Abstract
Background
Ginsenoside Rg3(S) and compound K (C-K) are pharmacologically active components of ginseng that promote human health and improve quality of life. The aim of this study was to produce Rg3(S) and C-K from ginseng extract using recombinant Lactococcus lactis.
Methods
L. lactis subsp. cremoris NZ9000 (L. lactis NZ9000), which harbors β-glucosidase genes (BglPm and BglBX10) from Paenibacillus mucilaginosus and Flavobacterium johnsoniae, respectively, was reacted with ginseng extract (protopanaxadiol-type ginsenoside mixture).
Results
Crude enzyme activity of BglBX10 values comprised 0.001 unit/mL and 0.003 unit/mL in uninduced and induced preparations, respectively. When whole cells of L. lactis harboring pNZBglBX10 were treated with ginseng extract, after permeabilization of cells by xylene, Rb1 and Rd were converted into Rg3(S) with a conversion yield of 61%. C-K was also produced by sequential reactions of the permeabilized cells harboring each pNZBgl and pNZBglBX10, resulting in a 70% maximum conversion yield.
Conclusion
This study demonstrates that the lactic acid bacteria having specific β-glucosidase activity can be used to enhance the health benefits of Panax ginseng in either fermented foods or bioconversion processes.
Keywords: β-glucosidase, bioconversion, compound K, ginsenoside Rg3(S), Lactococcus lactis
1. Introduction
Ginseng, the root of Panax ginseng Meyer, has been used as a traditional oriental medicine to treat various diseases for more than 2,000 years [1], [2]. The world ginseng market, including ginseng root and its processed products, is estimated to be worth United States (US) $2,084 million [3]. The majority of ginseng pharmacological effects are attributed to ginsenosides, which are triterpene saponins. Deglycosylated ginsenosides exhibit higher biological activity than glycosylated forms because of their smaller size, higher bioavailability, and better ability to penetrate the cell membrane [4].
Among various ginsenosides, Rg3(S), which is deglycosylated from Rd, exhibits a wide spectrum of pharmacological properties, including anticancer effects, as it has been shown to inhibit tumor cell invasion and metastasis [5]. Another attractive minor ginsenoside is compound K (C-K), which is deglycosylated from F2. While most ginsenosides are poorly absorbed from the gut, C-K is absorbed and therefore exhibits higher biological activities. Recently, in vitro and in vivo studies showed that C-K has multiple pharmacological activities, including anticarcinogenic, anti-inflammatory, antiallergic, antidiabetic, antiaging, and hepatoprotective effects [6]. C-K is used as the principal ingredient in some skin care products, because C-K is an effective antiwrinkle and antiaging agent for the skin [7]. However, the Rg3(S) and C-K content in ginseng is relatively low, and isolation of these valuable compounds from natural products is difficult [8], [9]. Thus, efficient methods to produce Rg3(S) and C-K are needed for their application as active pharmacological substances.
Production of ginsenosides has been carried out by various methods such as heat treatment [10], alkaline cleavage [11], mild acid hydrolysis [12], enzymatic conversion [13], and microbial conversion [14]. However, physicochemical methods show poor selectivity and often produce side reactions. By contrast, the enzymatic method is regarded as desirable for the production of minor ginsenosides because of its specificity in hydrolyzing sugar moieties at particular positions. Escherichia coli is the most frequently used microorganism for the expression of heterologous enzymes, because the system is well characterized, genetic information is available, and the cells have a fast growth rate along with the potential for high-density cultivation in inexpensive media [15]. However, a major disadvantage of this system is the concern about safety of its use as a food ingredient or medicine [2]. Meanwhile, lactic acid bacteria (LAB) exhibit a number of useful probiotic characteristics such as resistance to acid and bile, production of antimicrobial substances, ability to modulate intestinal microflora, and inability to generate endotoxins. Moreover, LAB have been given a “generally recognized as safe” status according to the US Food and Drug Administration and met the criteria for the designation of Qualified Presumption of Safety according to the European Food Safety Authority. Consequently, LAB microbial systems offer alternatives to E. coli, and are candidates for the production of bioresource materials.
We have previously developed a recombinant Lactococcus lactis subsp. cremoris (L. lactis) harboring pNZBgl expressing a heterologous β-glucosidase gene to produce an active minor ginsenoside F2. The nucleotide sequence of β-glucosidase gene (BglPm) was derived from Paenibacillus mucilaginosus, consisting of 1,260 bp and encoding 419 amino acids. The nucleotide sequence of BglPm was synthesized after codon optimization and cloned into pNZ8008, resulting in pNZBgl. The permeabilized L. lactis harboring pNZBgl resulted in a high conversion yield (74%) of F2 from the ginseng extract [16].
The aim of this study was to produce Rg3(S) and C-K with high conversion yields using a LAB system that is more suitable for use in the food industry, e.g., for preparation of yogurt, cheese, and kimchi. For this, the β-glucosidase gene (BglBX10) from Flavobacterium johnsoniae UW101T was transformed into L. lactis NZ9000 using the nisin controlled expression system. The BglBX10 gene, consisting of 2,445 bp and encoding 814 amino acids, belongs to the glycoside hydrolase family 3. This gene was cloned, and the recombinant enzyme that was overexpressed in E. coli was characterized [8]. The enzyme was reported to hydrolyze the outer and inner glucose moieties of ginsenoside Rb1 and Rd at C20 position of the aglycone into Rd and F2, respectively. After optimization of the expression conditions, the recombinant cells were used for the bioconversion of Rb1 and Rd into Rg3(S) within a relatively short period (≤ 24 h). Furthermore, the combination of two types of LAB cells (L. lactis expressing the BglBX10 gene from F. johnsoniae and L. lactis expressing the BglPm gene from P. mucilaginosus) was used for C-K production.
2. Materials and methods
2.1. Materials
Bacterial strains, plasmids, and primers used in this study are listed in Table 1. L. lactis NZ9000 and the pNZ8008 plasmid were used as the host and gene expression vector, respectively. L. lactis NZ9000 was grown in M17 medium (Difco, Detroit, MI, USA) supplemented with 0.5% glucose (GM17) at 30°C. E. coli MC1061 (MoBiTec, Goettingen, Germany) was grown in Luria-Bertani medium at 37°C under shaking conditions and used as the cloning host. For the selection of E. coli or L. lactis transformants, chloramphenicol (10 μg/mL) was added to the Luria-Bertani or GM17 medium. Gene expression in L. lactis was induced by nisin, which was prepared as follows: 2.5% nisin powder (Sigma-Aldrich, St. Louis, MO, USA) was dissolved in 0.05% acetic acid to obtain a final nisin concentration of 1 ng/mL. The protopanaxadiol-type ginsenoside mixture (PPDGM) was extracted from a ginseng root. The standard compounds of ginsenosides, Rb1, Rd, Rg3(S), F2, and C-K (≥ 98.0% purity), were purchased from Chengdu Biopurify Phytochemicals Ltd. (Chengdu, China).
Table 1.
Strains, plasmids, and primers used in this study
| Strains, plasmids, and primers | Relevant characteristics | Source |
|---|---|---|
| Strains | ||
| Lactococcus lactis subsp. cremoris NZ9000 | MG1363-derived, pepN::nisRK, expression host | MoBiTec |
| Escherichia coli MC1061 | araD139, Δ(ara, leu)7,697, ΔlacX74, galU−, galK–, hsr–, hsm+, strA, cloning host | MoBiTec |
| Plasmids | ||
| pGEX-bglBX10 | β-glucosidase BX10 template vector | [8] |
| pNZ8008 | PnisA, gus A, Cmr1), replicon of the rolling circle plasmid pSH71 | MoBiTec |
| pNZBglBX10 | pNZ8008 carrying the BglBX10 gene | This study |
| pNZBgl | pNZ8008 carrying the BglPm gene | [16] |
| Primers | ||
| For_EcoRI_BX10 | 5′-GACTCTGCAGGAATTCATGGCTTT AGTGATTTCGAATTTATC-3′ |
This study |
| Rev_HindIII_ BX10 | 5′-TGCACTCGAGAAGCTTTTAGTGG TGATGATGGTGATGTTGTTTTAACTCTTTAAATG-3′ |
This study |
Cmr indicates resistance to chloramphenicol
2.2. Construction of pNZBglBX10 and transformation into L. lactis
Based on sequence information in the National Center for Biotechnology Information genome database (http://www.ncbi.nlm.nih.gov), the BglBX10 gene from F. johnsoniae UW101T was amplified from pGEX-bglBX10 by polymerase chain reaction (PCR) using the specific primer set indicated in Table 1 [8]. The amplified fragment was inserted into corresponding sites in the expression plasmid pNZ8008 (4,953 bp), which was linearized by digestion with EcoRΙ and HindIII, resulting in pNZBglBX10 (5,572 bp). A schematic diagram of the recombinant plasmid pNZBglBX10 construction is shown in Fig. S1. For the transformation step, the recombinant pNZBglBX10 plasmid was electroporated into L. lactis NZ9000 as previously described [16]. The transformed cells were selected using the GM17 agar with chloramphenicol.
In addition, L. lactis harboring pNZBgl was used to produce C-K. This recombinant strain was constructed in our previous study by cloning the nucleotide sequence of the BglPm gene from P. mucilaginosus into pNZ8008 [16].
2.3. Expression and purification of recombinant pNZBglBX10
Recombinant L. lactis harboring pNZBglBX10 was cultivated in GM17 medium containing 10 μg/mL chloramphenicol at 30°C. The precultured cells (4%) were inoculated into 100 mL of fresh medium and cultured until the optical density (OD600nm) reached 0.4. Expression of the recombinant gene was induced with 1 ng/mL nisin, while uninduced cultures were used as a control. Then, the cells were grown for an additional 3 h at 30°C to express the β-glucosidase enzyme. Cells were collected by centrifugation at 10,000g for 5 min, and the pellets were resuspended in 50mM sodium phosphate buffer (pH 7.0). The cells were disrupted by sonication (on 50 s, off 10 s, 10 min), and the supernatant fraction was used as the crude enzyme. The enzyme fraction was purified using Ni-nitrilotriacetic acid (Ni-NTA) chromatography with a His Trap-FF column (GE Healthcare, Uppsala, Sweden) [16]. Expression and purification of β-glucosidase were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
2.4. Optimization of pNZBglBX10 expression in L. lactis and analysis of specific enzyme activity
To investigate the effect of nisin concentrations on β-glucosidase enzyme activity, L. lactis harboring pNZBglBX10 was induced by 0.5 ng/mL, 1 ng/mL, 5 ng/mL, 10 ng/mL, 20 ng/mL, and 50 ng/mL nisin when the O.D600nm reached 0.4. The enzyme activity of crude β-glucosidases was measured using p-nitrophenyl-β-D-glucopyranoside as a substrate. The crude enzyme (150 μL) was incubated at 37°C in 150 μL of the 50mM sodium phosphate buffer (pH 7.0) containing 4mM p-nitrophenyl-β-D-glucopyranoside. One unit of activity was defined as the amount of enzyme that produced 1 μmol of p-nitrophenyl in 1 min, as measured by a microplate reader at 405 nm. In addition, the effect of culture duration on enzyme activity was determined by culturing and harvesting cells at 1 h, 2 h, 3 h, 4 h, 5 h, 6 h, 12 h, and 24 h after nisin induction. Enzyme activity was measured using the method described above. After optimization of the expression conditions, the specific activities of crude and purified β-glucosidases were also determined. Protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA) with bovine serum albumin as the standard.
2.5. Bioconversion of PPDGM into Rg3(S) using recombinant L. lactis cells
To produce Rg3(S), three preparations (whole cells, cell lysates, and permeabilized cells) of recombinant L. lactis harboring pNZBglBX10 were used. After cultivation, whole cells were harvested by centrifugation at 10,000g for 10 min at 4°C and suspended in 50mM sodium phosphate buffer (pH 7.0). To prepare cell lysates, whole cells were disrupted by sonication, and the supernatant fraction was recovered after centrifugation. For permeabilized cells, 0.5% xylene was added to the whole cell preparations [17]. Then, the three types of cells (whole cells, cell lysates, and permeabilized cells, at a final concentration of 300 mg/mL) were reacted with 1% PPDGM in 50mM sodium phosphate buffer (pH 7.0) for 24 h. Samples were taken at 0 h, 6 h, 12 h, and 24 h intervals and centrifuged (10,000g, 2 min, 4°C) after boiling for 5 min. Both supernatant and residue fractions were extracted with 50% ethanol and the sum of the two fractions was used for analysis.
2.6. Bioconversion of major ginsenosides to C-K using recombinant L. lactis harboring pNZBgl and/or pNZBglBX10
To produce C-K, recombinant strains of L. lactis harboring pNZBgl [16] and/or pNZBglBX10 were used. Three methods were employed for the synthesis of ginsenoside C-K. The first method was a combined mode with reactions of mixed cells; after cultivation, the whole cells of both L. lactis (pNZBgl) and L. lactis (pNZBglBX10) were harvested (final concentration of cells: 100 mg/mL), and they were allowed to simultaneously react in 50mM sodium phosphate buffer (pH 7.0) with 1% PPDGM and 0.5% xylene for 36 h. The second method was a sequential mode in which two types of cells reacted one by one; L. lactis harboring pNZBgl (100 mg/mL) reacted in 50mM sodium phosphate buffer (pH 7.0) with 1% PPDGM and 0.5% xylene for 24 h, and when the ginsenoside Rb1 was almost fully converted to F2, L. lactis harboring pNZBglBX10 (100 mg/mL) was also added for an additional reaction for 48 h. The third method was a dual-plasmid mode, with the reaction of a recombinant cell harboring two plasmids (pNZBgl and pNZBglBX10); the two plasmids were simultaneously transformed into L. lactis which was confirmed by colony PCR using two specific primers for each β-glucosidase genes (BglPm and BglBX10) and the resulting recombinant cells (200 mg/mL) were reacted in the same conditions. Samples were taken at intervals of 0 h, 12 h, 18 h, 24 h, and 36 h for the combined mode and dual-plasmid mode, and at 0 h, 12 h, 18 h, 24 h, 36 h, 42 h, 48 h, 60, and 72 h for the sequential mode.
2.7. HPLC analysis
Ginsenoside [Rb1, Rd, F2, Rg3(S), and C-K] concentrations were quantified using an Agilent 1260 Infinity HPLC (Agilent Technology, Santa Clara, CA, USA) system equipped with a ZORBAX SB-C18 column (4.6 mm × 150 mm). Acetonitrile (Solvent A) and water (Solvent B) were used as the mobile phases. Gradient elution was performed, beginning with 70% Solvent A and 30% Solvent B for 0–5 min, and progressing to 70% Solvent A and 30% Solvent B for 5–15 min; 43% Solvent A and 57% Solvent B for 15–25 min; 30% Solvent A and 70% Solvent B for 25–30 min; and 30% Solvent A and 70% Solvent B for 30–40 min. The flow rate of the mobile phase was 0.8 mL/min, and it was monitored at 203 nm absorbance using a UV spectrophotometric detector. The bioconversion yields of the ginsenosides Rg3(S) and C-K were calculated as follows: the conversion yield of Rg3(S) (%) = ΔRg3(S)/(ΔRb1 + ΔRd) × 100, and the conversion yield of C-K (%) = ΔC-K/(ΔRb1 + ΔRd) × 100.
3. Results and discussion
3.1. Construction of recombinant L. lactis
There have been many previous reports that low levels of heterologous expression were caused by differences in synonymous codon usage between the expression and natural hosts, as well as because of the accumulation of rare codons during expression of the heterologous protein [18], [19]. To determine whether heterologous β-glucosidase could be expressed at a high level in L. lactis, the codon usage was analyzed. For this, the frequency of each codon (count per thousand) of BglBX10 gene and L. lactis genome were analyzed at http://www.bioinformatics.org/sms2/codon_usage.html and http://www.kazusa.or.jp/codon/, respectively. The codon usage was found to be similar in the two cases, except for CCG (proline), suggesting that codon usage differences would not significantly affect the β-glucosidase expression level. Therefore, the recombinant plasmid pNZBglBX10 was constructed by inserting the native BglBX10 gene and it was transformed into the host, L. lactis NZ9000.
3.2. Expression and characterization of the BglBX10 enzyme
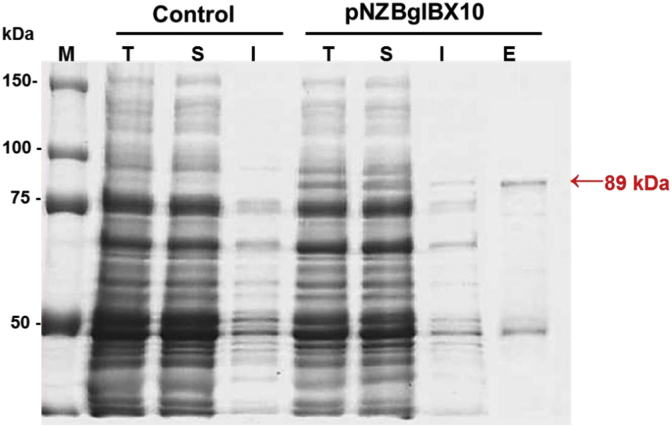
To measure the expression of the BglBX10 gene in L. lactis, SDS-PAGE analysis was conducted. As shown in Fig. 1, bands corresponding to the β-glucosidase protein were present in both the total and soluble fractions of the nisin-induced cell samples, whereas no bands were detected in the uninduced samples. The protein was purified to homogeneity using an Ni-NTA affinity chromatography column. The molecular weight of the recombinant protein was estimated to be 89 kDa by SDS-PAGE, which was similar to the theoretical value (89.3 kDa) calculated from its amino acid sequence.
Fig. 1.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis of BglBX10 expressed in Lactococcus lactis. Control, L. lactis harboring pNZBglBX10 without induction; pNZBglBX10, L. lactis harboring pNZBglBX10 with nisin induction. E, elution fraction after Ni-NTA purification; I, insoluble fraction of cell extracts; M, molecular weight marker; S, soluble fraction; T, total fraction.
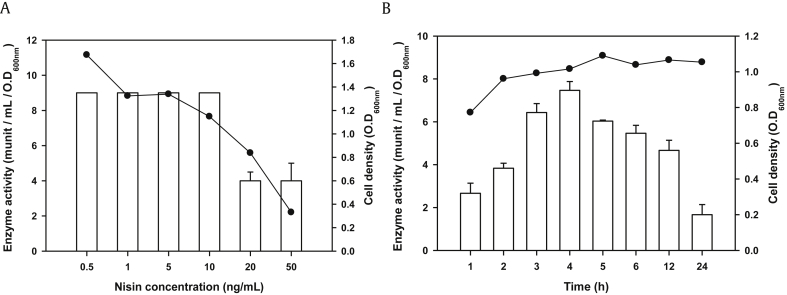
To investigate the effects of different nisin concentrations, L. lactis harboring pNZBglBX10 was induced with 0.5 ng/mL, 1 ng/mL, 5 ng/mL, 10 ng/mL, 20 ng/mL, and 50 ng/mL nisin in GM17 medium supplemented with chloramphenicol. As shown in Fig. 2A, β-glucosidase activity in the cytoplasmic fraction was similar (9 munit/mL/OD600nm) with nisin concentrations ranging from 0.5 ng/mL to 10 ng/mL. However, it decreased by half at higher nisin concentrations (20 ng/mL and 50 ng/mL) due to the inhibitory effect of nisin on microbial cell growth. Therefore, in the next part of this study, 1 ng/mL of nisin was used to induce enzyme expression in L. lactis. In addition, the effect of the postinduction period on enzyme activity was determined. As shown in Fig. 2B, the highest enzyme activity (7.5 munit/mL/OD600nm) was detected at 4 h after nisin induction; thereafter, the total enzyme activity decreased.
Fig. 2.
Changes in β-glucosidase activity of recombinant Lactococcus lactis harboring pNZBglBX10 (A) at different concentrations (0.5 ng/mL, 1 ng/mL, 5 ng/mL, 10 ng/mL, 20 ng/mL, and 50 ng/mL) of nisin and (B) at different culturing times (1 h, 2 h, 3 h, 4 h, 5 h, 6 h, 12 h, and 24 h) after induction by 1 ng/mL nisin.
In order to measure the specific activities of crude and purified preparations of β-glucosidase expressed in L. lactis harboring pNZBglBX10, cell-free extract was obtained after induction. The activity values of the BglBX10 enzyme in uninduced and induced cultures were 0.001 unit/mL and 0.003 unit/mL, respectively. The Ni-NTA-purified enzyme had an activity value of 0.002 unit/mL (Table 2). The specific activities of β-glucosidase in the crude extract and purified fraction were 0.002 units/mg and 0.02 units/mg, respectively.
Table 2.
β-Glucosidase activities in different cell-free extracts of Lactococcus lactis harboring pNZBglBX10 after induction and purification 1)
| Lactococcus lactis (pNZBglBX10) | Step | Volume (mL) | Activity (unit/mL) | Total activity (unit) | Protein (mg/mL) | Specific activity (unit/mg) | Yield (%) | Purification fold |
|---|---|---|---|---|---|---|---|---|
| Uninduced | Crude enzyme | 100 | 0.001 | 0.1 | 1.5 | |||
| Induced | Crude enzyme | 100 | 0.003 | 0.3 | 1.3 | 0.002 | 100 | 1 |
| Ni-NTA purification | 100 | 0.002 | 0.2 | 0.1 | 0.02 | 67 | 10 |
One unit of activity corresponds to the production of 1 μmol p-nitrophenyl (PNP)/min
3.3. Production of Rg3(S) using different preparations of recombinant cells
To produce Rg3(S) from PPDGM, purified from ginseng root, three different preparations of the recombinant cells (L. lactis harboring pNZBglBX10), namely whole cells, cell lysates, and permeabilized cells, were used. The concentrations of Rb1 and Rd in the 1% PPDGM solution were approximately 3.4mM and 2.4mM, respectively. As shown in Table 3, Rb1 was completely consumed by all three preparations, whereas no change was observed in the control fraction (uninduced cells). The ginsenoside Rg3(S) was produced after 5 h, and most conversions were completed within 24 h. A small amount of Rd, another major PPDGM component, remained after a 24 h reaction. The bioconversion yields of whole cells, cell lysates, and permeabilized cells were 44%, 58%, and 61%, respectively.
Table 3.
Concentrations of ginsenosides Rb1, Rd, and Rg3(S) and bioconversion yields from different preparations of recombinant cells
| Samples | Concentration (mM) |
Yield (%)1) | |||||
|---|---|---|---|---|---|---|---|
| Ginsenoside Rb1 |
Ginsenoside Rd |
Ginsenoside Rg3(S) |
|||||
| 0 h | 24 h | 0 h | 24 h | 0 h | 24 h | ||
| Control | 3.3 ± 0.03 | 3.3 ± 0.03 | 2.16 ± 0.05 | 2.16 ± 0.05 | 0.14 ± 0.08 | 0.14 ± 0.08 | 0 |
| Whole cells | 0.19 ± 0.02 | 0.28 ± 0.04 | 2.34 ± 0.04 | 44 | |||
| Cell lysates | 0.94 ± 0.00 | 3.70 ± 0.01 | 2.40 ± 0.01 | 58 | |||
| Permeabilized cells | 0.22 ± 0.05 | 0.73 ± 0.02 | 2.88 ± 0.05 | 61 | |||
The values in the table are averages with standard deviations determined from three independent experiments
Conversion yield (%) = ΔRg3(S)/(ΔRb1+ΔRd) × 100
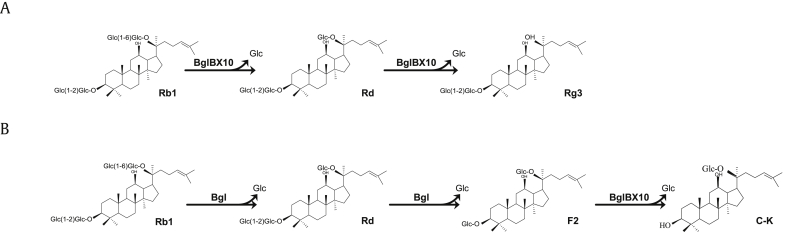
These results indicate that the enzyme was located mostly in the cytoplasm and could be released into the culture medium by manipulations such as sonication or xylene treatment. The use of whole cells has several advantages over the use of purified enzymes in many industrial bioconversion processes. However, reaction rates in whole cells are often very low because the cell envelope presents a permeability barrier for substrates and products [20]. To reduce the permeability barrier and obtain more enzymatically active whole cell preparations, cells can be permeabilized by ethanol or toluene. Permeabilization is an economical, easy, convenient, and safe process for enzymatic bioconversion and product formation [21]. In addition, the ginsenoside C-K was also produced during the bioconversion reactions. However, yields of bioconversion of PPDGM into C-K by whole cells, cell lysates, and permeabilized cells were very low (1%, 7%, and 5%, respectively), and BglBX10 could convert F2 into C-K when the F2 standard compound was used as a substrate (data not shown). This result suggests that BglBX10 is not suitable for C-K production. Taken together, the reaction pathway catalyzed by the BglBX10 enzyme included stepwise hydrolysis reactions of the outer and inner glucose moieties at positions C20 and C3 of ginsenoside Rb1. The reactions are as follows: Rb1 → Rd →Rg3(S) and F2→ C-K (Fig. 3).
Fig. 3.
Schematic presentation of the transformation pathways used to produce (A) Rg3(S) and (B) C-K and the structures of ginsenosides.
3.4. Production of C-K via the reactions of two recombinant cells
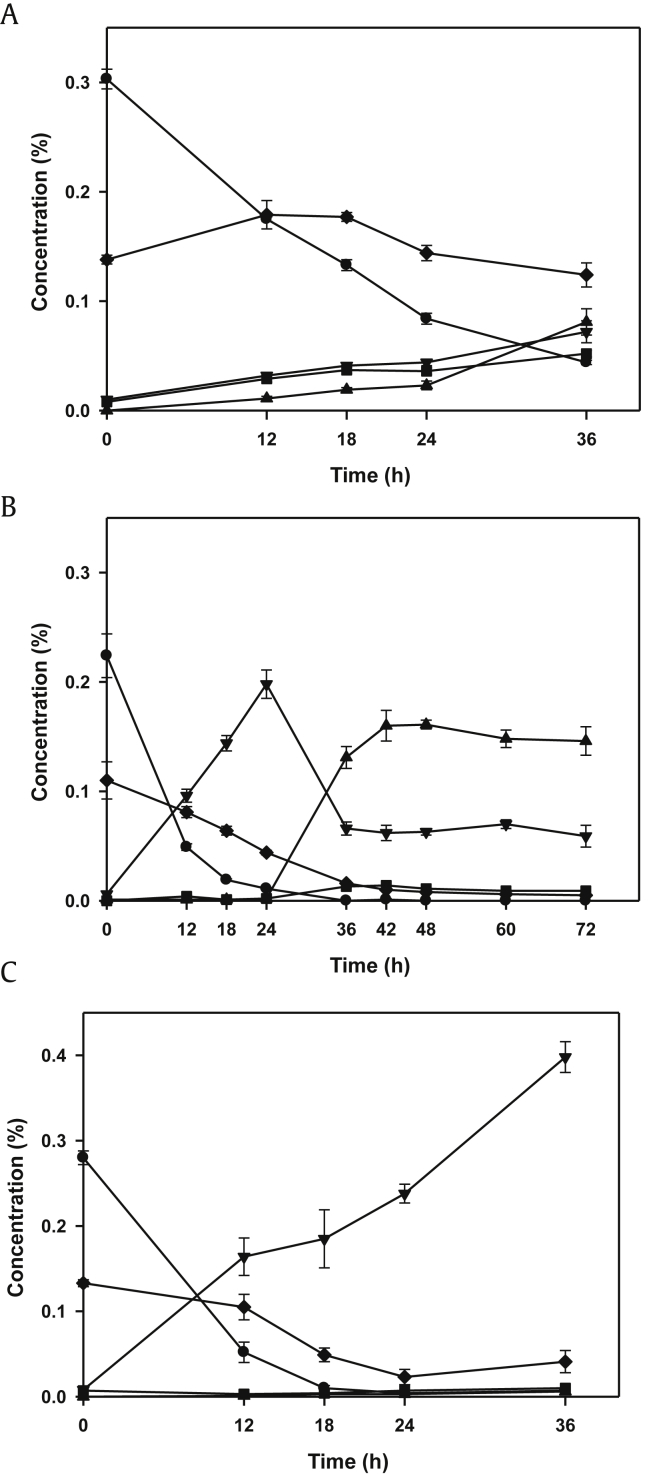
To achieve a higher conversion yield of C-K, additional recombinant cells of L. lactis harboring pNZBgl, which has β-glucosidase gene (BglPm) from P. mucilaginosus, were employed. Lysates of these cells have been shown to convert major ginsenosides into F2 with a conversion yield of 91% [16]. In this study, three modes of reactions using two recombinant cells were tested: the combined mode, the sequential mode, and the dual-plasmid mode. In the case of the combined two glucosidases mode, Rb1 was entirely consumed, and all of the three minor ginsenosides [Rg3(S), F2, and C-K] were produced from the beginning of the reaction, because the two major ginsenosides (Rb1 and Rd) were used competitively by the two enzymes (Fig. 4A). In the case of the sequential mode, Rb1 and Rd were completely converted into F2 within 24 h, and after the addition of the BglBX10 enzyme, F2 was almost converted into C-K within 12 h, and a small amount of Rg3(S) was synthesized (Fig. 4B). These results showed that the BglBX10 enzyme could hydrolyze F2 into C-K when there was enough F2 (Fig. 3B). In the case of the dual-plasmids mode, Rb1 and Rd were completely converted into F2 and the Rg3(S), and no C-K was formed (Fig. 4C). The bioconversion yields of F2, Rg3(S), and C-K were also different between modes. As shown in Table 4, the bioconversion yields of F2, Rg3(S), and C-K were 32%, 23%, and 53%, respectively, in the combined mode. By contrast, the bioconversion yields of F2, Rg3(S), and C-K were 25%, 5%, and 70%, respectively, in the sequential mode, with the bioconversion yield of C-K being the highest. Because F2 was fully synthesized as a final product in the dual-plasmid mode, the bioconversion yield of F2 was almost 100%.
Fig. 4.
Production of ginsenoside compound K (C-K) by the (A) combined mode, (B) sequential mode, and (C) dual-plasmid mode using recombinant Lactococcus lactis harboring pNZBgl and/or pNZBglBX10. C-K (▲); F2 (▼); Rb1 (●); Rd (◆);Rg3(S) (■).
Table 4.
Concentration of each ginsenoside and bioconversion yields from three reaction modes
| Modes | Ginsenoside Rb1 (mM) |
Ginsenoside Rd (mM) |
Ginsenoside F2 (mM) |
Ginsenoside Rg3(S) (mM) |
Ginsenoside C-K (mM) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 36 h | 0 h | 36 h | 0 h | 36 h | Yield (%)1) | 0 h | 36 h | Yield (%)2) | 0 h | 36 h | Yield (%)3) | |
| Combined mode | 2.73 (0.08) | 0.40 (0.02) | 1.43 (0.05) | 1.29 (0.12) | 0.12 (0.01) | 0.92 (0.13) | 32 | 0.10 (0.01) | 0.67 (0.04) | 23 | 0.00 (0.00) | 1.30 (0.20) | 53 |
| Sequential mode | 2.02 (0.31) | 0.00 (0.00) | 1.14 (0.31) | 0.17 (0.01) | 0.08 (0.00) | 0.84 (0.13) | 25 | 0.00 (0.00) | 0.17 (0.03) | 5 | 0.02 (0.02) | 2.11 (0.30) | 70 |
| Dual-plasmids mode | 2.52 (0.07) | 0.07 (0.03) | 1.38 (0.04) | 0.42 (0.13) | 0.10 (0.00) | 3.79 (0.21) | 100 | 0.09 (0.00) | 0.13 (0.02) | 1 | 0.00 (0.00) | 0.10 (0.02) | 3 |
The values in the table are averages determined from three independent experiments. Standard deviations are shown in brackets
Conversion yield (%) = ΔF2/(ΔRb1 + ΔRd) ×100
Conversion yield (%) = ΔRg3/(ΔRb1 + ΔRd) ×100
Conversion yield (%) = ΔC-K/(ΔRb1 + ΔRd) ×100
Production of C-K using β-glucosidase-active bacteria has been described in previous reports. Complete conversion took 48 h for Bifidobacterium sp. Int57, Bif. sp. SJ32, Aspergillus niger, and Aspergillus usamii; and 72 h for Leuconostoc mesenteroides DC102, Leuconostoc citreum LH1, and Lactobacillus paralimentarius LH4 [22], [23], [24], [25]. Compared to the above methods, the recombinant L. lactis developed in this study showed a superior ability to produce C-K with a fast reaction rate. The enzymes were also convenient to use in the whole cell system without a purification step.
The three reaction modes (the combined mode, the sequential mode, and the dual-plasmid mode) each have advantages and disadvantages: for the combined mode, all three minor ginsenoside compounds [F2, Rg3(S), and C-K] were synthesized from the beginning of the reaction, however, the production yield of C-K was only 50%; for the sequential mode, the production rate and yield of C-K were highest in the short term, which is useful for the efficient production of C-K; for the dual-plasmid mode, this system was simple using one recombinant cell, however, no C-K was formed. As shown in Fig. 3, the characteristics of the two glucosidase enzymes used in this study are different. The two enzymes both have β-1,2- and β-1,6-glucosidase activities, but the Bgl enzyme preferentially shows β-1,2-glucosidase over β-1,6-glucosidase activity [26], whereas the BglBX10 enzyme shows the reverse.
4. Conclusions
This study demonstrates that lysated or permeabilized L. lactis expressing the β-glucosidase gene can be used to efficiently produce the compound Rg3(S) from ginseng extract. The combined use of the two glucosidases enabled the efficient production of C-K. To our knowledge, this is the first report of a method to produce Rg3(S) and C-K with high conversion yields, using a system based on LAB. This food-grade microbial cell factory can be used to enhance the health benefits of Panax ginseng, in either fermented foods or bioconversion processes.
Conflicts of interest
All contributing authors declare no conflicts of interest.
Acknowledgments
This study was supported by the Intelligent Synthetic Biology Center of Global Frontier Project, funded by the Korean Ministry of Science, ICT, and Future Planning (2013M3A6A8073553), and a National Research Foundation of Korea (NRF) grant (2015R1A2A2A01007156) funded by the Korean government (MEST).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2017.04.007.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Chang K.H., Jee H.S., Lee N.K., Park S.H., Lee N.W., Paik H.D. Optimization of the enzymatic production of 20(S)-ginsenoside Rg3 from white ginseng extract using response surface methodology. New Biotechnol. 2009;26:181–186. doi: 10.1016/j.nbt.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 2.Park C.S., Yoo M.H., Noh K.H., Oh D.K. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl Microbiol Biotechnol. 2010;87:9–19. doi: 10.1007/s00253-010-2567-6. [DOI] [PubMed] [Google Scholar]
- 3.Baeg I.H., So S.H. The world ginseng market and the ginseng (Korea) J Ginseng Res. 2013;37:1–7. doi: 10.5142/jgr.2013.37.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim M.K., Lee J.W., Lee K.Y., Yang D.C. Microbial conversion of major ginsenoside Rb1 to pharmaceutically active minor ginsenoside Rd. J Microbiol. 2005;43:456–462. [PubMed] [Google Scholar]
- 5.Upadhyaya J., Kim M.J., Kim Y.H., Ko S.R., Park H.W., Kim M.K. Enzymatic formation of compound-K from ginsenoside Rb1 by enzyme preparation from cultured mycelia of Armillaria mellea. J Ginseng Res. 2016;40:105–112. doi: 10.1016/j.jgr.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X.D., Yang Y.Y., Ouyang D.S., Yang G.P. A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia. 2015;100:208–220. doi: 10.1016/j.fitote.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Shin K.C., Choi H.Y., Seo M.J., Oh D.K. Compound K production from red ginseng extract by β-glucosidase from Sulfolobus solfataricus supplemented with α-L-arabinofuranosidase from Caldicellulosiruptor saccharolyticus. PLoS One. 2015;10:e0145876. doi: 10.1371/journal.pone.0145876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J.K., Cui C.H., Liu Q., Yoon M.H., Kim S.C., Im W.T. Mass production of the ginsenoside Rg3(s) through the combinative use of two glycoside hydrolases. Food Chem. 2013;141:1369–1377. doi: 10.1016/j.foodchem.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 9.Liu C.Y., Zhou R.X., Sun C.K., Jin Y.H., Yu H.S., Zhang T.Y., Xu L.Q., Jin F.X. Preparation of minor ginsenosides C-Mc, C-Y, F2, and C-K from American ginseng PPD-ginsenoside using special ginsenosidase type-I from Aspergillus niger g.848. J Ginseng Res. 2015;39:221–229. doi: 10.1016/j.jgr.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park I.H., Piao L.Z., Kwon S.W., Lee Y.J., Cho S.Y., Park M.K., Park J.H. Cytotoxic dammarane glycosides from processed ginseng. Chem Pharm Bull. 2002;50:538–540. doi: 10.1248/cpb.50.538. [DOI] [PubMed] [Google Scholar]
- 11.Chen G.T., Yang M., Song Y., Lu Z.Q., Zhang J.Q., Huang H.L., Wu L.J., Guo D.A. Microbial transformation of ginsenoside Rb1 by Acremonium strictum. Appl Microbiol Biotechnol. 2008;77:1345–1350. doi: 10.1007/s00253-007-1258-4. [DOI] [PubMed] [Google Scholar]
- 12.Teng R.W., Li H.Z., Wang D.Z., Yang C.R. Hydrolytic reaction of plant extracts to generate molecular diversity: new dammarane glycosides from the mild acid hydrolysate of root saponins of Panax notoginseng. Helv Chim Acta. 2004;87:1270–1278. [Google Scholar]
- 13.Noh K.H., Oh D.K. Production of the rare ginsenosides compound K, compound Y, and compound Mc by a thermostable beta-glycosidase from Sulfolobus acidocaldarius. Biol Pharm Bull. 2009;32:1830–1835. doi: 10.1248/bpb.32.1830. [DOI] [PubMed] [Google Scholar]
- 14.Bae E.A., Han M.J., Choo M.K., Park S.Y., Kim D.H. Metabolism of 20(S)- and 20(R)-ginsenoside Rg3 by human intestinal bacteria and its relation to in vitro biological activities. Biol Pharm Bull. 2002;25:58–63. doi: 10.1248/bpb.25.58. [DOI] [PubMed] [Google Scholar]
- 15.Lee S.Y. High cell-density culture of Escherichia coli. Trends Biotechnol. 1996;14:98–105. doi: 10.1016/0167-7799(96)80930-9. [DOI] [PubMed] [Google Scholar]
- 16.Li L., Shin S.Y., Lee S.J., Moon J.S., Im W.T., Han N.S. Production of ginsenoside F2 by using Lactococcus lactis with enhanced expression of β-glucosidase gene from Paenibacillus mucilaginosus. J Agric Food Chem. 2016;64:2506–2512. doi: 10.1021/acs.jafc.5b04098. [DOI] [PubMed] [Google Scholar]
- 17.De León A., García B., Barba de la Rosa A.P., Villaseñor F., Estrada A., López-Revilla R. Periplasmic penicillin G acylase activity in recombinant Escherichia coli cells permeabilized with organic solvents. Process Biochem. 2003;39:301–305. [Google Scholar]
- 18.Roche E.D., Sauer R.T. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angov E., Hillier C.J., Kincaid R.L., Lyon J.A. Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host. PLoS One. 2008;3:e2189. doi: 10.1371/journal.pone.0002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo A., Liu Y., Furuta M., Fujita Y., Matsumoto T., Fukuda H. Preparation of high activity whole cell biocatalyst by permeabilization of recombinant flocculent yeast with alcohol. Enzyme Microb Technol. 2000;27:806–811. doi: 10.1016/s0141-0229(00)00304-5. [DOI] [PubMed] [Google Scholar]
- 21.Fontes E.A.F., Passos F.M.L., Passos F.J.V. A mechanistical mathematical model to predict lactose hydrolysis by beta-galactosidase in a permeabilized cell mass of Kluyveromyces lactis: validity and sensitivity analysis. Process Biochem. 2001;37:267–274. [Google Scholar]
- 22.Chi H., Ji G.E. Transformation of ginsenosides Rb1 and Re from Panax ginseng by food microorganisms. Biotechnol Lett. 2005;27:765–771. doi: 10.1007/s10529-005-5632-y. [DOI] [PubMed] [Google Scholar]
- 23.Quan L.H., Min J.W., Yang D.Y., Kim Y.J., Yang D.C. Biotransformation of ginsenoside Rb1 to prosapogenins, gypenoside XVII, ginsenoside Rd, ginsenoside F2, and compound K by Leuconostoc mesenteroides DC102. J Ginseng Res. 2011;35:344–351. doi: 10.5142/jgr.2011.35.3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quan L.H., Piao J.Y., Min J.W., Yang D.U., Lee H.N., Yang D.C. Bioconversion of ginsenoside Rb1 into compound K by Leuconostoc citreum LH1 isolated from kimchi. Braz J Microbiol. 2011;42:1227–1237. doi: 10.1590/S1517-838220110003000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quan L.H., Kim Y.J., Li G.H., Choi K.T., Yang D.C. Microbial transformation of ginsenoside Rb1 to compound K by Lactobacillus paralimentarius. Word J Microbiol Biotechnol. 2013;29:1001–1007. doi: 10.1007/s11274-013-1260-1. [DOI] [PubMed] [Google Scholar]
- 26.Cui C.H., Kim J.K., Kim S.C., Im W.T. Characterization of a ginsenoside-transforming β-glucosidase from Paenibacillus mucilaginosus and its application for enhanced production of minor ginsenoside F2. PLoS One. 2014;9:e85727. doi: 10.1371/journal.pone.0085727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.