Abstract
Background
Ginsenoside Rh2 has been known to enhance the activity of immune cells, as well as to inhibit the growth of tumor cells. Although the repertoire of genes regulated by Rh2 is well-known in many cancer cells, the epigenetic regulation has yet to be determined, especially for comprehensive approaches to detect methylation changes.
Methods
The effect of Rh2 on genome-wide DNA methylation changes in breast cancer cells was examined by treating cultured MCF-7 with Rh2. Pyrosequencing analysis was carried out to measure the methylation level of a global methylation marker, LINE1. Genome-wide methylation analysis was carried out to identify epigenetically regulated genes and to elucidate the most prominent signaling pathway affected by Rh2. Apoptosis and proliferation were monitored to examine the cellular effect of Rh2.
Results
LINE1 showed induction of hypomethylation at specific CpGs by 1.6–9.1% (p < 0.05). Genome-wide methylation analysis identified the “cell-mediated immune response”-related pathway as the top network. Cell proliferation of MCF-7 was retarded by Rh2 in a dose-dependent manner. Hypermethylated genes such as CASP1, INSL5, and OR52A1 showed downregulation in the Rh2-treated MCF-7, while hypomethylated genes such as CLINT1, ST3GAL4, and C1orf198 showed upregulation. Notably, a higher survival rate was associated with lower expression of INSL5 and OR52A1 in breast cancer patients, while with higher expression of CLINT1.
Conclusion
The results indicate that Rh2 induces epigenetic methylation changes in genes involved in immune response and tumorigenesis, thereby contributing to enhanced immunogenicity and inhibiting the growth of cancer cells.
Keywords: Breast cancer, CpG methylation, epigenetics, ginsenoside, Rh2
1. Introduction
Ginsenosides are the major active chemical constituents found in the root of Panax ginseng Meyer, which has been widely used as a traditional medicine in Eastern Asian countries to promote health [1]. Ginsenosides are known to have various pharmacological activities, including antiinflammatory, antiallergic, antifatigue, antistress, and anticancer activity [2]. So far, more than 100 ginsenosides have been identified from Panax species, most of which are categorized into four types of aglycone moieties: protopanaxadiol, protopanaxatriol, oleanolic acid, and ocotillol-type [1]. Rh2 is one of the converted ginsenosides that also includes K, Rg3, and Rh1, and it has shown stronger anticancer activity compared to the major ginsenosides, such as Rb1, Rb2, and Rg1 [3], [4]. The antiinflammatory effect of Rh2 was achieved by inhibiting the production of inflammatory cytokines through blocking the mitogen-activated protein kinase and nuclear factor (NF)-κB signaling pathways [5]. In the inhibitory pathway, Rh2 inhibited the lipopolysaccharide-induced production of nitric oxide, tumor necrosis factor (TNF)-α, interleukin (IL)-1β, and IL-6 [6]. Rh2 has been shown to have anticancer activity in a variety of cancer cells without having any remarkable toxicity [7], including in lung [8], liver [9], breast [10], prostate [11], and colorectal [12] cancer cells.
The molecular pathways affected by Rh2 in cancer cells were revealed to be mainly cell proliferation- and/or apoptosis-related ones. For example, Rh2 exerted anticancer activity targeting the IL-6-induced JAK2/STAT3 pathway in human colorectal cancer cells [12]. In glioblastoma cells, Rh2 suppressed viability of the cells via cell cycle arrest and downregulation of cell adhesion proteins [13]. In a study of HepG2 hepatoma cells, Rh2 suppressed proliferation while promoting apoptosis by downregulating β-catenin through activating GSK-3β [9]. For breast cancer cells, the major affected pathway was the mitogen-activated protein kinase /NF-κB pathway in MCF-7 cells [14]. As anticipated from the specific pathways, many genes affected by Rh2 are accordingly cancer cell proliferation- and/or apoptosis-related, including transcription factors, such as AP-1, E2F1, and c-Myc, and signaling kinases, such as cyclin D1, CDK, and matrix metalloproteinases.
Regulation of gene expression by altering the methylation level at the CpG of the promoter is a key epigenetic mechanism that many tumor suppressors and oncogenes adopt during tumor development [15], [16]. Representative tumor suppressors are PTEN, BRCA1, and p16, which are hypermethylated and thereby downregulated in cancer [17], [18], [19]. Meanwhile, oncogenes such as c-Myc, H-ras, and cyclin D2 are hypomethylated and upregulated in cancer [20], [21]. Although antiproliferation and other growth-related pathways and genes have been identified in Rh2-treated cancer cells, nothing is known about the methylation change of the specific genes.
In this study, to identify genes regulated by Rh2 via CpG methylation changes in cancer cells, the breast cancer cell line MCF-7 was treated with Rh2 and a genome-wide microarray-based methylation assay was carried out after confirming the ginsenoside's ability to change global methylation. The most prominent gene interaction networks and signaling pathways were constructed from the epigenetically affected genes.
2. Materials and methods
2.1. Cell culture and treatment with Rh2
The MCF-7 breast cancer cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (Gibco BRL, Carlsbad, CA, USA). The cultured cells were seeded in 60-mm culture dishes with 50% confluence 1 day before treatment of Rh2 (LKT Labs, St. Paul, MN, USA) The cells were treated with Rh2 at concentrations of 20μM and 50μM for 24 h and then harvested for further experiments.
2.2. Cell proliferation assay
Cells treated with Rh2 in a 60-mm culture dish were harvested using 0.05% trypsin-EDTA (Gibco BRL) and washed with phosphate buffered saline. To analyze cell proliferation, 2 × 103 cells were plated onto a 96-well plate, cultured for 8 d, and then stained with WST-8 from a Cell Counting Kit-8 (Dojindo, Kumamoto, Japan). OD450 was measured on a spectrophotometer to calculate cell density. For the colony formation assay, 1.3 × 104 cells were seeded onto 60-mm cell culture dishes. Following treatment with Rh2, cells were cultured for 2 wk, fixed with acetic acid/methanol (1:7), and stained with 0.1% crystal violet to count visible colonies and to measure the covered area.
2.3. Pyrosequencing
Measuring the methylation level at the CpG sites of the LINE1 and Alu sequence was carried out by pyrosequencing as described previously [22]. Briefly, 1 μg of genomic DNA was treated with bisulfite chromosomal DNA and the CpG-containing region was amplified by polymerase chain reaction (PCR). The methylation level was denoted by the percentage of methylated cytosines over the total of methylated and unmethylated cytosines after pyrosequencing. Sequencing was carried out for five independent samples treated with 20μM and 30μM Rh2, and assays were performed at least two times per sample.
2.4. Genome-wide methylation assay
The genome-wide methylation assay was carried out as described previously [23]. Briefly, 2 μg of chromosomal DNA isolated from a 60-mm culture dish was applied to an Illumina Infinium Human Methylation EPIC Beadchip (Illumina, San Diego, CA, USA) to detect genome-wide methylation. A methylation index (β) was calculated for each site, which is a variable ranging from 0 (no methylation) to 1 (100% methylation). The β value was then used to calculate a ratio of relative methylation between the control and Rh2-treated cells. The array data is accessible from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE93356.
2.5. Pathway analysis
The Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Redwood City, CA, USA) was used to perform functional categorization and pathway analysis of the gene pool affected by Rh2 treatment. Genes from the methylation array analysis were screened showing an false discovery rate (FDR)-adjusted p value < 0.05, Δβ ≥ 0.15, and |fold change| ≥ 1.5. The p value for each network was obtained by comparing the set of differentially methylated genes in Rh2-treated versus nontreated groups using Fisher's exact test, based on the hypergeometric distribution. The highest confidence functional network with the lowest p value was designated as the top network.
2.6. Real-time PCR
Total RNA isolation and reverse transcription were carried out as described previously [24]. Briefly, RNA from a 60-mm culture dish was eluted with 50 μL of distilled water and 2 μg was used for reverse transcription. Gene expression was measured by quantitative real time (RT)-PCR (qPCR) analysis using a Kapa SYBR Fast qPCR Kit (Kapa Biosystems, Woburn, MA, USA). A 1-μL aliquot of cDNA was used for PCR in triplicate. Gene expression was quantified according to the 2-ΔΔCt method after normalizing the RNA quantity to GAPDH. The primers adopted for the qPCR are listed in Table S1.
2.7. Statistical analysis
A Student t test was applied to detect differences in the mean expression levels for genes selected from the methylation array. All statistical analysis was carried out using SPSS for Windows, release 17.0 (SPSS Inc., Chicago, IL, USA). A p value < 0.05 was considered to be statistically significant.
3. Results
3.1. Rh induced global hypomethylation of the LINE1 sequence in MCF-7
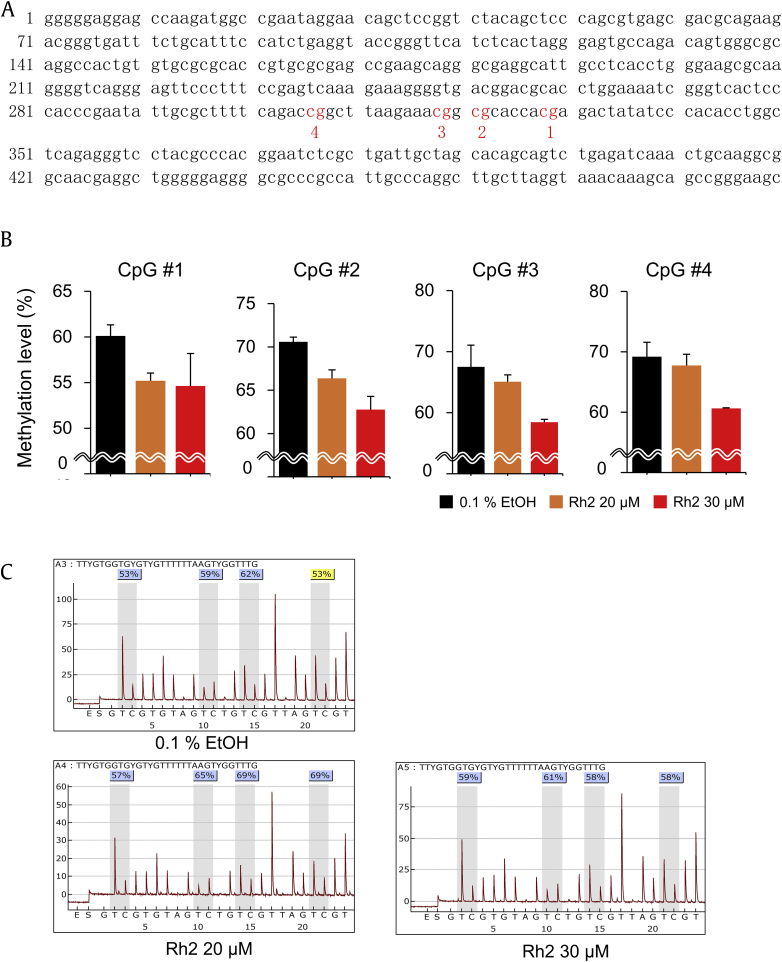
To examine the effect of Rh2 on the epigenetic profile of cancer cells, the breast cancer cell line MCF-7 was treated with the ginsenoside and then global as well as genome-wide methylation analyses were carried out. As global markers, LINE1 and Alu were selected because they were scattered on the chromosome in high copy numbers representing long- and short-sized repetitive elements, respectively. The methylation levels of specific CpG sites in the elements were determined by pyrosequencing, which could offer accurate estimation of the methylation levels at CpG loci (Fig. 1A). In LINE1, all the examined CpGs showed hypomethylation in the Rh2-treated cells compared to the nontreated cells. The decreased methylation level ranged from 1.6% to 5.0% at 20μM of Rh2 and from 5.5% to 9.1% at 30μM, showing dose-dependency (Figs. 1B and 1C). In contrast to LINE1, no significant change at the three examined CpG sites of Alu was observed regardless of Rh2 concentration (Fig. S1). These results indicate that Rh2 induced global hypomethylation but with element-specificity.
Fig. 1.
Effect of ginsenoside Rh2 on the global methylation level of LINE1 in MCF-7 cells. The methylation levels of the four CpGs on the LINE1 gene from the MCF-7 cells were determined by pyrosequencing after treatment with Rh2. (A) The LINE1 sequence adopted in this study [22]. The analyzed four CpGs are indicated in red and numbered. (B) The methylation level of the CpGs is shown on the bar graphs. Three independent treatments were performed and the values are given with mean ± standard error. (C) Representative pyrosequencing diagrams for 0μM, 20μM, and 30μM Rh2 treatment. EtOH, ethanol.
3.2. Rh2 changed methylation of genes involved in the cell-mediated immune response-related pathway
To interrogate the effect of Rh2 on the genome-wide methylation profile of the MCF-7 breast cancer cells, the methylation levels between Rh2-treated and nontreated MCF-7 cells were compared using a microarray covering 850K CpGs. 495 CpGs were filtered fitting our criteria: FDR < 0.05, Δβ ≥ 0.15, and |fold change| ≥ 1.5. A total of 355 CpGs were hypermethylated and 140 were hypomethylated in the Rh2-treated cells. The CpGs were scattered on the chromosomes with 74 being found in the promoter region, 18 in the coding region, and 213 in the intergenic region. The transcripts displaying the highest levels of altered methylation at the promoter CpG were SAMM50 (hypermethylated, Δβ = 0.63) and ST3GAL4 (hypomethylated, Δβ = –0.65). SAMM50 is a component of the SAM complex or the MINOS/MITOS complex, responsible for protein sorting or for the structural stability of mitochondrial cristae [25], [26]. ST3GAL4 is an enzyme accomplishing sialylation of N-glycans and its overexpression in cancer cells leads to altered glycosylation [27].
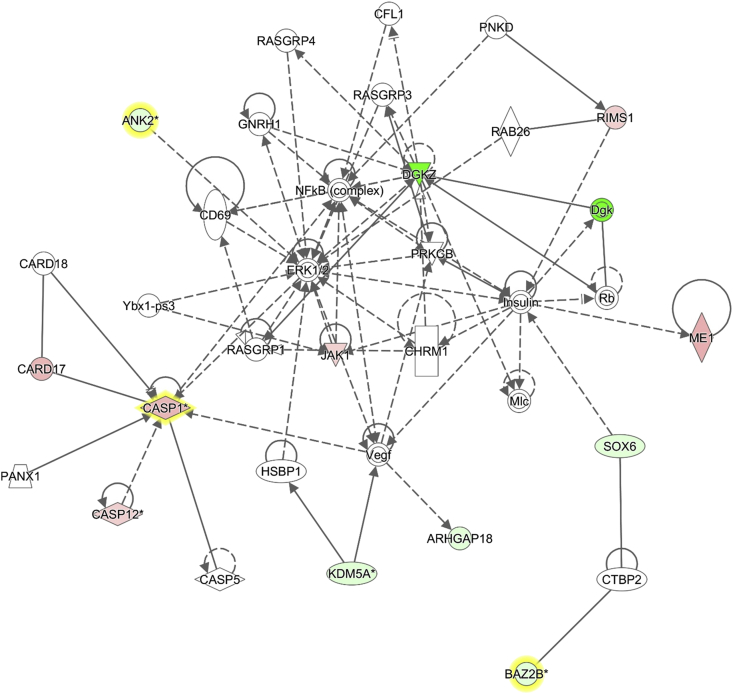
The 74 promoter CpGs, corresponding to 119 genes due to some promoters regulating more than two genes, were submitted to the IPA software tool to investigate functional interrelatedness. As a result, the “cell-mediated immune response”-related pathway emerged as the top network (Fig. 2). As Rh2 has been known to regulate many genes in the immune system such as CASP1 [28] and CARD17 [29] in the top network, the result may give us the insight that Rh2 regulates its immune-related target genes through, at least in part, an epigenetic mechanism.
Fig. 2.
Ginsenoside Rh2 epigenetically affects genes in the cell-mediated immune response pathway. The 850K microarray analysis identified 40 hypermethylated and 34 hypomethylated CpGs in promoter regions (Δβ ≥ 0.15 and |fold change| ≥ 1.5), and these were submitted for the construction of the highest confidence Ingenuity Pathway Analysis (IPA) network. The highest confidence network in the Rh2-treated MCF-7 cell was the “cell-mediated immune response” pathway. Shaded in red and green are those that are hypermethylated and hypomethylated, respectively, with intensity signifying the magnitude of the methylation change. Solid and dashed lines denote direct and indirect interactions, respectively.
3.3. Rh2 inhibited proliferation of MCF-7 through epigenetic regulation
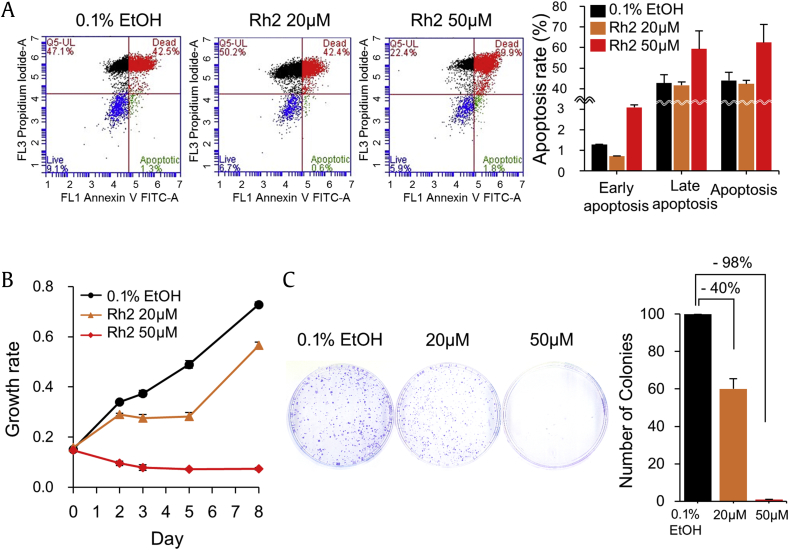
Rh2 has been known to increase apoptosis and inhibit cell growth in a few cancer cell lines [9], [10], [11]. We confirmed and detailed these effects by carrying out the experiments at two different Rh2 concentrations and found that 50μM was able to significantly induce early and late apoptosis of MCF-7 (Fig. 3A). At this concentration, cell growth was completely inhibited (Fig. 3B). Apoptosis was not induced at 20μM but cell proliferation was slightly inhibited. The effect of Rh2 on tumorigenicity was monitored by colony forming assay and the result showed a similar pattern to that observed for the cell growth inhibition with a 40% decrease at 20μM and a complete decrease at 50μM (Fig. 3C).
Fig. 3.
Ginsenoside Rh2 increased apoptosis and inhibited proliferation of MCF-7 cells. (A) MCF-7 cells were treated with Rh2 for 24 h and apoptosis was analyzed by FACS. The assay was performed in triplicate and the results are shown in a representative FACS diagram. The ratio of cells at apoptosis is represented by a bar graph with mean ± SE. (B) The effect of Rh2 on cell proliferation was analyzed using a Cell Counting Kit-8. Each sample was analyzed in triplicate, with the results shown as mean ± SE. (C) Colony formation was assayed on a 60-mm culture dish by staining the cells with crystal violet. The colonized areas are shown as a bar graph of the mean ± SE of three independent experiments. EtOH, ethanol.
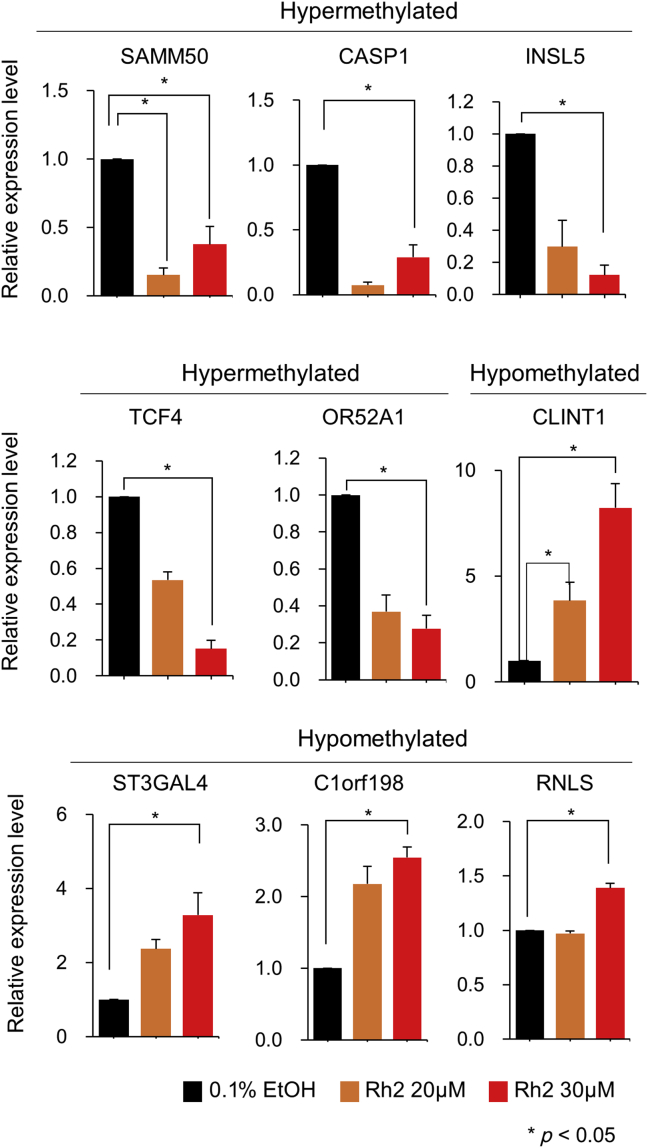
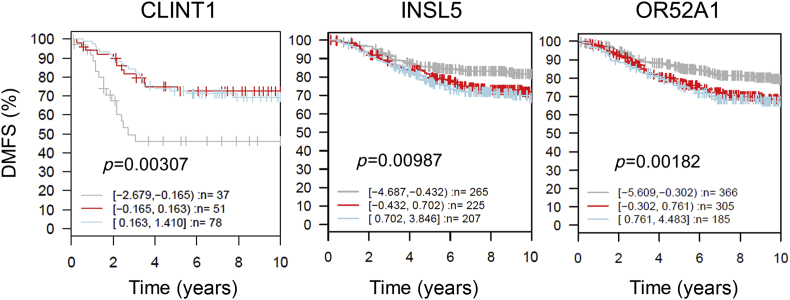
To see whether the methylation change by Rh2 accompanied any change of gene expression and eventually alteration of cell proliferation, we selected nine genes from the top 30 protein-coding genes (Table 1), five hypermethylated and four hypomethylated. Especially, CLINT1 [30] is known to be a tumor suppressor, and TCF4 [31] and INSL5 [32] are oncogenes or oncogenic markers. qPCR analysis revealed a strong opposite relationship between the levels of methylation and expression, i.e., hypermethylated genes were downregulated while hypomethylated genes were upregulated except for RNLS at 20μM of Rh2 (Fig. 4). For CLINT1, INSL5, and OR52A1, the relationship between expression and prognosis of breast cancer patients was investigated through the GOBO database and the results indicated a high rate of distant metastasis-free survival for patients with higher expression of CLINT1 while with lower expression of INSL5 and OR52A1 (p < 0.05) (Fig. 5).
Table 1.
Top 30 protein coding genes displaying differential methylation by Ginsenoside Rh2 in MCF-7 cells
| Symbol | Accession no. | Description | Δ β | Fold change |
|---|---|---|---|---|
| SAMM50 | NM_015380 | SAMM50 sorting and assembly machinery component | 0.632 | 3.175 |
| CCNDBP1 | NM_037370 | Cyclin D1 binding protein 1 | 0.362 | 8.048 |
| EYA1 | NM_172059 | EYA transcriptional coactivator and phosphatase 1 | 0.237 | 1.876 |
| CASP1 | NM_033294 | Caspase 1 | 0.2 | 2.373 |
| CARD17 | NM_001007232 | Caspase recruitment domain family member 17 | 0.2 | 2.373 |
| VWC2L | NM_001080500 | von Willebrand factor C domain containing protein 2-like | 0.198 | 2.902 |
| PXDNL | NM_144651 | Peroxidasin like | 0.188 | 1.898 |
| INSL5 | NM_005478 | Insulin like 5 | 0.185 | 2.073 |
| GALNT5 | NM_014568 | Polypeptide N-acetylgalactosaminyltransferase 5 | 0.182 | 2.651 |
| TCF4 | NM_001243235 | Transcription factor 4 | 0.18 | 2.019 |
| OR4L1 | NM_001004717 | Olfactory receptor family 4 subfamily L member 1 | 0.179 | 1.555 |
| ADAMTSL1 | NM_052866 | ADAMTS like 1 | 0.179 | 1.568 |
| VWA2 | NM_198496 | von Willebrand factor A domain containing 2 | 0.178 | 3.874 |
| MEI4 | NM_001282136 | Meiotic double-stranded break formation protein 4 | 0.176 | 2.034 |
| OR52A1 | NM_012375 | Olfactory receptor family 52 subfamily A member 1 | 0.173 | 1.696 |
| OVCH2 | NM_198185 | Ovochymase 2 (gene/pseudogene) | −0.183 | −2.626 |
| SDR39U1 | NM_020195 | Short chain dehydrogenase/reductase family 39U member 1 | −0.196 | −2.137 |
| BAZ2B | NM_001289975 | Bromodomain adjacent to zinc finger domain 2B | −0.198 | −1.783 |
| C6orf203 | NM_016487 | Chromosome 6 open reading frame 203 | −0.208 | −2.207 |
| OR9G4 | NM_001005284 | Olfactory receptor family 9 subfamily G member 4 | −0.238 | −1.728 |
| OR8U8 | NM_001013356 | Olfactory receptor family 8 subfamily U member 8 | −0.244 | −1.923 |
| OR8U1 | NM_001005204 | Olfactory receptor family 8 subfamily U member 1 | −0.244 | −1.923 |
| OR1L8 | NM_001004454 | Olfactory receptor family 1 subfamily L member 8 | −0.248 | −2.511 |
| LOC284798 | NR_027093 | Uncharacterized LOC284798 | −0.314 | −1.589 |
| SPOCK3 | NM_001040159 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 | −0.422 | −2.562 |
| RNLS | NM_001031709 | Renalase, FAD dependent amine oxidase | −0.481 | −4.75 |
| C1orf198 | NM_032800 | Chromosome 1 open reading frame 198 | −0.577 | −8.064 |
| CLINT1 | NM_014666 | Clathrin interactor 1 | −0.585 | −6.158 |
| DGKZ | NM_201533 | Diacylglycerol kinase zeta | −0.586 | −18.605 |
| ST3GAL4 | NM_006278 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | −0.654 | −15.594 |
Fig. 4.
Real time-polymerase chain reaction analysis of selected genes hypermethylated or hypomethylated by ginsenoside Rh2 in MCF-7 cells. Quantitative polymerase chain reaction analysis of selected genes showing altered methylation levels in response to Rh2 in MCF-7 cells. Cells were treated with 20μM and 30μM Rh2 and cultured for 24 h before harvesting. The control is 0.1% ethanol in the culture media used for dissolving Rh2. Each sample was analyzed in triplicate and the results are shown as the mean ± standard error.
Fig. 5.
Kaplan–Meier survival analysis of breast cancer patients with variable expression levels of CLINT1, INSL5, and OR52A1. Samples (n; sum of cases) were stratified into tertiles based on the expression level. Higher and medium CLINT1 expression is significantly associated with longer DMFS compared to lower expression. Lower expression of INSL5 and OR52A1 is associated with longer DMFS. DMFS stands for distant metastasis-free survival meaning the period until metastasis is detected. Samples were stratified into three quantiles based on gene expression, low (grey), medium (red), and high (sky-blue) with the log2 expression in the parenthesis. DMFS, distant metastasis-free survival.
4. Discussion
The aim of this study was to identify pathways epigenetically regulated by Rh2 and to examine the association between the epigenetic change and cancer cell proliferation. As a first step to check whether Rh2 has potential for inducing methylation changes, the genome-wide scattered LINE1 and Alu were examined. Notably, Rh2 induced hypomethylation of LINE1 but not Alu. This fact suggests a gene- or locus-specific action mode of Rh2 for epigenetic alteration. This kind of global marker-specific methylation change has been observed in a few cancers. For example, Alu methylation level but not LINE1 was associated with increased prostate cancer risk [33]. A characteristic chromatin structure imposed by unique histone modification was speculated to explain the differential methylation [34]. Furthermore, the existence of multiple subfamilies of the repetitive elements could also result in the type-specific methylation [35]. LINE1 has shown hypomethylation in various cancers, including breast, lung, and gastric cancer, possibly causing genomic instability, eventually leading cells to become cancerous [36]. Hypomethylation, however, was not the only cause of the development of cancer because hypermethylation of LINE1 was observed in melanoma [37]. The induction of hypomethylation by Rh2 does not seem to strengthen the cancerous state of MCF-7 because the cells rather showed higher apoptosis and a lower proliferation rate. Therefore, the hypomethylated LINE1 could be utilized as a global epigenetic marker to monitor whether Rh2 is able to induce an appropriate epigenetic change.
In the top IPA network, CASP12, CASP1, CARD17, ME1, JAK1, and RIMS1 are hypermethylated. CASP1, CASP12, and CARD17 are constituents of the so-called inflammasome, which has been known to play a central role in caspase-mediated inflammation. Activation of caspase-1 (CASP1) and subsequent processing of IL-1β are essential for initiation of the inflammatory response [28]. Previous investigations suggested that CARD17 can function as a negative regulator of CASP1 activity by binding the CARD domain of CASP1 [29]. CASP12 plays a major role in estrogen receptor stress-induced apoptosis and in the development of Alzheimer's disease [38]. Activation of the inflammasome by danger signals seems to play a diverse, and sometimes conflicting, suppressive or stimulatory role in cancer development and progression [39], [40]. Our results indicate that hypermethylation of CASP1 and its related genes by Rh2 would inactivate the inflammatory response, eventually leading to suppression of cancer development.
Malic enzyme 1 (ME1) links the glycolytic and citric acid cycles and is important for nicotinamide adenine dinucleotide phosphate production, glutamine metabolism, and lipogenesis. ME1 overexpression is associated with unfavorable prognoses in patients with various cancers, suggesting that ME1 is a poor prognostic predictor of carcinoma [41]. JAK1 is a pleiotropic signal transducer and is upregulated in various cancers [42], [43]. Hypermethylation of the two genes by Rh2 regulates the gene expression towards suppressing tumor development.
Hypomethylated genes in the top network are DGKZ, Dgk, ANK2, SOX6, KDM5A, ARHGAP18, and BAZZB. Among these, a few are notable because of being involved in cancer pathways. SOX6 has been described as a tumor suppressor in several cancers, inhibiting cell proliferation by stabilizing p53 protein and subsequently upregulating p21 [44]. KDM5A is an H3K4me3 demethylase required for NK cell activation by suppressing the suppressor SOCS1 [45].
The association between methylation and expression level for the genes appearing in the network is credited by the altered cellular activity and more specifically confirmed by qPCR for randomly selected genes from the list in Table 1, which showed altered methylation by Rh2. Furthermore, the tight association between the prognosis of breast cancer patients and the expression levels of the epigenetically regulated genes by Rh2 supplies clinical data showing that Rh2 can induce epigenetic changes of cancer-related genes.
The current study is limited by the absence of an assessment of genome-wide methylation to various concentration ranges of Rh2, even though two different concentrations were adopted for monitoring other events, including global marker methylation, cell activities, and gene expression. Experiments with a wider range of Rh2 concentrations will help pinpoint the precise concentration to achieve a desired epigenetic change in cultured cells and furthermore in applications such as in vivo animal models.
Taken together, Rh2 was proven to be able to induce hypomethylation of a global chromosomal element, LINE1. A genome-wide methylation analysis of the Rh2-treated MCF-7 cells identified “cell-mediated immune response”- and “cancer”-related pathways as the top networks. It is therefore suggested that the epigenetic methylation change by Rh2 accompanies gene expression change, leading to promotion of apoptosis and inhibition of proliferation for the cancer cells. The findings in the current study are, to the best of the authors' knowledge, the first observations on Rh2 via an epigenetic approach and could contribute to understanding the epigenetic mechanism of Rh2, how to stimulate immune response, and also how to inhibit cancer cell growth.
Conflicts of interest
The authors declare that they have no conflicts of interests.
Acknowledgments
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2016R1D1A1B01009235).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jgr.2017.05.003.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Jia L., Zhao Y. Current evaluation of the millennium phytomedicine–ginseng (I): etymology, pharmacognosy, phytochemistry, market and regulations. Curr Med Chem. 2009;16:2475–2484. doi: 10.2174/092986709788682146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen L.P. Ginsenosides chemistry, biosynthesis, analysis, and potential health effects. Adv Food Nutr Res. 2009;55:1–99. doi: 10.1016/S1043-4526(08)00401-4. [DOI] [PubMed] [Google Scholar]
- 3.Wang W., Zhao Y., Rayburn E.R., Hill D.L., Wang H., Zhang R. In vitro anti-cancer activity and structure-activity relationships of natural products isolated from fruits of Panax ginseng. Cancer Chemother Pharmacol. 2007;59:589–601. doi: 10.1007/s00280-006-0300-z. [DOI] [PubMed] [Google Scholar]
- 4.Wang C.Z., Du G.J., Zhang Z., Wen X.D., Calway T., Zhen Z., Musch M.W., Bissonnette M., Chang E.B., Yuan C.S. Ginsenoside compound K, not Rb1, possesses potential chemopreventive activities in human colorectal cancer. Int J Oncol. 2012;40:1970–1976. doi: 10.3892/ijo.2012.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi W.Y., Fu B.D., Shen H.Q., Wei Q., Zhang C., Song Z., Qin Q.Q., Li H.P., Lv S., Wu S.C. Sulfated derivative of 20(S)-ginsenoside Rh2 inhibits inflammatory cytokines through MAPKs and NF-kappa B pathways in LPS-induced RAW264.7 macrophages. Inflammation. 2012;35:1659–1668. doi: 10.1007/s10753-012-9482-1. [DOI] [PubMed] [Google Scholar]
- 6.Yi P.F., Bi W.Y., Shen H.Q., Wei Q., Zhang L.Y., Dong H.B., Bai H.L., Zhang C., Song Z., Qin Q.Q. Inhibitory effects of sulfated 20(S)-ginsenoside Rh2 on the release of pro-inflammatory mediators in LPS-induced RAW 264.7 cells. Eur J Pharmacol. 2013;712:60–66. doi: 10.1016/j.ejphar.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 7.Jeong M.K., Cho C.K., Yoo H.S. General and genetic toxicology of enzyme-treated ginseng extract: toxicology of ginseng Rh2. J Pharmacopuncture. 2016;19:213–224. doi: 10.3831/KPI.2016.19.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu F.Y., Shang W.Q., Yu J.J., Sun Q., Li M.Q., Sun J.S. The antitumor activity study of ginsenosides and metabolites in lung cancer cell. Am J Transl Res. 2016;8:1708–1718. [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Q., Shi X., Zuo G., Xiong W., Li H., Guo P., Wang F., Chen Y., Li J., Chen D.L. Anticancer effect of 20(S)-ginsenoside Rh2 on HepG2 liver carcinoma cells: Activating GSK-3beta and degrading beta-catenin. Oncol Rep. 2016;36:2059–2070. doi: 10.3892/or.2016.5033. [DOI] [PubMed] [Google Scholar]
- 10.Oh M., Choi Y.H., Choi S., Chung H., Kim K., Kim S.I., Kim D.K., Kim N.D. Anti-proliferating effects of ginsenoside Rh2 on MCF-7 human breast cancer cells. Int J Oncol. 1999;14:869–875. doi: 10.3892/ijo.14.5.869. [DOI] [PubMed] [Google Scholar]
- 11.Kim H.S., Lee E.H., Ko S.R., Choi K.J., Park J.H., Im D.S. Effects of ginsenosides Rg3 and Rh2 on the proliferation of prostate cancer cells. Arch Pharm Res. 2004;27:429–435. doi: 10.1007/BF02980085. [DOI] [PubMed] [Google Scholar]
- 12.Han S., Jeong A.J., Yang H., Bin Kang K., Lee H., Yi E.H., Kim B.H., Cho C.H., Chung J.W., Sung S.H. Ginsenoside 20(S)-Rh2 exerts anti-cancer activity through targeting IL-6-induced JAK2/STAT3 pathway in human colorectal cancer cells. J Ethnopharmacol. 2016;194:83–90. doi: 10.1016/j.jep.2016.08.039. [DOI] [PubMed] [Google Scholar]
- 13.Wanderi C., Kim E., Chang S., Choi C., Choi K. Ginsenoside 20(S)-protopanaxadiol suppresses viability of human glioblastoma cells via down-regulation of cell adhesion proteins and cell-cycle arrest. Anticancer Res. 2016;36:925–932. [PubMed] [Google Scholar]
- 14.Zhang J., Lu M., Zhou F., Sun H., Hao G., Wu X., Wang G. Key role of nuclear factor-kappaB in the cellular pharmacokinetics of adriamycin in MCF-7/Adr cells: the potential mechanism for synergy with 20(S)-ginsenoside Rh2. Drug Metab Dispos. 2012;40:1900–1908. doi: 10.1124/dmd.112.045187. [DOI] [PubMed] [Google Scholar]
- 15.Soes S., Daugaard I.L., Sorensen B.S., Carus A., Mattheisen M., Alsner J., Overgaard J., Hager H., Hansen L.L., Kristensen L.S. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience. 2014;1:367–374. doi: 10.18632/oncoscience.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahara T., Shibata T., Nakamura M., Yamashita H., Yoshioka D., Okubo M., Yonemura J., Maeda Y., Maruyama N., Kamano T. Association between IL-17A, -17F and MIF polymorphisms predispose to CpG island hyper-methylation in gastric cancer. Int J Mol Med. 2010;25:471–477. doi: 10.3892/ijmm_00000367. [DOI] [PubMed] [Google Scholar]
- 17.Schondorf T., Ebert M.P., Hoffmann J., Becker M., Moser N., Pur S., Gohring U.J., Weisshaar M.P. Hypermethylation of the PTEN gene in ovarian cancer cell lines. Cancer Lett. 2004;207:215–220. doi: 10.1016/j.canlet.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 18.Esteller M., Silva J.M., Dominguez G., Bonilla F., Matias-Guiu X., Lerma E., Bussaglia E., Prat J., Harkes I.C., Repasky E.A. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 19.Di Vinci A., Perdelli L., Banelli B., Salvi S., Casciano I., Gelvi I., Allemanni G., Margallo E., Gatteschi B., Romani M. p16(INK4a) Promoter methylation and protein expression in breast fibroadenoma and carcinoma. Int J Cancer. 2005;114:414–421. doi: 10.1002/ijc.20771. [DOI] [PubMed] [Google Scholar]
- 20.Luo J., Li Y.N., Wang F., Zhang W.M., Geng X. S-adenosylmethionine inhibits the growth of cancer cells by reversing the hypomethylation status of c-myc and H-ras in human gastric cancer and colon cancer. Int J Biol Sci. 2010;6:784–795. doi: 10.7150/ijbs.6.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oshimo Y., Nakayama H., Ito R., Kitadai Y., Yoshida K., Chayama K., Yasui W. Promoter methylation of cyclin D2 gene in gastric carcinoma. Int J Oncol. 2003;23:1663–1670. [PubMed] [Google Scholar]
- 22.Shigaki H., Baba Y., Watanabe M., Murata A., Iwagami S., Miyake K., Ishimoto T., Iwatsuki M., Baba H. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer. 2013;16:480–487. doi: 10.1007/s10120-012-0209-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park S.B., Kim B., Bae H., Lee H., Lee S., Choi E.H., Kim S.J. Differential epigenetic effects of atmospheric cold plasma on MCF-7 and MDA-MB-231 breast cancer cells. PLoS One. 2015;10:e0129931. doi: 10.1371/journal.pone.0129931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H., Lee S., Bae H., Kang H.S., Kim S.J. Genome-wide identification of target genes for miR-204 and miR-211 identifies their proliferation stimulatory role in breast cancer cells. Sci Rep. 2016;6:25287. doi: 10.1038/srep25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphries A.D., Streimann I.C., Stojanovski D., Johnston A.J., Yano M., Hoogenraad N.J., Ryan M.T. Dissection of the mitochondrial import and assembly pathway for human Tom40. J Biol Chem. 2005;280:11535–11543. doi: 10.1074/jbc.M413816200. [DOI] [PubMed] [Google Scholar]
- 26.Ott C., Ross K., Straub S., Thiede B., Gotz M., Goosmann C., Krischke M., Mueller M.J., Krohne G., Rudel T. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32:1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mereiter S., Magalhaes A., Adamczyk B., Jin C., Almeida A., Drici L., Ibanez-Vea M., Gomes C., Ferreira J.A., Afonso L.P. Glycomic analysis of gastric carcinoma cells discloses glycans as modulators of RON receptor tyrosine kinase activation in cancer. Biochim Biophys Acta. 2016;1860:1795–1808. doi: 10.1016/j.bbagen.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 28.Martinon F., Burns K., Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 29.Humke E.W., Shriver S.K., Starovasnik M.A., Fairbrother W.J., Dixit V.M. ICEBERG: a novel inhibitor of interleukin-1beta generation. Cell. 2000;103:99–111. doi: 10.1016/s0092-8674(00)00108-2. [DOI] [PubMed] [Google Scholar]
- 30.Dodd M.E., Hatzold J., Mathias J.R., Walters K.B., Bennin D.A., Rhodes J., Kanki J.P., Look A.T., Hammerschmidt M., Huttenlocher A. The ENTH domain protein Clint1 is required for epidermal homeostasis in zebrafish. Development. 2009;136:2591–2600. doi: 10.1242/dev.038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suryawanshi A., Manicassamy S. Tumors induce immune tolerance through activation of beta-catenin/TCF4 signaling in dendritic cells: A novel therapeutic target for cancer immunotherapy. Oncoimmunology. 2015;4:e1052932. doi: 10.1080/2162402X.2015.1052932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mashima H., Ohno H., Yamada Y., Sakai T., Ohnishi H. INSL5 may be a unique marker of colorectal endocrine cells and neuroendocrine tumors. Biochem Biophys Res Commun. 2013;432:586–592. doi: 10.1016/j.bbrc.2013.02.042. [DOI] [PubMed] [Google Scholar]
- 33.Barry K.H., Moore L.E., Liao L.M., Huang W.Y., Andreotti G., Poulin M., Berndt S.I. Prospective study of DNA methylation at LINE-1 and Alu in peripheral blood and the risk of prostate cancer. Prostate. 2015;75:1718–1725. doi: 10.1002/pros.23053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nusgen N., Goering W., Dauksa A., Biswas A., Jamil M.A., Dimitriou I., Sharma A., Singer H., Fimmers R., Frohlich H. Inter-locus as well as intra-locus heterogeneity in LINE-1 promoter methylation in common human cancers suggests selective demethylation pressure at specific CpGs. Clin Epigenetics. 2015;7:17. doi: 10.1186/s13148-015-0051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakshi A., Herke S.W., Batzer M.A., Kim J. DNA methylation variation of human-specific Alu repeats. Epigenetics. 2016;11:163–173. doi: 10.1080/15592294.2015.1130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J., Huang Q., Zeng F., Li W., He Z., Chen W., Zhu W., Zhang B. The prognostic value of global DNA hypomethylation in cancer: a meta-analysis. PLoS One. 2014;9:e106290. doi: 10.1371/journal.pone.0106290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Araujo E.S., Kashiwabara A.Y., Achatz M.I., Moredo L.F., De Sa B.C., Duprat J.P., Rosenberg C., Carraro D.M., Krepischi A.C. LINE-1 hypermethylation in peripheral blood of cutaneous melanoma patients is associated with metastasis. Melanoma Res. 2015;25:173–177. doi: 10.1097/CMR.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakagawa T., Zhu H., Morishima N., Li E., Xu J., Yankner B.A., Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 39.Muzes G., Sipos F. Inflammasome, inflammation and cancer: an interrelated pathobiological triad. Curr Drug Targets. 2015;16:249–257. doi: 10.2174/1389450115666141229154157. [DOI] [PubMed] [Google Scholar]
- 40.Guo W., Sun Y., Liu W., Wu X., Guo L., Cai P., Wu X., Wu X., Shen Y., Shu Y. Small molecule-driven mitophagy-mediated NLRP3 inflammasome inhibition is responsible for the prevention of colitis-associated cancer. Autophagy. 2014;10:972–985. doi: 10.4161/auto.28374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen D., Liu D., Tang J., Dong L., Liu Y., Tao Z., Wan J., Gao D., Wang L., Sun H. Malic enzyme 1 induces epithelial-mesenchymal transition and indicates poor prognosis in hepatocellular carcinoma. Tumour Biol. 2015;36:6211–6221. doi: 10.1007/s13277-015-3306-5. [DOI] [PubMed] [Google Scholar]
- 42.Liu D., Huang Y., Zeng J., Chen B., Huang N., Guo N., Liu L., Xu H., Mo X., Li W. Down-regulation of JAK1 by RNA interference inhibits growth of the lung cancer cell line A549 and interferes with the PI3K/mTOR pathway. J Cancer Res Clin Oncol. 2011;137:1629–1640. doi: 10.1007/s00432-011-1037-6. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M.X., Zhao X., Wang Z.G., Zhao W.M., Wang Y.S. Constitutive activation of signal transducer and activator of transcription 3 regulates expression of vascular endothelial growth factor in human meningioma differentiation. J Cancer Res Clin Oncol. 2010;136:981–988. doi: 10.1007/s00432-009-0743-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang J., Ding S., Duan Z., Xie Q., Zhang T., Zhang X., Wang Y., Chen X., Zhuang H., Lu F. Role of p14ARF-HDM2-p53 axis in SOX6-mediated tumor suppression. Oncogene. 2016;35:1692–1702. doi: 10.1038/onc.2015.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao D., Zhang Q., Liu Y., Li X., Zhao K., Ding Y., Li Z., Shen Q., Wang C., Li N. H3K4me3 demethylase Kdm5a is required for NK cell activation by associating with p50 to suppress SOCS1. Cell Rep. 2016;15:288–299. doi: 10.1016/j.celrep.2016.03.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.