Significance
This paper reveals the molecular logic for generating the basal methylation pattern in each individual following erasure of the gametic profile in the preimplantation embryo. The results show that transcription factors and the RNA polymerase complex play a major role in protecting recognized regions from de novo methylation by recruiting the H3K4 methylation machinery. Because methylation is stably maintained through development, this mechanism serves to perpetuate the activity state present in the early embryo. This model may also help explain how transient factors from the gametes may influence methylation patterns in the offspring and, thereby, contribute to intergenerational epigenetic inheritance.
Keywords: development, epigenetics, inheritance, histomodification
Abstract
Following erasure in the blastocyst, the entire genome undergoes de novo methylation at the time of implantation, with CpG islands being protected from this process. This bimodal pattern is then preserved throughout development and the lifetime of the organism. Using mouse embryonic stem cells as a model system, we demonstrate that the binding of an RNA polymerase complex on DNA before de novo methylation is predictive of it being protected from this modification, and tethering experiments demonstrate that the presence of this complex is, in fact, sufficient to prevent methylation at these sites. This protection is most likely mediated by the recruitment of enzyme complexes that methylate histone H3K4 over a local region and, in this way, prevent access to the de novo methylation complex. The topological pattern of H3K4me3 that is formed while the DNA is as yet unmethylated provides a strikingly accurate template for modeling the genome-wide basal methylation pattern of the organism. These results have far-reaching consequences for understanding the relationship between RNA transcription and DNA methylation.
In animals, the genome-wide DNA methylation pattern is initially erased in the early embryo and then reestablished in each individual at about the time of implantation. This is carried out by a process in which almost all of the DNA is subject to de novo methylation while CpG island-like regions are protected on the basis of underlying sequence motifs (1), but the biological logic and molecular mechanism of this process are still unknown. Analysis of these CpG-rich sites indicates that they are highly enriched for transcription start sites and characterized by the presence of binding motifs for many transcription factors (2). This close correlation suggested the possibility that protection from de novo methylation may actually be dictated by the binding of transcription complexes at recognized sites in the preimplantation embryo. In this work, we have used bioinformatic tools as well as genetic techniques to test this idea. The results of these experiments lead to a concept for how DNA methylation functions during development.
Results
Embryonic stem (ES) cells represent an excellent system for studying the process of global de novo methylation that takes place at the time of implantation. Although initially derived from the blastocyst stage of development, these cells harbor a DNA methylation pattern that is almost identical to the implantation-stage embryo (∼E6.5) (3, 4). Furthermore, unlike somatic cells in culture, ES cells constantly maintain the ability to actively de novo methylate newly introduced DNA sequences while at the same time protecting CpG islands (5–7). Since we were interested in characterizing the factors that may play a role in the formation of this pattern, we needed a model of the genomic landscape as it existed before de novo methylation. To this end, we took advantage of ES cells carrying knockouts (TKO) for all three DNA methylases, Dnmt1, Dnmt3a, and Dnmt3b, which are completely unmethylated at all CpG sites in the genome (8, 9). These cells, which have an epigenetic pattern similar, but not identical, to that of the preimplantation embryo (ICM) (4, 10) were chosen to scientifically evaluate the role of DNA methylation itself, independent of other variables.
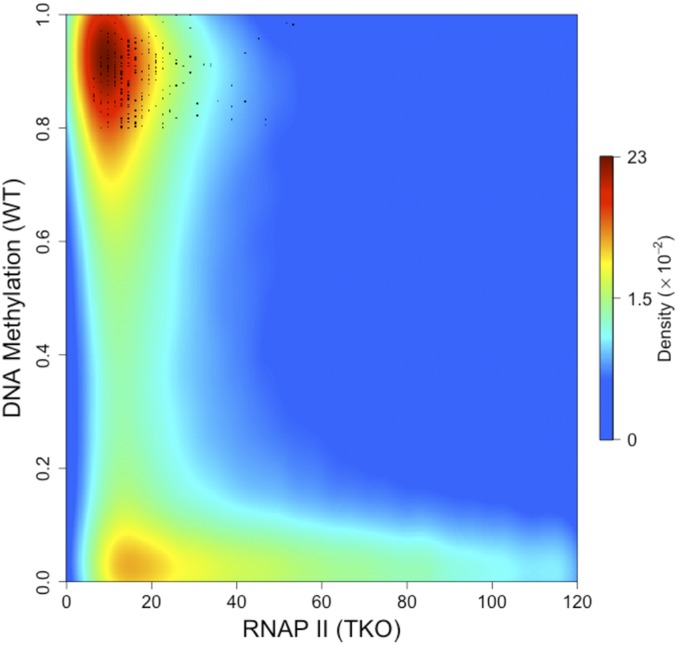
Using this dual cell-culture system, it was now possible to ask whether it is indeed positioning of the transcription machinery before implantation that determines what regions will be protected from subsequent de novo methylation. To this end, we used an antibody against RNA polymerase (RNAP II) to carry out ChIP-sequencing (Seq) in the unmethylated TKO ES cells and compared this pattern to the genome-wide levels of methylation observed in WT cells, which mimic the post-de novo state of implantation. Analysis employing both reduced representation bisulfite sequencing (RRBS), as well as whole genome bisulfite sequencing (WGBS) indicated that sites binding high concentrations of RNAP II remain largely unmethylated, while other low-binding regions in the genome undergo de novo methylation (Fig. 1 and SI Appendix, Figs. S1 and S2A). Consistent with this, sequences defined as CpG islands that are nonetheless fully methylated in WT ES cells were found to lack RNAP II binding in TKO, while non-CpG islands associated with RNAP in TKO cells largely remain unmethylated (SI Appendix, Fig. S2C). This was also validated by examining published data (11) on RNAP II binding in naive ground-state embryonic cells (grown in 2i medium), which provides an excellent model for the preimplantation ICM stage of development (SI Appendix, Fig. S2B). It should be noted that an analysis of all of the RNAP II-bound CpG islands in TKO cells indicated that, while most represent promoters of genes that are actually transcribed, over 20% do not generate RNA products in TKO cells as determined by RNA-Seq, suggesting that the presence of RNAP complex alone, even in the absence of active transcription, may be sufficient to mark these islands. Thus, as opposed to the idea that DNA methylation displaces transcription complexes, these results clearly demonstrate that it is the initial presence of RNA polymerase that actually serves to predict which sequences are ultimately protected from de novo methylation.
Fig. 1.
RNAP II binding predicts undermethylation. Density scatter plot of RNAP II binding (normalized ChIP-Seq reads) in TKO cells vs. percent DNA methylation (normalized RRBS) in WT ES cells. Note that regions bound to RNAP II are largely unmethylated in WT cells while unbound sites are highly methylated. Some unmethylated sites in the unbound fraction may reflect inefficiency of the RNAP II antibody ChIP. Sequences defined as CpG islands that are nonetheless methylated (>80%) in WT ES cells as well as four adult tissues (GSE60012) are marked as black dots. Data density is color-coded from blue (low) to red (high). Quantitative analysis indicates that the proportion of tiles that bind RNAP II (>40) but are methylated (>80%) in WT ES cells is 5%. The proportion of tiles not bound by RNAP II (<20), yet unmethylated (<20%) in WT ES cells was calculated to be 13%.
If it is indeed the presence of the RNA polymerase transcription complex that is responsible for keeping CpG islands from getting de novo methylated, we reasoned that it may be possible to protect any sequence from DNA modification by artificially recruiting this transcription complex to nonisland target loci. For this purpose, we developed a tethering system in ES cells using a plasmid vector expressing a fusion product containing the TATA-binding protein (TBP) linked to the yeast GAL4-binding domain (12). TBP is a general transcription factor that is known to initiate the recruitment of an RNAP II complex to begin transcription (13, 14). When introduced into ES cells by stable transfection, this vector was shown to be transcribed and to make the correct protein product (SI Appendix, Fig. S3). As a target for this system, we employed a second vector carrying the human CRYAA promoter linked in cis to multiple GAL4-binding sites (Fig. 2A).
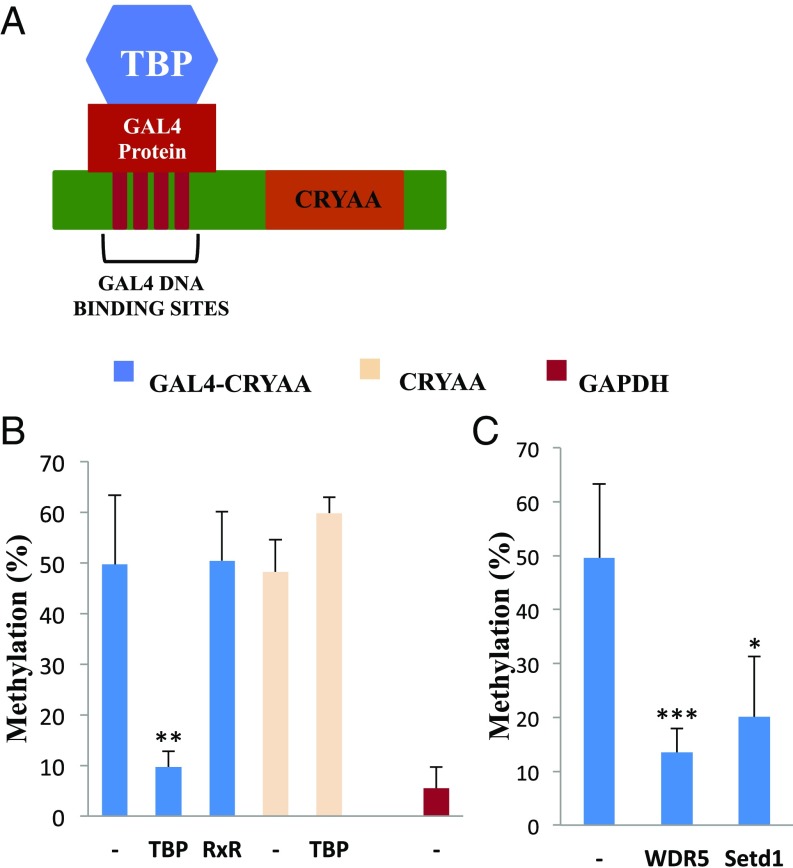
Fig. 2.
Tethering RNAP to a non-CpG island promoter. A vector carrying the human CRYAA promoter containing 4×GAL4 binding sites (A) was stably transfected (blue) into WT ES cells with or without (−) GAL4 binding domain fusion genes TBP and RXR (B) or WDR5 and Setd1 (C) and then analyzed for methylation by single molecule Bisulfite sequencing. Target controls include transfection of CRYAA without the GAL4 binding sites (yellow) and the GAPDH CpG-island promoter (red). Each bar represents results from three to six biological replicates. Statistical significance was determined by Student’s t test: *P < 0.05, **P < 0.005, ***P < 0.0005.
When inserted into WT ES cells by stable transfection, this target promoter (CRYAA) undergoes substantial de novo methylation as a function of time (SI Appendix, Fig. S4A), reflecting the fact that it is not recognized as a CpG island sequence. However, when this same vector is introduced into ES cells expressing TBP-GAL4, it is largely protected against the de novo reaction (Fig. 2B and SI Appendix, Fig. S4B), presumably as a result of fusion-protein binding at GAL4 sequences adjacent to the CRYAA promoter (SI Appendix, Fig. S5A). This mechanism appears to be specific, since protection was found to be strictly dependent on the presence of GAL4-binding sites and did not take place when tethering was carried out using a nonrelevant RXR fusion protein (Fig. 2B) which can clearly bind the promoter but has no effect on H3K4 methylation (SI Appendix, Fig. S5B). Furthermore, TBP-protection results were obtained (SI Appendix, Fig. S4C) using this same tethering system on another sequence (DAZ1), which normally undergoes de novo methylation in ES cells (2). Taken together, these results suggest that the artificial presence of TBP, which probably serves as a nucleus for the formation of an RNA polymerase complex, is sufficient to protect even non-CpG island sequences against de novo DNA methylation in ES cells. Current efforts are aimed at using the dCAS9 tethering system to prove this in vivo, as well.
ChIP studies in many systems have demonstrated that sites of transcription are packaged with nucleosomes containing H3K4me3, and it has been shown that this modification is catalyzed by the MLL1 complex, which is recruited together with the transcriptional machinery (15). Indeed, in keeping with this, ChIP analysis in our system showed that the mere tethering of TBP actually brings about H3K4 methylation of its CRYAA target sequence (SI Appendix, Fig. S5C). Biochemical studies indicate that the de novo methylation complex recognizes its DNA template through interactions with the lysine 4 residue on histone H3, and methylation of this site prevents this binding (16–20). With this in mind, we reasoned that the protection from de novo DNA methylation that comes about as a consequence of RNA polymerase binding may be mediated by the generation of H3K4me3 at these sites. To test this hypothesis, we generated a tethering vector that expresses the GAL4-binding domain fused to WDR5, a key protein that is an integral part of the Mll1 complex (15), and this was stably transfected into WT ES cells. When the GAL4-CRYAA target sequence was then introduced into these cells, it was found to be relatively protected from de novo methylation. Similar results were obtained in a tethering experiment using SETD1, another H3K4 methylase (15) (Fig. 2C).
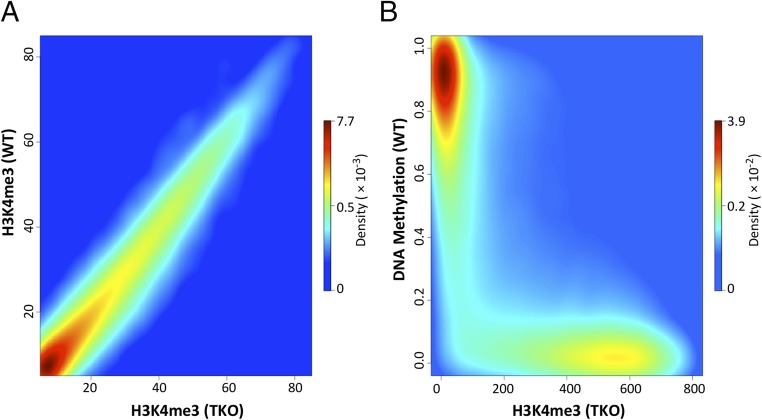
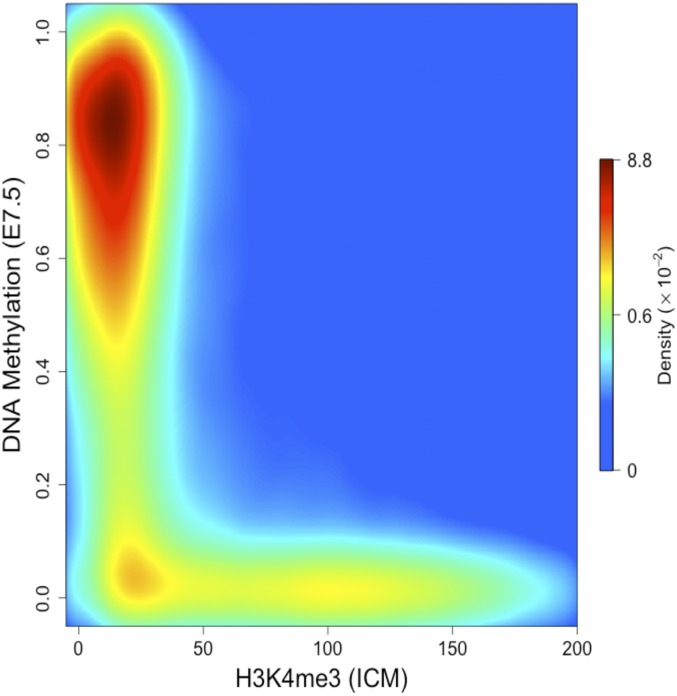
In light of these experiments, we thought it would be instructive to reexamine the relationship between the distribution of H3K4me3 and the pattern of DNA methylation in early embryonic cells. To this end, we carried out ChIP-Seq for H3K4me3 and compared its distribution in WT as opposed to TKO ES cells. Strikingly, the physical localizations and densities of H3K4me3 are almost identical in the two cell types, suggesting that this histone mark may already have been set up and present before the onset of DNA methylation (Fig. 3A and SI Appendix, Fig. S6A). Furthermore, the distribution of H3K4me3 in either TKO or naive ES cells (grown in 2i medium) turns out to be an excellent predictor for precisely defining the unmethylated DNA windows etched out during the generalized whole-genome de novo methylation reaction (Fig. 3B and SI Appendix, Fig. S6 B and C). To determine whether this is the case in vivo as well, we carried out ChIP-Seq of H3K4me3 in the ICM and found that this histone mark indeed serves as a template for whole-genome de novo methylation as seen in the early postimplantation embryo (E7.5) (Fig. 4). Thus, while the correlation between H3K4me3 and undermethylation of DNA is well known, our results suggest that the preestablished presence of a K4 histone methylase itself or the consequent methylation of H3K4 residues on local nucleosomes is actually capable of inhibiting de novo methylation of underlying DNA, even if the target sequence is not a genuine CpG island (Fig. 2C).
Fig. 3.
H3K4me3 predicts DNA undermethylation. (A) Density scatter plot of H3K4me3 (normalized ChIP-Seq reads) in TKO cells vs. H3K4me3 in WT ES cells. (B) Density scatter plot of H3K4me3 (normalized ChIP-Seq reads) in TKO cells vs. percent DNA methylation (normalized RRBS) in WT ES cells. Note that regions marked with H3K4me3 in TKO are unmethylated in WT ES cells. The density of dots is color-coded from blue (low) to red (high).
Fig. 4.
H3K4me3 predicts undermethylation in vivo. Density scatter plot of H3K4me3 (normalized ChIP-Seq reads) in ICM vs. percent DNA methylation (normalized RRBS) in 7.5-d embryos. Dot density is color-coded from blue (low) to red (high).
Discussion
Our results provide a way of understanding how the basic bimodal DNA methylation pattern of all cells is established in the early embryo. According to this model (Fig. 5), transient interactions (21) between cis-acting sequences and transacting factors in the preimplantation embryo indeed play a role in this process (5, 6). It appears to accomplish this by directly determining the binding pattern of transcription complexes on genomic DNA that, at this stage, is largely unmethylated. At the time of implantation, the entire genome is then subject to de novo methylation, but regions already primed for transcription and marked with H3K4me3 are protected, although they may not necessarily undergo active transcription (22). It should be noted that our data do not rule out the possibility that even sites not associated with RNA polymerase may be protected from generalized de novo DNA methylation, perhaps by virtue of other recognition factors (23) independently capable of recruiting H3K4 methylases (Fig. 2), as may be the case for polycomb-bound CpG island promoters (SI Appendix, Fig. S2D). This would be consistent with the observation that some nonpromoter-containing CpG-rich sequences remain unmethylated when introduced into embryonic cells (24). Once formed at the time of implantation, the bimodal methylation pattern is then largely maintained through all cell divisions despite the absence of the original factors that brought about de novo methylation in the first place (25).
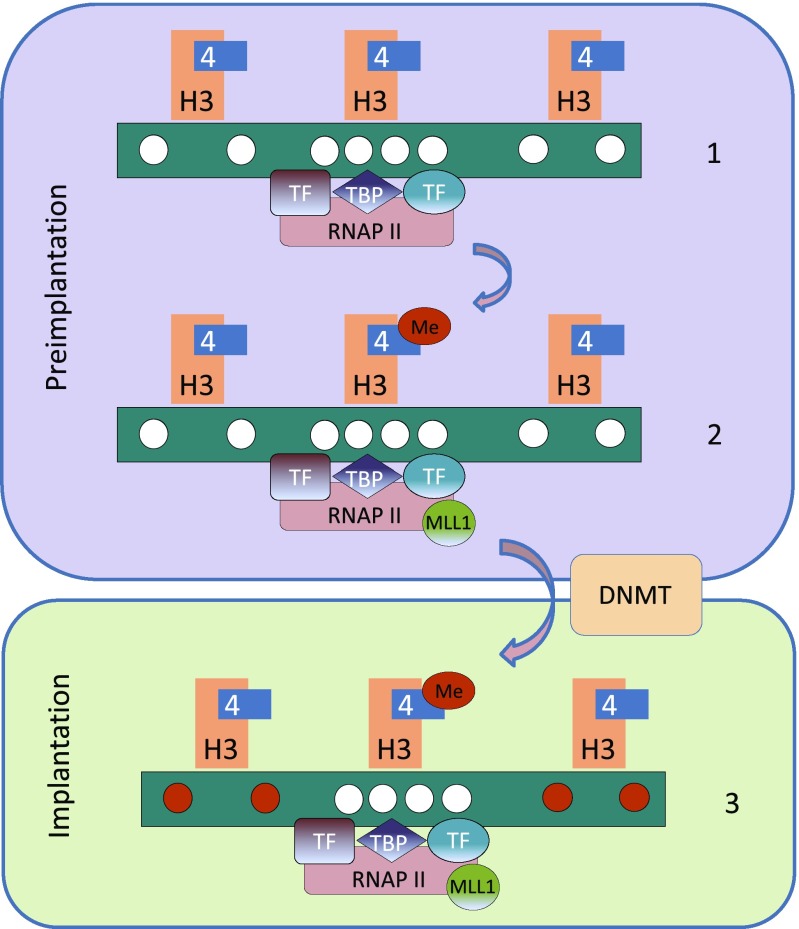
Fig. 5.
Mechanism of CpG island protection. Preimplantation, the genome is largely unmethylated (white circles) and RNAP II complexes are associated with sites of potential transcription by virtue of TBP as directed by transcription-factor recognition (1). RNAP II then recruits the MLL1 complex that brings about lysine 4 methylation of histone H3 within local nucleosomes (2). De novo methylation at the time of implantation (red circles) is carried out by the DNMT complex that can bind all regions of the DNA, as long as the local nucleosomes are not methylated at the H3 lysine 4 position (3).
Our results suggest that global DNA methylation at the time of implantation serves to provide an epigenetic memory of the early embryo transcription-complex map. Thus, while this modification does not itself forcefully initiate the repression of gene expression, it probably does serve as a stable mechanism for inhibiting gene activation in somatic cells where the transcription factor landscape may be much different from what was present in the early embryo. Good examples of genes that are subject to this process in vivo include many endogenous viral sequences as well as high profile tissue-specific genes (26, 27).
It appears that global methylation only occurs in the early embryo, and following implantation, all additional de novo events are probably targeted to specific sequences. Nonetheless, the general rules of this process remain the same, with methylation serving as a secondary mechanism intended to maintain repression at later stages of development. This is the case for the repression of pluripotency genes, such as Oct4, which is actually initiated by trans acting factors in the postimplantation embryo with modification only taking place at a later stage (28, 29), and the same is true for gene sequences subject to X inactivation in the female (25). Despite being a secondary event, the continued presence of methylation has been shown to play an important role in preventing reactivation (27, 30). In cancer, as well, abnormal de novo DNA methylation mostly takes place on polycomb-bound CpG-island promoters that are already repressed (31, 32), thereby converting the regulatory mode of these genes from initially being reversible to a state of stable silencing (33).
The general picture that emerges from these studies is that the addition of methyl groups to DNA can only take place on gene regions that are not selected for transcription and that this modification then serves to prevent activation, perhaps by decreasing their accessibility to protein factors (9, 34). A corollary to this idea is that for many genes, the removal of methylation at regulatory sequences may be necessary to turn on full expression, and experiments using Tet knockouts to prevent demethylation in ES cells (35) as well as in vivo during normal development (36, 37) now indicate that this is indeed the case.
The fact that the basal methylation pattern is established through interactions between cis-acting and early-embryonic transacting factors (5) that determine the positioning of transcription complexes on the DNA may have far reaching implications. Thus, while the expression of these regulatory proteins and perhaps noncoding RNA factors (38) may be largely intrinsic to the embryonic state, one can speculate that this composition may also be influenced by local environmental effectors, both in the early embryo as well as in the gametes. If this were the case, these interactions could provide a sophisticated mechanism for events that occur in the gametes to ultimately influence the overall methylation pattern of the offspring (39).
Methods
Cells.
Mouse WT (J1) and TKO (obtained from M. Okano, RIKEN, Japan) ES cells were grown in DMEM supplemented with 20% FCS, P/S, 1% sodium pyruvate, 0.2% mercaptoethanol and glutamine plus leukemia inhibitory factor. Cotransfection of DNA constructs was carried out using FuGENE (Roche) and subjected to G418 or puromycin selection to derive stable clones.
Mouse ICM cells were isolated from fertilized embryos in accordance with IACUC protocol B2010-141 “Transgenic Mouse Models of Human Disease/Transgenic Core.” To induce oocyte maturation and ovulation, females (3–5 wk of age) were given follicle-stimulating hormone (PMS) (5 IU/0.1 mL) on day 1 and, 46 h later, human CG was administered (5 IU/0.1 mL in PBS) and animals mated with stud male mice. Plugged mothers (3.5 d postcoitum) were killed by cervical dislocation, and the oviducts and uterine horns dissected for embryo harvest. Inner cell masses were isolated by immunosurgery as described (40). Embryos were pooled (>20,000 cells) before ChIP-Seq.
Constructs.
A plasmid containing TBP fused to the GAL4-binding domain under an SV40 promoter was obtained from K. Struhl, Harvard Medical School, Boston (41). The human CRYAA promoter was cloned into a plasmid vector carrying an upstream 4×GAL4-binding sequence. Coding sequences of Setd1 and RXR were cloned into a plasmid containing the GAL4 fusion vector by replacing the TBP sequence. WDR5-GAL4 under the CMV promoter was obtained from Joanna Wysocka, Stanford University School of Medicine, Stanford, CA (42).
For ChIP, cells were cross-linked and chromatin extracted and then sonicated to an average size of 500–2,000 bp. Immunoprecipitation was carried out using the ChIP assay kit as recommended by the manufacturer (Upstate Biotechnology). Antibodies (5 μg/10–30 μg DNA) were directed against the GAL4 DNA-binding domain (catalog no. 06-262; Millipore), histone H3K4me3 (catalog no. 39160; Active Motif) or RNAP II (catalog no. 17-620; Millipore). Incubation with the various antibodies was followed by Salmon Sperm DNA/Protein A Agarose (60 μL/10 μg DNA) precipitation to isolate the bound fraction. Because we usually precipitated <1% of the chromatin, PCR analysis of the bound fraction was compared with a 1:100 dilution of input DNA. Amplification was carried out by real-time PCR, and enrichment was determined after correction for the positive precipitation control and then normalized by setting the negative control to 1. ChIP-Seq was carried out using antibodies against RNAP II (MMS-128P; Antibodypedia) or H3K4me3 (Millipore 07–473 JBC 188194), as described (43). For ChIP-Seq, precipitations were carried out on two or three different samples and tested for maximum effectiveness by qPCR of selected sites before sequencing (single replicate). All data have been deposited in the Gene Expression Omnibus (GEO) under accession no. GSE98083.
Methylation Analysis.
Bisulfite conversion of genomic DNA was carried out using the EZH2 DNA Methylation Gold Kit (Zymo Research) according to the manufacturer’s instructions. PCR primers were designed using Methyl Primer Express Software v1.0 (https://www.thermofisher.com/us/en/home/brands/applied-biosystems.html). Two sets of primers 5′–3′ for CRYAA were used as follows:
FD1 TATTTTGGGGTTTTTTTATTTTAGT
BK1 AACCACCACCCTCTAAACAAC
FD2 TTTGTTGGTGGTATATAAAGGG
BK2 ATCCATATTCAACTTTAATACCCAT
Barcodes and adaptors were added to the primers and deep-sequenced using MiSeq.
Data Analysis.
ChIP-Seq reads were aligned to the mm9 genome assembly using MAQ and PCR duplicates as well as reads with low mapping quality (<30) were discarded. Read counts were binned into 25-bp regions and normalized by RPKM. ES cell DNA methylation data were downloaded from GSE11034 (RRBS) and GSE30202 (WGBS). E7.5 DNA methylation data were downloaded from GSE34864. CpGs with less than 10 reads were discarded, and average DNA methylation values were calculated for 100-bp regions. For comparison between DNA methylation and ChIP-Seq signals, DNA methylation values were compared with the strongest ChIP-Seq signal within 1,000 bp. For comparison between WT and TKO H3K4me3 signals, read counts were binned in 1,000-bp regions and normalized by RPKM.
To estimate the quality of our ChIP-Seq data, we compared the peaks obtained for H3K4me3 in both WT and TKO cells with published data (Series GSE23943) (11) and obtained >90% overlap. RNAP II in TKO compared with published data in naive ES cells (grown in 2i) (SI Appendix, Fig. S2B) yielded >70% overlap. Peak reads for both our ChIP-Seq data and from the literature for comparison (11) are shown in SI Appendix, Table S1.
Supplementary Material
Acknowledgments
This research was supported by grants from the European Research Council, the Israel Science Foundation, and the Israel Cancer Research Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98083).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804755115/-/DCSupplemental.
References
- 1.Long HK, King HW, Patient RK, Odom DT, Klose RJ. Protection of CpG islands from DNA methylation is DNA-encoded and evolutionarily conserved. Nucleic Acids Res. 2016;44:6693–6706. doi: 10.1093/nar/gkw258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Straussman R, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 3.Ludwig G, et al. Aberrant DNA methylation in ES cells. PLoS One. 2014;9:e96090. doi: 10.1371/journal.pone.0096090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Habibi E, et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell. 2013;13:360–369. doi: 10.1016/j.stem.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Brandeis M, et al. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 6.Lienert F, et al. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 7.Sabag O, et al. Establishment of methylation patterns in ES cells. Nat Struct Mol Biol. 2014;21:110–112. doi: 10.1038/nsmb.2734. [DOI] [PubMed] [Google Scholar]
- 8.Tsumura A, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 9.Domcke S, et al. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature. 2015;528:575–579. doi: 10.1038/nature16462. [DOI] [PubMed] [Google Scholar]
- 10.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marks H, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorris DR, Struhl K. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol Cell Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luse DS. The RNA polymerase II preinitiation complex. Through what pathway is the complex assembled? Transcription. 2014;5:e27050. doi: 10.4161/trns.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Fang J, Taatjes DJ, Nogales E. Structural visualization of key steps in human transcription initiation. Nature. 2013;495:481–486. doi: 10.1038/nature11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shilatifard A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otani J, et al. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noh KM, et al. Engineering of a histone-recognition domain in Dnmt3a alters the epigenetic landscape and phenotypic features of mouse ESCs. Mol Cell. 2015;59:89–103. doi: 10.1016/j.molcel.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo X, et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015;517:640–644. doi: 10.1038/nature13899. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg MV, et al. Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat Genet. 2017;49:110–118. doi: 10.1038/ng.3718. [DOI] [PubMed] [Google Scholar]
- 22.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 23.Hanna CW, et al. MLL2 conveys transcription-independent H3K4 trimethylation in oocytes. Nat Struct Mol Biol. 2018;25:73–82. doi: 10.1038/s41594-017-0013-5. [DOI] [PubMed] [Google Scholar]
- 24.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 26.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 27.Goren A, et al. Fine tuning of globin gene expression by DNA methylation. PLoS One. 2006;1:e46. doi: 10.1371/journal.pone.0000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epsztejn-Litman S, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enver T, Zhang JW, Papayannopoulou T, Stamatoyannopoulos G. DNA methylation: A secondary event in globin gene switching? Genes Dev. 1988;2:698–706. doi: 10.1101/gad.2.6.698. [DOI] [PubMed] [Google Scholar]
- 30.Feldman N, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 31.Keshet I, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 32.Nejman D, et al. Molecular rules governing de novo methylation in cancer. Cancer Res. 2014;74:1475–1483. doi: 10.1158/0008-5472.CAN-13-3042. [DOI] [PubMed] [Google Scholar]
- 33.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76:3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 34.Liu XS, et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247.e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawlaty MM, et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev Cell. 2014;29:102–111. doi: 10.1016/j.devcel.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlanski S, et al. Tissue-specific DNA demethylation is required for proper B-cell differentiation and function. Proc Natl Acad Sci USA. 2016;113:5018–5023. doi: 10.1073/pnas.1604365113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reizel Y, et al. Postnatal DNA demethylation and its role in tissue maturation. Nat Commun. 2018;9:2040. doi: 10.1038/s41467-018-04456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eidem TM, Kugel JF, Goodrich JA. Noncoding RNAs: Regulators of the mammalian transcription machinery. J Mol Biol. 2016;428:2652–2659. doi: 10.1016/j.jmb.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carone BR, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Solter D, Knowles BB. Immunosurgery of mouse blastocyst. Proc Natl Acad Sci USA. 1975;72:5099–5102. doi: 10.1073/pnas.72.12.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherwood RI, Chen TY, Melton DA. Transcriptional dynamics of endodermal organ formation. Dev Dyn. 2009;238:29–42. doi: 10.1002/dvdy.21810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 43.Busby M, et al. Systematic comparison of monoclonal versus polyclonal antibodies for mapping histone modifications by ChIP-seq. Epigenetics Chromatin. 2016;9:49. doi: 10.1186/s13072-016-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.