Abstract
Accumulating research has documented that microRNA-21 (miR-21) plays an important role in the development of human colorectal carcinoma (CRC). Our recent work also showed that antisense oligonucleotides (ASOs) against miR-21 can impair the growth of CRC cells in vitro. However, the potential role of miR-21 in gene therapy against CRC remains to be fully elucidated. Here, we further observed the effect of ASOs against miR-21 on the growth and metastasis of CRC in vivo using a xenograft model of human CRC. We found that ASOs could effectively inhibit the growth and metastasis of CRC in vivo, accompanied by downregulated expression of miR-21 and reduced transduction of the AKT and ERK pathway. Mechanically, global gene expression analysis showed that the expression of DUSP8, a novel target of miR-21, was upregulated in tumor mass. Furthermore, overexpression of DUSP8 could remarkably suppress the proliferation and migration of CRC cells in vitro. Finally, downregulation of DUSP8 could abrogate the effects of ASOs against miR-21 on the proliferation and migration of CRC cells, as well as altered transduction of the AKT and ERK signaling pathway. Together, these data suggest that ASOs against miRNAs are an attractive and potential therapeutic for the treatment of human CRC and warrant further development.
Keywords: miR-21, ASOs, colorectal carcinoma, growth, DUSP8
Introduction
Colorectal carcinoma (CRC), with an estimated 1.4 million cases and 693,900 deaths occurring worldwide, is the third most commonly diagnosed cancer in men and the second only to breast cancer in women.1 Even though the 5- and 10-year relative survival rates for patients with CRC have gradually improved, the side effects of regular treatment, including neuropathy, chronic diarrhea, bowel dysfunction, and so on, have a considerable impact on CRC patients in terms of quality of life and treatment compliance.2 Consequently, it has been necessary for clinical CRC patients to explore new reliable therapeutic strategies. Fortunately, molecular therapy, including techniques involving antisense oligonucleotides (ASOs), has been used to intervene in the growth and metastasis of CRC and thus represents an important tactic in gene therapy for clinical CRC.3, 4, 5, 6, 7 Furthermore, ASOs have recently undergone substantial development.8 Modification of DNA backbone chemistry has reduced toxicity, increased target engagement, and improved the destruction of DNA-RNA hybrids.9 For example, Tangudu et al.10 reported that upregulation of p53 using anti-miR-21 ASOs linked to rhodamine-labeled nanoparticles and could suppress a CRC model. Kasiri et al.11 found that ASOs mediated knockdown of HOXC13, a homeobox-containing gene that plays crucial roles in hair development and the origin of replication, and affected human CRC SW480 cell growth and apoptosis. Furthermore, Stolfi et al.12 revealed that inhibition of smad7 by ASOs reduces the in vitro proliferation of HCT116 cells and the in vivo growth of HCT116-derived xenografts. These studies indicate that ASO-based therapeutic strategy might be a promising approach for clinical therapy against CRC.
MicroRNA-21 (miR-21) is a distinct miRNA family member that plays a pivotal role in the context of pathology, especially induction of inflammatory response, and is involved in the progression of various cancers, including CRC.13, 14, 15, 16, 17, 18, 19, 20, 21 In CRC, accumulating evidence shows that miR-21 is a vital regulator in the development of CRC through regulation of the proliferation, invasion, and migration, as well as apoptosis, of CRC cells, and it has emerged as a novel potential therapeutic target in CRC. For instance, Mima et al.22 found that miR-21 expression level in CRC was associated with worse clinical outcomes, and this association was stronger in carcinomas expressing high levels, which played complex roles in immunity and inflammation in tumor progression. Furthermore, Li et al.23 reported that miR-21 was overexpressed in CRC cell lines and promoted the proliferation, migration, and invasion of these cells in vitro, which was associated with downregulation of Sec23A expression. Consistently, our recent evidence also showed that miR-21 ASO treatment could reduce the proliferation and migration of human CRC cells in vitro, accompanied by altered transduction of the AKT and ERK pathway.24 These studies indicate the critical role of miR-21 in the development, diagnosis, and clinical outcome and prognosis of CRC. Whether miR-21 might be used, however, and its effects on human CRC in vivo and as a potential target in the gene therapy against CRC remain to be further elucidated.
To address this issue, we used a pre-constructed eukaryotic vector encoding ASOs against miR-21 (termed p-miR-21-ASOs), which was injected locally into a human CRC model by using nude mice, establishing and observing its effects on the growth and metastasis of human CRC cells in vivo. Of note, we used the cDNA chip technique and found overexpression of DUSP8, a novel target molecule of miR-21, in tumor mass. Furthermore, overexpression of DUSP8 could remarkably suppress the proliferation and migration of CRC cells in vitro. Finally, downregulation of DUSP8 could abrogate the effects of ASOs against miR-21 on the proliferation and migration of CRC cells, as well as altered transduction of the AKT and ERK signaling pathway. Thus, this is the first study showing that ASOs against miR-21 can attenuate the growth and metastasis of CRC cells in vivo, closely correlated with upregulated expression of DUSP8, which may ultimately aid in the understanding of the development of CRC and the development of new gene therapeutic strategies against clinical CRC.
Results
ASOs against miR-21 Inhibited the Growth of Human CRC Cells In Vivo
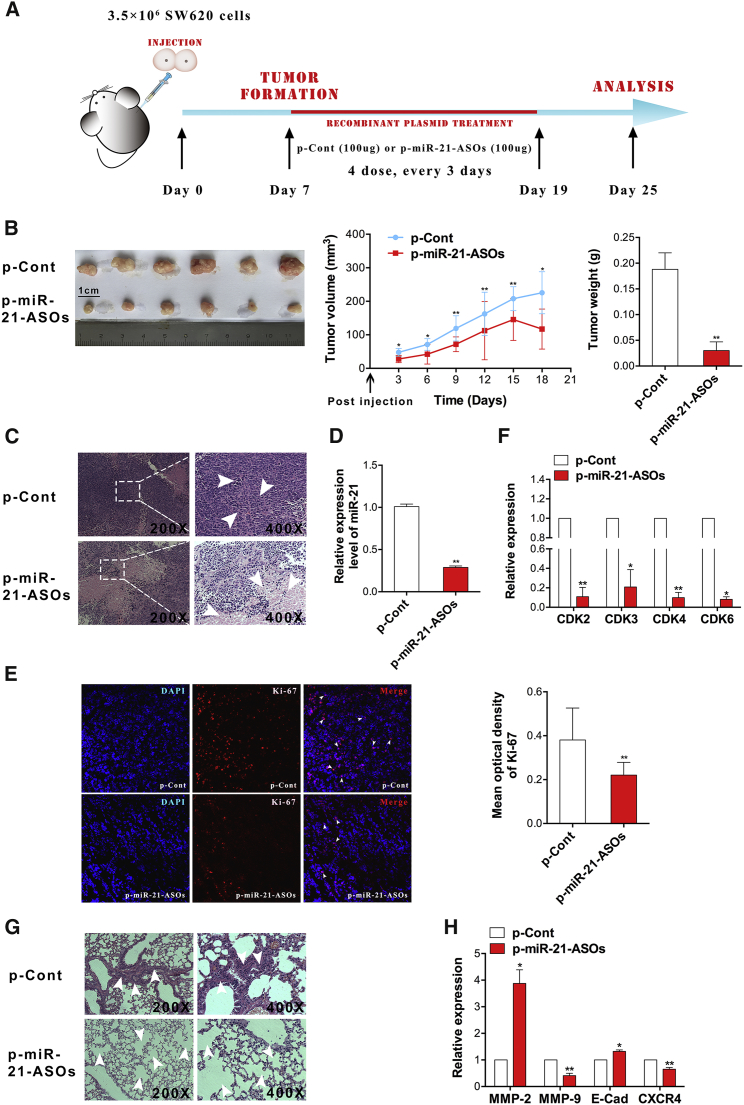
Our previous work showed that ASOs against miR-21 could reduce the proliferation and migration of human CRC cells in vitro.24 To verify the possible effect of ASOs against miR-21 on the growth and metastasis of human CRC cells in vivo, we designed a xenograft model of human CRC in nude mice to observe the effect of ASOs against miR-21 on the growth of tumor cells in vivo and to explore the potential molecular mechanism on the basis of global gene array analysis (Figures S1A and S1B). As shown in Figures 1A and 1B, both the volume and weight of tumor tissues in the p-miR-21-ASO group were reduced significantly compared with those in the p-Cont group (p < 0.05). H&E staining showed that there were many large areas of necrosis in tumor tissues in the p-miR-21-ASO group (Figure 1C). As expected, the expression of miR-21 was also much lower than in the p-Cont group (Figures 1D and S3D; p < 0.05), which was consistent with our previous work.24 Immunofluorescence assay demonstrated that the proliferation of SW620 tumor cells decreased significantly in the p-miR-21-ASO group (Figure 1E; p < 0.05). To confirm the impact of downregulation of miR-21 on the growth of cancer cells, we further detected the expression of cyclin-dependent kinases (CDKs), including CDK2, CDK3, CDK4, and CDK6. The data showed that the expression of these CDKs was downregulated remarkably (Figure 1F; p < 0.05). Finally, we also noticed that the apoptosis of cells in the p-miR-21-ASO group increased unmistakably in comparison with the p-Cont group (Figure S2).
Figure 1.
ASOs against miR-21 Inhibited the Growth and Metastasis of Human Colorectal Carcinoma Cells In Vivo
(A) Schematic diagram of the in vivo experiment. (B) Representative image, growth curve, and weight of tumors. (C) H&E staining of tumor tissues and (G) lung tissues was observed using a microscope (magnification 200× and 400× ). (D) Real-time PCR detected the expression of miR-21 in tumor tissues. (E) Expression of nuclear antigen Ki-67 in tumor tissues was analyzed using immunofluorescence assay, and the mean optical density of Ki-67 was calculated using ImageJ software (NIH). (F) Real-time PCR detected the expression of cyclin-dependent kinases (CDK2, CDK3, CDK4, and CDK6) and (H) metastasis-related molecules (MMP-2, MMP9, E-cadherin, and CXCR4) in tumor tissues. *p < 0.05 and **p < 0.01.
To further confirm the effect of ASOs against miR-21 on the growth of CRCs in vivo, we also observed the possible effect of ASOs against miR-21 on the growth of the CRC line HCT116 in vivo. As expected and as shown in Figures S3A and S3B, both the volume and weight of tumor tissues in the p-miR-21-ASO group were reduced significantly compared with those in the p-Cont group (p < 0.05). Moreover, immunofluorescence assay also showed that the proliferation of HCT116 tumor cells decreased significantly in the p-miR-21-ASO group (Figure S3C; p < 0.05), accompanied with the decreased level of miR-21 in tumor mass (Figure S3D; p < 0.05). Combining these data demonstrated that ASOs could clearly downregulate the expression of miR-21 in tumor tissues, which effectively inhibited the growth of CRC cells in vivo.
ASOs against miR-21 Suppressed the Metastasis of Human CRC Cells In Vivo
Our previous work showed that ASOs against miR-21 could impair the metastatic potential of CRC cell in vitro.24 To evaluate whether downregulated expression of miR-21 could affect the metastasis of human CRC cells in vivo, we examined metastatic lesions in lung tissue using H&E staining assay, which showed a more integral structure of alveolar space in the p-miR-21-ASO group (Figure 1G). Moreover, we also detected the expression of metastasis-related molecules, including CXCR4, E-cadherin, MMP-2, and MMP-9. The data showed that the expression of E-cadherin and MMP-2 were increased significantly, and MMP-9 and CXCR4 were decreased remarkably in the p-miR-21-ASO group (Figure 1H; p < 0.05). These findings demonstrated that ASOs against miR-21 also could suppress metastasis in lung tissue from the primary tumor of CRC in vivo.
ASOs against miR-21 Altered the Transduction of the AKT and ERK Signaling Pathway in CRC
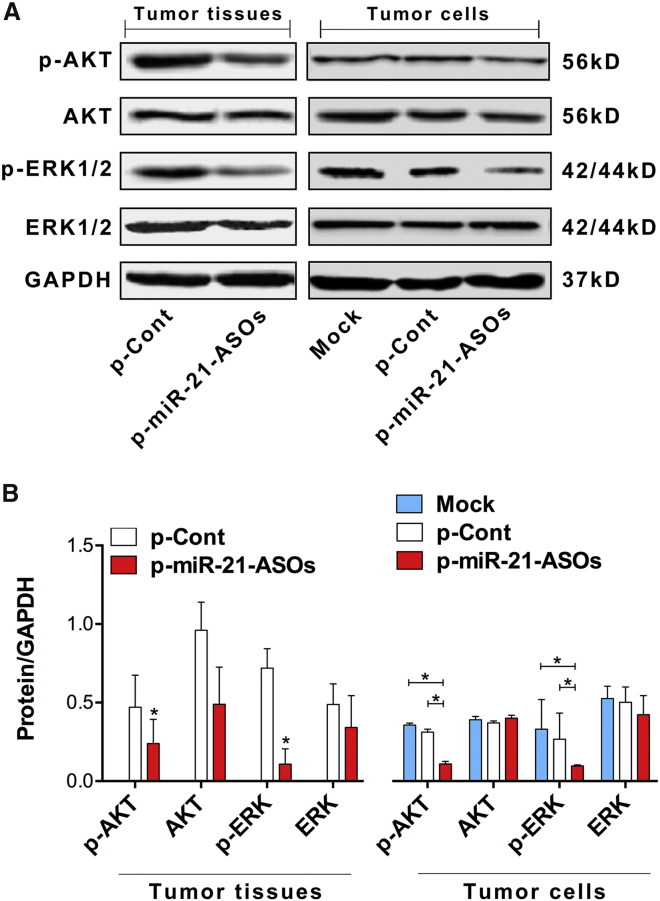
An increasing body of literature documents that miR-21 can regulate the growth of CRC cells through various signal pathways, such as the AKT and ERK pathway, which closely correlated with the proliferation and metastasis of CRC.25, 26, 27 Our previous work also showed that downregulated expression of miR-21 could inhibit the proliferation and migration of human CRC cells through the AKT and ERK pathway.24 Thus, to further investigate the effect of downregulated expression of miR-21 by ASOs on the growth and metastasis of CRC cells, we analyzed the expression level of phosphorylation of AKT (p-AKT) and ERK1/2 (p-ERK1/2) in tumor tissues derived from the p-miR-21-ASO group or p-Cont group. The data showed that there were no significant differences in the expression levels of both AKT and ERK between the p-miR-21-ASO group and the p-Cont group. Interestingly, the expression levels of p-AKT and p-ERK significantly decreased in the p-miR-21-ASO group (Figures 2A and 2B, left; p < 0.05). To confirm these findings, we also transiently transfected the recombined plasmid of p-miR-21-ASOs or p-Cont into SW620 in vitro and found that the expression levels of both p-AKT and p-ERK were reduced remarkably in the p-miR-21-ASO transfection group compared with those in p-Cont transfection group (Figures 2A and 2B, right; p < 0.05). These results suggested that downregulated expression level of miR-21 by ASOs could attenuate the growth of human CRC cells in vitro and in vivo by altering the transduction of the AKT and ERK signaling pathway, which was consistent with our previous findings.24
Figure 2.
ASOs against miR-21 Altered the Transduction of the AKT and ERK Signaling Pathway
(A) Human CRC SW620 cells were injected subcutaneously into the right flank of BALB/c nude mice (n = 10 per group). Seven days later, the plasmid of p-miR-21-ASOs (100 μg) or p-Cont (100 μg) was locally given by subcutaneous injection into the tumor tissues of nude mice four times every 3 days. Six days after the last injection, tumor tissues were collected. Protein levels of AKT, p-AKT, ERK, and p-ERK were detected using western blot, and the gray intensity value was calculated using ImageJ software. (B) Human CRC SW620 cells were transiently transfected with p-miR-21-ASOs (2.5 μg) or p-Cont (2.5 μg) in vitro. Forty-eight hours later, cells were collected, and protein levels of AKT, p-AKT, ERK, and p-ERK were detected using western blot and calculated. Representative data of three independent experiments are shown. Left: tumor tissues; right: tumor cells. *p < 0.05.
ASOs against miR-21 Altered Global Gene Expression of Tumor Tissue
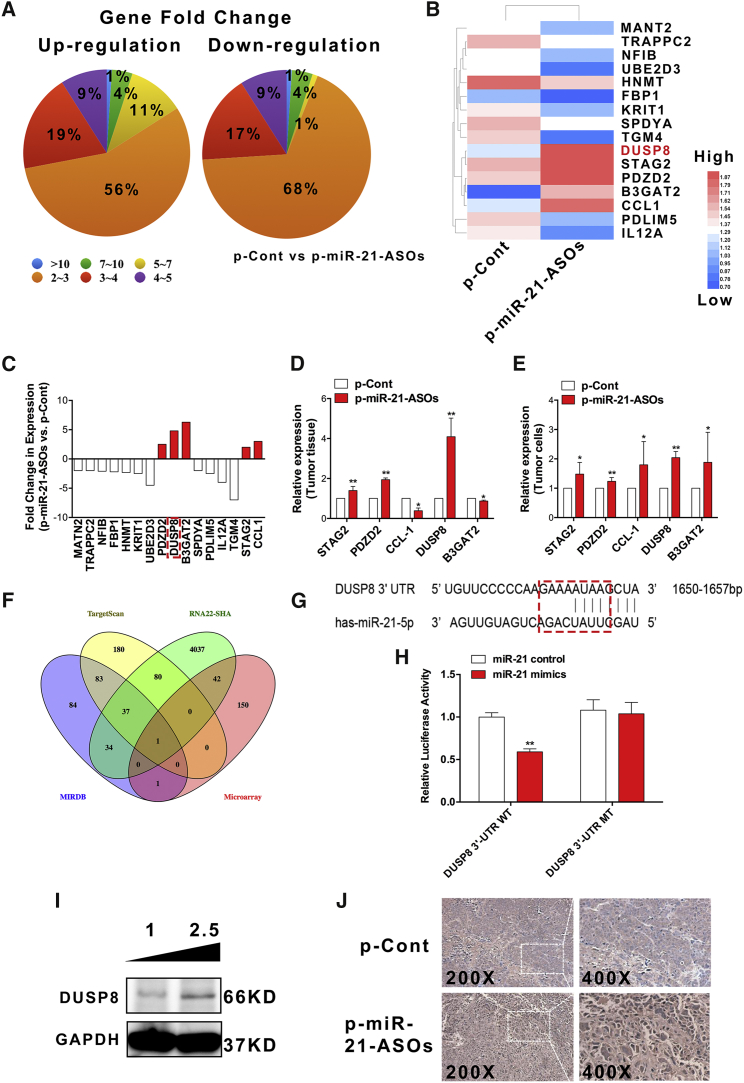
To investigate the underlying mechanism of the downregulated expression level of miR-21 by ASOs, the next experimental process was conducted (Figure S1B). We analyzed the global gene expression profiles in tumor tissues of SW620 cells between the p-miR-21-ASO group and the p-Cont group using gene expression microarray assay. The altered gene expression profiles in the p-miR-21-ASOs group are shown in a heatmap (Figures S4A and S4B). Given a 2-fold change and p < 0.05 (up and down) in differential expression as a cutoff, the number of altered genes was reduced to 1,726, 773 of which were upregulated and 953 downregulated (Figure 3A); 4-fold-changed genes are shown in Table S2. Gene Ontology (GO) analysis showed that the upregulation of differentially expressed genes were significantly enriched in the first 15 GO items in the three parts of a biological process, cell component, and molecular functions (Figure S4C).
Figure 3.
ASOs against miR-21 Altered Global Gene Expression of Tumor Tissue, and DUSP8 Was a New Target of miR-21
Human CRC SW620 cells were injected subcutaneously into the right flank of BALB/c nude mice (n = 10 per group). Seven days later, the plasmid of p-miR-21-ASOs (100 μg) or p-Cont (100 μg) was locally given by subcutaneous injection into the tumor tissues of nude mice four times every 3 days. Six days after the last injection, tumor tissues were collected. Global gene expression was analyzed and matched using cDNA microarray chip. (A) Gene fold change of all upregulation and downregulation genes. (B) Prediction of 18 target genes using MIRDB (http://www.mirdb.org) and TargetScan (http://www.targetscan.org) and (C) calculation of fold change in expression from microarray chip. (D) Expression of five genes, which were upregulated, was determined using real-time PCR. (E) Human CRC SW620 cells were transiently transfected with p-miR-21-ASOs (2.5 μg) or p-Cont (2.5 μg) in vitro. Forty-eight hours later, cells were collected, and expression of the indicated genes was detected using real-time PCR. (F) TargetScan, RNA22-SHA (https://cm.jefferson.edu/rna22/), and MIRDB had intersection elements with microarray chip using Venn analysis. (G) Putative miR-21-5p-binding sites in the 3′-UTR of human DUSP8 and (H) the result of dual luciferase reporter assay were shown. The protein level of DUSP8 in tumor tissues was analyzed using (I) western blot and (J) immunohistochemical staining (200× and 400×). *p < 0.05 and **p < 0.01.
To further elucidate the potential molecular mechanism through which downregulated expression level of miR-21 by ASOs affected the growth of CRC cells, we used TargetScan and MIRDB software to screen out 16 genes, 5 of which were upregulated, the putative target genes of miR-21 PDZD2, DUSP8, B3GAT2, STAG2, and CCL1, which are closely correlated with proliferation, apoptosis, and metastasis in tumor cells according to previous research28, 29, 30, 31, 32 (Figures 3B and 3C).
DUSP8 Is a New Direct Target of miR-21
To further investigate the putative target of miR-21, we verified the expression of these five predicted target genes on tumor tissues and tumor cells, respectively. Unexpectedly, real-time PCR assay showed that only DUSP8, one target among all predicted target genes of miR-21, was significantly upregulated both in tumor tissues in the p-miR-21-ASO group (Figure 3D; p < 0.05) and in the p-miR-21-ASO-transfected tumor cells (Figure 3E; p < 0.05). Interestingly, a spurious finding was that many predicted target genes of miR-21 from TargetScan, RNA22-SHA, and MIRDB had intersection elements with microarray chip, which indicated that DUSP8 was the only target molecule of miR-21 through Venn analysis (Figure 3F). Further analysis showed that miR-21 could directly bind to the 3′-UTR of DUSP8 mRNA (Figure 3G). Moreover, we performed western blot and immunohistochemistry assay to detect the expression of DUSP8 on protein levels in tumor tissues and cells and obtained similar results (Figures 3I and 3J). In addition, dual luciferase reporter assay showed that miR-21 could bind to the 3′-UTR of DUSP8 mRNA (Figure 3H; p < 0.05). Collectively, our data indicated that downregulated expression of miR-21 by ASOs could affect the growth of human CRC cells, which could be strongly linked to the upregulated expression of DUSP8, a new target molecule of miR-21.
Overexpression of DUSP8 Suppressed the Proliferation and Migration of Human CRC Cells In Vitro
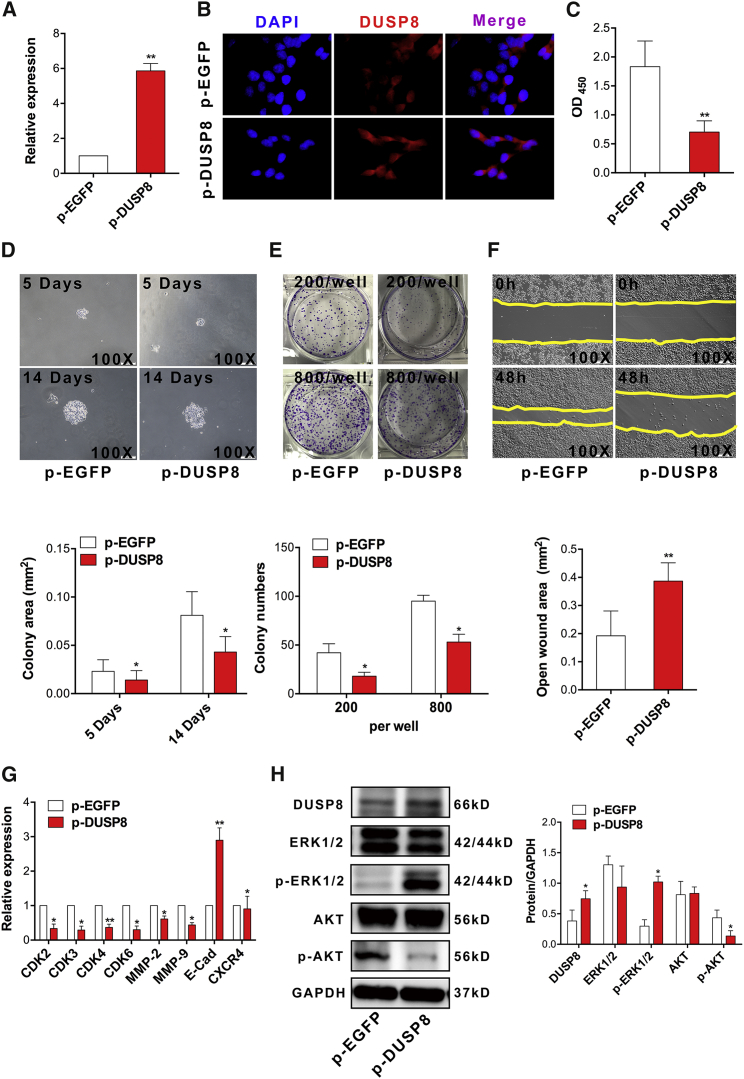
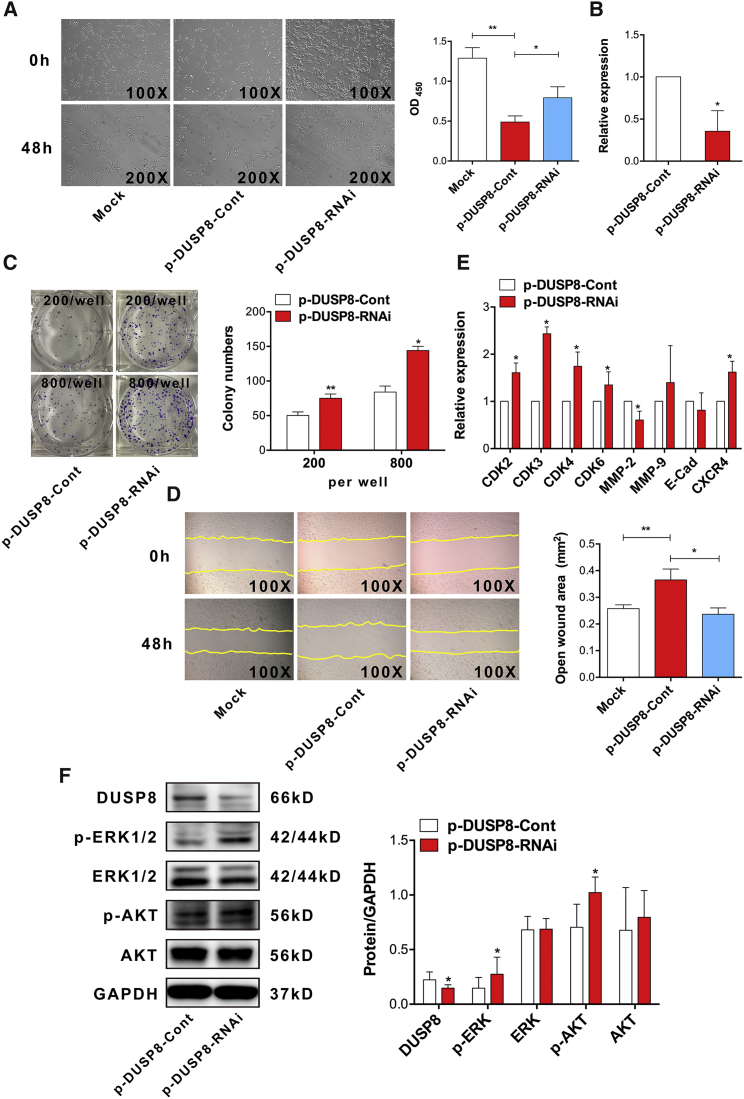
Previous works showed that DUSP8, a member of protein tyrosine phosphatase (PTP) family, played a vital part in the progression of cancers such as leukemia.33, 34 However, knowledge on the possible role of DUSP8 in the development of CRC is still limited. To explore whether DUSP8 plays a critical biological role in the proliferation and migration of CRC cells, we constructed a eukaryotic expression vector of DUSP8 (termed p-DUSP8) and then transiently transfected p-DUSP8 into CRC SW620 cells in vitro. As expected, real-time PCR assay showed that the expression level of DUSP8 significantly increased in the p-DUSP8 transfection group compared with the control group (Figure 4A; p < 0.05). Moreover, immunofluorescence assay produced a consistent result (Figure 4B). Importantly, CCK-8 assay showed that the proliferation of SW620 cells decreased significantly (Figure 4C; p < 0.05). Moreover, we found that the colony capability of single-cell and colony numbers was attenuated (Figures 4D and 4E; p < 0.05). Finally, wound-healing assay also showed that the migration ability of cells was abrogated (Figure 4F; p < 0.05).
Figure 4.
Overexpression of DUSP8 Suppressed the Proliferation and Migration of Human Colorectal Carcinoma Cells In Vitro
Human CRC SW620 cells were transiently transfected with p-DUSP8 (2.5 μg) or p-EGFP (2.5 μg) in vitro. Forty-eight hours later, (A) the relative expression of DUSP8 was determined using real-time PCR, (B) the protein level of DUSP8 was examined using immunofluorescence, and (C) the proliferation of cells was detected using the CCK-8 assay. (D) The colony capability of a single cell was observed using microscopy (magnification 100×) and the colony area (square millimeters) was calculated using ImageJ software. (E) Colon formation assay also was performed and calculated at the indicated time point. (F) Migration of SW620 cells was performed using wound-healing assay (magnification 100×), and the open wound area (square millimeters) also was calculated using ImageJ software. (G) Expression of indicated cyclin-dependent kinases and metastasis-related molecules was detected using real-time PCR. (H) Protein expression of DUSP8, ERK, p-ERK, AKT, and p-AKT also was detected by western blot, and the gray intensity value was calculated using ImageJ software. Representative data from three independent experiments are shown. *p < 0.05 and **p < 0.01.
Meanwhile, to further verify the role of DUSP8 in the proliferation and migration of CRC, we detected the expression of CDKs including CDK2, CDK3, CDK4, and CDK6, as well as metastasis-related molecules including MMP-2, MMP-9, E-cadherin, and CXCR4. The data showed that compared with those in the control group, the expression of all of these molecules (E-cadherin increased obviously) decreased significantly in the p-DUSP8 transfection group (Figure 4G; p < 0.05). Finally, we also analyzed the possible change in the transduction of the AKT and ERK pathway. Western blotting assay showed that the expression level of DUSP8 protein was significantly increased in the p-DUSP8 transfection group (Figure 4H; p < 0.05), consistent with our above data. Importantly, we noticed that the expression of p-AKT decreased remarkably, but p-ERK increased unmistakably (Figure 4H; p < 0.05). Combining these data indicated that DUSP8, as a new oncogene, could suppress the proliferation and migration of CRC cells by regulating the AKT/ERK signaling pathway.
Downregulation of DUSP8 Abrogated the Effects of ASOs against miR-21 on the Proliferation and Migration of Human CRC Cells
To further explore whether upregulation of DUSP8 contributed to the suppressive effect of ASOs against miR-21 on human CRC cells, we transiently co-transfected the plasmids of p-miR-21-ASOs and p-DUSP8-RNAi into SW620 cells and observed the possible change on cell proliferation and migration. As shown in Figure 5A, the proliferation of SW620 cells increased significantly in the p-miR-21-ASOs/p-DUSP8-RNAi co-transfection group compared with those in the p-miR-21-ASOs/p-DUSP8-Cont co-transfection group, in contrast to the above data. Moreover, we also determined the expression level of DUSP8, which was decreased unmistakably (Figure 5B; p < 0.05). Further analysis showed that the colony numbers and migration ability of SW620 cells in the p-miR-21-ASOs/p-DUSP8-RNAi co-transfection group were elevated significantly compared with those in the p-miR-21-ASOs/p-DUSP8-Cont co-transfection group (Figures 5C and 5D; p < 0.05). Consistently, real-time PCR showed that the expression of CDKs, including CDK2, CDK3, CDK4, and CDK6, was elevated significantly in the p-miR-21-ASOs/p-DUSP8-RNAi co-transfection group (Figure 5E; p < 0.05). Moreover, the expression of metastasis-related molecules including MMP-2 and CXCR4 was also elevated remarkably (Figure 5E; p < 0.05). Finally, we also analyzed the expression of p-AKT and p-ERK on SW620 cells in two groups. The data showed that the expression of DUSP8 on protein level decreased unmistakably (Figure 5F; p < 0.05). Importantly, the level of both p-AKT and p-ERK also significantly increased (Figure 5F; p < 0.05). Combing these results demonstrated that the upregulation of DUSP8 was critical for the effect of ASOs against miR-21 on the proliferation and migration of human CRC cells.
Figure 5.
Downregulation of DUSP8 Abrogated the Effects of ASOs against miR-21 on the Proliferation and Migration of Human Colorectal Carcinoma Cells In Vitro
(A) Human CRC SW620 cells were transiently co-transfected with p-miR-21-ASOs (2.0 μg) and p-DUSP8-RNAi (0.5 μg) or p-miR-21-ASOs (2.0 μg) and p-DUSP8-Cont (0.5 μg) in vitro. Forty-eight hours later, the morphology of cells was observed using microscopy (magnification 100× and 200×); the proliferation of cells was detected using the CCK-8 assay. (B) Real-time PCR detected the relative expression of DUSP8. (C) Colon formation assay also was performed and calculated at the indicated time point. (D) The migration of SW620 cells was performed using wound-healing assay (magnification 100×), and the open wound area (square millimeters) also was calculated using ImageJ software. (E) Expression of indicated cyclin-dependent kinases and metastasis-related molecules was detected using real-time PCR. (F) Protein expression of DUSP8, ERK, p-ERK, AKT, and p-AKT also were detected using western blot, and the gray intensity value was calculated using ImageJ software. Representative data from three independent experiments are shown. *p < 0.05 and **p < 0.01.
Discussion
In this study, the data first showed that ASOs against miR-21 could effectively inhibit the growth and metastasis of CRC in vivo. Notably, we further demonstrated that upregulation of DUSP8, a novel target of miR-21, contributed to the effects of downregulating the expression of miR-21 by ASOs on the growth and metastasis of CRC cells, accompanied by altered transduction of related signaling pathways, including the AKT and ERK pathway. Finally, overexpression of DUSP8 could remarkably suppress the proliferation and migration of CRC cells in vitro.
miR-21 was one of the first miRNAs detected in the human genome and has been associated with many types of cancers, including CRC.35, 36 More important, high miR-21 expression is associated with poor survival and poor therapeutic outcomes, and miR-21 also has been shown to correlate with prognosis in patients with stage II CRC.37, 38, 39 This previous research revealed that the expression level of miR-21 is closely associated with survival and prognosis in patients in different tumor-node-metastasis (TNM) stages. In our previous work, we found that ASOs against miR-21 could reduce the growth and metastasis of human CRC cells in vitro.24 In the present study, we extended our previous finding by demonstrating that ASOs could obviously abrogate the expression of miR-21 in tumor tissues and subsequently reduce the growth and metastasis of CRC cells in vivo, accompanied by altered expression of cell CDK factors and metastasis-related molecules such as CDK and MMP family members. These data suggest that miR-21-ASO can inhibit the growth and metastasis of CRC cells in vitro and in vivo. In line with our work, Li et al.40 reported that downregulation of the oncogenic miR-21 by ASOs resulted in upregulation of the tumor-suppressor genes PDCD4 and PTEN and the suppression of epithelial-mesenchymal transition, which inhibited the proliferation and reduced the clonal formation, migration, and invasion of pancreatic cancer cells in vitro. Moreover, Ross et al.41 described the preclinical evaluation of AZD4785, a high-affinity constrained ethyl-containing therapeutic ASO targeting KRAS mRNA. Systemic delivery of AZD4785 to mice bearing KRAS-mutant cell lines of non-small-cell lung cancer (NSCLC) using xenografts or patient-derived xenografts resulted in inhibition of KRAS expression in tumors and antitumor activity. Combing these data might highlight the promising prospect of ASOs against distinct miRNA molecules in cancer gene therapy. Therefore, further analysis of the distribution and potential toxicity of ASOs in vivo, which was not investigated in this study, might be of great value for the application of ASOs against miR-21 in gene therapy against clinical CRC.
Accumulating evidence shows that the molecular mechanism of miR-21 regulating proliferation and migration of CRC cells was very complicated. For instance, Ferraro et al.42 reported that miR-21 could regulate the metastasis of CRC cells through ITGB4 expression in vitro. Moreover, Peacock et al.43 found that miR-21 could control PDCD4 expression in CRC cells in vitro. In our previous work, we found that miR-21 could regulate the growth and metastasis of CRC cells through regulating PTEN expression.24 Unexpectedly, in the present study, global gene expression analysis showed that DUSP8, but not PTEN, was upregulated in CRC tumor tissue in vivo. Most important, downregulation of DUSP8 could abrogate the effect of ASOs against miR-21 on the growth and metastasis of CRC cells. These data demonstrate that DUSP8, a novel target of miR-21, contributes to the effects of ASOs against miR-21 expression in CRC. Similarly, Bleau et al.44 found that miR-146a targeted c-met and abolished CRC liver metastasis, and Xiong et al.45 reported that miR-7 regulated the growth of human lung cancer cells through PA28. In our previous work, we found that miR-7 could regulate the growth and metastasis of human lung cancer cells in vivo and in vitro through NUDFA4.46 Thus, we proposed that it reflects the complexity of miRNAs contributing to the development of cancers, and the different experimental setting in a distinct research study might also be responsible for this controversial phenomenon. Thus, in successive research work, further investigation of the connection between miR-21 and different targets in CRC is very important to illustrate the biological function of miR-21 in the development of CRC.
Dual-specificity phosphatase 8 (DUSP8), a novel PTP (a homolog of vaccinia virus H1 phosphatase gene clone 5 [hVH-5]), was expressed predominantly in the adult brain, heart, and skeletal muscle; it shared sequence similarity with a subset of PTPs that negatively regulate the activity of mitogen-activated protein kinases (MAPKs) through selective dephosphorylation.47 Recent studies showed that DUSP8 has broad substrate specificity aimed at p38MAPK, ERK1/2 MAPK, and JNK MAPK, which was involved in multiple biological processes, including cancers.48 Until now, the role of DUSP8 in the progression of cancers has been largely unknown.33, 34, 49, 50 Heminger et al.51 found that the expression of DUSP8 was induced by ionizing radiation and glycolytic inhibitor 2-deoxyglucose in human glioma cells, indicating that DUSP8 might be as a suppressor in cancer. However, Lim et al.52 reported that aberrant CpG methylation in DUSP8 was detected in ovarian cancer cell lines, and methylation of DUSP8 was observed in 15%–38% of primary ovarian tumors. Moreover, DUSP8 methylation was an independent predictor of favorable progression-free survival (PFS) and overall survival (OS), indicating that DUSP8 methylation, in a sense, is a favorable clinical outcome marker. In the present study, we found that overexpression of DUSP8 could obviously inhibit the proliferation and migration of CRC cells. Furthermore, overexpression of DUSP8 could significantly alter the transduction of the AKT and ERK signaling pathway, which was consistent with the effect of ASOs against miR-21 in CRC. These data suggest that DUSP8 might be a novel tumor suppressor gene in growth and metastasis of CRC cells. However, the expression of DUSP8 in clinical CRC and its potential connection with miR-21 expression, as well as the development of CRC, remain to be fully elucidated in the future, which was very helpful for the validation of the substantial role of the miR-21/DUSP8 axis in the development of CRC.
In conclusion, our study is the first to reveal that ASOs against miR-21 could significantly repress the growth and metastasis of human CRC in vivo, which was closely associated with upregulation of DUSP8, a novel target of miR-21, and altered transduction of related signaling pathways, including AKT and ERK (Figure 6). These data indicate that ASOs against miR-21 might be an ideal strategy for targeting therapy in human CRC, providing a preliminary experimental basis for gene therapy of miRNAs against clinical CRC.
Figure 6.
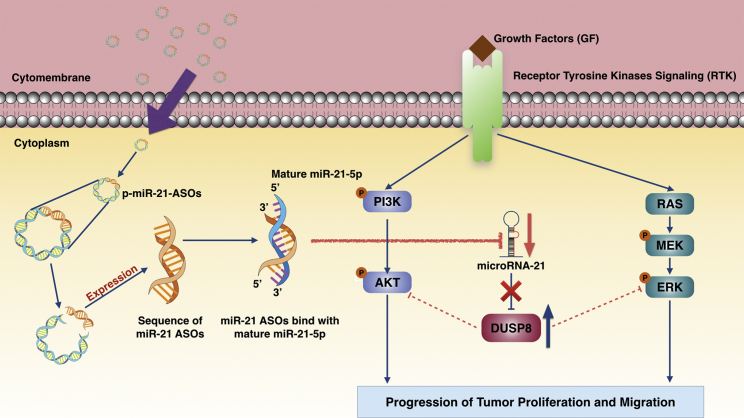
Proposed Model Representing the Regulation of Anti-tumor by ASOs against miR-21
The plasmid of antisense oligonucleotides (ASOs) against miR-21 entered tumor cell cytoplasm through the cytomembrane and expressed the sequence of miR-21 ASOs binding with mature miR-21-5p, which inhibited the expression of mature miR-21-5p. Downregulation of miR-21 endowed the upregulation of DUSP8 and subsequently attenuated the transduction of the AKT and ERK pathway, which repressed the progression of tumor proliferation and migration in vivo.
Materials and Methods
Eukaryotic Vector Construction
Plasmid pcDNA-6.2-miR-21-ASO (termed p-miR-21-ASO) was used per our previous description,24 and the map of p-miR-21-ASOs is shown in Figure S1A. The gene for DUSP8 (NM_004420.2) was expanded by PCR from human cDNA derived from FHC cells using a forward primer (CCGGAATTCATGGCTGGGGACCGGCTCCC) and a reverse primer (CGCGGATCCTCAGGACACCTCGATGACCT) and then subcloned into EcoR I and BamH I sites of pEGFP-N1 vector (Clontech, Kyoto, Japan). The shRNA sequence against human DUSP8 gene (NM_004420.2) was ligated into pLVX-shRNA1 vector (Clontech), termed p-DUSP8-RNAi. The Sh-DUSP8 sense strand was 5′-GATCCGCCATCGAGTTCATCGATAATTCAA- -GAGATTATCGATGAACTCGATGGTTTTTTACGCGTG-3′; the antisense strand was 5′-GCGGTAGCTCAAGTAGCTATTAAGTTCTCTAATAGCTACTTGAGCTACCAAAAAAT-GCGCACTTAA-3′. For negative control vector p-EGFP or p-DUSP8-Cont was used. All sequences were confirmed by sequencing.
Cell Culture and Transient Transfection
Human colon carcinoma cell line SW620 was obtained from the Shanghai Institutes for Biological Sciences Cell Resource Center (catalog number TCHu101, the latest identification in 2017) and cultured in Leibovitz’s L-15 medium (Life Technologies,Grand Island, NY, USA), containing 100 IU/mL penicillin, 100 μg/mL streptomycin, 20 mM glutamine, and 10% fetal bovine serum (FBS; GIBCO, Grand Island, NY, USA), incubated in a humidified incubator at 37°C without CO2. For transfection, cells were seeded at 80%–90% confluence and 18 hr later transiently transfected with indicated plasmids using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Cells were harvested 48 hr post-transfection for following experiments.
Xenograft Model Antitumor Assay in Nude Mice
For the in vivo study, athymic nude mice (BALB/C, 4–6 weeks old, female) were purchased from the Shanghai Laboratory Animal Research Center (Shanghai, China). Animals were handled in accordance with the Guidelines for the Care and Use of Laboratory Animals (Ministry of Health, People’s Republic of China, 1998), and all experimental procedures were in accordance with the ethical guidelines of Shanghai Medical Laboratory Animal Care and Use Committee (permit number 2013018). For the preparation of the subcutaneous xenograft model, cells were subcutaneously implanted into the right flanks of nude mice at 3.5 × 106 SW620 cell density (n = 20). Then, 7 days after tumor cell inoculation with confirmation of successful maturation of tumors, 20 mice were divided randomly into two groups (10 mice per group). A 100 μg plasmid of p-miR-21-ASOs or p-Cont was given locally by direct injection into the tumor tissues of nude mice at four times every 3 days. Tumor volume was calculated using the formula volume (mm3) = (length × width2)/2. After 18 days of treatment, all mice were sacrificed. Tumor tissues were peeled off and divided into two parts. One part was stored at −80°C for further experiments, and another section was fixed in formalin and embedded in paraffin, which was used for H&E staining.
RNA and miRNA Isolation and qRT-PCR
Total RNA was isolated from cell lines or tumor tissues with RNAiso Plus reagents (Takara, Ohtsu, Japan) according to the manufacturer’s instructions. The TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) and PrimeScripRT Reagent Kit (Takara, Kusatsu, Japan) were used for reverse transcription of miRNA-21 and indicated genes, respectively. qRT-PCR was performed to quantify mature miRNA-21 expression and mRNA expression using the miR-21 probe of TaqMan (Life Technologies) and SYBR Premix Ex Taq II (Takara, Kusatsu, Japan). Data collection was accomplished using the CFX96 Real-Time System (Bio-Rad). Relative expression level was normalized and calculated using the relative quantification (2−ΔΔCt) method. The indicated genes and their primer sequences are shown in Table S1.
Cell Proliferation Assay
Cells from each group were plated in 96-well plates at a density of 1 × 104 cells/well containing complete medium without any antibiotics. After incubating for 18 hr, cells were transfected with indicated plasmids using Lipofectamine 3000 for 48 hr. Cells were detected using Cell Counting Kit-8 (CCK8; Beyotime Institute of Biotechnology, Jiangsu, China). In brief, 10 μL CCK8 solution was added into each well and incubated at 37°C for 2 hr. Then absorbance was measured using a multifunctional microplate reader at 450 nm with 600 nm as a reference.
Gene Expression Microarray and Analysis
NanoDrop ND-1000 was used to measure RNA quantity and quality. RNA integrity was assessed using standard denaturing agarose gel electrophoresis. The Whole Human Genome Oligo Microarray is a broad view that represents all known genes and transcripts in the human genome. Sequences were compiled from a broad source survey and then verified and optimized by alignment to the assembled human genome. Sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technologies). Briefly, total RNA from each sample was linearly amplified and labeled with Cy3-UTP. The labeled cRNAs were purified using the RNeasy Mini Kit (QIAGEN). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured using NanoDrop ND-1000. One microgram of each labeled cRNA was fragmented by adding 11 μL 10 × blocking agent and 2.2 μL of 25 × fragmentation buffer, then heated at 60°C for 30 min, and finally 55 μL 2 × GE hybridization buffer was added to dilute the labeled cRNA. Hybridization solution (100 μL) was dispensed into the gasket slide and assembled to the gene expression microarray slide. The slides were incubated for 17 hr at 65°C in an Agilent hybridization oven. The hybridized arrays were washed, fixed, and scanned using the Agilent DNA Microarray Scanner (part number G2505C).
Agilent Feature Extraction software (version 11.0.1.1) was used to analyze acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX version 12.1 software package (Agilent Technologies). After quantile normalization of the raw data, genes that at least one of two samples had flags of detected (“all targets value”) were chosen for further data analysis. Differentially expressed genes with statistical significance between the two groups were identified using volcano plot filtering. Differentially expressed genes between the two samples were identified using fold change filtering. Hierarchical clustering was performed using R scripts. GO analysis was performed using the standard enrichment computation method. Global gene expression array data are available in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under accession number GEO: GSE109592.
Dual Luciferase Reporter Assay
By using the TargetScan database, a potential binding site of hsa-miR-21 was predicted at position 1650-1657 (5′-AAUAAGCU-3′) of DUSP8 3′-UTR. A fragment of 200 bp containing wild-type DUSP8 3′-UTR (5′-AAUAAGCU-3′) or a random mutation sequence of mutant DUSP8 3′-UTR (5′-AGAUUAGA-3′) was directly synthesized (Sangon, Shanghai, China). Two fragments were ligated to pEZX-FR02 reporter vector (GeneCopoeia, Rockville, MA, USA). Then, the wild-type (DUSP8 3′-UTR WT) or mutant reporter vector (DUSP8 3′-UTR MT) was co-transfected into 293T cells in 12-well plates with 100 nm miR-21 mimics or miR-21 mimics negative control by Lipofectamine 3000 (Invitrogen). The Dual-Luciferase Reporter Assay System (Promega) was used to detect the luciferase activity of Firefly and Renilla, and the ratio was normalized.
Plate Colony Formation Assay
Cells were trypsinized 48 hr post-transfection to prepare single-cell suspension and seeded in six-well plates (200 and 800 cells/well). Then the cells were cultured in a humidified atmosphere without CO2 at 37°C. The medium was changed every 5 days. Two weeks later, the cells were washed with PBS three times, immobilized with 4% paraformaldehyde for 20 min, and then stained with 0.2% crystal violet for 30 min. Next, the colonies were carefully rinsed with PBS until the background was clear. Thereafter the colony diameter and number were statistically analyzed. The colony formation rate was calculated as (number of colonies/number of incubating cells) × 100%.
Tissue Histopathology
Tumor tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and cut into 5-μm-thick sections. Sections were stained with H&E (Beyotime Institute of Biotechnology, Jiangsu, China), and images were taken using a light microscope (Olympus, Tokyo, Japan). All tumor tissues at original magnification 200× and 400× were examined for each sample.
In Vitro Wound-Healing Assay
Wound-healing assay was used to examine for cell migration and invasion. Cells were plated in 24-well plates at a density of 2 × 105 cells/well containing complete medium without any antibiotics and cultured for 18 hr. Then, cells from each well were transiently transfected indicated plasmids using Lipofectamine 3000 for 6 hr and wounded with pipette tips. Thereafter cells were carefully washed with PBS three times to remove cell debris, and the medium was refreshed with 0.1% FBS. The scratch areas were observed under a microscope with magnification 100×, and the migrated cells were counted 48 hr post-transfection.
Western Blot
Cells and tumor tissues were lysed with protein extraction reagent (KeyGEN BioTECH, Jiangsu, China) according to the manufacturer’s instructions, and total cell lysate protein samples were obtained. Then samples were equally loaded on 10% SDS-polyacrylamide gel, electrophoresed, and transferred to polyvinylidene difluoride membranes (Bio-Rad). After blocking with 5% BSA in Tris-buffered saline (TBS) with 0.1% Tween 20 (TBST) for 2 hr, the membranes were incubated with the primary antibodies rabbit-polyclonal anti-human DUSP8 (1:2,000; Abcam, Cambridge, UK), rabbit-monoclonal anti-human ERK (1:1,000; Cell Signaling Technology (Danvers, MA, USA)), rabbit-polyclonal anti-human p-ERK (1:2,000; Cell Signaling Technology), rabbit-monoclonal anti-human AKT (1:1,000; Cell Signaling Technology), rabbit-monoclonal anti-human p-AKT (1:2,000; Cell Signaling Technology), or rabbit-monoclonal anti-human GAPDH (1:2,000; Cell Signaling Technology) at 4°C overnight. After the overnight incubation with the primary antibodies, membranes were washed in TBST three times and subsequently probed with a secondary anti-rabbit Ab-conjugated to horseradish peroxidase (HRP) for 1 hr (1:2,000; Cell Signaling Technology). Finally, the signals were detected and analyzed using the chemiluminescence image system (Bio-Rad).
Immunofluorescence
Ki-67 (proliferating cell nuclear antigen) was used to assess the proliferation of tumor cells in tumor tissues after local injection of the plasmid of p-miR-21-ASOs or p-Cont. All frozen sections of tumor tissues were cut into 5-μm-thick slides. After washing with PBS three times and blocking with 10% normal goat serum for 30 min at room temperature and incubation with rabbit-monoclonal anti-human anti-Ki-67 antibody (1:100; Santa Cruz Biotechnology) at 4°C overnight, the sections were washed in PBS and incubated with a secondary antibody of Alexa Fluor 594-conjugated goat-anti-rabbit IgG (1:250; Invitrogen) for 1 hr in the dark at room temperature. After washing in PBS three times, the slides were counterstained, mounted with SlowFade Gold Antifade Reagent with DAPI (40,6-diamidino-2-phenylindole; Thermo Fisher Scientific), and left for 10 min in the dark at room temperature before examination by fluorescence microscopy (Zeiss Axioplan 2).
To detect the expression of DUSP8, cells were plated in 24-well plates with glass slides at a density of 2 × 105 cells/well containing complete medium without any antibiotics and cultured for 18 hr. Then, cells from each well were transiently transfected with indicated plasmids using Lipofectamine 3000 for 48 hr. After that, the cells were fixed, permeabilized and blocked with Image-it Fix-Perm Kit (Life Technologies), and incubated with rabbit-polyclonal anti-human DUSP8 (1:500; Abcam) at 4°C for overnight, followed by anti-rabbit IgG conjugated Alexa Fluor 647 (1:200; Cell Signaling Technology) for 1 hr. After washing in PBS three times, the slides were counterstained, mounted with Prolong Gold antifade reagent with DAPI (Invitrogen), and left overnight in the dark at room temperature. Cells were observed and photographed by confocal microscopy (Leica TCS SP8 X).
Tissue Immunohistochemistry
Immunohistochemical staining was performed following standard procedures. Tumor tissues were sliced into 5-μm-thick sections and fixed with formalin and embedded in paraffin. Then the slides were dewaxed, rehydrated, and fixed. Antigen epitope repairing was performed in 10 mmol/L citric acid buffer (pH 6.0), and endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol. After incubation with rabbit-polyclonal anti-human DUSP8 antibody overnight at 4°C, the sections were washed in PBS and incubated with a secondary antibody of polymer HRP conjugate (ZSGB-Bio, Beijing, China) for 1 hr. Finally, the sections were further incubated using the Liquid DAB Large-Volume Substrate-Chromogen System (ZSGB-Bio) and counterstained with hematoxylin. Slides were imaged under a light microscope (Olympus) at 200× and 400× magnification.
Statistical Analysis
All data are presented as mean ± SD from at least three independent experiments. Unpaired Student’s t test (two-tailed) for two groups or one-way ANOVA with Bonferroni’s correction for three or more groups was performed to evaluate statistical significance using GraphPad Prism 6 software. Differences were considered statically significant when p values were less than 0.05.
Author Contributions
T.D. and P.C. performed experiments, analyzed data, and wrote the paper. Y.Z., C.C., and J.Z. performed experiments and analyzed data. M.G. performed experiments. H.W. and Z.H. wrote the paper. L.X. conceived and designed experiments, analyzed data, and wrote the paper. All authors reviewed the paper.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
We thank the Department of Immunology, Zunyi Medical University, and the Special Key Laboratory of Gene Detection and Therapy of Guizhou Province for providing the experimental platform. This work was supported by the Applied Basic Research Major Project of Guizhou Province ([2015]2003), the Program for High-Level Innovative Talents of Guizhou Province (QKH-RC-2016-4031), the National Natural Science Foundation of China (31760258), the Project of Guizhou Provincial Department of Science and Technology (QKH-JC-2018-1428), the Program for New Century Excellent Talents in University, Ministry of Education of China (NCET-12-0661), and the Program for Excellent Young Talents of Zunyi Medical University (15ZY-001).
Footnotes
Supplemental Information includes Supplemental Materials and Methods, four figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.09.004.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Miller K.D., Siegel R.L., Lin C.C., Mariotto A.B., Kramer J.L., Rowland J.H., Stein K.D., Alteri R., Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J. Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 3.Storm E.E., Durinck S., de Sousa e Melo F., Tremayne J., Kljavin N., Tan C., Ye X., Chiu C., Pham T., Hongo J.A. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature. 2016;529:97–100. doi: 10.1038/nature16466. [DOI] [PubMed] [Google Scholar]
- 4.Ye L., Jiang T., Shao H., Zhong L., Wang Z., Liu Y., Tang H., Qin B., Zhang X., Fan J. MiR-1290 is a biomarker in DNA-mismatch-repair-deficient colon cancer and promotes resistance to 5-fluorouracil by directly targeting hMSH2. Mol. Ther. Nucleic Acids. 2017;7:453–464. doi: 10.1016/j.omtn.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yun J., Mullarky E., Lu C., Bosch K.N., Kavalier A., Rivera K., Roper J., Chio I.I., Giannopoulou E.G., Rago C. Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science. 2015;350:1391–1396. doi: 10.1126/science.aaa5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrasco R.A., Stamm N.B., Marcusson E., Sandusky G., Iversen P., Patel B.K. Antisense inhibition of survivin expression as a cancer therapeutic. Mol. Cancer Ther. 2011;10:221–232. doi: 10.1158/1535-7163.MCT-10-0756. [DOI] [PubMed] [Google Scholar]
- 7.Conde J., Oliva N., Zhang Y., Artzi N. Local triple-combination therapy results in tumour regression and prevents recurrence in a colon cancer model. Nat. Mater. 2016;15:1128–1138. doi: 10.1038/nmat4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin A.A. Targeting therapeutic oligonucleotides. N. Engl. J. Med. 2017;376:86–88. doi: 10.1056/NEJMcibr1613559. [DOI] [PubMed] [Google Scholar]
- 9.Scoles D.R., Meera P., Schneider M.D., Paul S., Dansithong W., Figueroa K.P., Hung G., Rigo F., Bennett C.F., Otis T.S., Pulst S.M. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature. 2017;544:362–366. doi: 10.1038/nature22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tangudu N.K., Verma V.K., Clemons T.D., Beevi S.S., Hay T., Mahidhara G., Raja M., Nair R.A., Alexander L.E., Patel A.B. RNA interference using c-Myc-conjugated nanoparticles suppresses breast and colorectal cancer models. Mol. Cancer Ther. 2015;14:1259–1269. doi: 10.1158/1535-7163.MCT-14-0970. [DOI] [PubMed] [Google Scholar]
- 11.Kasiri S., Ansari K.I., Hussain I., Bhan A., Mandal S.S. Antisense oligonucleotide mediated knockdown of HOXC13 affects cell growth and induces apoptosis in tumor cells and over expression of HOXC13 induces 3D-colony formation. RSC Advances. 2013;3:3260–3269. doi: 10.1039/C2RA22006G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolfi C., De Simone V., Colantoni A., Franzè E., Ribichini E., Fantini M.C., Caruso R., Monteleone I., Sica G.S., Sileri P. A functional role for Smad7 in sustaining colon cancer cell growth and survival. Cell Death Dis. 2014;5:e1073. doi: 10.1038/cddis.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F., Fang Q., Chen C., Zhou L., Li H., Yin Z., Wang Y., Zhao C.X., Xiao X., Wang D.W. Recombinant adeno-associated virus-mediated delivery of microRNA-21-3p lowers hypertension. Mol. Ther. Nucleic Acids. 2018;11:354–366. doi: 10.1016/j.omtn.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie T., Huang M., Wang Y., Wang L., Chen C., Chu X. MicroRNAs as regulators, biomarkers and therapeutic targets in the drug resistance of colorectal cancer. Cell. Physiol. Biochem. 2016;40:62–76. doi: 10.1159/000452525. [DOI] [PubMed] [Google Scholar]
- 15.Mohammadi A., Mansoori B., Baradaran B. The role of microRNAs in colorectal cancer. Biomed. Pharmacother. 2016;84:705–713. doi: 10.1016/j.biopha.2016.09.099. [DOI] [PubMed] [Google Scholar]
- 16.Hu Y., Wang C., Li Y., Zhao J., Chen C., Zhou Y., Tao Y., Guo M., Qin N., Ren T. MiR-21 controls in situ expansion of CCR6+ regulatory T cells through PTEN/AKT pathway in breast cancer. Immunol. Cell Biol. 2015;93:753–764. doi: 10.1038/icb.2015.37. [DOI] [PubMed] [Google Scholar]
- 17.Pfeffer S.R., Yang C.H., Pfeffer L.M. The role of miR-21 in cancer. Drug Dev. Res. 2015;76:270–277. doi: 10.1002/ddr.21257. [DOI] [PubMed] [Google Scholar]
- 18.Sheedy F.J. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front. Immunol. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasti A., Mehrazma M., Madjd Z., Keshtkar A.A., Roudi R., Babashah S. Diagnostic and prognostic accuracy of miR-21 in renal cell carcinoma: a systematic review protocol. BMJ Open. 2016;6:e009667. doi: 10.1136/bmjopen-2015-009667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrović N. miR-21 might be involved in breast cancer promotion and invasion rather than in initial events of breast cancer development. Mol. Diagn. Ther. 2016;20:97–110. doi: 10.1007/s40291-016-0186-3. [DOI] [PubMed] [Google Scholar]
- 21.Nedaeinia R., Sharifi M., Avan A., Kazemi M., Rafiee L., Ghayour-Mobarhan M., Salehi R. Locked nucleic acid anti-miR-21 inhibits cell growth and invasive behaviors of a colorectal adenocarcinoma cell line: LNA-anti-miR as a novel approach. Cancer Gene Ther. 2016;23:246–253. doi: 10.1038/cgt.2016.25. [DOI] [PubMed] [Google Scholar]
- 22.Mima K., Nishihara R., Yang J., Dou R., Masugi Y., Shi Y., da Silva A., Cao Y., Song M., Nowak J. MicroRNA MIR21 (miR-21) and PTGS2 expression in colorectal cancer and patient survival. Clin. Cancer Res. 2016;22:3841–3848. doi: 10.1158/1078-0432.CCR-15-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C., Zhao L., Chen Y., He T., Chen X., Mao J., Li C., Lyu J., Meng Q.H. MicroRNA-21 promotes proliferation, migration, and invasion of colorectal cancer, and tumor growth associated with down-regulation of sec23a expression. BMC Cancer. 2016;16:605. doi: 10.1186/s12885-016-2628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao Y.J., Li Y.J., Zheng W., Zhao J.J., Guo M.M., Zhou Y., Qin N.L., Zheng J., Xu L. Antisense oligonucleotides against microRNA-21 reduced the proliferation and migration of human colon carcinoma cells. Cancer Cell Int. 2015;15:77. doi: 10.1186/s12935-015-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai W., Bai Y., Hebda L., Zhong X., Liu J., Kao J., Duan C. Calcium deficiency-induced and TRP channel-regulated IGF1R-PI3K-Akt signaling regulates abnormal epithelial cell proliferation. Cell Death Differ. 2014;21:568–581. doi: 10.1038/cdd.2013.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y., Xiao X., Zhang C., Yu W., Guo W., Zhang Z. Melatonin synergizes the chemotherapeutic effect of 5-fluorouracil in colon cancer by suppressing PI3K/AKT and NF-κB/iNOS signaling pathways. J. Pineal Res. 2016;62(2) doi: 10.1111/jpi.12380. [DOI] [PubMed] [Google Scholar]
- 27.Peng H., Dara L., Li T.W., Zheng Y., Yang H., Tomasi M.L., Tomasi I., Giordano P., Mato J.M., Lu S.C. MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology. 2013;57:2299–2313. doi: 10.1002/hep.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tam C.W., Cheng A.S., Ma R.Y., Yao K.M., Shiu S.Y. Inhibition of prostate cancer cell growth by human secreted PDZ domain-containing protein 2, a potential autocrine prostate tumor suppressor. Endocrinology. 2006;147:5023–5033. doi: 10.1210/en.2006-0207. [DOI] [PubMed] [Google Scholar]
- 29.Cotsiki M., Oehrl W., Samiotaki M., Theodosiou A., Panayotou G. Phosphorylation of the M3/6 dual-specificity phosphatase enhances the activation of JNK by arsenite. Cell. Signal. 2012;24:664–676. doi: 10.1016/j.cellsig.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 30.He H., Nilsson C.L., Emmett M.R., Marshall A.G., Kroes R.A., Moskal J.R., Ji Y., Colman H., Priebe W., Lang F.F., Conrad C.A. Glycomic and transcriptomic response of GSC11 glioblastoma stem cells to STAT3 phosphorylation inhibition and serum-induced differentiation. J. Proteome Res. 2010;9:2098–2108. doi: 10.1021/pr900793a. [DOI] [PubMed] [Google Scholar]
- 31.Solomon D.A., Kim T., Diaz-Martinez L.A., Fair J., Elkahloun A.G., Harris B.T., Toretsky J.A., Rosenberg S.A., Shukla N., Ladanyi M. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das S., Sarrou E., Podgrabinska S., Cassella M., Mungamuri S.K., Feirt N., Gordon R., Nagi C.S., Wang Y., Entenberg D. Tumor cell entry into the lymph node is controlled by CCL1 chemokine expressed by lymph node lymphatic sinuses. J. Exp. Med. 2013;210:1509–1528. doi: 10.1084/jem.20111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bermudez O., Pagès G., Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am. J. Physiol. Cell Physiol. 2010;299:C189–C202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- 34.Peti W., Page R. Molecular basis of MAP kinase regulation. Protein Sci. 2013;22:1698–1710. doi: 10.1002/pro.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shenouda S.K., Alahari S.K. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 36.Yu Y., Kanwar S.S., Patel B.B., Oh P.S., Nautiyal J., Sarkar F.H., Majumdar A.P. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis. 2012;33:68–76. doi: 10.1093/carcin/bgr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen B.S., Jørgensen S., Fog J.U., Søkilde R., Christensen I.J., Hansen U., Brünner N., Baker A., Møller S., Nielsen H.J. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin. Exp. Metastasis. 2011;28:27–38. doi: 10.1007/s10585-010-9355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J.X., Song W., Chen Z.H., Wei J.H., Liao Y.J., Lei J., Hu M., Chen G.Z., Liao B., Lu J. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol. 2013;14:1295–1306. doi: 10.1016/S1470-2045(13)70491-1. [DOI] [PubMed] [Google Scholar]
- 39.Schetter A.J., Leung S.Y., Sohn J.J., Zanetti K.A., Bowman E.D., Yanaihara N., Yuen S.T., Chan T.L., Kwong D.L., Au G.K. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Chen Y., Li J., Zhang Z., Huang C., Lian G., Yang K., Chen S., Lin Y., Wang L. Co-delivery of microRNA-21 antisense oligonucleotides and gemcitabine using nanomedicine for pancreatic cancer therapy. Cancer Sci. 2017;108:1493–1503. doi: 10.1111/cas.13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross S.J., Revenko A.S., Hanson L.L., Ellston R., Staniszewska A., Whalley N., Pandey S.K., Revill M., Rooney C., Buckett L.K. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl. Med. 2017;9:9. doi: 10.1126/scitranslmed.aal5253. [DOI] [PubMed] [Google Scholar]
- 42.Ferraro A., Kontos C.K., Boni T., Bantounas I., Siakouli D., Kosmidou V., Vlassi M., Spyridakis Y., Tsipras I., Zografos G., Pintzas A. Epigenetic regulation of miR-21 in colorectal cancer: ITGB4 as a novel miR-21 target and a three-gene network (miR-21-ITGΒ4-PDCD4) as predictor of metastatic tumor potential. Epigenetics. 2014;9:129–141. doi: 10.4161/epi.26842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peacock O., Lee A.C., Cameron F., Tarbox R., Vafadar-Isfahani N., Tufarelli C., Lund J.N. Inflammation and MiR-21 pathways functionally interact to downregulate PDCD4 in colorectal cancer. PLoS ONE. 2014;9:e110267. doi: 10.1371/journal.pone.0110267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bleau A.M., Redrado M., Nistal-Villan E., Villalba M., Exposito F., Redin E., de Aberasturi A.L., Larzabal L., Freire J., Gomez-Roman J., Calvo A. miR-146a targets c-met and abolishes colorectal cancer liver metastasis. Cancer Lett. 2018;414:257–267. doi: 10.1016/j.canlet.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Xiong S., Zheng Y., Jiang P., Liu R., Liu X., Qian J., Gu J., Chang L., Ge D., Chu Y. PA28gamma emerges as a novel functional target of tumour suppressor microRNA-7 in non-small-cell lung cancer. Br. J. Cancer. 2014;110:353–362. doi: 10.1038/bjc.2013.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei L., Chen C., Zhao J., Wang H., Guo M., Zhou Y., Luo J., Zhang J., Xu L. Targeted expression of miR-7 operated by TTF-1 promoter inhibited the growth of human lung cancer through the NDUFA4 pathway. Mol. Ther. Nucleic Acids. 2017;6:183–197. doi: 10.1016/j.omtn.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martell K.J., Seasholtz A.F., Kwak S.P., Clemens K.K., Dixon J.E. hVH-5: a protein tyrosine phosphatase abundant in brain that inactivates mitogen-activated protein kinase. J. Neurochem. 1995;65:1823–1833. doi: 10.1046/j.1471-4159.1995.65041823.x. [DOI] [PubMed] [Google Scholar]
- 48.Nunes-Xavier C., Romá-Mateo C., Ríos P., Tárrega C., Cejudo-Marín R., Tabernero L., Pulido R. Dual-specificity MAP kinase phosphatases as targets of cancer treatment. Anticancer. Agents Med. Chem. 2011;11:109–132. doi: 10.2174/187152011794941190. [DOI] [PubMed] [Google Scholar]
- 49.Hunter T., Cooper J.A. Protein-tyrosine kinases. Annu. Rev. Biochem. 1985;54:897–930. doi: 10.1146/annurev.bi.54.070185.004341. [DOI] [PubMed] [Google Scholar]
- 50.Keyse S.M. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 51.Heminger K., Jain V., Kadakia M., Dwarakanath B., Berberich S.J. Altered gene expression induced by ionizing radiation and glycolytic inhibitor 2-deoxy-glucose in a human glioma cell line: implications for radio sensitization. Cancer Biol. Ther. 2006;5:815–823. doi: 10.4161/cbt.5.7.2812. [DOI] [PubMed] [Google Scholar]
- 52.Lim S., Green J.A., Wong H., VanderBurg M.E., Crook T. DUSP7 and DUSP8 promoter hypermethylations: predictors of clinical outcomes in advanced epithelial ovarian carcinoma. J. Clin. Oncol. 2007;25(18_suppl):5501. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.