Abstract
Modeling and visualization of the cellular mesoscale, bridging the nanometer scale of molecules to the micrometer scale of cells, is being studied by an integrative approach. Data from structural biology, proteomics, and microscopy are combined to simulate the molecular structure of living cells. These cellular landscapes are used as research tools for hypothesis generation and testing, and to present visual narratives of the cellular context of molecular biology for dissemination, education and outreach.
Graphical Abstract

Introduction
Current research is building a bridge that spans from the nanoscale of molecular biology to the microscale of cellular biology. Experimental and computational methods are extending this bridge from both sides. Methods of integrative structural biology are revealing structures of larger and larger functional assemblies, such as the nuclear pore or even entire organelles 1. Imaging methods in cell biology, including cryoelectron tomography and single molecule microscopy, are probing entire cells at finer and finer levels 2, 3. However, the center of this bridge, the cellular mesoscale, is still largely the realm of interpretation. Modeling and illustration of this level of scale, where inanimate matter becomes life, is helping to make this bridge more concrete, to improve our understanding, and to identify regions that require additional study 4.
The goal of this work is to create accurate depictions of the molecular structure of cells, integrating data from structural biology, biochemistry, and cell biology. Mesoscale models have shown utility both in the research community and beyond. Some workers are using these models as scientific tools, for instance, to quantify the effects of crowding in diffusion or to reconcile interpretations of micrographic observations. Other workers use these as thinking tools, to develop and explore hypotheses that will then be subjected to experiment. These depictions are also essential elements in education and outreach, providing context for discussions of molecular and cellular topics, and impressing on viewers the complexity and interconnectedness of the subcellular world. In this review, we will discuss some of the technical and aesthetic aspects of creating mesoscale imagery, as well as applications in research and in education.
Historical
Artistic depictions of the inner structure of cells have been used since the development of microscopy, both to document scientific observations and as figures in textbooks. After a long history of drawing and redrawing by generations of artists, an iconic form has emerged, typically showing a generic “cell” with a spherical nucleus in the center, surrounded by small ovoid mitochondria, a sheet-like endoplasmic reticulum, and perhaps a single Golgi. Most often, the only molecular details included in these figures are ER-associated ribosomes and nuclear pores, often drawn considerably larger than the actual relative size to make them visible.
Soon after the first electron micrographs of cells were obtained, the first integrated views with molecular detail were created. For example, illustrations depicted the “microtrabecular lattice,” a hypothetical structure of the cytoplasm organized by cytoskeletal filaments 5, 6, and Srere presented an illustration of mitochondrial membranes 7. In 1991, several mesoscale illustrations were created for the best-studied organism of the time, Escherichia coli, integrating atomic structures of molecules, proteomics information from 2D electrophoresis, and a variety of other sources 8. This illustrative mesoscale work has continued to other systems, ranging from HIV 9 to a panorama from the nucleus to cell surface of a T-cell 10. Currently, artistic illustrations and animations of the cellular mesoscale are widely used for education and outreach (described in more detail below).
Building on this illustrative tradition, mesoscale modeling and visualization are now moving into three dimensions, becoming a tool for research. The increased availability of data at all levels has made this possible, including the use of electron microscopy to determine atomic structures of very large assemblies, the high resolution currently achieved in cryoelectron tomography of entire cells, and advances in genomics, proteomics and interactomics. The results are semiquantitative models of large portions of cells. Notable examples include models of a synaptic vesicle 11, a synaptic bouton 12, HIV 13, and bacterial nucleoids 14, 15. By all expectations, the grand challenge of creating a predictive model of an entire eukaryotic cell is within reach given our rapidly improving experimental and computational abilities 16.
Technical Concerns
Modeling and depiction of the cellular mesoscale is necessarily an integrative process, since there currently are no direct methods for observing the atomic structure of cells. At a minimum, a few types of information are needed:
Cellular Ultrastructure. This is most often obtained from electron microscopy, such as negative-stained thin sections for cross-sectional mesoscale models and tomograms for 3D mesoscale models 2, 3. Typically, these experimental data are segmented to yield a description of the ultrastructure of membranes and location of large molecular assemblies. Techniques such as template-free structure detection 17 promise to extend this segmentation to finer and finer scales.
Molecular Composition. Genomic information can provide a comprehensive list of proteins and nucleic acids that are synthesized, and proteomic data can define the proteins and estimates of their abundance for different compartments in the cell. For example, a recent immunofluorescence study created a “cell map” for ~12,000 proteins in a human cell 18.
Molecular Structure. Atomic structures are available for tens of thousands of biomolecules through the Worldwide Protein Data Bank (www.wwpdb.org), and homology modeling allows prediction of structures that have not been determined for the organism of interest. The recent radical improvements in cryoelectron microscopy are a significant advance, providing structures for very large and complex assemblies that were not tractable with other structural methods 1.
Molecular Interactions. Higher-order molecular interactions are still an area of intense research, both to define functional interactomes 19 and to determine or model structures of assemblies 20, 21.
Generation of mesoscale models for visualization is being approached in many ways, depending on the audience and goals of the model. Traditional artistic methods, including hand-drawn images and digitally-produced images and animations, are still most commonly used for images in education and outreach settings (Figure 1a). This is typically a laborious process of generating models for each of the molecules, then placing them manually to generate the scene 22. However, artistic approaches have a great advantage, since all the tricks used by artists can be employed: manual placement allows building of scenes that highlight salient features and minimize occlusion, and portions of the scene (such as molecules not directly related to the story) may be excluded. With experimentally-generated models, we don’t typically have as many of these options.
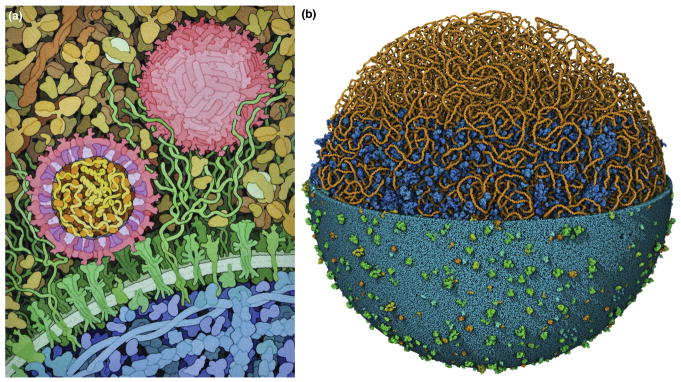
Figure 1.
Mesoscale illustrations. (a) Hand-drawn illustration of Zika virus recognizing a target cell, created for educational outreach at the RCSB Protein Data Bank (http://pdb101.rcsb.org). (b) Three-dimensional model of a full mycoplasma cell. The DNA is modeled using lattice-based methods 29, the membrane with LipidWrapper 28, and the remaining molecules with CellPACK 30. The model is visualized in CellView 31, using selective clipping to progressively reveal the interior molecules (center) and only the DNA (top). Mycoplasma illustration created by Ludovic Autin.
Several methods have been developed to assist with the process of designing a mesoscale scene. Plugins such as MolecularMaya (https://clarafi.com/tools/mmaya), ePMV 23, and BioBlender (http://www.bioblender.org) assist with the import, modeling and animation of atomic structures within popular 3D animation packages. The GraphiteLifeExplorer 24 streamlines the construction of complex assemblies with proteins and nucleic acids, allowing users to define 3D paths for DNA and other flexible chains, and populate the scene with associated protein structures. CellPAINT (http://cellpaint.scripps.edu) uses the approach of a digital painting program: a palette of molecules, each with characteristic properties for interaction, is used to paint a 2.5 D mesoscale scene, and a temperature slider can add diffusive motion to the resulting scene.
While these methods allow users to control the placement of elements in a scene, automated stochastic methods for generating and simulating mesoscale models are also becoming available 4, 25. Typically, these define the ultrastructure of the environment, then randomly populate it with molecules based on defined composition, concentration, and rules of interaction. Several laboratories are working on detailed atomic models of bacterial cytoplasm, which are used in Brownian dynamics simulations to explore the consequences of macromolecular crowding on cellular function 26, 27. Specialized methods have been developed for large structures in the cells. For example, LipidWrapper 28 uses a tiling approach to populate a membrane with lipid structures taken from detailed molecular dynamics simulations, and bacterial nucleoids have been approached using lattice methods 29 and in an ambitious hierarchical approach that starts with coarse grain models and progressively refines the resolution to give an atomic model 14, 15. CellPACK 30 brings these tools together to build mesoscale models of entire cells (Figure 1b).
Given their size and complexity, representation of these stochastic models poses a number of challenges, so specialized tools are being built to visualize them. For example, CellVIEW 31 provides a variety of selecting, clipping and coloring tools to help simplify these scenes and highlight features of interest. Rendering of these models poses a great challenge, since the datasets are extremely large. GPUs and instancing, where the structure of each type of molecule is stored only once and then repeated throughout the scene, are emerging as the techniques of choice 32, 33. Methods are also being developed to assist with navigation and comprehensibility. These include control of hierarchical level of detail, custom clipping planes and advanced lighting models (such as ambient occlusion) 34.
Building a Narrative
Mesoscale modeling is all about context. In research, mesoscale models are being developed to allow simulation of molecular processes in their cellular context, exploring the consequences of crowding, compartmentation and copy number on biomolecular function, and presenting these consequences in dissemination of the research. In science outreach and education, mesoscale imagery allows viewers to see biomolecular processes in the context of their cellular environment. Effective mesoscale modeling, both for research and for outreach, requires careful thought to design a narrative that captures the salient features of this context. The landscapes allow us to link data from multiple sources into a coherent integrated story.
Weak points in this hierarchy of data will compromise the narrative. One area of intensive current work is the development of methods to quantify these fuzzy regions and use them to define confidence measures in the resultant mesoscale models. For example, current work in integrative modeling is using Bayesian inference to create probabilistic models that are consistent with a body of heterogeneous data 35, 36. There is also a great need to develop effective methods to represent levels of certainty in mesoscale imagery. Combination of detailed and schematic representations is one simple, and readily comprehensible, approach (Figure 2). Embedded citation within a visualization is also being explored as a way to add information on sources to images 37.
Figure 2.
Illustration from the educational portal of the RCSB Protein Data Bank (http://pdb101.rcsb.org), showing six atomic structures of proteins from Ebola virus, along with an artistic conception of their place in the entire virion. Schematic circles show the approximate volume of portions that were not determined in the atomic structures.
There are multiple ways of representing scientific data, and in science dissemination and outreach, researchers need to select ways of displaying their data that tell a clear, coherent story. Fortunately, we can draw on a variety of existing narrative devices, ranging from textbooks to classics of literature and film. These techniques build on a rich tradition of communication practices and are as diverse as the people telling them and the audiences for which they are designed. The complexity of mesoscale subjects poses a few unique challenges for the design of narratives.
The first issue is to highlight a story from within a sea of competing molecules. Most commonly, much of the extraneous detail is simply omitted, creating imagery that only includes molecules of interest. If the crowded nature of the cellular environment is a part of the story, an introductory image may be presented. For instance, molecular biology textbooks often include a single figure in introductory sections showing the crowded nature of the cell, then use simplified images for the following sections that explore specific topics. Coloring and labels can also help viewers parse out the central molecules in a scene, and scenes may be designed such that the central players occupy prominent positions in the scene, with minimal occlusion by other molecules. Cross-sectional metaphors may also be used to clip away portions that occlude areas of interest. For example, Figure 1a shows a traditional cross section, similar to the way samples are prepared for microscopy, and Figure 1b employs several selective clipping planes to reveal the inner structure of the mesoscale model 34.
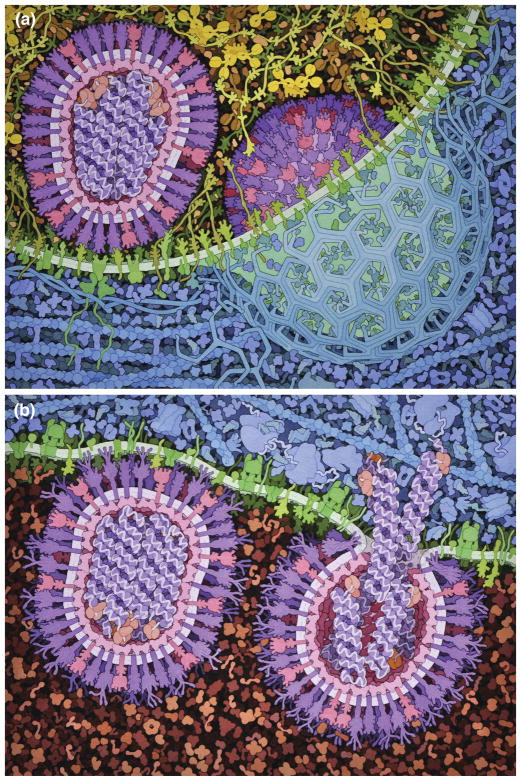
Viewers also must navigate large differences in scale, and large differences in time, for many processes 22. Mesoscale processes span from picoseconds to seconds and from nanometers to microns. In static images, these may be captured visually in multiples. For instance, Figure 3 shows two processes in influenza infection. Two separate panels show processes of entry and release that are separated by a large distance in time and space, and each panel includes two snapshots of the virus at progressive stages of each local process.
Figure 3.
Snapshots of the influenza life cycle, created as part of educational materials being developed at the Center for BioMolecular Modeling. Major steps occur at distant regions in the cell, so they are shown in separate panels: (a) entry at the cell membrane; (b) exit from the endosome. Within each panel, two viruses show separate substeps: (a) attachment at left and endocytosis at right; (b) on the left, hemagglutinin conformational change and insertion into the endosome membrane, and on the right, fusion with the membrane and release of viral RNA into the cytoplasm.
In animations, all manner of pans, zooms, and cuts may be employed to transition between scenes at different temporal and spatial scales. For this, we need to continuously change the representation to highlight features of the current scale level, showing atoms when we’re looking at enzymatic reactions and shifting to smoother representations as we move to assemblies and above. Pioneering animations such as Charles and Ray Eames “Powers of Ten” (http://www.eamesoffice.com/the-work/powers-of-ten/) and Nelson Max’s continuous zoom from DNA atoms to a chromosome for “The Universe: We Are Born of Stars” 38 presented effective visual solutions to these transitions. Drew Berry’s animation of cellulose structure (Figure 4) employs many visual techniques to make the narrative clear, including consistent use of color, smooth transition of the level of detail, and design of the scene to include recognizable features that bridge scale levels. Today, methods such as CellVIEW are implementing these transitional techniques in visualization software, automatically picking the appropriate coloration and level of detail for the current view 31.
Figure 4.
An animation by Drew Berry presents a continuous zoom from atoms to an entire plant. Four frames from the mesoscale portion of the film are shown here: nanoscale views of (a) cellulose and (b) cellulose synthase; (c,d) microscale views of the cell wall, with chloroplasts visible through a translucent cell membrane. Images kindly provided by Drew Berry.
Many properties of the biological mesoscale are non-intuitive, with aspects that are foreign to our familiar experience. Diffusive motion is perhaps the most pervasive feature to be considered. Animations of the mesoscale address this at all levels. Brownian dynamics simulations of bacterial cytoplasm depict accurate models of random diffusive motion, allowing detailed prediction of biophysical properties, but making functional transitions hard to predict and depict. On the other hand, animations for education and outreach (described below) typically work at longer time scales, script molecules to follow story-driven trajectories and add various amounts of random motion to give the impression of diffusion.
Animation and Interaction
Animation is the most effective approach for exploring the dynamic nature of the mesoscale 22, 39. In research, short animated segments of a simulation are invaluable for debugging, because small errors in dynamics methods are often readily apparent when viewing the motion. Animated segments are also commonly used for dissemination, often as supplementary files in publication and to present key dynamic processes in presentations.
Animation is widely used for mesoscale dissemination, education and outreach. Animation is a natural narrative device, with a strong tradition of cinematic techniques that guide viewers through a story. In addition, we can leverage an extensive toolbox of software developed by the entertainment industry. The mesoscale poses a few challenges to traditional methods of animation. The complexity and crowded nature of the mesoscale may present problems. Animation often takes an immersive approach, placing the viewer in the middle of the environment, surrounded on all sides by molecules or ultrastructure. This gives an exciting sense of immediacy and presence to the animation, but can pose pedagogical problems with occlusion or perspective distortion. Animators may employ simplifications to address these potential problems, sometimes omitting molecules that aren’t of functional interest, or choosing viewpoints that are some distance from the subject, allowing an unobstructed view.
The diffusive motion of the cellular mesoscale also poses challenges. Storytelling often requires a coherent series of events, following a process step-by-step, which is almost impossible to extract from an unbiased dynamics simulation. Animators typically employ a variety of tricks to make the action follow a story. Most commonly, traditional keyframe animation is used to script the story, and then stochastic motion is layered on top of this scripted motion. Looking to movies presented at the Clarifi showcase (https://clarafi.com/showcase/), animators take very different approaches to this balance of random and directed motion.
Two leaders in the field have found a sweet spot that balances scientific accuracy and comprehensibility. Drew Berry’s work incorporates extensive stochastic motions while following scripted biomolecular processes. See, for example, his iconic presentations of basic genetic mechanisms in “Body Code” (https://www.wehi.edu.au/wehi-tv/body-code). Janet Iwasa’s animations of the HIV life cycle often employ larger stochastic motions of entire molecules, since they depict processes that occur over longer time scales (http://scienceofhiv.org). One trick that may be employed for these complex trajectories is to create a model, disassemble it with a dynamics simulation, and then play the process in reverse. As Berry describes, even this presents challenges: “It works to reduce the sense of agency but is very tricky to plan and implement for animation production.” This technique was used, for example, for an animation of assembly of the apoptosome (https://www.youtube.com/watch?v=DR80Huxp4y8).
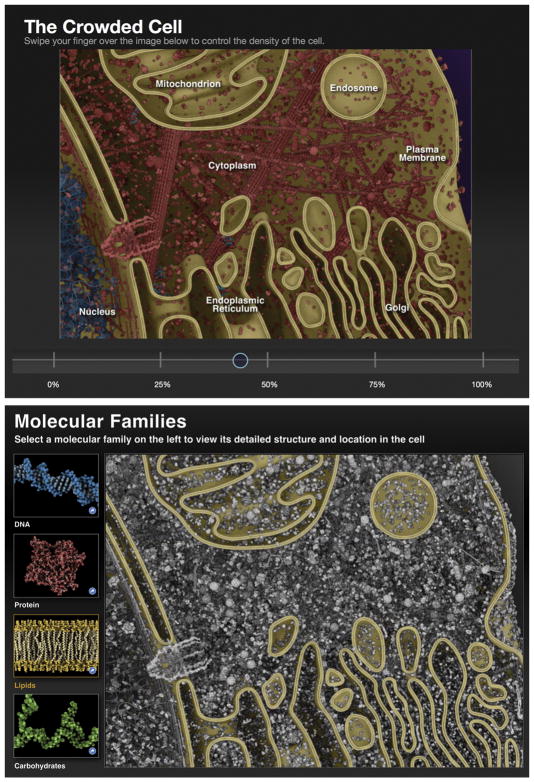
Animations are typically scripted, allowing the creator to guide us through a topic, but also necessarily limiting the way we interact with the scene. Methods for interactive display can add an exploratory aspect, adding to the pedagogical value of mesoscale visualizations and providing entertaining ways to address the complexity of these scenes. One effective approach is to segment scenes into functional portions, then provide interactive tools to display portions selectively. For example, Gael McGill designed a series of interactive figures for “EO Wilson’s Life on Earth” to allow students to explore the types of molecules displayed in a molecular scene, and the crowding of the scene (Figure 5). Image maps provide another intuitive mode of interaction: users click on portions of the scene, and detailed information is provided. This is used effectively in a series of virus illustrations by Visual Science, such as a collaborative illustration of influenza virus that captures the current state of structural knowledge (https://visual-science.com/projects/influenza/illustration/). As part of an e-book on biological energetics, image maps were created for paintings of mitochondria and chloroplasts, and were use as a navigation device to integrate interactive visualization and animations across an entire educational site 40. CellPAINT (http://cellpaint.scripps.edu) extends interactivity to the construction of mesoscale imagery. It is modeled after familiar digital painting programs, with a collection of molecular brushes to paint onto mesoscale canvas (Figure 6). New approaches to the human-computer interface, including virtual reality and 3D printing, are also being explored to enhance navigation and presence in mesoscale visualization.
Figure 5.
Interactive figures from “EO Wilson’s Life on Earth.” In “The Crowded Cell,” viewers use a slider to add molecules to the scene, exploring the crowded nature of the cell. In “Molecular Families”, viewers selectively color molecules from the families shown at left. Here, “Lipids” have been chosen. Images kindly provided by Gael McGill, Digizyme Inc.
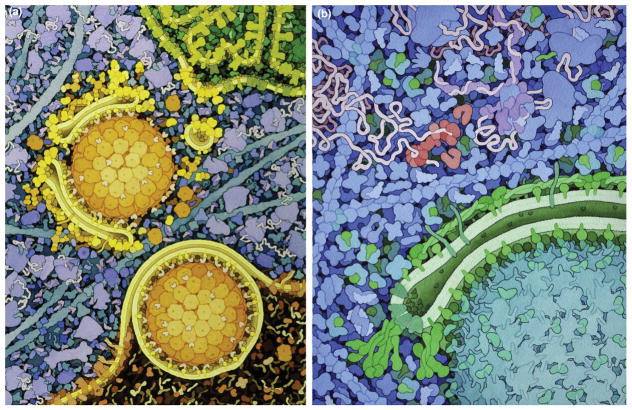
Figure 6.
CellPAINT (http://cellpaint.scripps.edu) uses the approach of a digital painting program to create mesoscale illustrations. (a) The interface has a palette of molecules, and tools for painting them into the scene. In this screen capture, the user is drawing a membrane based on an electron micrograph. (b) A completed scene with HIV (green), blood serum (oranges and yellows), and the surface of a T-cell (blue and magenta).
Molecular Landscapes in Research
In mesoscale research, we search for hypotheses that link molecular structure with cellular function. Our goal is to build a model that captures enough of the experimental observations to yield scientific insight into a mesoscale process. As part of this process, we need to validate each step of the hierarchy: from atomic structure/interaction, to how ultrastructures are populated with these molecular structures, and finally to the cellular properties that emerge. This is currently a field in growth phase, with methods being developed as fast as applications are discovered that need them 4, 16, 25, 41. Two trends are stimulating the success of mesoscale research: integrative structural biology and coarse-grain computational modeling.
Integrative structural biology combines experimental data from multiple levels of resolution to build atomic models of large assemblies. Notable examples include a plethora of structures showing the action of ribosomes 42 and nuclear pore models of ever increasing detail 43. This integration is continually extending to larger levels of scale, for example, to interpret features in data from cryoelectron tomography 44 and fluorescence microscopy 30.
Coarse-grain computational models explore mesoscale properties, such as diffusion and cellular ultrastructure, using simplified representations of the molecular components. These are particularly effective because the level of detail may be chosen to complement the system, while keeping computational demands to achievable limits. For example, bead models of nucleic acids may be used to study everything from the role of entropy in partition of bacterial chromosomes 45 to the ultrastructure of entire eukaryotic chromosomes 46.
Mesoscale imagery is also used as a thinking tool, to capture the current state of knowledge in a field, and for dissemination of this state of knowledge. For example, a series of illustrations of the molecular processes of autophagy were created in a collaboration between artist and scientist 47 (Figure 7). The first illustrations, created in 2006, included many approximations for molecular size and interaction. In the following years, as more experimental data were obtained, the illustrations gained more detail. Similarly, Janet Iwasa is creating animations of the HIV life cycle in collaboration with researchers, integrating the enormous body of experimental data that has been gathered (Figure 8). For these types of applications, the modeling and visualization must have a sufficient level of scientific rigor to be used in professional research settings.
Figure 7.
Illustrations of the “cytoplasm to vacuole targeting” (Cvt) process of autophagy, created in a collaboration with Daniel Klionsky. (a) The first illustration was created in 2006 and includes simple representations of the many proteins orchestrating the formation of the phagophore. (b) In 2014, new data allowed more detailed representation of the proteins and complexes.
Figure 8.
Frame from “Egress” by Janet Iwasa, created in collaboration with HIV researchers in the CHEETAH Center. The animation integrates data from the field of HIV structural biology to show the complex spatial interactions and ordering of molecules in the budding process. This frame is near the end of the process, when a helical complex of ESCRT-III (in turquoise near the bottom) is pinching off the new virus from the cell surface. Image kindly provided by Janet Iwasa.
Molecular Landscapes in the Classroom
Well-designed mesoscale visualizations address scale, proportion, and quantity within a cellular scene, as well as the relationship of structure and function. These concepts have been identified as central themes for science education in the Crosscutting Concepts in Next Generation Science Standards 48. Students most often encounter mesoscale concepts and imagery in texts, but effective resources are increasingly breaking through the limitations of the page, including the interactive, animated approach of “EO Wilson’s Life on Earth,” many animations that are freely available online, and innovative approaches to new media such as augmented reality and 3D printing.
Illustrators portraying complex molecular interactions for educational contexts face a daunting challenge. If they oversimplify the content, students think that the cell is “simple” and miss the nuanced complexities. If all the details are provided, novices become overwhelmed and can easily give up 49, 50. Vygotsky’s theory of the zone of proximal development suggests that learning is optimal when content is just beyond the learner’s reach, but attainable with appropriate scaffolding 51. If details are oversimplified and there is not a little challenge, optimal learning conditions are not achieved, as demonstrated in a study on cellular illustrations of signaling networks 52.
Bruner introduced the concept of spiraled learning, in which novices are introduced to the basics of a topic, then spiral back to the same concepts repeatedly throughout the curriculum, digging ever deeper each time they visit the topic 53. This approach can be employed effectively either by providing increasingly more complex visualizations over time, or by teaching students how to “mine” visualizations multiple times to uncover additional layers of complexity and meaning. This revisiting of topics repeatedly in the curriculum also improves learning retention 54, 55. In our hands, mesoscale illustrations have been particularly useful in this context. They present multiple processes, all occurring simultanously in the context of their cellular environment. Using a single illustration, students can explore multiple levels of scale, for example, starting with the multisubunit structure of ATP synthase, then exploring its location and orientation in the mitochondrial membrane, then jumping back to individual molecules to explore the respiratory electron transport chain, and so on.
Recent science education reforms have focused on ‘thinking like a scientist’ with a concomitant ability to apply knowledge in new contexts 56, 57. Unfortunately, many students memorize without understanding, then quickly forget after being tested. To overcome this tendency to compartmentalize information, then forget it, students must be provided opportunities to use the information and organize it into larger “schema” 58. This is the basis for constructivism, in which students come to their own understanding by working through the content and making sense of it themselves 59.
In developing conceptual understanding, it is essential to make connections between seemingly disparate ideas. Carefully crafted illustrations and animations serve as visual organizers of information, unifying themes that are often presented in the classroom in a compartmentalized fashion. Why, for instance, would nuclear proteins be found in the cytoplasm? Students are encouraged to think about where proteins are made, and then to address the question: How do nuclear proteins know they should go to the nucleus? Such cognitive dissonance leads to deeper thinking and richer questions 60. Visualization of these deeper connections assists in scaffolding student understanding 61, 62.
Although molecular animations can readily demonstrate complex processes in a temporal fashion at the mesoscale, they may introduce detailed interactions at such a rapid pace that the novice learner doesn’t have time to process and internalize the information. Experts often assume that novices “see” the same details that experts see; most educators can provide amusing (and even tragic) examples that demonstrate discrepant interpretations 63. Many studies have demonstrated that passive learning is ineffective 64, whereas active learning increases learning and retention 65. Educators choosing to incorporate molecular animations and illustrations in their classrooms must create activities in which students are fully engaged and actively building their own understanding of the concepts. Some suggested approaches based on our own work with mesoscale illustrations are provided in Table 1. These ideas may be implemented using readily available materials. For example, the Center for BioMolecular Modeling has created grayscale coloring pages and molecular flashcards to accompany a cellular panorama landscape (Figure 9).
Table 1.
Engaging students in interpreting mesoscale landscapes
| Activity | Description | Examples | Applications |
|---|---|---|---|
| What is it? | Identifying cell structures | Endoplasmic reticulum, Golgi body, ribosomes, cytoskeleton | Color the endomembrane system. Which structures have a double membrane and why? |
| Where’s Waldo? | Locating specific structures | RNA polymerase, ribosome, ATP synthase | Explore cellular compartmentalization and why structures have differing concentrations throughout the cell |
| Where in the cell are we? | Identify all locations of various structures | DNA, ATP synthase | Why is the structure in one location and not another? Or in two different locations? |
| Where does this process occur? | Identify key players in various processes and identify their location | Protein synthesis, energy metabolism | Why is mRNA necessary? |
| Compare and contrast | Prokaryotic vs. eukaryotic cells | RNA polymerase, ribosomes, ATP synthase | Why does eukaryotic RNA polymerase have a long tail? |
Figure 9.

A combination of grayscale and color is used to create molecular flashcards that can be used in a number of active learning activities. Here, eukaryotic RNA polymerase with its unstructured C-terminus is identified in the nucleus.
There is even greater value in engaging students in the process of creating visualizations. As a part of the Connecting Researchers, Educators and Students (CREST) Project, undergraduates were involved with researchers and educators in exploring a research topic, then creating instructional materials to introduce current research in the classroom. Undergraduates conducted background research to create molecular landscapes exploring angiogenesis 66 and endocytosis (http://cbm.msoe.edu/includes/modules/coatedPit/coatedPit.html). Students determined the types of cell and what portion of the cells to display to tell the molecular story. They decided which proteins to include, as well as their shape, size, location and concentration. They discovered discrepancies in the literature and worked with researchers to develop a rationale for selecting one interpretation over another 66. In this hands-on approach, students gained personal experience in the process of integrating data into a coherent vision of the topic.
Perspective
Klaus Schulten referred to modeling and simulation as a “computational microscope” to explore molecules in their living context 67. This microscope may be focused at many levels, from molecular dynamics simulations of individual proteins to continuum models of molecular diffusion in entire cells. Amazingly, the advance of experimental imaging techniques such as cryoelectron tomography and fluorescence microscopy are rapidly putting us out of business, as scientists view the cellular mesoscale directly. In some senses, mesoscale modeling is giving us an exciting preview of what we will see as experimental mesoscale science continues to sharpen and improve.
Mesoscale visualization is playing perhaps its most pervasive role in the dissemination of science. These images, animations and interactives allow students and the public to understand the connections between different disciplines--how study of an individual protein can give us insights into our own health and welfare. Mesoscale imagery, such as the popular “Inner Life of the Cell” movie (Figure 10), are inspiring a new generation of scientists to explore the mesoscale mysteries of life.
Figure 10.
Frame from “Inner Life of the Cell,” showing the kinesin transporting a vesicle along a microtubule. Image kindly provided by Michael Astrachan. ‘Inner Life of the Cell,’ © 2006 President and Fellows of Harvard College. Created by Alain Viel, PhD and Robert Lue, PhD in collaboration with XVIVO, LLC.
Highlights.
Mesoscale illustrations integrate data from molecular and cellular biology.
Methods for building and visualizing 3D mesoscale models are being developed.
Mesoscale illustrations provide context in molecular biology education.
Acknowledgments
This work is supported by R01-GM120604 from the National Institutes of Health (DSG), DBI-1338415 to the RCSB from the National Science Foundation, the National Institutes of Health, the US Department of Energy (DSG), and DUE 1725940 from the National Science Foundation and R25OD023723 from NIH-SEPA (TH and MAF). TSRI publication #29685.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annu Rev Biochem. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Harush K, Maimon T, Patla I, Villa E, Medalia O. Visualizing cellular processes at the molecular level by cryo-electron tomography. J Cell Sci. 2010;123:7–12. doi: 10.1242/jcs.060111. [DOI] [PubMed] [Google Scholar]
- 3.Beck M, Baumeister W. Cryo-electron tomography: Can it reveal the molecular sociology of cells in atomic detail? Trends Cell Biol. 2016;26:825–837. doi: 10.1016/j.tcb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 4.Im W, Liang J, Olson A, Zhou HX, Vajda S, Vakser IA. Challenges in structural approaches to cell modeling. J Mol Biol. 2016;428:2943–2964. doi: 10.1016/j.jmb.2016.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter KR, Tucker JB. The ground substance of the living cell. Sci Am. 1981;244:56–67. doi: 10.1038/scientificamerican0381-56. [DOI] [PubMed] [Google Scholar]
- 6.Clegg JS. Properties and metabolism of the aqueous cytoplasm and its boundaries. Amer J Physiol. 1984;246:R133–R151. doi: 10.1152/ajpregu.1984.246.2.R133. [DOI] [PubMed] [Google Scholar]
- 7.Srere PA. The structure of the mitochondrial inner membrane-matix compartment. Trends Biochem Sci. 1982;7:375–378. [Google Scholar]
- 8.Goodsell DS. Inside a living cell. Trends Biochem Sci. 1991;16:203–206. doi: 10.1016/0968-0004(91)90083-8. [DOI] [PubMed] [Google Scholar]
- 9.Torbett BE, Goodsell DS, Richman DD. The future of HIV-1 therapeutics: Resistance is futile? Springer; New York, NY: 2015. [Google Scholar]
- 10.Goodsell DS. Eukaryotic cell panorama. Biochem Mol Biol Educ. 2011;39:91–101. doi: 10.1002/bmb.20494. [DOI] [PubMed] [Google Scholar]
- 11.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J, Wieland F, Jahn R. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelm BG, Mandad S, Truckenbrodt S, Krohnert K, Schafer C, Rammner B, Koo SJ, Classen GA, Krauss M, Haucke V, Urlaub H, Rizzoli SO. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- 13.Johnson GT, Goodsell DS, Autin L, Forli S, Sanner MF, Olson AJ. 3D molecular models of whole HIV-1 virions generated with cellPACK. Faraday Discuss. 2014;169:23–44. doi: 10.1039/c4fd00017j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hacker WC, Li S, Elcock AH. Features of genomic organization in a nucleotide-resolution molecular model of the Escherichia coli chromosome. Nucl Acids Res. 2017;45:7541–7554. doi: 10.1093/nar/gkx541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildirim A, Feig M. High-resolution 3D models of Caulobacter crescentus chromosome reveal genome structural variability and organization. Nucl Acids Res. 2018 doi: 10.1093/nar/gky141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singla J, McClary KM, White KL, Alber F, Sali A, Stevens RC. Opportunities and challenges in building a spatiotemporal multi-scale model of the human pancreatic beta cell. Cell. 2018;173:11–19. doi: 10.1016/j.cell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu M, Beck M, Alber F. Template-free detection of macromolecular complexes in cryo electron tomograms. Bioinformatics. 2011;27:i69–76. doi: 10.1093/bioinformatics/btr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, Backstrom A, Danielsson F, Fagerberg L, Fall J, Gatto L, Gnann C, Hober S, Hjelmare M, Johansson F, Lee S, Lindskog C, Mulder J, Mulvey CM, Nilsson P, Oksvold P, Rockberg J, Schutten R, Schwenk JM, Sivertsson A, Sjostedt E, Skogs M, Stadler C, Sullivan DP, Tegel H, Winsnes C, Zhang C, Zwahlen M, Mardinoglu A, Ponten F, von Feilitzen K, Lilley KS, Uhlen M, Lundberg E. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 19.Petrey D, Honig B. Structural bioinformatics of the interactome. Annu Rev Biophys. 2014;43:193–210. doi: 10.1146/annurev-biophys-051013-022726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kundrotas PJ, Zhu Z, Vakser IA. GWIDD: A comprehensive resource for genome-wide structural modeling of protein-protein interactions. Hum Genomics. 2012;6:7. doi: 10.1186/1479-7364-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosca R, Ceol A, Aloy P. Interactome3D: adding structural details to protein networks. Nat Methods. 2013;10:47–53. doi: 10.1038/nmeth.2289. [DOI] [PubMed] [Google Scholar]
- 22.McGill G. Molecular movies … Coming to a lecture near you. Cell. 2008;133:1127–1132. doi: 10.1016/j.cell.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Johnson GT, Autin L, Goodsell DS, Sanner MF, Olson AJ. ePMV embeds molecular modeling into professional animation software environments. Structure. 2011;19:293–303. doi: 10.1016/j.str.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hornus S, Levy B, Lariviere D, Fourmentin E. Easy DNA modeling and more with GraphiteLifeExplorer. PLoS One. 2013;8:e53609. doi: 10.1371/journal.pone.0053609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingolfsson HI, Arnarez C, Periole X, Marrink SJ. Computational ‘microscopy’ of cellular membranes. J Cell Sci. 2016;129:257–268. doi: 10.1242/jcs.176040. [DOI] [PubMed] [Google Scholar]
- 26.McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu I, Mori T, Ando T, Harada R, Jung J, Sugita Y, Feig M. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. eLife. 2016;5:e19274. doi: 10.7554/eLife.19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durrant JD, Amaro RE. LipidWrapper: an algorithm for generating large-scale membrane models of arbitrary geometry. PLoS Comput Biol. 2014;10:e1003720. doi: 10.1371/journal.pcbi.1003720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodsell DS, Autin L, Olson AJ. Lattice models of bacterial nucleoids. J Phys Chem B. 2018 doi: 10.1021/acs.jpcb.1027b11770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson GT, Autin L, Al-Alusi M, Goodsell DS, Sanner MF, Olson AJ. cellPACK: a virtual mesoscope to model and visualize structural systems biology. Nat Methods. 2015;12:85–91. doi: 10.1038/nmeth.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Music M, Autin L, Parulek J, Viola I. cellVIEW: a tool for illustrative and multi-scale rendering of large biomolecular datasets. In: Buhler KLL, John NW, editors. Eurographics Workshop on Visual Computing for Biology and Medicine. The Eurographics Association; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindow N, Baum D, Hege HC. Interactive rendering of materials and biological structures on atomic and nanoscopic scale. Comput Graph Forum. 2012;31:1325–1334. [Google Scholar]
- 33.Falk M, Krone M, Ertl T. Atomistic visualization of mesoscopic whole-cell simulations using ray-casted instancing. Comput Graph Forum. 2013;32:195–206. [Google Scholar]
- 34.Le Muzic M, Mindek P, Sorger J, Autin L, Goodsell D, Viola I. Visibility equalizer cutaway visualization of mesoscopic biological models. Comput Graph Forum. 2016;35:161–170. doi: 10.1111/cgf.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rieping W, Habeck M, Nilges M. Inferential structure determination. Science. 2005;309:303–306. doi: 10.1126/science.1110428. [DOI] [PubMed] [Google Scholar]
- 36.Viswanath S, Bonomi M, Kim SJ, Klenchin VA, Taylor KC, Yabut KC, Umbreit NT, Van Epps HA, Meehl J, Jones MH, Russel D, Velazquez-Muriel JA, Winey M, Rayment I, Davis TN, Sali A, Muller EG. The molecular architecture of the yeast spindle pole body core determined by Bayesian integrative modeling. Mol Biol Cell. 2017;28:3298–3314. doi: 10.1091/mbc.E17-06-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jantzen SG, Jenkinson J, McGill G. Transparency in film: increasing credibility of scientific animation using citation. Nat Methods. 2015;12:293–297. doi: 10.1038/nmeth.3334. [DOI] [PubMed] [Google Scholar]
- 38.Max N. DNA animation, from atom to chromosome. J Mol Graph. 1985;3:69–71. [Google Scholar]
- 39.Iwasa JH. Bringing macromolecular machinery to life using 3D animation. Curr Op Struct Biol. 2015;31:84–88. doi: 10.1016/j.sbi.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 40.Batiza AF, Gruhl M, Zhang B, Harrington T, Roberts M, LaFlamme D, Haasch MA, Knopp J, Vogt G, Goodsell D, Hagedorn E, Marcey D, Hoelzer M, Nelson D. The effects of the SUN project on teacher knowledge and self-efficacy regarding biological energy transfer are significant and long-lasting: results of a randomized controlled trial. CBE Life Sci Educ. 2013;12:287–305. doi: 10.1187/cbe.12-09-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amaro RE, Mulholland AJ. Multiscale methods in drug design bridge chemical and biological complexity in the search for cures. Nat Rev Chem. 2018;2:0148. doi: 10.1038/s41570-018-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frank J. Whither ribosome structure and dynamics research? (A perspective) J Mol Biol. 2016;428:3565–3569. doi: 10.1016/j.jmb.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, Chemmama IE, Pellarin R, Echeverria I, Shivaraju M, Chaudhury AS, Wang J, Williams R, Unruh JR, Greenberg CH, Jacobs EY, Yu Z, de la Cruz MJ, Mironska R, Stokes DL, Aitchison JD, Jarrold MF, Gerton JL, Ludtke SJ, Akey CW, Chait BT, Sali A, Rout MP. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 2018;555:475–482. doi: 10.1038/nature26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oikonomou CM, Chang YW, Jensen GJ. A new view into prokaryotic cell biology from electron cryotomography. Nat Rev Microbiol. 2016;14:205–220. doi: 10.1038/nrmicro.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci U S A. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosa A, Zimmer C. Computational models of large-scale genome architecture. Int Rev Cell Mol Biol. 2014;307:275–349. doi: 10.1016/B978-0-12-800046-5.00009-6. [DOI] [PubMed] [Google Scholar]
- 47.Goodsell DS, Klionsky DJ. Artophagy: The art of autophagy--the Cvt pathway. Autophagy. 2010;6:3–6. doi: 10.4161/auto.6.1.10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.NRC. A framework for K-12 science education: practices, crosscutting concepts, and core ideas. The National Academies Press; Washington, DC: 2012. [Google Scholar]
- 49.Cook MP. Visual representations in science education: The influence of prior knowledge and cognitive load theory on instructional design principles. Sci Educ. 2006;90:1073–1091. [Google Scholar]
- 50.Cook M, Wiebe EN, Carter G. The influence of prior knowledge on viewing and interpreting graphics with macroscopic and molecular representations. Sci Educ. 2008;92:848–867. [Google Scholar]
- 51.Vygotsky LS. Mind in society: The development of higher psychological processes. Harvard Univ. Press; Cambridge, MA: 1978. [Google Scholar]
- 52.Kramer IM, Dahmani HR, Delouche P, Bidabe M, Schneeberger P. Education catching up with science: Preparing students for three-dimensional literacy in cell biology. CBE Life Sci Educ. 2012;11:437–447. doi: 10.1187/cbe.12-06-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruner JS. The Process of Education. Harvard Univ. Press; Cambridge, MA: 1960. [Google Scholar]
- 54.Dempster FN. The spacing effect: A case study in the failure to apply the results of psychological research. Am Psychologist. 1988;43:627–634. [Google Scholar]
- 55.Donovan JJ, Radosevich DJ. A meta-analytic review of the distribution of practice effect: Now you see it, now you don’t. J Appl Psychol. 1999;84:795–805. [Google Scholar]
- 56.AAAS. Vision and change in undergraduate biology education: A call to action. American Association for the Advancement of Science; Washington, DC: 2011. [Google Scholar]
- 57.NSTA. NGSS for all students. National Science Teachers Association; Arlington, VA: 2015. [Google Scholar]
- 58.Bartlett FC. Remembering: A Study in Experimental and Social Psychology. Cambridge University Press; Cambridge, England: 1932. [Google Scholar]
- 59.Smith JP, III, diSessa AA, Roschelle J. Misconceptions reconceived: A constructivist analysis of knowledge in transition. J Learning Sci. 1994;3:115–163. [Google Scholar]
- 60.Festinger L. A Theory of Cognitive Dissonance. Stanford Univ. Press; Stanford, CA: 2001. [Google Scholar]
- 61.Hartman H. Human Learning and Instruction. New York, NY: City College of New York; 2002. Scaffolding & cooperative learning; pp. 23–69. [Google Scholar]
- 62.Offerdahl EG, Arneson JB, Byrne N. Lighten the load: Scaffolding visual literacy in biochemistry and molecular miology. Cell Biol Educ. 2017;16:es1. doi: 10.1187/cbe.16-06-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dries DR, Dean DM, Listenberger LL, Novak WRP, Franzen MA, Craig PA. An expanded framework for biomolecular visualization in the classroom: Learning goals and competencies. Biochem Mol Biol Educ. 2016;45:69–75. doi: 10.1002/bmb.20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arum R, Roksa J. Academically Adrift: Limited Learning on College Campuses. University of Chicago Press; Chicago, IL: 2011. [Google Scholar]
- 65.Freeman S, Eddy SL, McDonough M, Smith MK, Okoroafor N, Jordt H, Wenderoth MP. Active learning increases student performance in science, engineering, and mathematics. Proc Natl Acad Sci USA. 2014;111:8410–8415. doi: 10.1073/pnas.1319030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Span EA, Goodsell DS, Ramchandran R, Franzen MA, Herman T, Sem DS. Protein structure in context: The molecular landscape of angiogenesis. Biochem Mol Biol Educ. 2013;41:213–223. doi: 10.1002/bmb.20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee EH, Hsin J, Sotomayor M, Comellas G, Schulten K. Discovery through the computational microscope. Structure. 2009;17:1295–1306. doi: 10.1016/j.str.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]