermal unfolding process stabilizes folding intermediates of LDH as soluble micro-β-aggregates with amyloid-like properties. Size and heterogeneity of the oligomers formed in this process was dependent on the polyP chain length, with longer chains forming smaller, more homogenous complexes. This ability of polyP to stabilize thermally unfolded proteins even upon exposure to extreme temperatures, appears to contribute to the observed resistance of E. coli towards severe heat shock treatment. These results suggest that the working mechanism of polyP is the same for both soluble and amyloidogenic proteins, with the ultimate outcome likely being determined by a combination of polyP chain length and the client protein itself. They help to explain how polyP can simultaneously function as general stress protective chaperone and instigator of amyloidogenic processes in vivo.
Introduction
Polyphosphate is an extremely simple molecule composed of as few as three and as many of 1,000 inorganic phosphates linked by high-energy phosphoanhydride bonds. Highly conserved and universally found in all organisms studied, polyP has likely been present for billions of years [1]. PolyP appears to play key roles in a large number of seemingly unrelated biological processes, ranging from stress protection, biofilm formation and virulence in bacteria, to blood clotting, apoptosis and mTOR activation in eukaryotes [2–5]. The wide range of biological functions together with the extremely simple nature of polyP has long intrigued researchers from different fields and raised the obvious question as to how polyP works. The molecular mechanisms that have been previously proposed to contribute to polyP’s protective role during stress conditions ranged from energy storage and metal chelation to transcriptional regulation of stress responses [1, 6]. Recent work from our lab now showed that bacteria lacking the polyPsynthesizing enzyme polyphosphate kinase (PPK) are exquisitely oxidative stress sensitive. In response to physiological oxidants such as hypochlorous acid (i.e. bleach), bacteria lacking polyP accumulate large amounts of aggregated proteins, despite the bacteria’s attempts to compensate for the absence of polyP with the upregulation of protein-based chaperones [7, 8]. In vitro studies confirmed these results and showed that polyP binds unfolded proteins and prevents the formation of toxic protein aggregates [7]. Counterintuitively, we also discovered that presence of polyP promotes the transition of amyloidogenic proteins (e.g., α-synuclein, bacterial CsgA) into a β-sheet conformation, thereby accelerating the transition from presumably toxic oligomers into non-toxic amyloid fibrils [9]. These studies revealed a previously unrecognized connection between polyP and amyloid diseases, and served to explain how polyP stimulates processes such as biofilm formation, which depend on bacterial amyloid fibril formation [5, 9].
To investigate how polyP can possibly fulfill both functions, serving as an aggregationpreventing reagent for thermally unfolded proteins such as luciferase, and as fibril-stimulating agent for amyloidogenic proteins, we decided to investigate the influence of polyP on the thermal unfolding of lactate dehydrogenase (LDH), a commonly used protein folding model. We discovered that polyP stabilizes thermally unfolded intermediate(s) of LDH in form of small, defined-size oligomers. These oligomers, whose size decreases with increasing polyP chain length, exert amyloid-like features, including increased thioflavin T binding and surface hydrophobicity, characteristic of β-aggregates. These results suggest that the basic working mechanism of polyP is the same for both soluble and amyloidogenic proteins. It is of note that a substantial proportion of LDH intermediates stabilized by polyP remain refolding competent even upon exposure to near-boiling temperatures and reactivate once non-stress conditions are restored and a chaperone folding system in provided. We found that this unique ability of polyP to maintain protein solubility and refolding competence even upon incubation at extreme temperatures appears to contribute to the thermoprotective effect of polyP in bacteria.
Results
PolyP maintains lactate dehydrogenase soluble at near-boiling temperatures
To investigate how polyP maintains solubility of thermally unfolded protein, we used lactate dehydrogenase (LDH) from rabbit muscle and monitored its temperature-mediated changes in secondary structure by far-UV-circular dichroism spectroscopy. LDH, which is tetrameric and primarily α-helical, rapidly unfolds upon incubation at temperatures above 55°C, making it a popular in vitro chaperone client (Figure 1A-C). Thermal unfolding of LDH is irreversible, consistent with the formation of insoluble, pelletable LDH aggregates (Figure 1D) [10]. This situation changed completely, however, when we heated LDH in the presence of 1 mM polyP (concentration given in Pi units) of various chain lengths, ranging from 14mers (polyP14) to 300mers (polyP300). Although LDH incubated in the presence of polyP started to unfold at similar temperatures than LDH alone, the protein did not completely unfold. Instead, we found that LDH adopts an apparently thermostable intermediate conformation at about 60°C, which does not change its secondary structure even upon subsequent heating to 85°C (Figure 1A). Analysis of the far-UV-CD spectrum revealed that the thermally unfolded intermediate(s) of LDH that accumulate at temperatures above 60°C are highly enriched for β-sheets compared to native LDH (Figures 1B, C) and, according to spin down experiments, soluble (Figures 1D, E). The βsheet composition of the intermediates formed in the presence of polyP was maintained upon cooling the samples to RT (Figures 1B, C), suggestive of an apparently stable conformation. In contrast, LDH incubated in the absence of polyP lacked much of its original signal (Figures 1B, C). Our finding that LDH undergoes an α-helical to β-sheet transition upon thermal denaturation was reminiscent of studies with the regulatory subunit of protein kinase PKA or ovalbumin, which both adopt soluble so-called β-aggregates during thermal unfolding [11, 12]. In contrast to PKA and ovalalbumin, however, soluble LDH intermediates were only populated in the presence of polyP. No significant difference in thermal transition or the secondary structure of the LDH unfolded intermediates was observed when different polyP chain lengths were used, suggesting that at this polyP concentration, chain length does not alter the polyP effects (Figure 1A-E). These results suggested that polyP stabilizes partially structured thermal unfolded intermediate(s) of LDH, making them potentially amendable to further biophysical characterization.
Figure 1. PolyP stabilizes thermally unfolded intermediates of LDH.
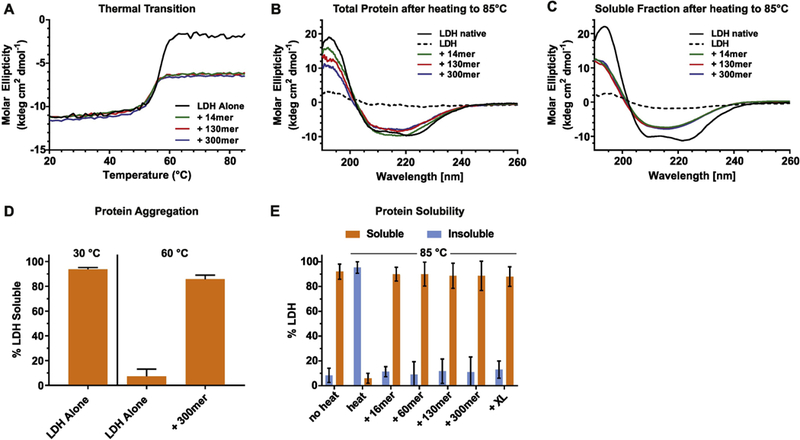
(A) LDH (5.7 μM) was heated in the spectrophotometer in the absence (black) or in the presence of 1 mM polyP14 (green), polyP130 (red), or polyP300 (blue) using a heating rate of 1°C/min. The molar ellipticity at 222 nm was monitored. Far-UV-CD spectra of each sample from A) were recorded either immediately after the incubation at 85°C (B) or upon removal of the aggregates by centrifugation (C). Spectra were recorded at RT. Native LDH was used as control (black solid line). The dotted line represents heated LDH in the absence of polyP. At 20°C, presence of polyP had no apparent influence on the secondary structure of native LDH. (D) Percent soluble LDH after heating LDH (5.7 μM) in the absence or presence of 1 mM polyP300 to 60°C using a heating rate of 1°C/min. No significant difference in solubility was observed when shorter polyP-chains were used. The amount of soluble protein at 20°C was set to 100%. (E) Percent soluble LDH after heating LDH (5.7 μM) in the absence or presence of 1 mM polyP using different chain lengths. Samples were incubated in a thermomixer at 20°C and heated to 85°C (~10 min). The amount of soluble protein at 20°C was set to 100%.
PolyP forms stable complexes with unfolding intermediates of LDH
Our results suggested that polyP either remains stably associated with the thermally unfolded intermediates of LDH upon subsequent cooling to RT, or that the LDH intermediates, once stabilized by polyP, remain in a soluble conformation even in the absence of polyP. To first determine whether polyP remains associated with thermally unfolded LDH, we prepared LDHpolyP300 complexes as before, and analyzed them as well as mixtures of native LDH +/− polyP300 by size exclusion chromatography. We tested each fraction of the eluates for both protein abundance using absorbance at 280 nm (Figure 2A, solid lines), and polyP content, using DAPI staining (Figure 2A, dashed lines) [13]. We did not test LDH heated in the absence of polyP since most of the protein aggregated during the incubation. Native LDH tetramers eluted in one discrete peak, independent of the presence or absence of polyP300 (Figure 2A, compare blue and black solid lines). Similarly, no significant differences between the elution profiles of polyP300 alone or polyP in the presence of native LDH were observed (Figure 2A, compare green and blue dashed lines). In contrast, however, LDH heated in the presence of polyP300 eluted substantially earlier and in a wide peak (Figure 2A, red solid line). The elution profile of polyP300 was similarly shifted to earlier fractions, suggesting that LDH and polyP300 remain associated (Figure 2A, dashed red line).
Figure 2. PolyP forms apparently stable complexes with thermally unfolded LDH.
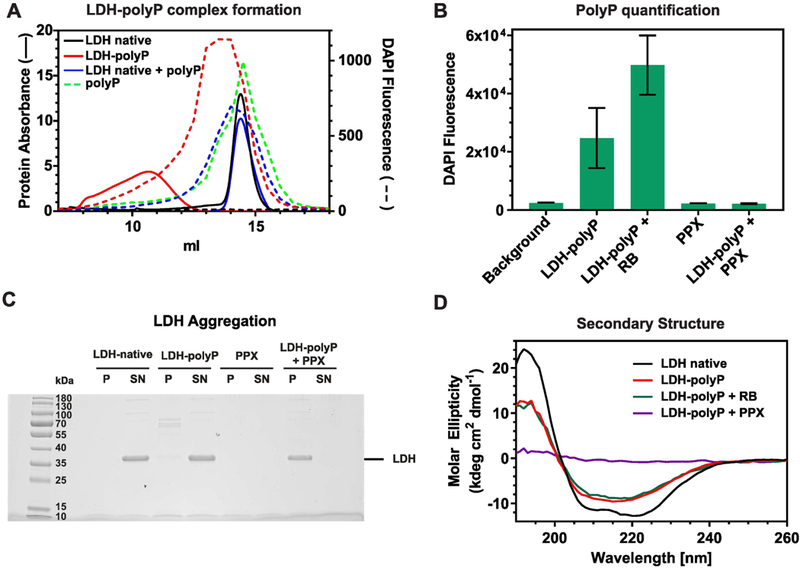
(A) LDH (5.7 μM) was heated in the absence or presence of 1 mM polyP300 (red) to 70°C using a heating rate of 1°C/min. Samples were spun down and the soluble supernatant was analyzed by size exclusion chromatography. Samples of unheated, native LDH in the absence (black) or presence of polyP (blue) as well as polyP in the absence of LDH (green) were used as controls. Protein abundance in the eluates was determined by absorbance (solid lines) and polyP content was measured by using DAPI staining (dashed lines). LDH heated in the absence of polyP completely aggregated and was therefore not tested. (B-D) Complexes between LDH and polyP300 (LDH-polyP) were prepared as before, cooled down to RT and either analyzed directly or upon incubation with 300 nM ScPPX for 30 min at 37°C to hydrolyze polyP. (B) DAPI fluorescence measurements were conducted to determine polyP levels before and after incubation with 300 nM ScPPX. ScPPX reaction buffer (RB) was added as control, and PPX alone was tested as control as well. (C) Samples containing native LDH or pre-formed LDHpolyP complexes were either treated with nothing, RB or 300 nM ScPPX for 30 min at 37°C. After centrifugation, the insoluble pellets as well as the soluble supernatants were analyzed on SDS-PAGE and visualized by Coommassie staining. PPX alone was tested as control. (D) Far- UV-CD spectra of native LDH (black line) or LDH-polyP complexes in the absence of additives (red line), in the presence of reaction buffer (RB) (green line) or upon treatment with 300 nM ScPPX for 30 min at 37°C and subsequent centrifugation. All spectra were recorded at RT.
To directly test whether LDH once thermally unfolded in the presence of polyP300 remains soluble even upon removal of polyP, we added the exopolyphosphatase PPX from Saccharomyces cerevisiae (ScPPX) to the LDH-polyP300 complexes to hydrolyze polyP [7]. Analysis of the remaining polyP levels in the incubation reaction revealed the complete digestion of polyP after 30 min at 37°C when ScPPX was present (Figure 2B). To test whether thermally unfolded LDH remains soluble and partially folded even upon removal of polyP, we centrifuged the samples and analyzed both soluble supernatant and insoluble pellet. While the majority of native LDH and LDH-polyP complexes were soluble, treatment of LDH-polyP complexes with PPX caused LDH to aggregate and partition into the insoluble pellet (Figure 2C). Secondary structure analysis of the supernatant by CD revealed a complete loss of signal in the PPX-treated sample, consistent with the aggregation of LDH upon digestion of polyP (Figure 2D).
Thermally unfolded LDH-polyP complexes form defined-sized oligomers
Our size exclusion chromatography experiments suggested that polyP and thermally unfolded LDH form soluble, higher molecular weight complexes (Figure 2, solid red line). To obtain more detailed insights into the size distribution of the LDH-polyP complexes and assess whether polyP chain length affects oligomer size, we used sedimentation velocity analytical ultracentrifugation (SV-AUC). We prepared LDH-polyP complexes using polyP chain lengths between 14 Pi and > 700 Pi (polyPXL) by rapidly heating the samples to 85°C (~10 min), and compared their sedimentation properties with that of native LDH incubated in the absence and presence of polyP14 or polyPXL. Native LDH (MW 35 kDa per monomer) sedimented primarily as tetramer [14], and its sedimentation behavior was unaffected by the presence of polyP (Figure 3A, insert). In contrast, however, LDH-polyP complexes sedimented in discrete peaks, with the shorter polyP14 chains causing a much wider range and larger size distribution than the longer poly3P300 or polyP XL chains (Figure 3A). The average size of oligomers appeared to be directly correlated with the length of polyP-chains, with longer polyP chains generating smaller, more homogenous oligomers. Analysis of the frictional ratio, which relates to the shape of the complexes, versus the sedimentation coefficient, which relates to their size revealed that LDHpolyP14 complexes are larger yet more globular compared to LDH-polyPXL-complexes, which form smaller, more elongated oligomeric structures resulting in a larger frictional ratio (Figure 3B). These results suggest that the longer polyP-chains (i.e., polyPXL and polyP300) are more effective than shorter polyP chains in protecting LDH oligomers from forming non-specific interactions with other LDH oligomers.
Figure 3. LDH-polyP complexes form defined-size oligomers.
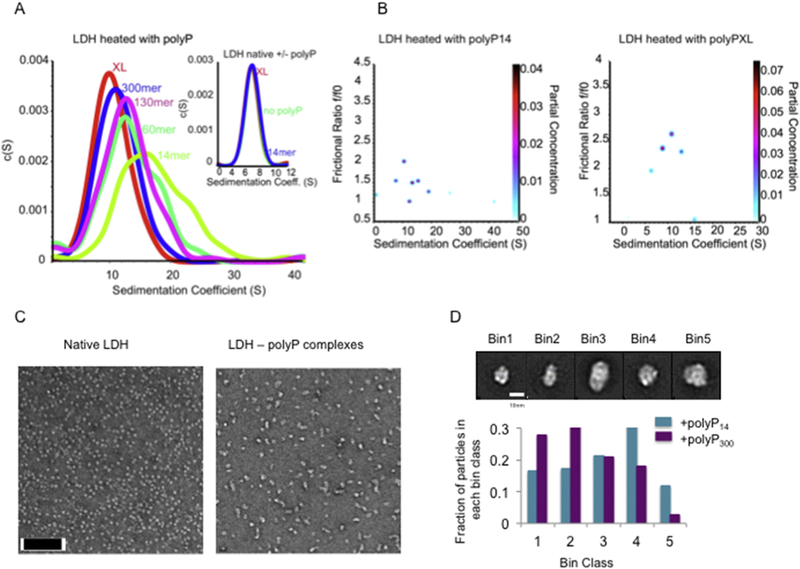
(A) Two-dimensional sedimentation analysis (2DSA) of a sedimentation velocity experiment. LDH (5.7 μM) was heated in the presence of 1 mM polyP chains from 20°C to 85°C in the thermomixer (duration: ~10 min). The samples were analyzed by analytical ultracentrifugation. Insert: Sedimentation behavior of native LDH with and without polyP14 or polyPXl. (B) Twodimension genetic analysis (GA) plots of f/f0 versus sedimentation coefficient. (C) Samples prepared for (A) were analyzed by transmission electron microscopy. An image of native LDH as well as an image of LDH-polyP130 complexes is shown as illustration (scale bar: 100 nm). (D) Representative reference-free class averages seen in both samples were selected as templates for 5 size bins (top; scale bar 10 nm). The total set of 2D class averages for each condition was subjected to reference-based matching and the total particles belonging to each bin were tabulated for both conditions (bottom).
To directly visualize the complexes that form between thermally unfolded LDH and polyP-chains of various lengths, we prepared LDH-polyP14 and LDH-polyPXL-complexes using the same rapid heating rate as before (i.e., transition from 20°C to 85°C in about 10 min), and analyzed the particles in the soluble supernatant by transmission electron microscopy (TEM). The native LDH preparation was highly homogeneous and revealed globular shaped particles of about 90 × 90 Å likely representing the native tetramer (Figure 3C). Both LDH-polyP14 and LDH-polyPXL preparations contained a range of defined oligomers with both globular and elongated shapes. The smallest soluble LDH-polyP complexes, which were found primarily in the presence of the longer polyP chains, appeared to have dimensions of about 100 × 100 Å or 100 × 140 Å, only slightly larger than native LDH. In addition, larger oligomers were also identified. To determine if there is a change in the size distribution following incubation with the two different chain lengths, we analyzed about 6500 particles each of the LDH-polyP14 and LDH-polyPXL complexes and performed 2D classification analysis. Five of the resulting 2D averages that represent the majority of particles based on size and shape were then selected and used as templates for reference matching. The particles were subsequently split into these five discrete size “bins” (Figure 3D, top). Analysis of the particle distribution in the LDH-polyP14 and LDH-polyPXL agreed well with our ultracentrifugation data, confirming that longer polyP chains stabilize smaller oligomeric structures of LDH intermediates, while shorter polyP chains form larger more elongated oligomeric structures (Figure 3D, bottom).
Biophysical characterization of soluble LDH-polyP complexes
Our finding that LDH-polyP complexes, once formed by heating the samples to temperatures >60°C are soluble and stable at RT, provided us with the unique ability to determine their biophysical characteristics. To assess the structural properties of thermally unfolded LDH in complex with polyP compared to native LDH, we monitored tryptophan fluorescence (Figure 4A), analyzed surface hydrophobicity via bis-ANS binding [15] (Figure 4B) and assessed the cross β-sheet character by thioflavin T (ThT) fluorescence [16] (Figure 4C). As observed before, neither one of these three biophysical features changed when native LDH was incubated with polyP, confirming that polyP does not interact with properly folded LDH (Figure 4, dashed lines). The tryptophan fluorescence spectra of native LDH showed a maximum around 340 nm (Figure 4A), consistent with the notion that four of the six tryptophan residues in each LDH monomer are surface exposed (Figure 4D). Moreover, native LDH neither bound significant amounts of bis-ANS nor thioflavin T, agreeing with its soluble and predominantly α-helical character (Figures 4B, C). In contrast, however, the thermally unfolded intermediate(s) of LDH that were stabilized by polyP behaved distinctly different from native LDH. We observed a substantial fluorescence quench as well as a marked blue shift in the tryptophan spectra, suggesting that some of the previously exposed tryptophan(s) become buried within hydrophobic regions likely as a result of oligomer formation (Figure 4A). Furthermore, the LDH-polyP complexes showed substantially increased surface hydrophobicity as determined by bis-ANS binding, which might explain their tendency to aggregate once polyP is removed (Figure 4B). Finally, and fully consistent with the idea that LDH intermediates stabilized by polyP form soluble β-aggregates, we found substantial binding to thioflavin T, a compound whose fluorescence is known to increase upon interacting with cross-β-sheet fibrils, such as present in amyloid fibrils like αsynuclein [16](Figure 4C). The ThT positive signal of LDH-polyP complexes suggested a high degree of structural similarity between insoluble amyloid-fibrils and soluble polyP-associated LDH unfolded intermediates. Since regions in proteins that appear to be particularly prone to undergo cross-β-sheet formation are buried, unstructured or short helices that connect segments of β-sheets [17], we used the prediction program TANGO to identify potential regions in LDH that might be involved in β-aggregation [18]. We found the highest predictive value for two short helices immediately adjacent to a 6-stranded parallel β-sheet, which is in very close proximity to the corresponding β -sheet of the other second subunit (Figure 4D). Both helices contain surface exposed tryptophan residues in native LDH.
Figure 4. Biophysical properties of LDH-polyP intermediates.
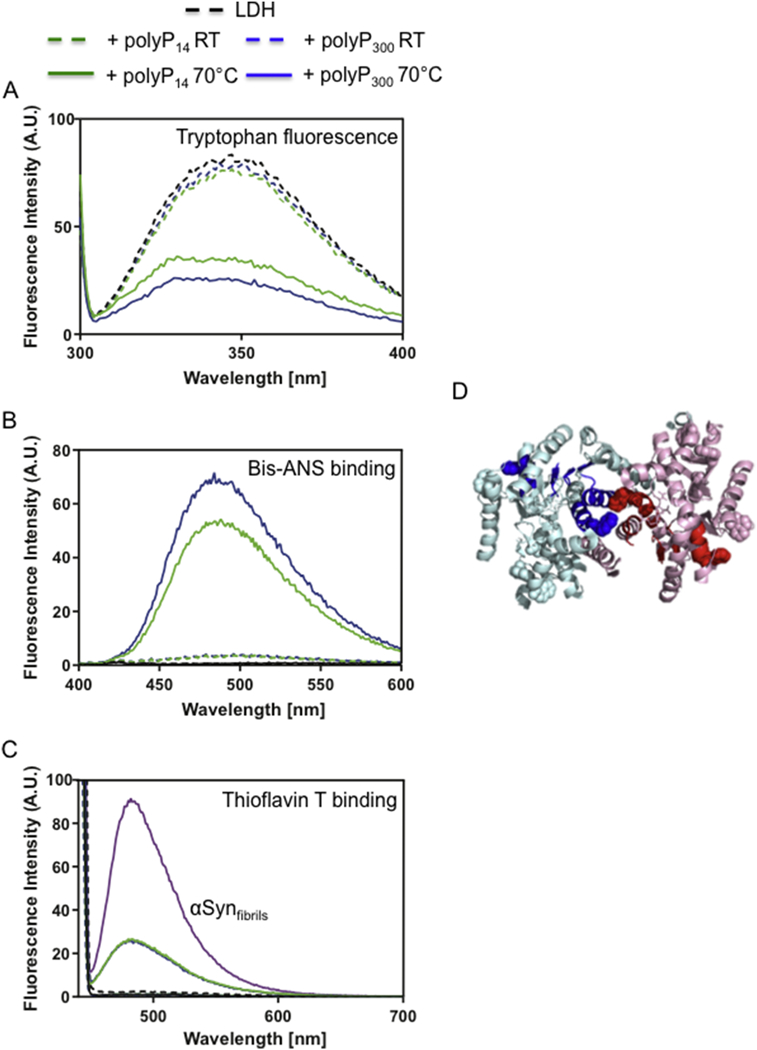
LDH (5.7 μM) was heated in the absence or presence of 1 mM polyP16 (solid green) or polyP300 (solid blue) to 70°C using a heating rate of 1°C/min. Native LDH in the absence (black) or presence of 1 mM polyP16 (dashed green) or polyP300 (dashed blue) were used as control. The heated samples were spun down, and the protein concentration of the soluble supernatant was determined. (A) Fluorescence spectra of 5 μM of each sample were recorded. (B) To assess surface hydrophobicity, each sample was adjusted to a concentration of 0.5 μM LDH and supplemented with 16.5 μM bis-ANS. (C) To determine thioflavin T (ThT) binding, each sample was adjusted to a concentration of 5.7 μM LDH and supplemented with 10 μM ThT. As positive control, 5.7 μM α-synuclein fibrils incubated with 10 μM ThT was used. LDH heated in the absence of polyP completely aggregated and was not tested. (D) Two of the four subunits (cyan and pink) in rabbit muscle LDH structure (pdb: 3H3F) are shown. The regions predicted by TANGO to undergo β-aggregation are depicted in blue and red, respectively. Tryptophan residues are indicated as balls.
PolyP maintains LDH refolding-competent upon incubation at near-boiling temperatures
To test whether polyP is able to maintain thermally unfolded LDH refolding competent upon incubation at severe heat shock temperatures, we heated LDH to either 60°C or 85°C with or without polyP, cooled the sample to RT and analyzed refolding in the absence or presence of combinations of ScPPX and the chaperone system DnaK-DnaJ-GrpE (DnaKJE). This chaperone system consist of the ATP-dependent chaperone DnaK, the co-chaperone DnaJ and the nucleotide exchange factor GrpE [19]. We reasoned that hydrolysis of polyP by ScPPX might be necessary to release LDH from the complex while the chaperone system, which is known to use ATP to support refolding of thermally unfolded proteins both in vitro and in vivo [20], might be required for promoting the refolding of LDH upon shift to non-stress temperatures. Importantly, we found that presence of polyP not only maintained LDH refolding-competent upon heating to 60°C (Figure 5A) but also upon incubation of the enzyme at near boiling temperatures (Figure 5B). These results demonstrate the unique ability of polyP to stabilize unfolded proteins upon incubation at non-physiologically high temperatures. LDH refolding upon return to non-stress conditions was absolutely dependent on the presence of polyP during the thermal unfolding, and the presence of the DnaKJE-ATP system during the subsequent incubation at RT (Figure 5A). In the absence of either component, LDH failed to refold. Addition of fresh polyP together with the DnaKJE-ATP during the refolding did not further improve the reactivation yield, indicating that polyP acts primarily during the unfolding reaction. To test whether hydrolysis of polyP during the refolding increases the yield of reactivated LDH, we added ScPPX during the refolding (Figure 5C). However, contrary to our assumption we found that ScPPX increasingly inhibited the extent of LDH refolding. These results suggest that the ScPPX-mediated release of LDH, which causes LDH aggregation according to our earlier size exclusion studies (Figures 2C, D), out-competes the kinetically slow DnaKJE-mediated refolding of LDH, and imply that the DnaKJE-system can refold client proteins in the presence of polyP. In summary, our in vitro studies indicated that polyP is exceptionally well-suited for maintaining solubility of thermolabile proteins, preventing their irreversible aggregation and promoting their refolding upon subsequent return to non-stress temperatures.
Figure 5. PolyP binding maintains thermally unfolded LDH refolding-competent.
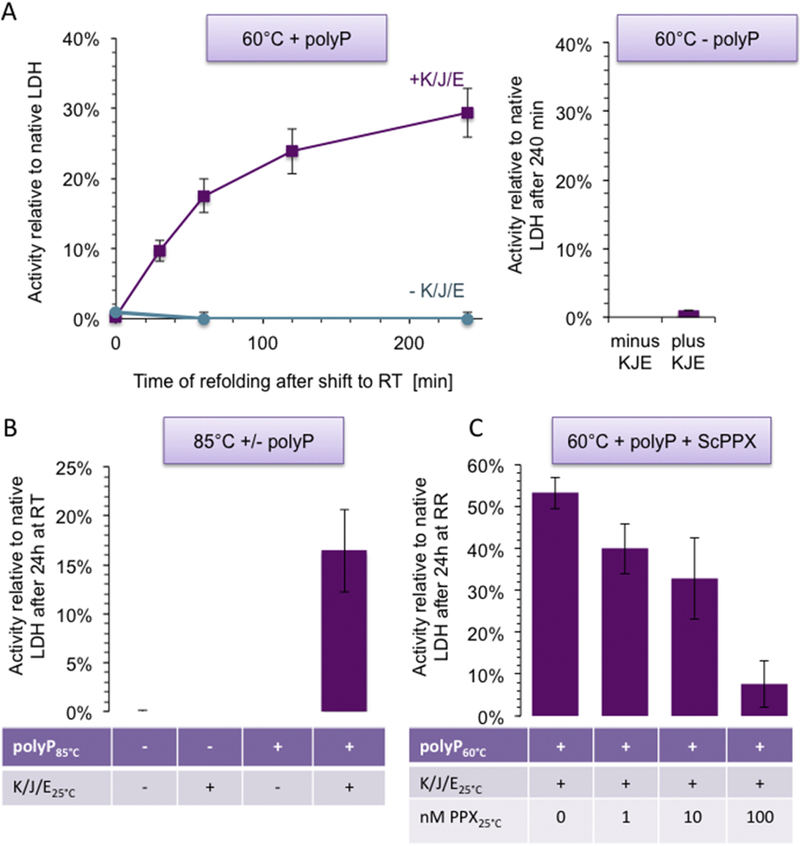
(A) LDH (5.7 μM) was incubated in the presence (left panel) or absence (right panel) of 1 mM polyP130 at 60°C for 10 min. Immediately after the incubation, the samples were diluted 1:29 (0.2 μM final LDH concentration) into 10 mM KPi buffer (pH 7.5) containing 2 mM Mg-ATP, 50 mM KCl +/− 2 μM DnaK, 0.4 μM DnaJ, and 0.2 μM GrpE (KJE). Aliquots from these samples were taken at various time points, diluted 1:40 into the LDH activity assay and the initial rates of the reaction were recorded. (B) Reactivation of LDH after heating the sample from 20°C – 85°C with or without polyP130, and dilution into refolding buffer +/− KJE. (C) Relative activity of LDH heated with polyP130 at 60°C (see (A) for details) and refolded with KJE in the presence of 0, 1, 10, or 100 nM ScPPX.
PolyP protects E. coli against 70°C heat shock treatment
To test whether the protective effects of polyP towards high temperature-induced protein unfolding/aggregation in vitro plays any role in the survival of bacteria at high temperature, we conducted survival assays using the uropathogenic UPEC wild-type and the polyphosphate kinase ppk deletion mutant upon a brief exposure to 70°C during mid-log growth. Both strains grew similarly at 37°C before the heat shock treatment. However, after a short exposure to extreme heat shock, wild-type E. coli was able to quickly recover while the ppk deletion strain took substantially longer before it resumed growth (Figure 6A), resulting in significant lower survival (Figure 6A, Insert). Analysis of the intracellular polyP levels agreed with this result and revealed that exposure of the wild-type strain to a 1 min heat treatment at 70°C was sufficient to trigger polyP levels that were about 1/3 of the highest known levels of polyP in E. coli, typically achieved by nutrient-shift (Figure 6B). As expected, no significant amount of polyP was determined in the ppk deletion strain. Unfortunately, we were unable to reproducibly observe significant differences in total protein aggregation between wild-type and ppk deletion strain. This, however, is likely due to the experimental setup, which required us to conduct an extremely brief heat treatment (1–2 min) to avoid excessive killing, followed by a 60-min recovery of the bacteria. However, it is well known that accumulation of unfolded proteins triggers the expression of heat shock proteins [21]. We therefore decided to use the expression levels of select heat shock genes (i.e. ibpA, ibpB, dnaK) before and after heat shock treatment as proxy for the amount of unfolded proteins present in the cells (Figure 6C). In support of our in vitro results, we found that the ppk deletion strain responds to the heat shock treatment with a significantly higher upregulation of heat shock gene expression compared to wild-type cells, suggesting that cells attempt to compensate for the absence of polyP through increased chaperone production (Figure 6C). Although we cannot exclude that other protective effects of polyP are also involved in heat shock resistance, these results strongly suggest that the chaperoning effect of polyP contributes to high temperature survival of bacteria.
Figure 6. PolyP protects UPEC strain UTI89 against high temperature treatment.
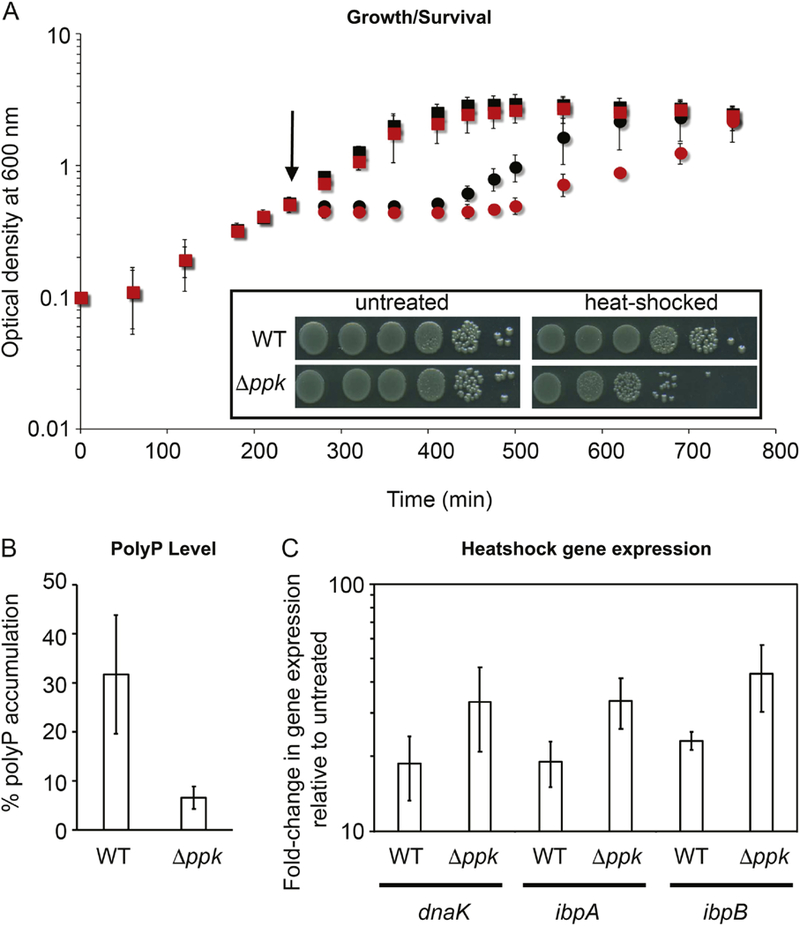
(A) UTI89 wild-type (black) and Δppk cells (red) were grown at 37°C in MOPS-glucose media. At mid-log phase (A600=0.4–0.5), cells were either left untreated (squares) or transferred into pre-warmed flasks (indicated by the arrow) at 70°C for 1 min (circles). Cells were then allowed to recover at 37°C. Growth was recorded for 8.5 h post heat-stress treatment. Insert: Survival of UTI89 wildtype and Δppk cells after 1 min heat treatment at 70°C and subsequent recovery at 37°C for 60 min. Wild-type and Δppk cells were serial-diluted and spot-titered onto LB agar plates and incubated for 15h at 37°C. (B) The accumulation of polyP after 1 min at 70°C and subsequent recovery at 37°C for 60 min was determined and normalized to the polyP accumulation in nutrient-starved cells. (C) Heat shock gene (dnaK, ibpA, ipbB) expression changes in response to brief high temperature treatment. Exponentially growing UTI89 wild-type and Δppk cells in MOPS-glucose medium (OD600 ≈ 0.4–0.5) were either left untreated or exposed to 70°C for 1 min with subsequent recovery for 15 min at 37°C. Gene expression was normalized to the expression of the housekeeping gene rrsD (encoding 16S rRNA), and fold-changes were calculated relative to the expression of each gene in the respective untreated UTI89 strain using the □□CT Experiments were performed independently at least three times.
Discussion
Polyphosphate is a universally conserved and widely distributed compound, whose molecular functions have remained largely enigmatic. In this study, we now demonstrated that polyP, when added at physiologically relevant low millimolar concentrations, is sufficient to stabilize LDH against further unfolding and irreversible aggregation. The unfolding intermediate(s) of LDH that form when polyP is present, are thermostable up to 85°C, form discrete-sized small, soluble oligomers, and show an inherent ability to refold and gain activity once the temperature is reduced and an appropriate chaperone folding system is added. In contrast to native LDH, which is a predominantly α-helical protein, we found that the structural features of the soluble intermediates were distinctly different, revealing substantially increased surface hydrophobicity and pronounced thioflavin T-positive signatures, suggestive of an amyloid-like cross β-sheet character. These results agreed with our previous findings, which showed that polyP stabilizes the β-sheet rich conformation of amyloidogenic proteins, and suggest a very similar mechanism of polyP interaction with non-amyloidogenic proteins, such as LDH.
Over the past years, a number of different non-amyloidogenic proteins have been shown to adopt β-cross sheet features under the appropriate unfolding conditions. Examples include apomyoglobin, a purely α-helical, compactly folded proteins, which undergoes cross β-sheet fibril formation upon incubation in boric acid at pH 9.0, 65°C [22] or the regulatory subunit of PKA, which is a mixed α-helix / β-sheet protein that forms soluble cross β-sheet oligomers upon thermal denaturation [11]. These results led to the proposal that many (if not most) polypeptides have an intrinsic propensity to adopt a cross β-sheet conformation [23]. By extension, this conclusion implied that cross-β-sheet conformations are less influenced by the amino acid side chain composition but by stabilizing interactions between neighboring main chains [22]. We therefore analyzed the LDH sequence using the β-aggregation prediction program TANGO, and identified a distinct region in LDH, where partial unfolding combined with temperature-induced structural fluctuations might promote the transient formation of cross β-sheet structures. We now propose that presence of polyP stabilize these β-sheet structures, thereby preventing higher-order aggregation processes and maintaining the intermediates as small soluble oligomers. PolyP might therefore be an appropriate tool to stabilize and subsequently characterize other protein unfolding intermediates that adopt cross β-sheet character, and potentially aid in determining whether cross β-sheet formation is indeed a generic feature of polypeptides [23].
One of the most intriguing aspects of polyP action deals with the question as to how such a highly charged and simple polymer can recognize and bind so many different protein folding intermediates. We found no evidence to support the notion that polyP binds to native LDH or that presence of polyP triggers the formation of β-sheet structures in LDH under non-denaturing conditions. These results make physiological sense since they exclude the possibility that endogenous polyP targets native proteins to undergo presumably inactivating structural changes. Instead, our data suggest that some partial unfolding and/or secondary structure rearrangement must occur before polyP interacts with proteins. Given its highly charged character, we can likely exclude the possibility that polyP interacts with hydrophobic regions of proteins. Interactions with polar and charged side chains are much more likely, however, they would not necessarily explain polyP’s preference to bind/stabilize protein folding intermediates in a β-sheet rich structure [7, 9]. It is feasible, however, that polyP undergoes electrostatic interactions with the main chain, potentially binding in register with the β-strands. Future highresolution structure analyses such as solid state NMR are necessary to answer this intriguing question. In either case, our study suggests that polyP’s basic working mechanism with soluble versus amyloidogenic proteins is the same, despite the differences in outcome. In fact, presence of (non-physiologically) high levels of polyP has been shown to also increase amyloid solubility and prevent fibril formation [9], suggesting that the ratio between polyP-conferred surface charge and hydrophobicity is ultimately the deciding factor whether soluble oligomers or insoluble fibrils form.
Previous work revealed that the efficacy of polyP in protecting non-amyloidogenic proteins against irreversible aggregation or stimulating amyloidogenic proteins to form fibrils depends on the chain length, with longer chains of polyP being significantly more effective [7, 9]. At the polyP concentration that we used in this study, we did not find any obvious differences in the effect of polyP chain lengths on either the temperature transition of LDH or the secondary structure of the thermal unfolded intermediate(s). We did, however, observe a clear correlation between the size of the LDH-polyP oligomers and the length of polyP chains, with the longer polyP chains forming substantially smaller complexes with LDH than the shorter chains. It now remains to be tested whether shorter polyP chains have a lower binding affinity. This would explain why they are less able to effectively compete with other LDH oligomers for interactions. PolyP chain length has been shown to vary immensely in nature, with microorganisms generally harboring very long >500Pi chains [1]. Based on our results, these polyP species seem to be well-suited to protect against stress-induced protein aggregation. However, more work needs to be done in order to study the dynamics of the complex formation and structure depending on the polyP chain length.
The stress-protective role of polyP has long been recognized [1]. However, the precise role that polyP played under these conditions was far from clear. Early work revealed a stationary growth defect in polyP-depleted bacteria that became particularly pronounced when the bacteria were exposed to oxidative or heat stress [24]. The finding that overexpression of the stress regulator RpoS rescued the growth defects suggested a synergistic interaction between polyP and the RpoS response [24]. In contrast, our studies were conducted in exponentially growing bacteria, where RpoS levels are generally low and not thought to be involved in oxidative or heat stress responses [25], implying that the mechanism by which polyP protects bacteria against these stress treatments is likely independent of RpoS. Instead, our results suggested that the accumulation of polyP in high temperature-treated E. coli cells protects bacterial proteins against irreversible aggregation, thereby promoting cell survival. It appears that the cytoprotective role of polyP becomes most critical under extreme protein-unfolding stress conditions, such as very high temperatures as tested in this study, or upon exposure to fast acting protein unfolding oxidants such as hypohalous acids [8]. Under these conditions, the capacity of canonical chaperones might simply become overwhelmed and/or the translational responses might not be fast enough to appropriately deal with protein unfolding. Future studies are needed to reveal what mechanism triggers the observed accumulation of polyP during high heat treatment, and the precise fate of unfolded client proteins upon return to non-stress conditions. Our in vitro studies and in vivo aggregation results suggest that polyP maintains the protein unfolded intermediates refolding competent until non-stress conditions have been restored, upon which canonical chaperones can refold the proteins. Based on these results, we propose that polyP serves as a general chaperone that protects bacteria against fast-acting protein-unfolding stress conditions.
Material and Methods
Reagents and Proteins
Polyphosphate stocks of different lengths were generously provided by Dr. Toshikazu Shiba (Regenetiss, Japan). Long chain polyP (>800 Pi; polyPXL) was provided by the Morrissey lab [26]. The polyP concentrations provided in the text refer to the concentrations of [Pi]. DnaK, DnaJ, GrpE and ScPPX were purified as described [7, 27]. L-Lactate Dehdrogenase (LDH) from rabbit muscle (Roche) was dialyzed into 50 mM KPi with 50 mM NaCl, pH 7.5, concentrated to about 300 μM with Amicon centrifugation filters and stored at 80°C. All other chemical reagents were purchased from Sigma-Aldrich or Fisher.
Thermal unfolding of LDH and preparation of LDH-polyP complexes
To determine the effects of polyP on the thermal unfolding of LDH, and form complexes between thermally unfolded LDH and polyP, 5.7 μM LDH was incubated in 10 mM KPi (pH 7.5) with or without 1 mM polyP. The respective polyP chain lengths used in this study ranged from 14 Pi units (polyP14) to extra-long chains (polyPXL), and are indicated in the figure legends. Complexes between LDH and polyP were formed by various means and are indicated in the figure legend; i) heating samples in the circular dichroism (CD) spectrophotometer equipped with a Peltier, starting at 20°C and using a heating rate of 1°C/min or 10°C/min; ii) heating the samples in an Eppendorf Benchtop incubator, either by incubating the samples for 10 min at 60°C or by incubating the samples at 20°C and heating them to 85°C (~10 min). For spectroscopic studies, size exclusion analysis, SV-AUC and TEM, the samples were spun down at 16,000 x g for 30 min at 4°C. The soluble supernatants were taken and the protein concentration was analyzed using the published extinction coefficient of LDH at 43,680 M−1cm-1. Samples were immediately analyzed.
Digestion of polyP by Saccharomyces cerevisiae PPX (ScPPX)
To digest polyP, LDH-polyP complexes were incubated with 300 nM ScPPX in reaction buffer (10 mM Tris/HCl, pH 7.5, 0.5 mM MgCl2, 5 mM ammonium acetate) for 30 min at 37°C. PPX digestion was verified by diluting samples 1:100 into 10 μM DAPI, 10 mM KPi, and 1 mM EDTA (pH 7.5). DAPI fluorescence was measured at an excitation wavelength of 415 nm and emission wavelength of 550 nm.
Spin-down assays
To analyze the solubility of LDH after heating, 5.7 μM LDH incubated in the presence of absence of 1 mM polyP in 10 mM KPi (pH 7.5) was heated as indicated. Afterwards, the samples were spun down at 16,000 x g for 30 min at 4°C. The soluble supernatants were transferred into a separate tube, and the insoluble pellets were resuspended in 10 mM KPi. Samples were supplemented with reducing 5-fold Laemmli buffer and loaded onto an SDS-PAGE (Bio-Rad). The gels were stained with Coommassie blue, and the bands were quantified with ImageJ.
Circular Dichroism spectroscopy
To monitor changes in the secondary structure of LDH during thermal unfolding, 5.7 μM LDH was incubated in 10 mM KPi (pH 7.5) with or without 1 mM polyP. Far-UV circular dichroism (CD) spectra were taken from 190 nm to 260 nm in a J-810 spectrophotometer (Jasko) at the indicated temperatures. Protein melting curves were recorded in the same way, except that the CD signal at 222 nm was monitored as the sample was heated (heating rate: 1°C/min).
Size exclusion chromatography
Samples containing 5.7 μM of LDH and polyP of various chain lengths were prepared by heating the samples to 70°C using a heating rate of 1°C/min. 200 μl of sample volume was applied onto a 24 ml Superdex 200 column (GE Healthcare) that had been equilibrated with 10 mM KPi, 50 mM NaCl (pH 7.5). Samples were run at a flow rate of 0.2 ml per minute at a temperature of 4°C. Protein absorbance was used to determine protein elution. The fractions were tested for polyP by DAPI staining [13].
Sedimentation velocity analytical ultracentrifugation (SV-AUC) measurements
LDH-polyP complexes were prepared as described, heating 5.7 μM LDH and 1 mM of different length poyP chains to 85°C using a rate of 10°C/min. SV-AUC was carried out using 420 μl sample loaded into two-sector Epon centerpieces with 1.2 cm path-length in an An60 Ti rotor in a Beckman Optima XL-I analytical ultracentrifuge, and run at 22°C. Measurement were completed in intensity mode. A rather large sedimentation coefficient distribution of 15 to 35 S was observed for the LDH-polyP complexes. In order to maximize the resolution of the size-distribution, all samples were spun at 30,000 and 20,000 rpm, respectively. Sedimentation of LDH was monitored continuously by absorbance at 280 nm, where polyP exhibits no absorbance. All SVAUC data were analyzed using UltraScan 3 software, version 3.5 and fitting procedures were completed on XSEDE clusters at the Texas Advanced Computing Center (Lonestar, Stampede) through the UltraScan Science Gateway (https://www.xsede.org/web/guest/gateways-listing). The partial specific volume (vbar) of the LDH-polyP complex was estimated within UltraScan III based on the LDH protein sequence [28]. Raw intensity data were converted to pseudoabsorbance by using the intensity of the air above the meniscus as a reference and edited. Next, 2-dimensional sedimentation spectrum analysis (2DSA) was performed [29] to subtract time-invariant noise and the meniscus was fit using ten points in a 0.05-cm range. Arrays were fit using an S range of 1–40 S, an f/f0 range of 1–4 with 64 grid points for each, 10 uniform grid repetitions and 400 simulation points. 2DSA was then repeated at the determined meniscus to fit radially invariant and time-invariant noise together using ten iterations. The 2DSA solution was refined by a genetic algorithm (GA), which uses an evolutionary based approach using random cross-over, mutations and deletion operations to alter the solute characteristics of the 2DSA solutes to eliminate false positive solutions [30].
Tryptophan fluorescence
LDH-polyP complexes were formed as described before. Samples were diluted to a final concentration of 0.5 μM LDH in 10 mM KPi (pH 7.5). Fluorescence was measured in a Hitachi 4500 fluorescence spectrophotometer at 30°C. The samples were excited at 285 nm and emission was measured from 300 to 400 nm using slit widths of 5 nm and PMT voltage of 700 V.
Bis-ANS fluorescence
LDH-polyP complexes were described as before. Samples were diluted to a final concentration of 0.5 μM LDH, 10 mM KPi (pH 7.5). Then, 16.5 μM of 4,4’-dianilino-1,1’binaphthyl-5,5’-disulfonic acid (bis-ANS) was added. The samples were incubated at 30°C and excited in the fluorescence spectrophotometer at 370 nm. Emission was measured at 400 nm to 600 nm using slit widths of 5 nm and a PMT voltage of 700 V.
Thioflavin T fluorescence
LDH-polyP complexes were described as before except that 10 μM LDH and 1.75 mM polyP60 or polyP130 was used for complex preparation. The soluble supernatant was diluted to 5.71 μM LDH and incubated with 10 μM thioflavin T at 30°C. Samples were excited in the fluorescence spectrophotometer at 440 nm at 30°C and emission was measured from 470 nm to 700 nm, using slit widths of 5 nm each and a PMT voltage of 700 V. As positive control, 5.7 μM preformed α-synuclein fibrils incubated with 10 μM thioflavin T were used.
Negative stain electron microscopy
LDH-polyP complexes were described as before. The samples were negatively stained with 0.75% uranyl formate (pH 5.5–6.0) on thin carbon layered 400-mesh copper grids (Pelco) as described [31] . Micrograph images were collected on a Tecnai T12 transmission electron microscope (FEI) equipped with a LaB6 filament operated at 120 kV. Images were recorded at 50,000X magnification with 2.2 Å/pixel spacing and a 1.0–1.5 μM defocus on a 4k × 4k CCD camera (Gatan). Contrast transfer parameters for the collected micrographs were then calculated using the CTFFIND4 program [32], and particles were selected using the EMAN2 autoboxing protocols in the E2boxer program [33]. Subsequent particle extraction, phase flipping and reference free 2D classification protocols were performed within RELION 1.4 software package [34]. Five class averages representing different particle sizes were selected as templates or size “bins” for reference-based classification of the total 35 generated class averages into bins using routines in the SPIDER software package [35]. The particles that belonged to each class average were used to estimate the total number of particles that belonged to each bin.
LDH activity assay
The LDH activity assay was adapted from Sigma Aldrich. In short, 0.1 mM NADH, 2 mM sodium pyruvate, and 0.3 mg/mL bovine serum albumin were freshly added to 100 mM NaPi, pH 7.5 at RT. To start the reaction, 5 nM LDH was added, and the absorbance decrease at 340 nm over time was measured in a Cary WinUV Bio50 UV spectrophotometer. LDH activity was determined as the initial change in absorbance. Addition of polyP had no effect on native LDH activity.
LDH-inactivation and reactivation assay
5.7 μM LDH in 10 mM KPi (pH 7.5) was prepared with or without 1 mM polyphosphate. Samples were heated at the indicated temperatures. After the incubation, the samples were diluted 1:29 (final LDH concentration 0.2 μM) into buffer containing 2 mM Mg-ATP, 50 mM potassium chloride, 10 mM KPi (pH 7.5) with or without 2 μM DnaK, 0.4 μM DnaJ and 0.2 μM GrpE and/or 1, 10 or 100 nM ScPPX at RT. The samples were incubated at RT and aliquots were taken at defined time points to monitor LDH activity.
Heat treatment and phenotypical analysis of UPEC strains
Wild-type UTI89 and UTI89Δppk strains were grown aerobically at 37°C in 50 ml potassium morpholinopropanesulfonate (MOPS) minimal medium (Teknova, Inc.) containing 0.2% w/v glucose, 1.32 mM potassium phosphate and 10 μM thiamine. When the cultures reached an optical density at 600 nm (OD600) of 0.4 – 0.5, 13 ml of cells were transferred into pre-warmed flasks and incubated at either 37°C or 70°C. After 1 min at 70°C, the flask was transferred to 37°C and growth was recorded every 30 to 60 min for 8.5 h post heat treatment by measuring OD600. To monitor survival, cells were serial-diluted and spot-titered onto LB agar plates after 60 min of recovery and incubated at 37°C for 15 h.
Extraction and quantification of polyP
UTI89 wild-type and UTI89Δppk strains were cultivated and treated as described before. Changes in intracellular polyP levels upon short-term exposure to heat were quantified as previously described [8]. The fold-change in polyP was compared to the fold-change in polyP observed in wild-type UTI89 grown under nutrient deprivation, a condition known to induce polyP accumulation [36].
Quantitative real-time PCR
UTI89 wild-type and UTI89Δppk strains were cultivated and treated as described before prior to gene expression analysis by RT-PCR with the only modification that transcription was stopped with methanol after only 15 min of recovery at 37°C post treatment at 70°C for 1 min. RNA was prepared using the NucleoSpin RNA kit (Macherey & Nagel) and DNA-free™ kit (Ambion). PrimeScript 1st strand cDNA Synthesis Kit (Takara) was used to generate cDNA, and RT-PCRs were set up with SYBR(R) GreenER™ qRT-PCR mix (Invitrogen) and a Mastercycler(R) realplex2 real-time PCR system (Eppendorf). Expression ratios were calculated compared with expression of each gene in untreated UTI89 and UTI89Δppk cultures, respectively, by the 2ΔΔCT method [37] and normalized to expression of rrsD (encoding 16S rRNA), expression of which did not change under the conditions tested. Primers used for RT- PCR analysis were: ibpAfw: 5’-ACAACCAGAGCCAGAGTAATGG-3’; ibpArev: 5’TATCCTGGGCGGTAATTTCC-3’; ibpBfw: 4’-AAAGCGACGATAACCACTACCG-3’; ibpBrev: 5’-CGTAAAGCTCAGGCTAAAAGGC-3’; rrsDfw 5′-AAG AAC TTA CCT GGT CTT GAC ATC-3′; rrsDrev 5′-CAG TTT ATC ACT GGC AGT CTC CTT-3′.
Highlights.
-
◻
PolyP stabilizes thermal unfolding intermediates of LDH as soluble oligomers of defined size and shape
-
◻
PolyP-LDH intermediates exert features of amyloid-like β-aggregate
-
◻
PolyP maintains refolding competence of LDH upon incubation at near-boiling temperatures
-
◻
PolyP production protects bacteria against extreme temperature stress
Acknowledgements
We thank Lihan Xie for the purification of ScPPX, and Wilhelm Voth for the purification of the DnaK/DnaJ and GrpE proteins. We thank Toshikazu Shiba (Regentiss) and J. Morrissey for providing us with different chain length polyPs. This work was supported by the National Institute of Health Grants GM065318 and GM116582 to U.J. J.-U. D. was supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (DA 1697/1–1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rao NN, Gomez-Garcia MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78:605–47. [DOI] [PubMed] [Google Scholar]
- [2].Morrissey JH. Polyphosphate: a link between platelets, coagulation and inflammation. Int J Hematol. 2012;95:346–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kornberg A, Rao NN, Ault-Riche D. Inorganic polyphosphate: a molecule of many functions. Annu Rev Biochem. 1999;68:89–125. [DOI] [PubMed] [Google Scholar]
- [4].Wang L, Fraley CD, Faridi J, Kornberg A, Roth RA. Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proc Natl Acad Sci U S A. 2003;100:11249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, et al. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2000;97:9636–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gray MJ, Jakob U. Oxidative stress protection by polyphosphate--new roles for an old player. Curr Opin Microbiol. 2015;24:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gray MJ, Wholey WY, Wagner NO, Cremers CM, Mueller-Schickert A, Hock NT, et al. Polyphosphate is a primordial chaperone. Mol Cell. 2014;53:689–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Groitl B, Dahl JU, Schroeder JW, Jakob U. Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol Microbiol. 2017;106:335–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cremers CM, Knoefler D, Gates S, Martin N, Dahl JU, Lempart J, et al. Polyphosphate: A Conserved Modifier of Amyloidogenic Processes. Mol Cell. 2016;63:768–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lee GJ, Pokala N, Vierling E. Structure and in vitro molecular chaperone activity of cytosolic small heat shock proteins from pea. J Biol Chem. 1995;270:10432–8. [DOI] [PubMed] [Google Scholar]
- [11].Dao KK, Pey AL, Gjerde AU, Teigen K, Byeon IJ, Doskeland SO, et al. The regulatory subunit of PKA-I remains partially structured and undergoes beta-aggregation upon thermal denaturation. PLoS One. 2011;6:e17602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Azakami H, Mukai A, Kato A. Role of amyloid type cross beta-structure in the formation of soluble aggregate and gel in heat-induced ovalbumin. J Agric Food Chem. 2005;53:1254–7. [DOI] [PubMed] [Google Scholar]
- [13].Allan RA, Miller JJ. Influence of S-adenosylmethionine on DAPI-induced fluorescence of polyphosphate in the yeast vacuole. Can J Microbiol. 1980;26:912–20. [DOI] [PubMed] [Google Scholar]
- [14].Dams T, Ostendorp R, Ott M, Rutkat K, Jaenicke R. Tetrameric and octameric lactate dehydrogenase from the hyperthermophilic bacterium Thermotoga maritima. Structure and stability of the two active forms. Eur J Biochem. 1996;240:274–9. [DOI] [PubMed] [Google Scholar]
- [15].Das KP, Surewicz WK. Temperature-induced exposure of hydrophobic surfaces and its effect on the chaperone activity of alpha-crystallin. FEBS Lett. 1995;369:321–5. [DOI] [PubMed] [Google Scholar]
- [16].Biancalana M, Makabe K, Koide A, Koide S. Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies. J Mol Biol. 2009;385:1052–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Soldi G, Bemporad F, Chiti F. The degree of structural protection at the edge beta-strands determines the pathway of amyloid formation in globular proteins. J Am Chem Soc. 2008;130:4295–302. [DOI] [PubMed] [Google Scholar]
- [18].Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequencedependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–6. [DOI] [PubMed] [Google Scholar]
- [19].Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–55. [DOI] [PubMed] [Google Scholar]
- [20].Mayer MP, Rudiger S, Bukau B. Molecular basis for interactions of the DnaK chaperone with substrates. Biol Chem. 2000;381:877–85. [DOI] [PubMed] [Google Scholar]
- [21].Arsene F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55:3–9. [DOI] [PubMed] [Google Scholar]
- [22].Fandrich M, Fletcher MA, Dobson CM. Amyloid fibrils from muscle myoglobin. Nature. 2001;410:165–6. [DOI] [PubMed] [Google Scholar]
- [23].Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–90. [DOI] [PubMed] [Google Scholar]
- [24].Rao NN, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol. 1996;178:1394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Smith SA, Baker CJ, Gajsiewicz JM, Morrissey JH. Silica particles contribute to the procoagulant activity of DNA and polyphosphate isolated using commercial kits. Blood. 2017;130:88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U. Identification of a redox-regulated chaperone network. EMBO J. 2004;23:160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Demeler B, Brookes E, Nagel-Steger L. Analysis of heterogeneity in molecular weight and shape by analytical ultracentrifugation using parallel distributed computing. Methods Enzymol. 2009;454:87–113. [DOI] [PubMed] [Google Scholar]
- [29].Brookes E, Cao W, Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 2010;39:405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brookes EaD B Genetic Algorithm Optimization for obtaining accurate Molecular Weight Distributions from Sedimentation Velocity Experiments In: Wandrey CaC H, editor. Progress in Colloid and Polymer Science: Springer; 2006. p. 78–82. [Google Scholar]
- [31].Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. [DOI] [PubMed] [Google Scholar]
- [34].Scheres SH. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415:406–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Dahl JU, Gray MJ, Bazopoulou D, Beaufay F, Lempart J, Koenigsknecht MJ, et al. The antiinflammatory drug mesalamine targets bacterial polyphosphate accumulation. Nat Microbiol. 2017;2:16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]


