Abstract
Background
Attenuated measles virus (MV) strains are promising agents currently being tested against solid tumors or hematologic malignancies in ongoing phase I and II clinical trials; factors determining oncolytic virotherapy success remain poorly understood, however.
Methods
We performed RNA sequencing and gene set enrichment analysis to identify pathways differentially activated in MV-resistant (n = 3) and -permissive (n = 2) tumors derived from resected human glioblastoma (GBM) specimens and propagated as xenografts (PDX). Using a unique gene signature we identified, we generated a diagonal linear discriminant analysis (DLDA) classification algorithm to predict MV responders and nonresponders, which was validated in additional randomly selected GBM and ovarian cancer PDX and 10 GBM patients treated with MV in a phase I trial. GBM PDX lines were also treated with the US Food and Drug Administration–approved JAK inhibitor, ruxolitinib, for 48 hours prior to MV infection and virus production, STAT1/3 signaling and interferon stimulated gene expression was assessed. All statistical tests were two-sided.
Results
Constitutive interferon pathway activation, as reflected in the DLDA algorithm, was identified as the key determinant for MV replication, independent of virus receptor expression, in MV-permissive and -resistant GBM PDXs. Using these lines as the training data for the DLDA algorithm, we confirmed the accuracy of our algorithm in predicting MV response in randomly selected GBM PDX ovarian cancer PDXs. Using the DLDA prediction algorithm, we demonstrate that virus replication in patient tumors is inversely correlated with expression of this resistance gene signature (ρ = –0.717, P = .03). In vitro inhibition of the interferon response pathway with the JAK inhibitor ruxolitinib was able to overcome resistance and increase virus production (1000-fold, P = .03) in GBM PDX lines.
Conclusions
These findings document a key mechanism of tumor resistance to oncolytic MV therapy and describe for the first time the development of a prediction algorithm to preselect for oncolytic treatment or combinatorial strategies.
Oncolytic virotherapy relies on virus replication within tumor cells to mediate direct cell killing of tumor cells and/or activation of antitumor immunity. Although individual cases of patients benefiting from oncolytic virotherapy have been documented and one immuno-virotherapy product recently received regulatory approval, success and preclinical promise have not always been reproducible (1–3). There is an urgent need to identify factors beyond virus receptor expression that are involved in restriction of viral replication in treated tumors of individual patients.
Attenuated measles virus (MV) strains of the Edmonston vaccine lineage enter cells through three known receptors: CD46, SLAM, and Nectin-4 (4–7). MV represents an appealing oncolytic platform due to the tumor selectivity, bystander killing effect, amenability to genetic engineering, and the excellent safety record observed in several clinical trials (8). Ongoing clinical trials utilize MV derivatives for the treatment of several tumor types, such as glioblastoma (GBM; NCT00450814), multiple myeloma (NCT00450814), ovarian cancer (OvCa; NCT02364713), head and neck cancer (NCT01846091), breast cancer (NCT01846091), and mesothelioma (NCT01503177). In the context of early evidence of biological activity in these trials, we aimed to identify specific mechanisms within tumor cells that can directly restrict virus replication (1,9,10).
RNA sequencing technology and the use of pharmacogenomics can provide an effective tool to identify gene variants and expression signatures that correlate with drug response and metabolism (11–15). To our knowledge, such an approach has not been used to optimize efficacy in virotherapy trials. We hypothesized that oncolytic MV replication in GBM cells is determined by restriction mechanisms following viral entry. In this study, we sought to analyze predictors for replication of oncolytic MV in GBM and OvCa tumors through the use of an in-depth expression analysis of differentially activated pathways between MV-permissive and -resistant primary patient-derived xenograft (PDX) lines. The goal of this study is to identify mechanisms of oncolytic MV resistance that can be used to preselect patients who are likely to respond to MV therapy and develop combinatorial strategies that can circumvent resistance.
Methods
An additional description of the methods can be found in the Supplementary Methods (available online).
MV Antitumor Effect In Vitro
GBM cells were infected at multiplicities of infection (MOIs) of 0.1 and 1.0 with MV strains diluted in Opti-MEM (Life Technologies, Carlsbad, CA). MTT proliferation assay (ATCC, Rockville, MD) was used to determine the cell viability at 24 hours to 10 days post–MV inoculation. Samples were run in triplicate wells in 96-well plates. Cells inoculated with inactivated MV were used as controls.
Patients and Patient Samples
All studies involving patients were approved by the Mayo Clinic Institutional Review Board (Mayo Clinic IRB#06-004440). Written informed consent was obtained for all patients. Patients with recurrent GBM tumors were treated by stereotactic injection of MV-CEA in an ongoing phase I clinical trial at Mayo Clinic (NCT00390299; Rochester, MN). Patients included in this analysis derived from cohort B and received intratumoral injection of the virus on day 1 via a catheter. On day 5, the tumor was resected, and a second dose was administered to the tumor-infiltrated parenchyma surrounding the resection cavity. Tumor tissue derived from 10 consecutive study patients was included in this analysis. RNA was isolated, and gene expression was analyzed from tumor tissue prior to MV therapy, described in the Supplementary Methods (available online).
Statistical Analysis
Statistical analysis was performed using Microsoft Excel, GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA), and R (16). Student t tests were utilized to test the difference in comparisons between the two groups, including virus production, MV entry as measured by green fluorescent protein (GFP), IFN-α/β production, interferon-stimulated gene (ISG) expression, and MV replication (17). Differences in expression between the MV-resistant and MV-permissive groups were evaluated using unequal-variance t tests. P values of less than .05 were considered statistically significant. All statistical tests were two-sided.
Kaplan-Meier survival curves and log-rank tests were used to obtain median survival times and compare the survival of the animal groups treated with active or inactive MV (18).
The GeneCodis online resource (http://genecodis.cnb.csic.es/) was utilized to perform pathway enrichment analysis between the MV-resistant group (GBM6, GBM150 and GBM39) and the MV-permissive group (GBM43 and GBM64) (19,20); 19 835 genes were used as an input into GeneCodis. We used a custom script in the R programming language to combine fold-change values and P values with functional grouping of proteins (16). Additional information is described in detail in the Supplementary Methods (available online).
Diagonal linear discriminant analysis (DLDA) was used as the tool to build a class prediction statistical model (21). The DLDA model was generated using the 22 ISG gene panel identified in the pathway enrichment analysis. As described in Figure 2, a comparison of MV-resistant and MV-permissive cells revealed the constitutive activation of the IFN pathway, characterized by elevated expression of 22 genes (P < .05 and fold-change > 1.5). Using this gene panel, the DLDA model yields coefficients for each gene and a constant, which can be used to generate a DLDA score for prospective samples to predict response. To accommodate expression data from multiple gene expression platforms, expression data were log2-transformed then standardized to a mean of 0.0 and standard deviation of 1.0. Additional information on the DLDA model is described in the Supplementary Methods (available online).
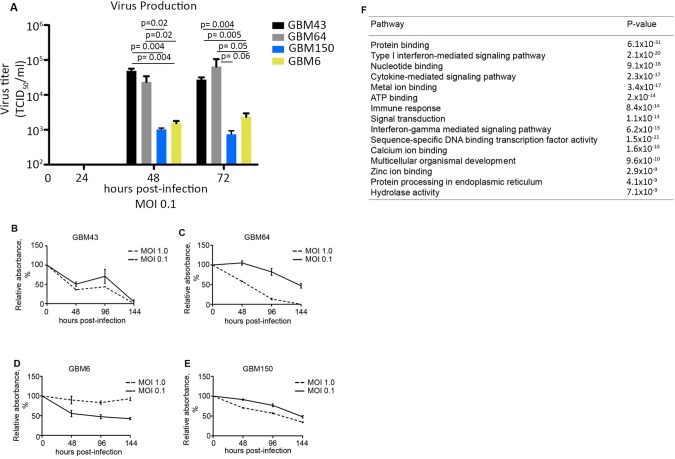
Figure 2.
Gene-set enrichment analysis (GSEA) identifies important interferon-stimulated genes differentially expressed between measles virus (MV)–resistant and permissive glioblastoma (GBM) lines. A) Additional primary GBM lines were infected with MV at a multiplicity of infection (MOI) of 0.1, and virus production was measured by titration on Vero cells 24–72 hours postinfection. B–E) Cell viability was measured by MTT assay in GBM64 and GBM43 cells following infection with MV-NIS (MOI = 1.0 and 0.1; n = 3). F) Gene expression for GBM64, GBM43, GBM150, and GBM6 cells was measured by RNA-Seq. Cells were grouped according to their response to MV. The resistant group consists of GBM39, GBM6, and GBM150, while the permissive group consists of GBM43 and GBM64. GSEA was performed on the MV-resistant and -permissive groups to identify pathways differentially activated between the two groups (expression of 19 835 genes was included in the analysis). The top 15 pathways overrepresented in the resistant group are listed; 2.14 × 10-20). In vitro experiments were repeated two or more times. A two-sided Student t test was used to determine statistically significant differences. Data are presented as the mean and SD (A–E). GBM = glioblastoma; MOI = multiplicity of infection.
Pearson coefficients were obtained to evaluate the correlation between virus replication in patient samples and the DLDA scores using R (22). Heat maps were used to visually display the standardized expression data. The colors relate to the overall range of the expression data. The dendrograms associated with each heat map show the clustering of both samples and genes based on hierarchical clustering using complete linkage with the distance metric 1 correlation between the samples or genes, respectively (23).
Results
MV Infection in Primary GBM PDX Lines
We evaluated MV replication in MV-resistant (GBM39) and MV-permissive (GBM12) primary patient-derived GBM xenograft lines (24). These lines are derived from resected human GBM patient tumors propagated as xenografts, and following orthotopic implantation, they have been shown to maintain the molecular characteristics and invasiveness of their human tumor of origin (25,26). MV production in GBM12 cells reached 6.8×104 tissue culture infection dose 50% per mL (TCID50/mL) at 72 hours with complete destruction of the cell monolayer (Figure 1, A and C). In comparison, virus production in GBM39 cells was only 1.8×102 TCID50/mL (387-fold lower infectious progeny, P < .001), with more than 47.0% (SD = 3.9%) of GBM39 cells remaining alive seven days postinfection (Figure 1, A and B). Orthotopic in vivo models demonstrated that athymic nude mice implanted with GBM12 cells responded to MV therapy with a 37-day prolongation in median survival (194% increase). In comparison, the MV-treated GBM39 mouse group demonstrated only a six-day prolongation in median survival (Figure 1D).
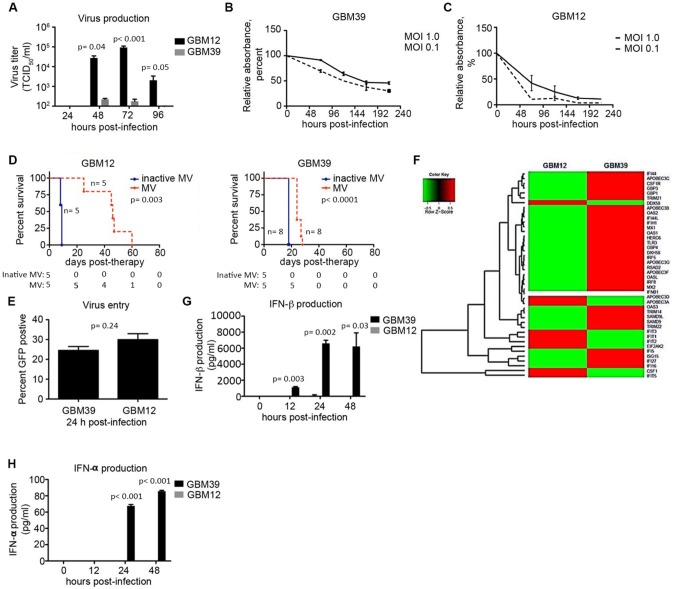
Figure 1.
Constitutive expression of interferon-stimulated genes measured in measles virus (MV)–resistant glioblastoma (GBM) cells. A) GBM12 and GBM39 cells were infected with MV-NIS (multiplicity of infection [MOI] = 0.1) and virus production measured at 24–96 hours (n = 3). B and C) GBM12 and GBM39 cells were infected with MV-NIS (MOI = 1.0 and 0.1), and cell viability was assessed by MTT assay (n = 3). D) Athymic nude mice were implanted orthotopically with GBM12 or GBM39 cells and received MV therapy (1 × 105 TCID50/mL) once every three days for a total of three or five doses, respectively. E) GBM12 and GBM39 cells were infected with MV-GFP at an MOI of 1.0, and virus entry efficiency was measured by GFP-positive cells assessed by flow cytometry at 24 hours postinfection (n = 2). F) RNA was extracted from uninfected GBM12 and GBM39 cells, and gene expression was analyzed by RNA-Seq. A heat map illustrates the expression of several antiviral interferon-stimulated genes overexpressed in GBM39 cells relative to GBM12. G and H) GBM39 and GBM12 cells were untreated or infected with MV-NIS (MOI = 5), and supernatant was harvested at 12, 24, and 48 hours postinfection (n = 2). IFN-α and IFN-β concentrations were measured by enzyme-linked immunosorbent assay. In vitro experiments were repeated two or more times. A two-sided Student t test was used to determine statistically significant differences. Kaplan-Meier survival curves and log-rank tests were used to obtain median survival times and compare survival between animal groups. Data are presented as the mean and standard deviation (A–C, E, G, and H). GBM = glioblastoma; MOI = multiplicity of infection; MV = measles virus.
We next evaluated MV entry into the GBM cells using MV encoding a green fluorescent protein (GFP) reporter (27). MV enters GBM12 and GBM39 cells with a similar efficiency (mean [SD] = 29.9 [1.4]%; mean [SD] = 24.5 [3.0]%, respectively; P = .24) (Figure 1E). Additionally, both GBM12 and GBM39 cells have similar levels of CD46, SLAM, and Nectin-4 entry receptor expression, suggesting a mechanism of postentry restriction (Supplementary Figure 1A, available online). Therefore, we performed RNA-Seq analysis on uninfected GBM12 and GBM39 cells. The constitutive expression of interferon-stimulated genes emerged as a prominent difference between these lines (Figure 1F;Supplementary Table 1, available online). ISGs are the effector proteins in the interferon pathway and play a critical role in the mammalian antiviral defense system (28). Importantly, constitutive expression of the ISGs was independent of IFN-β production (Figure 1G).
Infection with MV (MOI 5) induces production of IFN-β in GBM39 cells (mean [SD] = 6600 [383.3] pg/mL) 24 hours postinfection, whereas production is delayed and statistically significantly lower in GBM12 cells upon infection (mean [SD] = 92.6 [131.0] pg/mL, P = .002) (Figure 1G). Additionally, infection led to production of IFN-α by 48 hours (85.7 pg/mL) in GBM39 cells, whereas IFN-α was not detectable in infected GBM12 cells up to 48 hours postinfection (Figure 1H). In line with these results, ISG expression is induced upon MV infection in both lines (Supplementary Figure 2, available online). These results indicate that resistance to the virus is not due to differences in receptor expression or baseline interferon levels, but rather to an ISG-mediated preexisting antiviral state in GBM39 cells. Importantly, induction of ISGs with exogenous IFN-β in permissive cells prior to infection leads to MV resistance (Supplementary Figure 1B, available online).
Constitutive Type I Interferon Pathway Activation in MV-Resistant GBM Cells
We performed RNA-Seq analysis using RNA isolated from athymic mice orthotopically implanted with 41 primary orthotopic GBM models. Expression data for GBM12 were excluded from this analysis due to poor RNA quality and low gene coverage. From this analysis, we identified four additional permissive and resistant GBM models deriving from human patients. GBM43 and GBM64 cells supported MV replication (>2.0×104 TCID50/mL), whereas replication in GBM6 and GBM150 cells was statistically significantly lower (<1.6×103 TCID50/mL, P < .05) (Figure 2A;Supplementary Table 2, available online). In line with these differences in MV replication, 100% of GBM43 and GBM64 cells were effectively killed upon MV infection, whereas 42.5% (SD = 4.7%) and 34.3% (SD = 1.2%) of GBM6 and GBM150 cells survived seven days postinfection, respectively (Figure 2B-E).
Using expression data from these lines, we performed gene set enrichment analysis to identify differential pathway activation between the resistant (GBM39, 6, and 150) and permissive lines (GBM43 and 64). Constitutive activation of the type I Interferon pathway in the resistant group was the second most differentially activated pathway (P = 2.14×10-20) (Figure 2F;Supplementary Table 3, available online). We focused on the type I interferon pathway due to the known antiviral functions of this pathway (29). Twenty-two genes involved in the interferon pathway are differentially expressed (fold-change > 1.5, P < .05 for all) between the resistant and permissive groups and were used as the gene signature. Activation of the IFN pathway remains an important difference between MV-resistant and -permissive cells also when using more stringent criteria (fold-change > 2, P < .05 for all) (Supplementary Table 3, available online).
IFN Activation as a Predictive Marker for Response to Oncolytic MV Therapy
The diagonal linear discriminant analysis (DLDA) class prediction algorithm was generated using the 22 gene signature (13). The training data set for the DLDA model includes standardized gene expression values for the 22 ISGs in the three resistant (GBM39, 6, 150) and two permissive (GBM43, 64) PDX lines. Next, we measured gene expression in the remaining 35 GBM PDX lines for the 22 ISGs (Figure 3A;Supplementary Table 4, available online). DLDA scores for test samples are calculated using this specific gene list and corresponding coefficient for each gene (Supplementary Table 5, available online). Test set DLDA scores above and below 0.0 are classified as resistant and permissive, respectively. Additionally, we performed unsupervised hierarchical clustering for the 35 samples using the 22 gene signature (Figure 3A).
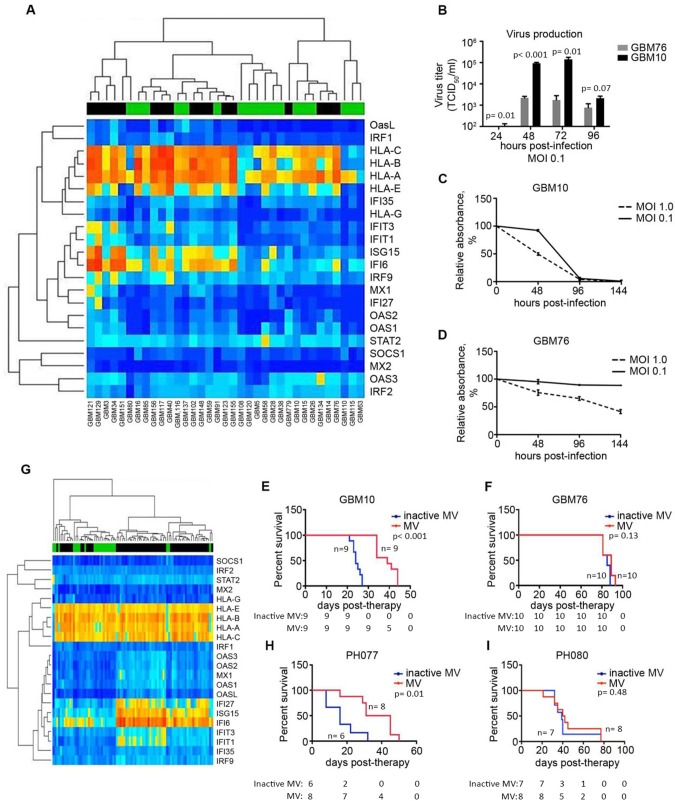
Figure 3.
Application of the diagonal linear discriminant analysis (DLDA) algorithm in primary glioblastoma (GBM) and ovarian cancer (OvCa) xenografts. A) Expression of the 22-gene signature was analyzed and illustrated in a heat map consisting of 35 primary GBM lines (predicted responders and nonresponders are indicated by a green or black bar, respectively). B) Representative lines classified as permissive (GBM10) or resistant (GBM76), based on the DLDA analysis, were infected with MV-NIS (multiplicity of infection [MOI] = 0.1), and virus production was measured by titration on Vero cells 24–96 hours postinfection (n = 3). C and D) GBM10 and GBM76 cells were infected with MV-NIS (MOI = 1.0 and 0.1), and cell viability was measured by MTT assay (n = 3). E and F) Athymic nude mice were implanted orthotopically with either GBM10 or GBM76 cells. Mice were treated with MV-NIS (1 × 105 TCID50/mL) once every three days for a total of three (GBM10) or five (GBM76) doses. G) The DLDA scoring algorithm was applied in gene expression data obtained from primary patient-derived OvCa tumors passaged in mice. H) Low interferon-stimulated gene (ISG)–expressing OvCa line, PH077, and (I) a high-ISG expressing line, PH080, were implanted intraperitoneally in SCID beige mice. In vitro experiments were repeated two or more times. A two-sided Student t test was used to determine statistically significant differences. Kaplan-Meier survival curves and log-rank tests were used to obtain median survival times and compare survival between animal groups. Data are presented as the mean and SD (B–D). GBM = glioblastoma; MOI = multiplicity of infection; MV = measles virus.
Of the 35 primary GBM PDX lines tested, 18 were predicted to be responders. MV production in a randomly selected line assigned to the responders (GBM10) reached high levels of virus production (1.4×105 TCID50/mL), whereas MV production in another randomly selected line designated as a nonresponder (GBM76) only reached 2.2×103 TCID50/mL at 72 hours (Figure 3B). In line with the decreased virus production, cell killing was statisitically significantly impaired in the resistant line relative to the predicted responder (Figure 3, C and D). Furthermore, MV therapy in athymic nude mice implanted with GBM10 cells statistically significantly improved median survival, as compared with inactivated MV (median survival = 39 days and 24 days, respectively, hazard ratio [HR] = 24.24, 95% confidence intervals [CI] = 5.69 to 103.2, P < .001), whereas athymic nude mice implanted with MV-resistant GBM76 cells demonstrated no statistically significant difference in median survival (85 vs 89 days, HR = 3.79, 95% CI = 0.67 to 21.46, P = .13) (Figure 3, E and F).
Similar analysis and DLDA predictions were generated for 86 OvCa PDXs (Figure 3G;Supplementary Table 6, available online). SCID beige mice implanted with a randomly selected responder (PH077) resulted in a statistically significant 22-day prolongation in median survival for inactivated MV vs MV therapy groups (16 vs 38 days, respectively, HR = 7.49, 95% CI = 1.62 to 34.54, P = .01). In comparison, SCID beige mice implanted with the randomly selected nonresponder (PH080) had no benefit from MV therapy, as compared with inactivated virus (41 vs 39 days, respectively, HR = 1.58, 95% CI = 0.45 to 5.55, P = .48) (Figure 3, H and I).
ISG Expression in MV-Treated GBM Patients
To further validate constitutive IFN activation as an indicator for response to MV therapy, we examined tumors of GBM patients treated with MV in an ongoing phase I clinical trial (NCT00390299). Virus replication in treated patients was analyzed five days post-therapy in the first 10 patients consecutively treated in arm B of the trial and ranged from nondetectable to 6.0×107 genome copies/µg RNA (Figure 4A).
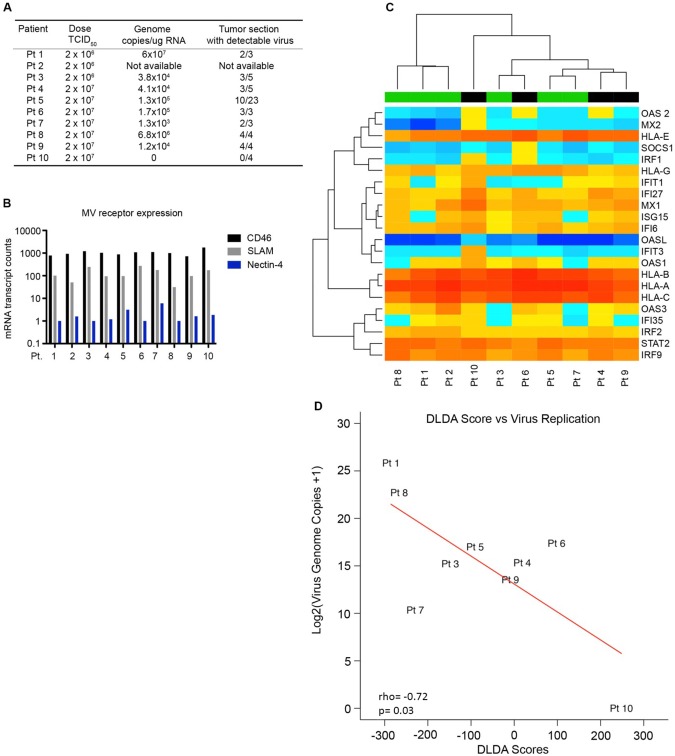
Figure 4.
Expression of the interferon gene signature in MV-treated GBM patients. A) GBM patients were treated with MV-CEA (2 × 106 patients 1–3 or 2 × 107 TCID50 for patients 4–10) on day 0, and tumors were resected on day 5. RNA was extracted from multiple regions of the treated tumors, and virus replication was assessed by quantitative real-time polymerase chain reaction for viral genome copies. B) RNA was extracted from the primary tumors of GBM patients scheduled to receive MV therapy in an ongoing phase I trial. Gene expression was assessed by Nanostring analysis using a custom gene panel that assessed the interferon-stimulated gene (ISG) profile and virus entry receptor expression. C) Expression of the 22 ISG panel is depicted in the heat map. The diagonal linear discriminant analysis (DLDA) scoring system was applied to the patients, and classification was assigned to each individual patient (black bar = resistant, green bar = permissive). D) DLDA scores calculated for each patient were assessed for correlation to virus replication measured from the treated tumors. Pearson coefficients were utilized to evaluate the correlation between virus replication and DLDA scores. DLDA = diagonal linear discriminant analysis; MV = measles virus.
Gene expression was measured in the 10 MV-treated patients using a custom Nanostring gene panel (Supplementary Table 7, available online). Expression of the MV entry receptors was similar among the 10 treated patients, suggesting a mechanism of postentry restriction (Figure 4B). Utilizing gene expression of the 22 gene signature, we applied the DLDA scoring system in 10 consecutive GBM patients treated in arm B of the study (NCT00390299) (Figure 4C;Supplementary Table 8, available online). An increase in the DLDA score (elevated ISG expression) was inversely correlated with the virus replication measured in the patient’s tumor (ρ = –0.717, P = .03) (Figure 4D). The patient with the highest DLDA score (greater than 150) had no detectable viral replication in the tumor, whereas the two patients with the lowest DLDA scores (less than –250) had the highest level of viral replication (6.8×106 and 6.0×107 MV genome copies/µg of RNA) recovered from the tumor. Interestingly, patient 1 had the lowest DLDA score and the highest level of virus replication detected, despite receiving one of the lower viral doses (2×106 TCID50) (Figure 4, A and D). The remaining six patients had moderate DLDA scores (–250 to +150) and intermediate levels of virus replication (1.3×103 to 1.7×105 MV genome copies/µg of RNA). Overall, these results are consistent with our hypothesis that ISG expression can determine MV permissiveness and serve as a screening tool.
Effects of JAK1/2 Inhibition in MV-Resistant GBM Lines
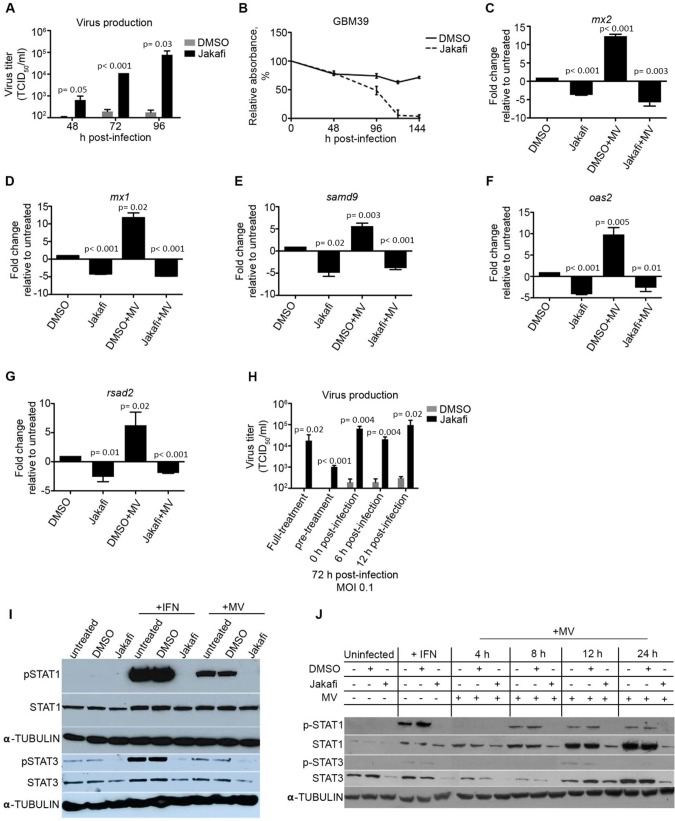
Upon viral infection, the host antiviral defense mechanism triggers IFN production and signaling through JAK1/Tyk2 to activate STAT1/2, which associates with IRF9 to form the ISGF3 complex that activates ISG expression (30). Therefore, we explored the use of pharmacological agents to overcome resistance through inhibition of the JAK/STAT signaling pathway with the US Food and Drug Administration (FDA)–approved JAK1/2 inhibitor ruxolitinib (Jakafi). Virus production was statistically significantly increased at 48 hours postinfection and nearly 1000-fold greater by 96 hours in Jakafi-treated cells (P = .03) (Figure 5A). The increased virus production and spread also led to increased cell killing in Jakafi-pretreated cells relative to control-treated cells (Figure 5B;Supplementary Table 2, available online). A similar result was observed in four additional resistant and permissive lines (Supplementary Figure 3, A and B, available online).
Figure 5.
Inhibition of JAK/STAT signaling in MV resistant primary GBM lines. A) Resistant glioblastoma (GBM) 39 cells were treated with the JAK1 inhibitor Ruxolitinib (Jakafi; 3 µM) or DMSO for 48 hours prior to infection. Treated cells were infected with MV-NIS (multiplicity of infection [MOI] = 0.1), and virus production was measured at 48, 72, and 96 hours postinfection (n = 3). B) MV-mediated cell killing was assessed in GBM39 cells pretreated with Jakafi (3 µM) or DMSO control by MTT assay (n = 3). C–G) RNA was extracted from GBM39 cells under the indicated conditions, and interferon-stimulated gene (ISG) expression was assessed by quantitative real-time polymerase chain reaction (n = 2, run in technical triplicates). Pretreatment with Jakafi decreased baseline ISG expression, as well as ISG induction upon MV infection. H) Protein was extracted from GBM39 cells and (I) GBM12 cells under the indicated conditions, and p-STAT1/3 activation was measured by immunoblot. J) Jakafi (3 µM) was added at different points during the course of infection (MOI = 0.1), and virus production was assessed at 48 hours postinfection (n = 3). In vitro experiments were repeated two or more times. A two-sided Student t test was used to determine statistically significant differences. Data are presented as the mean +/- SD (A–H). MV = measles virus.
Relative to control-treated cells, expression of ISGs, such as RSAD2, OAS2, MX1, MX2, and SAMD9, were statistically significantly decreased in Jakafi-treated cells (Figure 5, C–G). Furthermore, pretreatment with Jakafi blocked ISG induction upon MV infection (MOI 5), whereas ISG expression was statistically significantly increased upon infection in the control-treated cells.
We next investigated the signaling molecules involved in mediating ISG expression. Interestingly, STAT3 is constitutively activated in the uninfected cells, whereas activated STAT1 was not detected. Phosphorylation of both STAT1 and STAT3 was induced by four hours postinfection; however, this process was inhibited in the presence of Jakafi (Figure 5I). In MV replication–permissive cells, a similar response was observed, but it peaked eight to 12 hours postinfection (Figure 5J;Supplementary Figure 4, available online). We next investigated the timing of JAK/STAT signaling inhibition that would be required to reverse restriction. Jakafi administered up to 12 hours postinfection is sufficient to restore MV production to levels similar to a full course of Jakafi treatment (Figure 5H), suggesting that inhibition of the antiviral response is critical for propagating infection in resistant lines. Additionally, we challenged resistant cells with a recombinant MV encoding the wild-type phosphoprotein (MV-GFP-Pwt). The phosphoprotein gene of wild-type MV encodes the P/V/C proteins and plays an important role in inhibiting IFN induction and signaling (31–33). Infection with MV-GFP-Pwt leads to a statistically significant decrease in ISG induction relative to infection with MV-GFP (P = .02) and successfully replicates in resistant GBM39 cells (Supplementary Figure 3, B and C, and Supplementary Table 2, available online). These results highlight the importance of utilizing the basal ISG expression profile as a reliable indicator for responsiveness to MV therapy.
Discussion
Although the interferon pathway is an effective defense mechanism against viral infection, tumor cells are frequently defective in this pathway. Differential activation of the IFN signaling pathway has been employed as an argument supporting tumor selectivity of oncolytic viruses (34–36). The impetus of the work presented in this manuscript was the observation that despite adequate levels of virus receptor expression enabling viral entry, there was variability in MV replication, permissiveness, and antitumor effect in patient-derived xenografts. An association between constitutive interferon activation and reduced infection was observed in GBM and OvCa PDXs. We report for the first time the development of a DLDA algorithm to predict permissiveness to viral replication in human tumors. Chemical inhibition of JAK1/2, a critical component of ISG signaling, which can reverse the primed state and/or the antiviral response upon infection, sensitized virus-resistant cells to MV infection, further supporting our conclusions.
These results have several clinical implications for oncolytic virotherapy. First, a defective IFN system in tumor cells is one of the main arguments for the use of oncolytic viruses to selectively target tumor cells (34,37,38). Our data demonstrate innate variations in interferon response antiviral defense mechanisms within tumors that can have a statistically and biologically significant impact on MV replication. Importantly, several of the ISGs identified are known to restrict other oncolytic viruses currently being tested, such as vesicular stomatitis virus, HSV-1, Vaccinia, and Newcastle Disease Virus (39–45). Second, our DLDA scoring–based algorithm identified three tiers of GBM patients: low scores (<–250) identified patients with tumors extremely permissive to MV replication, and high scores (>150) correlated with MV-resistant tumors, while a third class of patients who had moderate DLDA scores (–250 to +150) had intermediate levels of virus replication. The level of replication reached in these patients would still be adequate to allow them to benefit from the immunostimulatory potential of oncolytic cell death, however. Thus, these patients could be excellent candidates for combinatorial strategies, for example, using the FDA-approved JAK1/2 inhibitor ruxolitinib, checkpoint inhibitors, such as monoclonal antibodies targeting CTLA-4 and PD-L1, or recombinant MV strains encoding immunomodulatory genes (46–50). We and others have recently demonstrated that even low levels of oncolytic viral replication are adequate for the synergistic effect of virotherapy with anti-PD1 inhibition to be materialized (50). A schematic proposal of how this algorithm could be applied for optimal patient selection in clinical trials is provided as Supplementary Figure 5 (available online).
While our study is able to validate the predictive capabilities of our classification system, according to ISG expression, our validation in patients is limited by the number of GBM patients who have been treated with oncolytic MV. We are in the process of validating these results in additional GBM and ovarian cancer patients treated with MV strains in other clinical trials (3), as well as prospectively utilizing the DLDA algorithm we developed to enrich measles virotherapy trials for patients likely to respond. Additional mechanistic studies are planned to characterize the specific role of JAK1 vs JAK2 inhibition on MV infection response and develop clinical combinatorial strategies.
Our results represent the first successful effort to associate tumor molecular characteristics with viral replication in PDXs and clinical trial patients alike. A recently reported trial attempted to identify predictors of response to oncolytic virotherapy in patient samples (51). The identified survival-related neuronal subtype signature appeared to be prognostic rather than predictive because it lacked the mechanistic rationale that could explain an association with virotherapy effect; it was not shown to be associated with viral replication and could not predict outcome at the individual patient level. In our study, we developed a unique scoring system that was able to accurately predict the extent of viral replication at the individual patient level. The data were validated in ovarian and GBM avatars; therefore, our algorithm is predictive across multiple tumor types. Additionally, given the role of ISGs in determining replication across many different oncolytic platforms, this predictive gene signature could have broad translational and clinical implications for the field of oncolytic virotherapy (39–45).
Funding
This work was supported in part by National Institutes of Health (NIH) grants P50 CA108961, P50 CA136393, R01 CA154348, and R01 CA200507, The Richard M. Schulze Family Foundation, and the Siebens Foundation.
Notes
Affiliations of authors: Department of Molecular Medicine (CK, IDI, IA, EG), Department of Health Sciences Research (SKA, AAL, MJM, ALO), Department of Radiation Oncology (MAS, JNS), Department of Laboratory Medicine and Pathology (CG, MEJ, JJ), Division of Medical Oncology (SMG, MAB, PH, SJW, EG), Cancer Biology, Mayo Clinic Florida, Jacksonville, FL (EAT), Department of Neurological Surgery (IFP), and Genome Analysis Core, Medical Genome Facility (JJ), Mayo Clinic, Rochester, MN.
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
We thank B. Carlson, J. Kachergus, E. Heinzen, A. Nair, and X. Hou for technical support. We thank P. Devaux for helpful discussion regarding experimental design. The data from this study are available upon request from the corresponding author.
Author contributions: CK, II, and EG conceived the study, designed experiments, and interpreted data. KA, AL, CK, and EG designed and performed computational analysis. MM, KA, JJ, AO, and JW processed and analyzed RNA-Seq analysis. CK, II, IA, CP, SG, and MB performed experiments. MJ, IP, PH, CG, MS, JS, and EG contributed to the generation and maintenance of the patient-derived xenografts and collection of patient samples. CK and EG wrote the manuscript. All authors have reviewed and approved the manuscript.
Supplementary Material
References
- 1. Russell SJ, Federspiel MJ, Peng KW, et al. Remission of disseminated cancer after systemic oncolytic virotherapy. Mayo Clinic Proc. 2014;897:926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andtbacka RH, Kaufman HL, Collichio F, et al. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol. 2015;3325:2780–2788. [DOI] [PubMed] [Google Scholar]
- 3. Galanis E, Atheron PJ, Maurer MJ, et al. Oncolytic measles virus expressing the sodium iodide symporter to treat drug-resistant ovarian cancer. Cancer Res. 2015;751:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muhlebach MD, Mateo M, Sinn PL, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;4807378:530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dorig RE, Marcil A, Chopra A, et al. The human Cd46 molecule is a receptor for measles-virus (Edmonston strain). Cell. 1993;752:295–305. [DOI] [PubMed] [Google Scholar]
- 6. Tatsuo H, Ono N, Tanaka K, et al. SLAM (CDw150) is a cellular receptor for measles virus. Nature 2000;4066798:893-897. [DOI] [PubMed] [Google Scholar]
- 7. Noyce RS, Bondre DG, Ha MN, et al. Tumor cell marker PVRL4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 2011;78:e1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Msaouel P, Opyrchal M, Domingo Musibay E, et al. Oncolytic measles virus strains as novel anticancer agents. Exp Opin Biol Ther. 2013;134:483–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galanis E, Hartmann LC, Cliby WA, et al. Phase I trial of intraperitoneal administration of an oncolytic measles virus strain engineered to express carcinoembryonic antigen for recurrent ovarian cancer. Cancer Res. 2010;703:875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Galanis E, Atherton PJ, Maurer MJ, et al. Oncolytic measles virus expressing the sodium iodide symporter to treat drug-resistant ovarian cancer. Cancer Res. 2015;751:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brandes AA, Franceschi E, Tosoni A, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;2613:2192–2197. [DOI] [PubMed] [Google Scholar]
- 12. Salonga D, Danenberg KD, Johnson M, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;64:1322–1327. [PubMed] [Google Scholar]
- 13. Hess KR, Anderson K, Symmans WF, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;2426:4236–4244. [DOI] [PubMed] [Google Scholar]
- 14. Del Rio M, Molina F, Bascoul-Mollevi C, et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J Clin Oncol. 2007;257:773–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;35127:2817–2826. [DOI] [PubMed] [Google Scholar]
- 16.Team RC. R: A language and environment for statistical computing. https://www.R-project.org/. Accessed March 7, 2018.
- 17. Gosset WS. The probable error of a mean. Biometrika. 1908;61:1–25. [Google Scholar]
- 18. Kaplan E, Meier P.. Non-parametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- 19. Nogales-Cadenas R, Carmona-Saez P, Vazquez M, et al. GeneCodis: Interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37(Web Server issue):W317–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A.. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40(Web Server issue):W478–W483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dudoit S, Fridlyand J.. A prediction-based resampling method for estimating the number of clusters in a dataset. Genome Biol. 2002;37:RESEARCH0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pearson K. Notes on regression and inheritance in the case of two parents. Proc R Soc Lond. 1895;58:240–242. [Google Scholar]
- 23. Ling RL. A computer generated aid for cluster analysis. Commun ACM. 1973;166:355. [Google Scholar]
- 24. Sarkaria JN, Carlson BL, Schroeder MA, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12(7 Pt 1):2264–2271. [DOI] [PubMed] [Google Scholar]
- 25. Gupta SK, Kizilbash SH, Carlson BL, et al. Delineation of MGMT hypermethylation as a biomarker for veliparib-mediated temozolomide-sensitizing therapy of glioblastoma. J Natl Cancer Inst. 2016;1085:djv369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giannini C, Sarkaria JN, Saito A, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-oncology. 2005;72:164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duprex WP, McQuaid S, Hangartner L, et al. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J Virol. 1999;7311:9568–9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schoggins JW, MacDuff DA, Imanaka N, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;5057485:691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schneider WM, Chevillotte MD, Rice CM.. Interferon-stimulated genes: A complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Silvennoinen O, Ihle JN, Schlessinger J, et al. Interferon-induced nuclear signalling by Jak protein tyrosine kinases. Nature. 1993;3666455:583–585. [DOI] [PubMed] [Google Scholar]
- 31. Devaux P, Priniski L, Cattaneo R.. The measles virus phosphoprotein interacts with the linker domain of STAT1. Virology. 2013;444(1–2):250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Devaux P, Cattaneo R.. Measles virus phosphoprotein gene products: Conformational flexibility of the P/V protein amino-terminal domain and C protein infectivity factor function. J Virol. 2004;7821:11632–11640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakatsu Y, Takeda M, Ohno S, et al. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J Virol. 2008;8217:8296–8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stojdl DF, Lichty B, Knowles S, et al. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med. 2000;67:821–825. [DOI] [PubMed] [Google Scholar]
- 35. Noser JA, Mael AA, Sakuma R, et al. The RAS/Raf1/MEK/ERK signaling pathway facilitates VSV-mediated oncolysis: Implication for the defective interferon response in cancer cells. Mol Ther. 2007;158:1531–1536. [DOI] [PubMed] [Google Scholar]
- 36. Critchley-Thorne RJ, Simons DL, Yan N, et al. Impaired interferon signaling is a common immune defect in human cancer. Proc Natl Acad Sci U S A. 2009;10622:9010–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He LF, Gu JF, Tang WH, et al. Significant antitumor activity of oncolytic adenovirus expressing human interferon-beta for hepatocellular carcinoma. J Gene Med. 2008;109:983–992. [DOI] [PubMed] [Google Scholar]
- 38. Jenks N, Myers R, Greiner SM, et al. Safety studies on intrahepatic or intratumoral injection of oncolytic vesicular stomatitis virus expressing interferon-beta in rodents and nonhuman primates. Hum Gene Ther. 2010;214:451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Staeheli P, Pavlovic J.. Inhibition of vesicular stomatitis virus mRNA synthesis by human MxA protein. J Virol. 1991;658:4498–44501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Broberg EK, Hukkanen V. Immune response to herpes simplex virus and y134.5 deleted HSV vectors. Current Gene Ther. 2005;55:523–530. [DOI] [PubMed] [Google Scholar]
- 41. Guerra S, Caceres A, Knobeloch KP, et al. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;47:e1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shi HX, Yang K, Liu X, et al. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol. 2010;3010:2424–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Patel MR, Jacobson BA, Ji Y, et al. Vesicular stomatitis virus expressing interferon-beta is oncolytic and promotes antitumor immune responses in a syngeneic murine model of non-small cell lung cancer. Oncotarget. 2015;632:33165–33177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Escobar-Zarate D, Liu YP, Suksanpaisan L, et al. Overcoming cancer cell resistance to VSV oncolysis with JAK1/2 inhibitors. Cancer Gene Ther. 2013;2010:582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moerdyk-Schauwecker M, Shah NR, Murphy AM, et al. Resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus: Role of type I interferon signaling. Virology. 2013;4361:221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stewart CE, Randall RE, Adamson CS.. Inhibitors of the interferon response enhance virus replication in vitro. PLoS One. 2014;911:e112014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Engeland CE, Grossardt C, Veinalde R, et al. CTLA-4 and PD-L1 checkpoint blockade enhances oncolytic measles virus therapy. Mol Ther. 2014;2211:1949–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zamarin D, Holmgaard RB, Subudhi SK, et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci Translat Med. 2014;6226:226ra32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iankov ID, Allen C, Federspiel MJ, et al. Expression of immunomodulatory neutrophil-activating protein of Helicobacter pylori enhances the antitumor activity of oncolytic measles virus. Mol Ther. 2012;206:1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hardcastle J, Mills L, Malo CS, et al. Immunovirotherapy with measles virus strains in combination with anti-PD-1 antibody blockade enhances antitumor activity in glioblastoma treatment. Neuro-oncology. 2017;194:493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cloughesy TF, Landolfi J, Hogan DJ, et al. Phase 1 trial of vocimagene amiretrorepvec and 5-fluorocytosine for recurrent high-grade glioma. Sci Translat Med. 2016;8341:341ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.