Significance
Fragile X syndrome (FXS) is the most common form of inherited intellectual disability and is a leading genetic cause of autism. The mammalian target of rapamycin (mTOR) complex 1 cascade is a central regulator of protein translation, cell growth, proliferation, survival, and autophagy. Findings in the present study demonstrate that autophagy and protein degradation via the autophagy/lysosomal pathway are reduced in hippocampal neurons of Fmr1-KO mice, a model of human FXS. We show that excess mTOR activity is causally related to decreased autophagy, which induces spine defects, exaggerated synaptic plasticity, and impaired cognition in Fmr1-KO mice. These findings increase our understanding of the etiology of FXS and suggest components of the autophagy pathway as promising targets for amelioration of FXS in humans.
Keywords: fragile X syndrome, autophagy, cognition, autism, mTOR
Abstract
Fragile X syndrome (FXS) is the most frequent form of heritable intellectual disability and autism. Fragile X (Fmr1-KO) mice exhibit aberrant dendritic spine structure, synaptic plasticity, and cognition. Autophagy is a catabolic process of programmed degradation and recycling of proteins and cellular components via the lysosomal pathway. However, a role for autophagy in the pathophysiology of FXS is, as yet, unclear. Here we show that autophagic flux, a functional readout of autophagy, and biochemical markers of autophagy are down-regulated in hippocampal neurons of fragile X mice. We further show that enhanced activity of mammalian target of rapamycin complex 1 (mTORC1) and translocation of Raptor, a defining component of mTORC1, to the lysosome are causally related to reduced autophagy. Activation of autophagy by delivery of shRNA to Raptor directly into the CA1 of living mice via the lentivirus expression system largely corrects aberrant spine structure, synaptic plasticity, and cognition in fragile X mice. Postsynaptic density protein (PSD-95) and activity-regulated cytoskeletal-associated protein (Arc/Arg3.1), proteins implicated in spine structure and synaptic plasticity, respectively, are elevated in neurons lacking fragile X mental retardation protein. Activation of autophagy corrects PSD-95 and Arc abundance, identifying a potential mechanism by which impaired autophagy is causally related to the fragile X phenotype and revealing a previously unappreciated role for autophagy in the synaptic and cognitive deficits associated with fragile X syndrome.
Fragile X syndrome (FXS) is the most common heritable form of intellectual disability and is a leading genetic cause of autism (1–4). Patients with FXS exhibit a complex and debilitating neurological phenotype including impaired cognition and social interactions, delayed speech, hyperactivity, emotional lability, attentional deficits, resistance to change, seizures, sleep disorders, gross motor delays, hypersensitivity to sensory stimuli, autistic behaviors, and autonomic dysfunction (1–4). In FXS, expansion of a poly-CGG trinucleotide repeat located in the 5′ UTR of the fragile X mental retardation 1 (Fmr1) gene from ∼50 to >200 repeats results in hypermethylation and epigenetic silencing of the Fmr1 gene, causing FXS (5–7). Fragile X mental retardation protein (FMRP), the gene product of Fmr1, is an RNA-binding protein implicated in cognitive dysfunction and autism that tightly regulates the trafficking, localization, and translation of a vast number of neuronal mRNAs critical to neural development, synaptic plasticity, and dendritic spine architecture (1, 8–10). An effective treatment for fragile X in humans is still lacking.
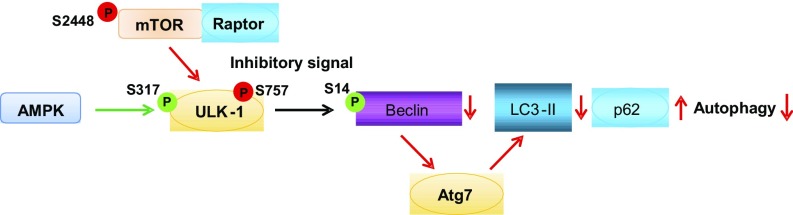
Mammalian target of rapamycin (mTOR) is a serine/threonine kinase that integrates external cues and is a central regulator of protein synthesis, cell growth, proliferation, survival, and autophagy (11–14). Overactivation of mTOR complex 1 (mTORC1) at hippocampal synapses is a phenotypic feature of fragile X humans, mice, and flies (7, 13, 15, 16). In neurons, mTORC1 is strategically positioned at pre- and postsynaptic sites where it promotes local protein translation (6–9) and serves as a brake on autophagy (11–14). Autophagy is a catabolic process that plays a pivotal role in synaptic remodeling and memory formation (17–19). Upon activation, mTOR phosphorylates Unc-51–like autophagy-activating kinase 1 (ULK-1) at S757, a target of mTORC1 and a well-established antiautophagy site (Fig. 1) (20). This, in turn, sequesters ULK-1 away from AMP kinase (AMPK). AMPK phosphorylates and activates ULK-1 at S317. Upon activation, ULK-1 promotes phosphorylation and activation of Beclin-1 at S14, a critical step in the nucleation phase of autophagy (21). Beclin-1 promotes lipidation of LC3-I to generate its lipidated form LC3-II, enabling elongation of the limiting membrane and the formation of autophagosomes (22). Autophagosomes act via protein adaptors such as p62 to deliver proteins and organelles to lysosomes for degradation.
Fig. 1.
Schematic of mTOR and autophagy signaling. Upon activation, mTOR phosphorylates ULK-1 at S757, a target of mTORC1 and a well-established antiautophagy site. This, in turn, sequesters ULK-1 away from AMPK, which phosphorylates and activates ULK-1 at S317. Phosphorylation of ULK-1 at S317 promotes phosphorylation and activation of Beclin-1 at S14, a critical step in the nucleation phase of autophagy. Beclin-1 promotes lipidation of LC3-I to generate its lipidated form LC3-II, enabling elongation of the limiting membrane and formation of autophagosomes. Lipidation of LC3-I is an ubiquitin-like reaction that requires Atg7. Autophagosomes act via protein adaptors such as p62 to deliver proteins and organelles to lysosomes for degradation.
Whereas it is well established that protein translation is elevated in neurons of fragile X mice (7, 10, 13–15, 23, 24), a role for suppressed autophagy and protein degradation is, as yet, unclear. The present study was undertaken to examine the possibility that autophagy, which is negatively regulated by phosphorylation of mTOR on S2448 [(25), but see ref. 26], is reduced in hippocampal neurons from fragile X mice. We focused on the hippocampus, which is central to cognition (27, 28). Using a combination of molecular, electrophysiological, and behavioral approaches, we found that autophagy is reduced in hippocampal neurons of fragile X mice and that activation of autophagy rescues, at least in part, aberrant dendritic spine morphology, metabotropic glutamate receptor-dependent long-term depression (mGluR-LTD), and cognition in fragile X mice. Moreover, activation of autophagy largely corrects the abundance of the postsynaptic density protein PSD-95 and the activity-regulated cytoskeletal-associated synaptic proteins Arc/Arg3.1 implicated in spine structure and synaptic plasticity. In summary, these findings document a causal relation between decreased autophagy and the synaptic and cognitive deficits in FXS and identify components of the autophagy pathway as potential therapeutic targets for amelioration of the symptoms of FXS.
Results
Autophagy Is Down-Regulated in Hippocampal Neurons of Fragile X Mice.
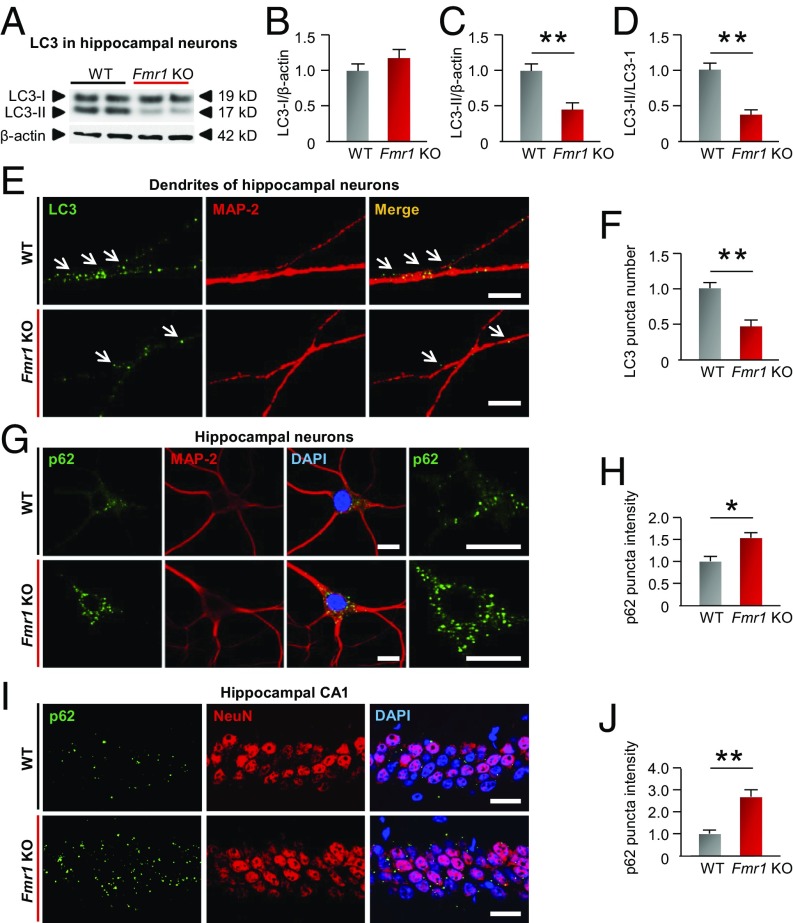
Initiation of macroautophagy starts with autophagosome formation, which depends on the lipidation of LC3-I to generate its lipidated counterpart LC3-II. Upon lipidation, LC3-II localizes to the phagophore membrane, enabling elongation of the limiting membrane to enclose the substrate and form mature LC3-II+ autophagosomes (17–19). We assessed autophagosome formation by two different assays. First, we measured the abundance of LC3-I and LC3-II (Fig. 2 A–C) and the LC3-II/I ratio (Fig. 2 A and D) in neurons from fragile X and WT mice. LC3-II abundance and the LC3-II/I ratio were markedly diminished in hippocampal neurons from Fmr1-KO mice, indicating reduced autophagosome synthesis. Second, we examined the distribution of LC3 in neurons by immunofluorescence. In neurons, autophagosomes form mainly in dendrites and are transported to the cell body for degradation (18). Punctate LC3 immunofluorescence in dendrites affords a measure of newly formed autophagosomes (Fig. 2E) (18, 29). LC3 puncta were markedly reduced in dendrites of Fmr1-KO vs. WT neurons, consistent with reduced autophagosome formation (Fig. 2F).
Fig. 2.
Autophagy is down-regulated in hippocampal neurons of fragile X mice. (A) Representative Western blots of whole-cell lysates from primary cultures of hippocampal neurons at DIV 14 from WT and Fmr1-KO mice were probed for LC3-I and LC3-II. β-Actin was used as a loading control. (B–D) Summary data showing LC3-I (B), LC3-II (C), and the LC3-II/I ratio (D) (normalized to the corresponding WT values). (E) Green immunofluorescence (LC3+ puncta) marks autophagosomes (arrows) localized to MAP-2–labeled dendrites (red) in neurons from KO and WT mice. (Scale bars, 10 µm.) (F) Summary data showing number of green (LC3+) puncta colocalized with red (MAP2+) in dendrites (normalized to WT group). (G) Images show immunolabeling of p62 together with MAP-2 to mark neurons. p62 staining at higher magnification is shown in Right panels. (Scale bars, 15 µm.) (H) Summary data (bar graphs) showing fluorescence intensity (normalized to WT values). (I) Five-week-old male WT and Fmr1-KO mice were killed, and frozen sections of brain were subjected to immunolabeling with an antibody to p62 together with an antibody to NeuN to mark neurons. (Scale bars, 25 µm.) (J) Summary data show the calculated intensity (normalized to corresponding WT values). *P < 0.05, **P < 0.01. Significance was calculated by a two-tailed Student’s t test; n = 5 experiments involving independent batches of neurons cultured from different litters in B–D, F, and H. At least 50 neurons were analyzed for each treatment group in F and H. n = 4 mice for J. Values reflect mean ± SEM.
We next measured autophagic flux, a functional readout of lysosomal degradation (17–19, 30). Autophagosomes deliver proteins and organelles to lysosomes for degradation by means of cargo adaptor proteins, such as p62 (18). When the final degradation step is reduced, p62 accumulates. We examined the abundance of p62 in primary cultures of hippocampal neurons from Fmr1-KO and WT mice. p62 was markedly increased in neurons from Fmr1-KO vs. WT mice (Fig. 2 G and H), primarily in the somata, consistent with the notion that the degradation of substrate by the autophagy/lysosomal pathway is markedly reduced. Similar results were observed in CA1 neurons assessed in brain sections from Fmr1-KO and WT mice (Fig. 2 I and J). Collectively, these findings indicate that autophagic flux is down-regulated in neurons of Fmr1-KO mice.
mTORC1 Activity Is Enhanced in Hippocampal Neurons from Fragile X Mice.
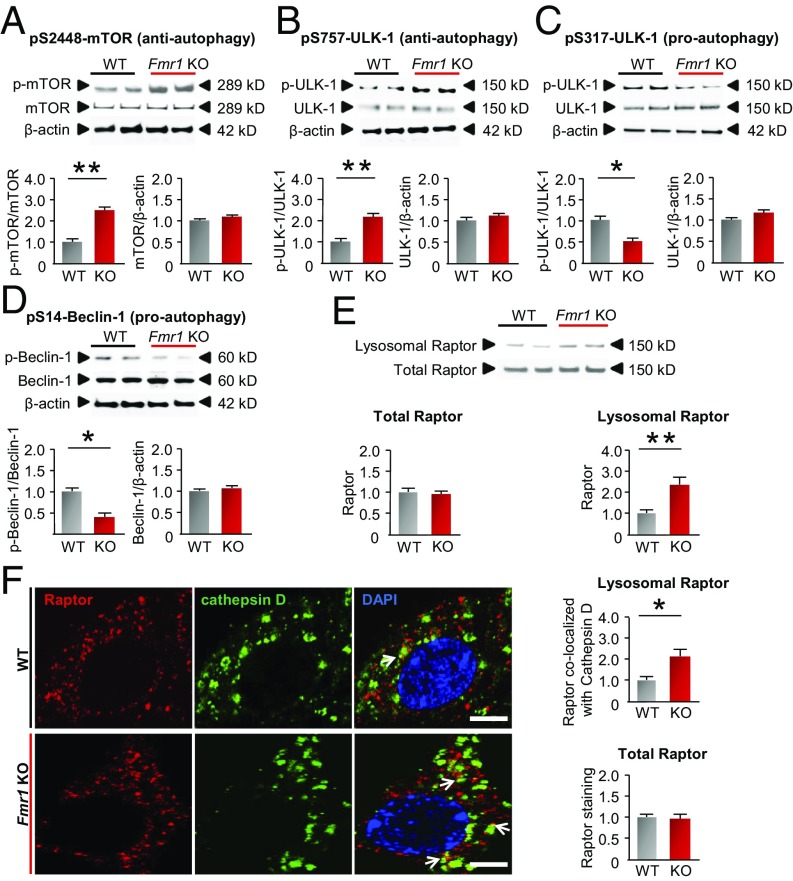
The results thus far suggest that autophagosome formation and autophagic flux are reduced in hippocampal neurons from fragile X mice but do not address the mechanisms underlying this deficit. Overactivation of mTORC1 in hippocampal neurons is a phenotypic feature common to FXS and other autism-like disorders (15, 23). Hippocampal neurons from Fmr1-KO mice exhibit elevated phosphorylation of mTOR at S2448, indicating overactivation of mTOR (Fig. 3A). mTOR-dependent phosphorylation of ULK-1 at S757, an antiautophagy site, prevents assembly of ULK-1 with scaffolding protein Atg13 and adhesion protein FIP200 (20) and halts the initiation of autophagy. Phosphorylation of ULK-1 at S757 was markedly increased in Fmr1-KO vs. WT neurons, consistent with down-regulated autophagy, with little or no change in ULK-1 abundance (Fig. 3B). In response to cellular stress, ULK-1 is phosphorylated at S317 by AMPK to induce the initiation of autophagy (31). Consistent with this, phosphorylation of ULK-1 at S317 was decreased in neurons from KO vs. WT mice (Fig. 3C). Moreover, phosphorylation and activation of Beclin-1 at S14, a downstream target of ULK-1, was markedly reduced in neurons from KO vs. WT mice, with little or no change in protein abundance (Fig. 3D). Loss of Beclin-1 phosphorylation halts initiation of autophagy (21). These findings indicate that overactivated mTORC1 signaling imposes a brake on autophagy in the hippocampal neurons of Fmr1-KO mice.
Fig. 3.
mTORC1 activity and downstream signals are dysregulated in Fmr1-KO mice. Primary neurons were cultured from the hippocampus of WT and Fmr1-KO mice at E18. (A–D) Cell lysates were probed with antibodies to components of upstream signaling pathways that regulate autophagy and protein abundance, and phosphorylation status was assessed by Western blot analysis with phospho-specific antibodies. (A) Phosphorylation of mTOR at S2448 (antiautophagy site). (B) Phosphorylation of ULK-1 at S757 (antiautophagy site). (C) Phosphorylation of ULK-1 at S317 (proautophagy site). (D) Phosphorylation of Beclin-1 at S14 (proautophagy site). (Upper) Representative Western blots. (Lower) Bar graphs show summary data (values were normalized to the corresponding WT group). β-Actin was used as loading control. (E) Lysosome fractions and whole-cell lysates were collected from the hippocampi of 5-wk-old male WT and Fmr1-KO mice. Raptor protein abundance was assessed in whole-cell lysates (total Raptor) and in lysosomal fractions (lysosomal Raptor) by means of Western blot analysis. Summary data show total and lysosomal Raptor (normalized to the corresponding values for WT); n = 5 WT and 5 KO mice. (F) Lysosomal Raptor localization was shown by immunolabeling of Raptor together with a lysosomal marker, cathepsin D, in primary hippocampal neurons cultured from WT and Fmr1-KO mice at DIV 14. Bar graphs show the calculated intensities of colocalized puncta and total Raptor (normalized to WT). (Scale bars, 8 µm.) *P < 0.05, **P < 0.01. Significance was calculated by two-tailed Student’s t test; n = 4 experiments involving independent batches of neurons cultured from different litters. At least 30 neurons were included for each group in F. Values reflect mean ± SEM.
Raptor, an obligatory component of mTORC1, is a critical binding partner of mTOR that facilitates recruitment of substrates such as S6K and is required for the localization of mTORC1 to lysosomes, where it imposes a brake on autophagy by sequestering ULK-1 (32, 33). We examined the translocation of Raptor to the lysosomal surface, where it plays a critical role in suppression of autophagy. Whereas Raptor abundance did not differ significantly in whole-cell lysates from the hippocampus, it was markedly increased in the lysosomal fraction isolated from the hippocampus of Fmr1-KO vs. WT mice, indicating increased mTORC1 activity (Fig. 3E). Cathepsin D is a lysosomal aspartyl protease and is widely used as a marker of lysosomes (34). Localization of Raptor to lysosomes marked by cathepsin D was elevated in hippocampal neurons from Fmr1-KO vs. WT mice, as assessed by immunolabeling of neurons (Fig. 3F). Thus, localization of mTORC1 to lysosomes is elevated, enforcing the brake imposed by mTOR on autophagy, in neurons from Fmr1-KO mice.
RNAi-Mediated Depletion of Raptor Rescues Autophagy in Fragile X Mice.
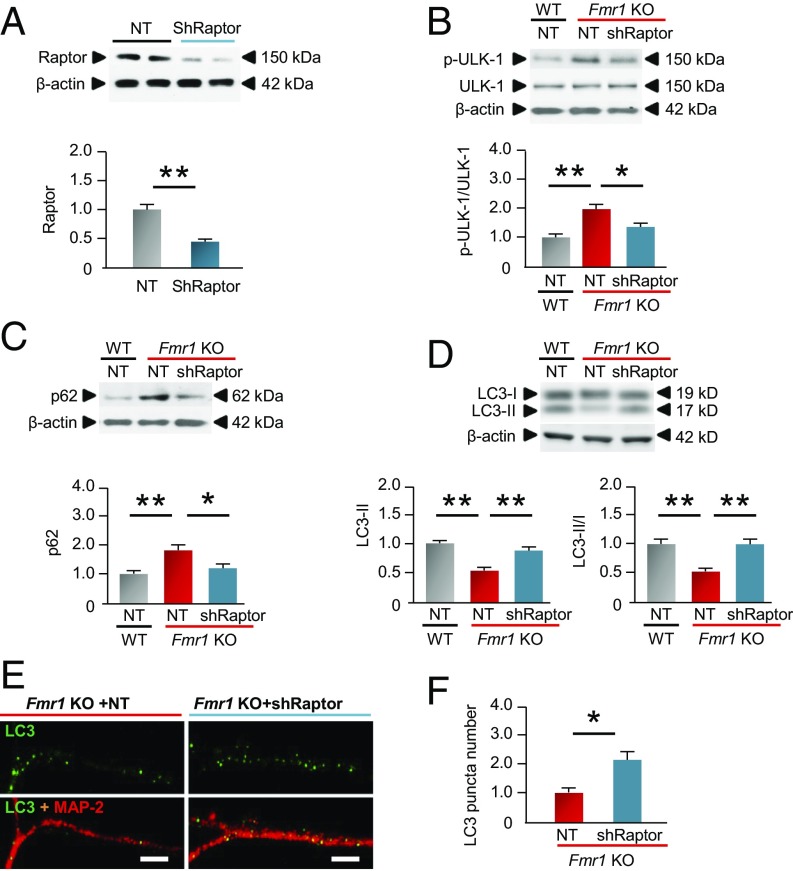
The results thus far indicate that elevated mTORC1 activity and localization of Raptor to lysosomes are associated with decreased autophagy but do not address a causal relation between them. Toward this end, we expressed shRNA to Raptor (shRaptor) directly into the hippocampal CA1 of intact mice and in hippocampal neurons by means of the lentivirus expression system (to suppress mTORC1) and assessed autophagic flux by p62 accumulation and autophagosome formation. Nontargeting (NT) shRNA, which does not recognize any known vertebrate sequence, served as a negative control. Validation experiments performed in vivo revealed that shRaptor injected directly into the CA1 of WT mice reduced Raptor to ∼30% of that of WT mice expressing NT shRNA (Fig. 4A). We reasoned that if mTORC1 suppresses autophagy, then knockdown of Raptor, which inhibits mTORC1 activity, would disinhibit autophagy. We examined the impact of shRaptor on biochemical markers of autophagy in hippocampal neurons cultured from Fmr1-KO mice. shRaptor reduced phosphorylation of ULK-1 at S757 (Fig. 4B) and the abundance of the cargo adaptor protein p62 (Fig. 4C), elevated LC3-II abundance and the LC3-II/I ratio (Fig. 4D), and increased the number of LC3+ autophagosomes in hippocampal neurons from Fmr1-KO mice (Fig. 4 E and F). These findings strongly suggest that knockdown of Raptor increases autophagy and that overactivated mTORC1 is causally related to the reduced autophagy observed in Fmr1-KO mice.
Fig. 4.
Raptor knockdown activates autophagy in neurons from Fmr1-KO mice. (A) We injected lentivirus carrying nontargeting shRNA (NT, negative control) or shRNA to Raptor (shRaptor) bilaterally directly into the hippocampal CA1 of 5-wk-old male WT mice. One week later, we assessed the ability of shRaptor to suppress Raptor abundance in whole-cell lysates of CA1 by Western blot analysis. (Upper) Representative Western blot. (Lower) Summary data show Raptor abundance in WT mice expressing NT shRNA or shRaptor (normalized to the value for WT mice expressing NT shRNA). (B–D) Primary hippocampal neurons cultured from WT and Fmr1-KO mice (DIV 14) were transduced with lentivirus carrying either NT shRNA (negative control) or shRaptor. Hippocampal neurons cultured from WT mice expressing NT shRNA were used as a control. Three days after transduction, the phosphorylation status of ULK-1 at Ser757 (antiautophagy site) and p62 and LC3-I and II abundance were assessed in whole-cell lysates by means of Western blot analysis. (Upper) Representative Western blots. (Lower) Summary data showing p-S757-ULK-1 (B), p62 protein (C), and the LC3-II and LC3-II/I ratio (normalized to the corresponding values for neurons from WT mice expressing NT shRNA) (D). (E) Immunolabeling of LC3+ autophagosomes (green) in dendrites marked by MAP-2 (red). (Scale bars, 10 µm.) (F) Summary data showing the number of LC3 puncta in dendrites from KO mice expressing NT or Raptor shRNA (normalized to the corresponding value for KO mice expressing NT shRNA). β-Actin was used as a loading control. *P < 0.05, **P < 0.01. Statistics were calculated by two-way ANOVA with Tukey’s test. n = 4–5 WT mice in each group for A. Four independent experiments involving separate batches of neurons cultured from different litters were analyzed in B–F. At least 50 neurons were analyzed for each treatment group in F. Values reflect mean ± SEM.
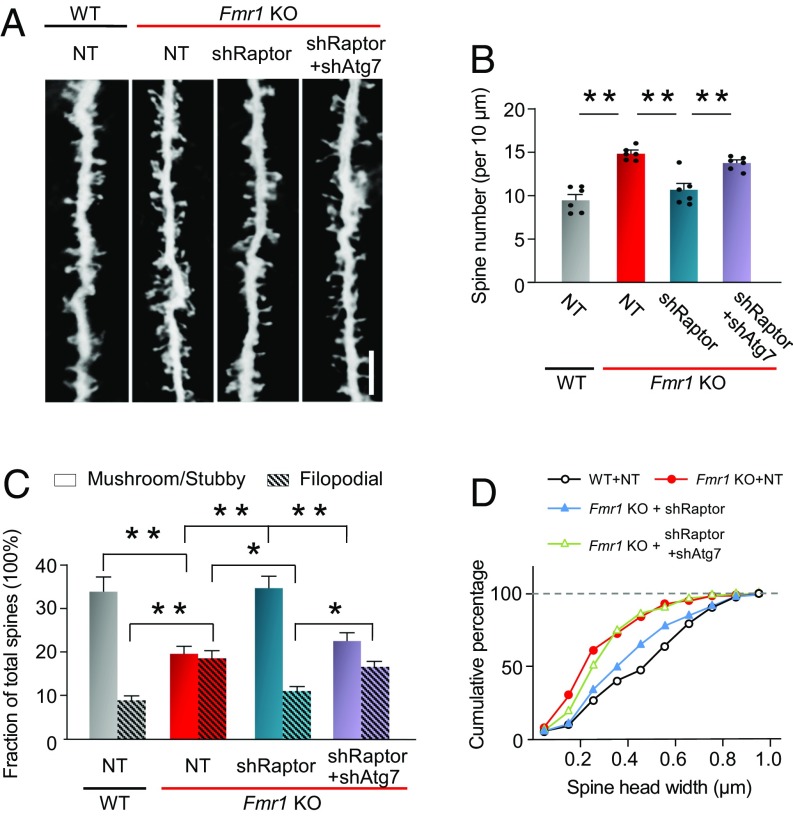
Activation of Autophagy Ameliorates Aberrant Spine Morphology of Fmr1-KO Mice.
Hippocampal neurons of fragile X patients and Fmr1-KO mice exhibit an excess of dendritic spines (27, 35, 36). To examine a role for impaired autophagy in aberrant spine morphology, we assessed the impact of shRaptor, which suppresses mTORC1 and activates autophagy, alone and in the presence of shAtg7, which suppresses autophagy, on spine structure and density. Validation experiments performed in vivo revealed that shRaptor injected directly into the CA1 of Fmr1-KO mice reduced Raptor to ∼42% of that of WT mice expressing NT shRNA; injection of shRaptor together with shAtg7 directly into the CA1 reduced Raptor protein to ∼50% of that of WT mice expressing NT shRNA (SI Appendix, Fig. S1A). We first transduced Fmr1-KO mice and WT littermates with lentivirus carrying GFP and shRaptor or NT shRNA (negative control) directly into the hippocampal CA1 and 7 d later impregnated brains with Golgi and assessed dendritic spine morphology (Fig. 5A). Untransfected CA1 neurons (not illustrated) and GFP+ CA1 neurons from Fmr1-KO mice expressing NT shRNA exhibited a higher density of spines than WT littermates (Fig. 5 A and B, compare bar 2 with bar 1), in corroboration of work published by us (37) and by others (27, 35, 38). Delivery of shRaptor directly into the CA1 of Fmr1-KO mice corrected spine density on dendrites of pyramidal neurons to nearly that of WT mice expressing NT shRNA (Fig. 5 A and B, compare bar 3 with 1). Suppression of mTORC1 not only disinhibits autophagy but also reduces protein translation, which may independently affect spine morphology and soma body size (35, 38, 39), and can potentially inhibit cell growth, proliferation, and ribosome biogenesis. To distinguish the contribution of autophagy to the rescue of spine morphology, we next delivered lentivirus carrying shRaptor to suppress mTORC1 together with shRNA directed to Atg7 (40–42), an E1-like ligase essential for autophagosome formation, directly into the CA1 of Fmr1-KO mice to selectively block autophagy (43). Validation experiments performed in vivo revealed that shAtg7 administered together with shRaptor reduced Atg7 protein in CA1 to ∼24% of that of WT mice expressing NT shRNA (SI Appendix, Fig. S1B). Expression of shAtg7 together with shRaptor partially reversed the rescue of spine density by shRaptor, consistent with the concept that rescue by shRaptor occurs, at least in part, via activation of autophagy (Fig. 5 A and B, compare bar 4 with bar 3).
Fig. 5.
Activation of autophagy corrects aberrant spine morphology in Fmr1-KO mice. Five-week-old male Fmr1-KO mice were bilaterally injected in the hippocampal CA1 with lentiviruses expressing GFP with NT shRNA, shRaptor alone, or shRaptor together with shRNA to Atg7 (shRaptor + shAtg7). Age-matched WT male mice expressing NT shRNA were used as control. One week after injection, brains were subjected to Golgi staining, and the morphology of all spines located on apical dendrites on CA1 pyramidal neurons was analyzed. (A) Representative images of spine morphology. (B) Spine number per 10 μm of dendrite. (C) Analysis of stubby/mushroom and filopodial spine fractions. (D) The head diameter was defined as the maximum width of the spine head, and spines were classified according to their head diameter at intervals of 0.1 µm. Values at each interval were calculated, and the cumulative frequency distribution was plotted. Each point in B represents data from an individual mouse. Values are shown as mean ± SEM. *P < 0.05, **P < 0.01. Statistics were calculated by two-way ANOVA with Tukey’s test; n = 6 mice for each group. Five neurons per mouse and 200 10-µm segments on the apical dendrite from the cell body to a distance of 130 μm along primary, secondary, and tertiary branches of CA1 pyramidal neurons were analyzed per group.
Next we examined the impact of autophagy on spine maturation by classifying spines as stubby and mushroom-shaped (mature) or spindly, filopodial-like protrusions (immature), together with measurement of spine-head width. Untransfected CA1 neurons (not illustrated) and GFP+ CA1 neurons expressing NT shRNA from Fmr1-KO mice exhibited a marked decrease in the percentage of mushroom or stubby spines and a marked increase in the percentage of long, filopodial-like protrusions relative to that of WT littermates (Fig. 5C, compare bars 3 and 4 with bars 1 and 2), in corroboration of refs. 27, 28, 36, and 37. shRaptor restored the percentage of mature spines to nearly WT levels (Fig. 5C, compare bars 5 and 6 with bars 1 and 2). The rescue afforded by shRaptor was largely reversed by shRNA to Atg7, a key component of autophagy, indicating that the rescue of spine morphology by shRaptor occurs largely via activation of autophagy (Fig. 5C, compare bars 7 and 8 with bars 5 and 6). Finally, shRaptor corrected the smaller head width observed in Fmr1-KO vs. WT mice (Fig. 5D), an effect significantly reduced by shAtg7 (Fig. 5D). Thus, activation of autophagy corrected, at least in part, abnormalities in spine structure associated with Fmr1-KO mice.
Activation of Autophagy Corrects mGluR-LTD at CA1 Synapses of Fragile X Mice.
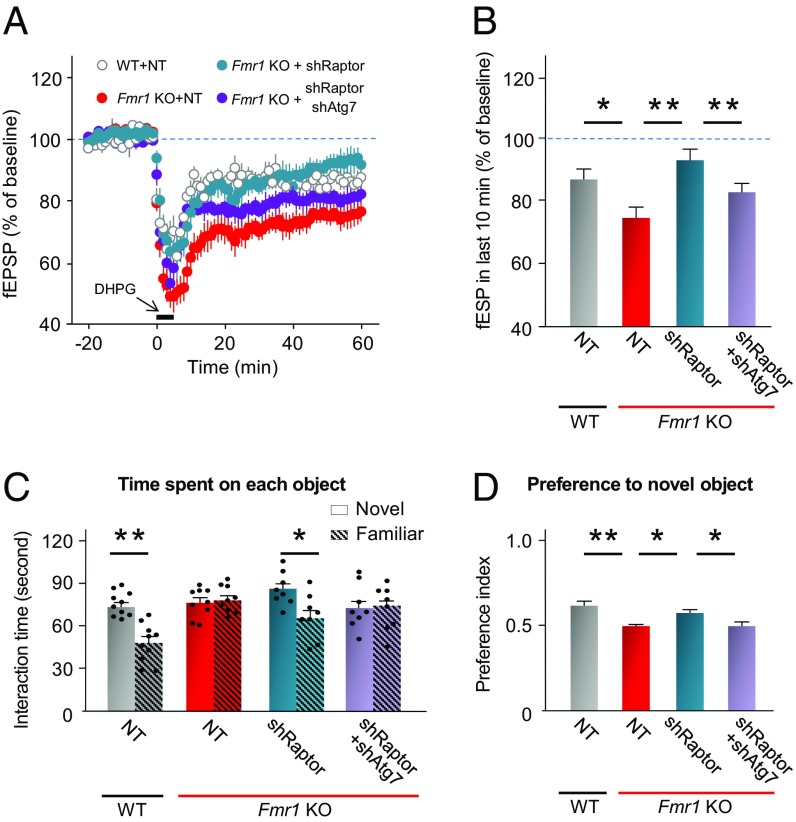
Fmr1-KO mice display exaggerated protein-synthesis independent mGluR-LTD at Schaffer collateral-to-CA1 pyramidal cell (Sch-CA1) synapses (15, 27, 44–46). Activation of group I mGluRs (mGluR1/5) with the selective agonist S-3,5-dihydroxyphenylglycine (DHPG) elicits a form of homosynaptic LTD (chemical mGluR-LTD) of synaptic transmission at Sch-CA1 pyramidal cell synapses that is independent of NMDA receptor activation (47, 48). We applied DHPG (100 μM, 5 min) to organotypic hippocampal slices from 4-wk-old WT and Fmr1-KO mice cultured for 48 h (Fig. 6A). In WT slices, DHPG produced a robust depression of the evoked field potential (fEPSP) slope to 87.2 ± 3.3% of the pre-DHPG baseline, assessed at 50 min after the induction of LTD (n = 6) (Fig. 6 A and B), consistent with reports by us and by others (15, 44). In contrast, in slices from Fmr1-KO mice, DHPG produced a greater LTD (depression of fEPSP to 75.0 ± 3.2% of baseline at 50 min, n = 6, P < 0.05 Fmr1-KO vs. WT) (Fig. 6A). To examine whether impaired autophagy contributes to exaggerated mGluR-LTD, we examined mGluR-LTD at CA1 synapses in hippocampal slices cultured from Fmr1-KO mice expressing shRaptor in the absence or presence of shAtg7. shRaptor restored mGluR-LTD to a value near that observed in slices from WT mice expressing NT shRNA (negative control). Whereas shRaptor corrected mGluR-LTD at CA1 synapses of Fmr1-KO mice to a level near that observed in WT mice, the rescue was partially reversed in slices from Fmr1-KO mice expressing shRaptor together with shAtg7 (Fig. 6 A and B), indicating that activation of autophagy is important to the rescue.
Fig. 6.
Autophagy ameliorates exaggerated mGluR-LTD and deficit in cognition in fragile X mice. (A and B) Organotypic hippocampal slices from 4-wk-old male WT and Fmr1-KO mice were cultured for 48 h. Fmr1-KO slices were transfected with lentivirus carrying NT shRNA, shRaptor, or shRaptor + shAtg7. WT slices expressing NT shRNA were used as control. Seventy-two hours after transfection, DHPG-induced mGluR-LTD was examined. (A) Time course of mGluR-LTD. (B) Mean values of the slope of the fEPSP from 50 to 60 min after initiation of DHPG application. Values are shown as mean ± SEM. *P < 0.05, **P < 0.01. Statistics were calculated by two-way ANOVA with Tukey’s test; n = two or three slices per mouse, and n = 6 mice per group. (C and D) Five-week-old male Fmr1-KO mice were bilaterally injected in the hippocampal CA1 with lentiviruses expressing: control NT shRNA, shRaptor, or shRaptor + shAtg7. Age-matched WT male mice expressing NT shRNA were used as control. One week after virus injection, visual memory was assessed by a novel object recognition task. (C) Time spent exploring novel and familiar objects in the test session. (D) The preference index for the novel object was calculated as the time spent exploring the novel object divided by the total time exploring the novel and familiar objects. Values are shown as mean ± SEM. *P < 0.05, **P < 0.01. Statistics were calculated by two-way ANOVA with Tukey’s test; n = 10 mice for WT NT group, n = 9 mice for Fmr1-KO NT group, n = 8 mice for the Fmr1-KO shRaptor group, and n = 8 mice for the Fmr1-KO shRaptor + shAtg7 group. Each point in C represents data from an individual mouse.
Activation of Autophagy Rescues Impaired Cognition in Fmr1-KO Mice.
Fmr1-KO mice display deficits in visual memory (46, 49, but see ref. 50). We next examined the impact of autophagy on cognition in Fmr1-KO mice. To this end, we delivered lentivirus carrying NT shRNA, shRaptor, or shRaptor together with shAtg7 directly into the CA1 of WT and Fmr1-KO mice and 7 d later subjected mice to the novel object recognition assay (Fig. 6 C and D). The novel object recognition task assesses visual memory and takes advantage of the innate tendency of WT mice to spend more time exploring a novel object than a familiar one. Whereas WT mice expressing NT shRNA spent more time exploring the novel object (73.5 ± 3.2 s with the novel object vs. 48.0 ± 4.3 s with the familiar object), Fmr1-KO mice expressing NT shRNA spent approximately equal times exploring the novel and familiar object (novel object, 76.1 ± 3.3 s vs. familiar object, 77.8 ± 2.5 s) (Fig. 6C), indicating, in corroboration of findings of others (27, 36), that Fmr1-KO mice had no preference for the novel object (Fig. 6D). Fmr1-KO mice expressing shRaptor spent more time exploring the novel object than the familiar object (Fig. 6C), indicating a preference for the novel object comparable to that of WT mice (Fig. 6D). shRaptor, which suppresses mTORC1 activity and thereby disinhibits autophagy, rescued visual memory in Fmr1-KO mice. Fmr1-KO mice expressing shRaptor together shAtg7 spent approximately equal time exploring novel and familiar objects (Fig. 6C), indicating little preference for the novel object (Fig. 6D). Thus, shAtg7, which inhibits autophagy, reversed, at least in part, the rescue by shRaptor, as is consistent with a mechanism in which increased autophagy is important for the rescue of cognition by shRaptor.
Reduced Protein Degradation via the Autophagy/Lysosomal Pathway Is Critical to Fragile X Pathology.
The results thus far indicate that activation of autophagy in neurons lacking FMRP rescues spine morphology, mGluR-LTD, and cognitive defects in Fmr1-KO mice but do not address the mechanism by which this occurs. A mechanism implicated in the defects in spine morphology, exaggerated mGluR-LTD at CA1 synapses, and impaired cognition associated with fragile X is the overabundance of synaptic proteins in neurons (1, 8, 9). The correct amounts of proteins at synaptic sites are maintained by a fine balance between protein synthesis and protein degradation. In neurons, autophagy plays an important role in protein degradation and the structural remodeling of synapses (51). Molecules such as p62, which bind cargo and components of the autophagic machinery, recognize and bind ubiquitinated proteins, enabling their engulfment by autophagosomes targeted for degradation (17). Reduced autophagy in brains from humans diagnosed with autism is associated with an accumulation of ubiquitinated proteins (41).
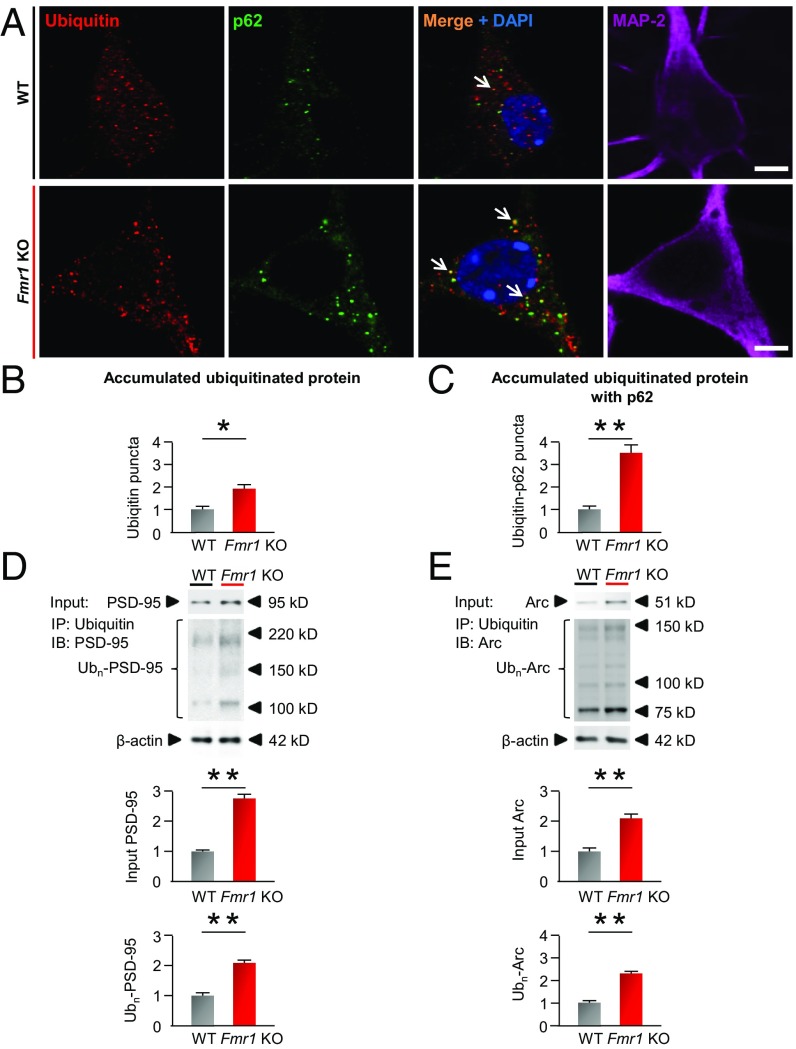
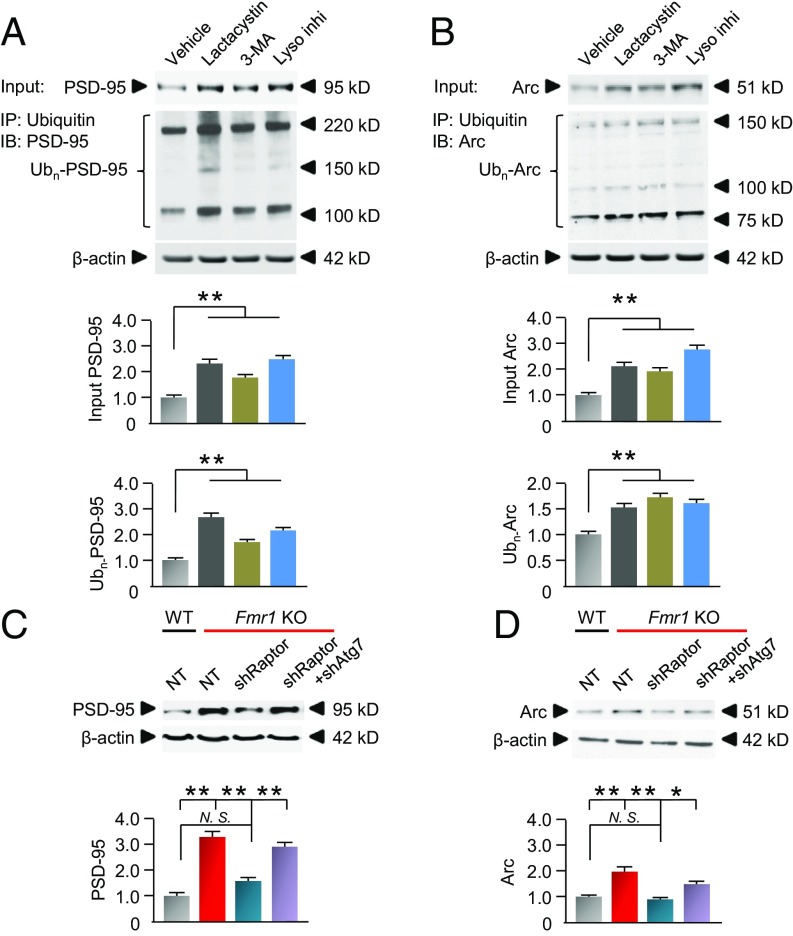
Hippocampal neurons from Fmr1-KO mice exhibited an accumulation of ubiquitinated protein aggregates (Fig. 7 A and B), which colocalized with p62 (Fig. 7 A and C), relative to that of WT neurons (Fig. 7 A–C), indicating that autophagy is involved in this accumulation of ubiquitinated proteins. A hypothesis under consideration is that a subset of synaptic proteins such as Arc and PSD-95 is degraded not only by the ubiquitin-based proteasomal degradation pathway but also by the autophagy/lysosomal pathway. Indeed, neurons from Fmr1-KO mice exhibited markedly elevated abundance of both total and ubiquitinated PSD-95 (Fig. 7D) and Arc (Fig. 7E), indicating that ubiquitinated PSD-95 and Arc accumulate and are not degraded efficiently in Fmr1-KO neurons. In corroboration of work by others (52), treatment of WT neurons with the proteasomal inhibitor lactacystin increased the abundance of both total and ubiquitinated PSD-95 (Fig. 8A) and Arc (Fig. 8B). Treatment of neurons with an inhibitor of autophagosome formation, 3-methyladenine (3-MA), and/or the lysosomal inhibitor leupeptin with ammonium chloride also induced striking accumulation of both total and ubiquitinated PSD-95 (Fig. 8A) and Arc (Fig. 8B), consistent with the concept that not only the proteasomal pathway but also the autophagy/lysosomal pathway is important to the degradation of at least a subset of synaptic proteins.
Fig. 7.
Accumulation of ubiquitinated proteins in hippocampal neurons from fragile X mice. (A) Primary cultures of hippocampal neurons (DIV 14) from WT and Fmr1-KO mice were immunolabeled with antibodies against ubiquitin, p62, and the neuronal marker MAP-2. (Scale bars, 10 µm.) (B and C) Bar graphs show density of ubiquitin puncta per soma (B) and density of puncta colabeled for p62 and ubiquitin per soma (C), each normalized to the corresponding WT value. (D and E) Protein in whole-cell lysates of primary hippocampal neurons from WT and Fmr1-KO mice was immunoprecipitated with an antibody to ubiquitin. Whole-cell lysates (Input) and immunoprecipitates (IP) were probed for PSD-95 (D) and Arc (E). (Top) Representative blots. (Middle) Summary data for input. (Bottom) Summary data for precipitate. All summary data were normalized to the corresponding values for vehicle-treated neurons. *P < 0.05, **P < 0.01. Statistics were calculated by a two-tailed Student’s t test. n = 4 independent experiments from different batches of neurons; a minimum of 50 neurons per batch for each treatment group were analyzed in B and C. Values reflect mean ± SEM.
Fig. 8.
Synaptic proteins PSD-95 and Arc are degraded via the autophagy/lysosomal pathway. (A and B) Primary cultures of hippocampal neurons were treated with vehicle, 5 μM lactacystin, 2.5 mM 3-MA, or 10 mM NH4Cl with 50 μM leupeptin (Lyso inhi) for 6 h. Lysates were extracted and immunoprecipitated with an antibody to ubiquitin. Whole-cell lysates (Input) and immunoprecipitates (IP) were probed for PSD-95 (A) and Arc (B). (Top) Representative blots. (Middle) Summary data for input. (Bottom) Summary data for precipitate. All summary data were normalized to vehicle. (C and D) Primary cultures of hippocampal neurons from WT and Fmr1-KO mice were transfected with lentiviruses expressing control NT shRNA, shRaptor, or shRaptor + shAtg7. WT neurons expressing NT shRNA served as control. Seventy-two hours after transfection, lysates were subjected to Western blots and probed for PSD-95 (C) and Arc (D). (Upper) Representative blots. (Lower) Summary data (normalized to vehicle). β-Actin served as loading control. *P < 0.05, **P < 0.01, N. S., no significant difference. Statistics were calculated by two-way ANOVA with Tukey’s test. All data were calculated from four independent experiments from different batches of neurons. Values reflect mean ± SEM.
Neurons from Fmr1-KO mice exhibited markedly elevated PSD-95 and Arc protein abundance relative to that of WT neurons. This is significant in that PSD-95 is a synaptic scaffolding protein which, when improperly degraded, contributes to the overabundance of immature spines in hippocampal neurons of Fmr1-KO mice (52), and Arc is an immediate early gene that interacts with dynamin and endophilin to facilitate endocytosis of AMPA receptors and, when dysregulated, is implicated in aberrant mGluR-LTD at CA1 synapses and memory consolidation in Fmr1-KO mice (53–55). In Fmr1-KO neurons, shRaptor, which inhibits translation and activates autophagy, reduced PSD-95 and Arc protein abundance to levels near those of WT neurons (Fig. 8 C and D). Coexpression of shRaptor with shAtg7 in Fmr1-KO neurons partially diminished the reduction in PSD-95 and Arc afforded by shRaptor, consistent with a substantial role for the autophagy/lysosomal pathway in the degradation of these synaptic proteins.
The results thus far indicate that PSD-95 and Arc are degraded, at least in part, by the autophagy/lysosomal pathway but do not provide anatomical evidence of autophagy-mediated degradation. Toward this end, we imaged PSD-95 and Arc in LC3+ autophagosomes in dendrites of WT and Fmr1-KO hippocampal neurons. RNAi-mediated knockdown of Raptor increased the number of LC3+ autophagosomes, consistent with activation of autophagy, and reduced PSD-95 and Arc puncta in dendrites of hippocampal neurons (SI Appendix, Figs. S2 and S3). Moreover, colocalization of PSD-95 and Arc puncta with LC3 puncta was increased (SI Appendix, Figs. S2 and S3), consistent with targeting of PSD-95 and Arc to the autophagy/lysosomal pathway for degradation. In Fmr1-KO neurons expressing shRaptor together with shAtg7, the number of LC3+-stained puncta was greatly diminished, PSD-95 and Arc abundance was increased, and colocalization of PSD-95 and Arc puncta with LC3 puncta was diminished, consistent with a decrease in autophagy-dependent degradation (SI Appendix, Figs. S2 and S3). shRNA to Atg7 with shRaptor increased PSD-95 and Arc puncta to nearly that in Fmr1-KO mice expressing NT shRNA while reducing the colocalization of PSD-95 and Arc puncta with LC3 puncta, indicative of reduced autophagy-dependent degradation (SI Appendix, Figs. S2 and S3). These findings strongly suggest that PSD-95 and Arc are degraded, at least in part, via the autophagy/lysosomal pathway.
Discussion
In this study we demonstrate that autophagic flux, a functional readout of autophagy, and biochemical markers of autophagy are decreased in hippocampal neurons of Fmr1-KO mice. We further show that in neurons lacking FMRP, mTORC1, which imposes a brake on autophagy, is overactivated and reduces autophagy. RNAi-mediated depletion of Raptor, a critical binding partner of mTOR and a defining component of mTORC1, activates autophagy in Fmr1-KO neurons and thereby rescues, at least in part, aberrant spine morphology, synaptic function, and cognition in Fmr1-KO mice. Rescue by shRaptor was significantly reversed by shRNA directed to Atg7, a key component of the autophagy/lysosomal pathway (Fig. 1). These findings establish a causal relation between down-regulated autophagy and cognitive defects in fragile X mice. We also show that the synaptic scaffolding protein PSD-95, critical to spine morphology, and the immediate early gene Arc, critical to mGluR-LTD at CA1 synapses, are degraded, at least in part, via the autophagy/lysosomal pathway. Whereas it is well established that elevated translation contributes to elevated PSD-95 and Arc (2), these findings document an additional role for decreased protein degradation. Although there are undoubtedly numerous other proteins that are dysregulated due to reduced autophagy in hippocampal neurons of fragile X mice, these findings provide a potential mechanism by which down-regulated autophagy can lead to aberrant spine structure and synaptic plasticity at CA1 synapses and, by inference, cognition in fragile X mice.
We propose a model in which autophagy is critical for maintaining the correct balance between protein synthesis and degradation at the synapse in hippocampal neurons of WT mice. In neurons lacking FMRP, overactivation of mTOR suppresses autophagy, which in turn leads to reduced degradation of a subset of synaptic proteins. Synaptic proteins such as PSD-95 and Arc are elevated, leading to an overabundance of filopodial-like spines, exaggerated mGluR-LTD, and, by inference, impaired cognition. Acute knockdown of Raptor, a binding partner of mTOR critical to mTORC1 activity, activates autophagy, thereby enabling autophagy-dependent degradation of synaptic proteins such as PSD-95 and Arc. This, in turn, corrects spine morphology, mGluR-LTD at CA1 synapses, and, by inference, cognition in Fmr1-KO mice. These findings identify a previously unappreciated role for autophagy in the aberrant synaptic plasticity and cognition observed in fragile X mice and suggest drugs that activate autophagy as a potential therapeutic strategy to ameliorate the sensory and cognitive deficits associated with fragile X.
Whereas Huber and coworkers (52) report that the E3 ligase Mdm2 ubiquitinates PSD-95 and, together with myocyte enhancer factor-2 (Mef2) and protocadherin10 (Pcdh10), targets PSD-95 to the proteasome for degradation, thereby enabling synapse elimination assessed by recording of miniature EPSCs, we show that PSD-95 is degraded by both the proteasomal and autophagy/lysosomal pathways and that down-regulated autophagy is causally related to elevated PSD-95 and aberrant spine morphology assessed by imaging of Golgi-impregnated neurons. Our findings suggest that autophagy is a mechanism by which MEF2 might promote synapse elimination in WT neurons. Whereas Sulzer and coworkers (41) show that overactivated mTOR signaling suppresses autophagy in the brain of Tsc1+/− and Tsc2+/− mice during postnatal development and that reduced autophagy is causally linked to impaired pruning of spines on dendrites of cortical layer V pyramidal neurons and autism-relevant behaviors in these mice, we report that reduced autophagy is causally related to an overabundance of long, filopodial-like spines on dendrites of hippocampal neurons, exaggerated mGluR-LTD at CA1 synapses, and impaired cognition in fragile X mice.
It is widely accepted that elevated protein translation in the hippocampus of Fmr1-KO mice is causally related to major fragile X phenotypes (7, 10, 13–15, 23, 24) and that blunting of protein translation can rescue aberrant spine morphology, synaptic function, and cognitive deficits in fragile X mice (27). In Fmr1-KO mice, knockout of p70 ribosomal S6 kinase, a downstream effector of mTORC1 and regulator of protein translation, rescues aberrant morphology of dendritic spines on hippocampal neurons and cognitive deficits (27). Reduction of eIF4E phosphorylation by genetic deletion of MNK1 or MNK2 in Fmr1-KO mice rescues aberrant spine morphology, enhanced mGluR-LTD, and behavioral deficits (56). Inhibition of ERK1/2 in the hippocampus or its downstream effector RSK in cortex corrects elevated protein synthesis in Fmr1-KO mice and reduces the susceptibility of the mice to audiogenic seizures (57, 58). Genetic ablation and acute knockdown of cytoplasmic polyadenylation element-binding protein 1 (CPEB1) in Fmr1-KO mice ameliorates exaggerated mGluR-LTD and spine defects at CA1 synapses in the hippocampus, aberrant social behaviors, and impaired working memory (59).
In the present study, we find that a substantial fraction of the rescue afforded by shRaptor is attenuated by coadministration of shRNA to the autophagy factor Atg7, suggesting that not only elevated protein translation but also reduced protein degradation via the autophagy/lysosomal pathway contributes to synaptic phenotypes in FXS. In the case of some Fmr1-KO phenotypes, coadministration of shAtg7 appeared to reverse the rescue afforded by shRaptor, possibly because, in addition to its striking inhibition of autophagy, shAtg7 also directly or indirectly affects protein translation (60, 61). For example, macroautophagy degrades components of the translational machinery, including factors involved in the initiation and elongation of protein translation (60, 61). Thus, pharmacological inhibition or RNAi-mediated acute knockdown of autophagy would be expected to increase protein translation, although it decreases substrate availability for protein synthesis (60, 61). In this way, shAtg7 not only reverses the activation of autophagy but also promotes translation. This may explain why, in several experiments, coadministration of shAtg7 appears to substantially reverse the rescue afforded by shRaptor. Our findings are consistent with a scenario in which both elevated protein translation and reduced autophagy contribute to the cognitive deficits associated with FXS.
Autophagy is a key regulator of cell growth, differentiation, and survival (17–19). In neurons, autophagy plays a pivotal role in synaptic remodeling, synaptic plasticity, and memory formation (18, 51). In the present study, we identify a mechanism by which reduced autophagy is causally related to the overabundance of dendritic spines on hippocampal neurons in a mouse model of FXS. Our findings, together with those of Sulzer and coworkers, add autism spectrum disorders (ASDs) to the growing list of brain disorders in which down-regulated autophagy is an important factor.
In summary, the present study links aberrant mTORC1 and autophagy signaling to cognitive deficits of Fmr1-KO mice. These findings reveal a role for autophagy as a therapeutic target for FXS. Given that aberrant mTORC1 activity has been noted in different populations of neurons at different stages of development of Fmr1-KO mice, reduced autophagy in brain regions other than hippocampus may also serve as therapeutic targets for the amelioration of the symptoms associated with fragile X. Importantly, aberrant mTORC1 signaling is implicated not only in FXS but also in other disorders such as Rett syndrome, neurofibromatosis type 1, and tuberous sclerosis complex (13, 62) in which mTOR signaling is dysregulated. Thus, findings in the present study indicate components of the autophagy pathway as therapeutic targets not only for FXS but also other for ASDs.
Materials and Methods
Animals.
FVB.129P2-Pde6b+ Tyrc-ch Fmr1tm1Cgr/J (Fmr1-KO) mice and FVB.129P2-Pde6b+ Tyrc-ch/AntJ (WT) mice were obtained from The Jackson Laboratory as described (25). All mice were housed in a standard, pathogen-free animal facility with a 12-h/12-h light/ dark cycle, and only male mice were used. Genotyping was performed using standard PCR techniques as described (63). Animal protocols were approved by the Institutional Animal Care and Use Committee of the Albert Einstein College of Medicine.
Vectors and Lentivirus.
Oligonucleotides with the following nucleotide sequences were cloned for Raptor shRNA: CCGGGCCCGAGTCTGTGAATGTAATCTCGAGATTACATTCACAGACTCGGGCTTTTTG (64) and Atg7 shRNA: GATCCCCGCAGCTCATTGATAACCATTTCAAGAGAATGGTTATCAATGAGCTGCTTTTTC (43). The NT shRNA sequence ATCTCGCTTGGGCGAGAGT was used as control. Raptor and NT shRNAs were cloned into the pGIPZ vector containing GFP. Atg7 shRNA was cloned to the pLenti vector carrying mCherry. High-titer lentiviral stocks were produced by calcium phosphate–mediated transfection of HEK293T cells with viral plasmids together with helper plasmids and were purified via ultracentrifugation as described (65–68). Viral titers were determined by transducing HeLa cells. GFP and mCherry fluorescence was examined by flow cytometry (Becton Dickinson LSR II flow cytometer) 72 h after transduction. Final virus titer was diluted to 2 × 106 transducing units/μL.
Cell Culture, Viral Transduction, and Immunocytochemistry.
Primary cultures of hippocampal neurons were prepared from embryonic day 18 (E18) mice and were maintained in Neurobasal medium with B-27 supplementation and GlutaMAX (Invitrogen) as described (37). Neuronal cultures were maintained for 14 d before use. For immunocytochemistry, neurons were fixed with 4% paraformaldehyde (PFA), blocked with serum from the appropriate species, and subjected to reaction with primary antibodies, followed by reaction with Alexa Fluor 488-, 555-, or 647-conjugated secondary antibodies (Invitrogen). DAPI staining was used to reveal all cells in the section. At least three coverslips per group and multiple areas per slide selected on a random basis were used for counting analysis. Consecutive z-section images were acquired with a Leica SP5 confocal microscope (63× objectives). Images taken at 0.6-µm depth intervals were averaged four times and processed using ImageJ software (NIH). Labeled neurons were selected randomly for quantification. The integrated puncta intensity for a given neuron was quantified and normalized to the area of the cell body. To make comparisons between experiments involving different batches of neurons or different cohorts of animals, we used the same immunochemistry procedure and included all control and treatment groups in each preparation. Moreover, laser settings of the microscope were uniform across all preparations.
In Vivo Virus Injection.
Mice were placed in a stereotaxic frame (Kopf Instruments), anesthetized with 4% isoflurane, and maintained in anesthesia with 1.5% isoflurane as described (58, 65, 66). Before injection, the incision site was scrubbed with Betadine (Purdue), and a small burr hole was drilled in the skull. Concentrated viral solution (0.5 μL) was injected into the right and left CA1 by means of a 10-µL Hamilton syringe with a 26-gauge needle driven by a Quintessential Stereotaxic Injector (Stoelting Company) at a flow rate of 0.2 µL/min. The injection site was defined by the following coordinates: 2 mm posterior to bregma, 1.5 mm below the surface of the skull, and 1.8 mm lateral to the midline (65, 66). The needle was left in place for an additional 2 min and then was withdrawn gently. The incision was closed with cyanoacrylate glue. After injection, animals were placed in a heated cage to recover.
Golgi Staining, Spine Morphology, Immunolabeling, and Histology.
To image spine morphology, we used an FD Rapid Golgi stain Kit (FD Neurotechnologies) as described (37). In brief, mouse brains were collected, quickly rinsed, immersed in Golgi impregnation solution, which includes fixation, and stored in the dark at room temperature for 2 wk. Brains were then transferred and stored in solution C for 72 h at 4 °C and were cut into 200-mm-thick sections by means of a Leica cryostat at −20 °C. Sections were transferred to cold SuperFrost Plus microscope slides (Fisher Scientific), rinsed, dehydrated, and cleared according to the manufacturer’s instructions. Spines on apical dendrites of hippocampal CA1 pyramidal neurons were imaged by means of an Olympus brightfield microscope using Neurolucida software (MBF Bioscience) with a 100× oil immersion lens. Dendritic spine density was determined by counting the total number of spines along the apical dendrite from the soma to a 130-μm distance on primary, secondary, and tertiary branches. Spine-head width was measured, and spines were classified as filopodial-like or mushroom-like in all imaged neurons by means of a categorization macro in Neurolucida software, which excludes branched and detached spines, as described (27, 37).
Immunochemistry was performed on frozen brain sections from fragile X and WT mice as described. Mice were anesthetized and transcardially perfused with 4% PFA, and brains were removed, postfixed for 24 h, and infiltrated with 20–30% sucrose. Brain sections (20 µm thick) were blocked with serum from the appropriate species, treated with primary antibodies, and then reacted with Alexa Fluor 488 or 555 secondary antibodies (Invitrogen). Naive IgG of the appropriate species was used as a negative control. DAPI staining was used to reveal all cells in brain sections. Images were acquired using a confocal microscope.
Organotypic Slice Culture and Electrophysiological Recordings.
Organotypic hippocampal slices were cultured from Fmr1-KO mice and WT littermates at 4 wk of age and were transduced with lentivirus carrying NT shRNA, shRaptor, or shRaptor together with shRNA to Atg7 at day in vitro (DIV) 3 and were recorded at DIV 6–8 as described (27, 44, 45). The investigator was blind to the genotype and treatment. Baseline slope was calculated from the mean values recorded for 20 min before drug application. LTD was induced by a single application of 100-µM DHPG for 5 min and was measured as the change in the response at 50–60 min after initiation of drug application. Slices were visualized using an infrared differential interference contrast Zeiss microscope. Data were acquired with a Multiclamp 700B amplifier (Molecular Devices) and analyzed by Igor Pro software (WaveMetrics).
Immunoprecipitation.
Primary neurons were homogenized in ice-cold lysis buffer as described (15). Homogenates were incubated with anti-ubiquitin (1:100) (Santa Cruz) antibody and were gently shaken overnight at 4 °C. Supernatant with antibody was added to a slurry of IgG bound to agarose beads (Protein A/G, Pierce) and was incubated with rocking at 25 °C for 2 h. Efficiency of the immunoprecipitation was determined by comparing the abundance of immunoprecipitated protein in the supernatant and in wash fractions obtained from the procedure.
Lysosome Isolation and Western Blotting.
Lysosomes were isolated from mouse hippocampus by means of a Lysosome Enrichment Kit (Thermo Scientific). Briefly, tissues were homogenized and centrifuged at 850 × g for 2 min. Pellets were isolated, vortexed, resuspended, and centrifuged according to the manual. Supernatant fractions were subjected to gradient ultracentrifugation at 145,000 × g for 2 h at 4 °C. Lysosomal fractions were collected, resuspended in PBS buffer, and centrifuged at 12,000 × g for 5 min at 4 °C; lysosomal proteins were collected from the supernatant.
Hippocampal tissue was homogenized in lysis buffer supplemented with proteinase inhibitors and centrifuged at 12,000 × g for 10 min at 4 °C to collect whole-cell proteins in the supernatant. Primary neurons were lysed and centrifuged as above, except that centrifugation was for 5 min. Protein concentrations were measured, and Western blots were performed as described (69). Band densities were normalized to β-actin (as a loading control) and then to WT values. Band densities were quantified using ImageJ software.
Primary antibodies used for Western blotting included rabbit anti–LC3-I/II (Novus), rabbit anti-p62 (MBL), rabbit anti-Raptor (Cell Signaling), rabbit anti-Arc (Cell Signaling), rabbit anti–PSD-95 (Millipore), mouse anti-mTOR (Cell Signaling), rabbit anti–p-mTOR (S2448) (Cell Signaling), rabbit anti–p-ULK-1 (S757) (Cell Signaling), rabbit anti–p-ULK-1 (S317) (Cell Signaling), rabbit anti–ULK-1 (Cell Signaling), rabbit anti–p-Beclin-1 (S14) (Cell Signaling), rabbit anti–Beclin-1 (Cell Signaling), mouse anti-ubiquitin (Enzo), rabbit anti-Atg7 (Cell Signaling), and mouse anti–β-actin (Sigma). Antibodies for LC3-I/II, p62, Raptor, Arc, PSD-95, and ubiquitin were used for immunocytochemistry with primary antibodies of chicken anti–MAP-2 (Millipore) and rat anti-cathepsin D (R&D Systems). Primary antibodies of p62 and mouse anti-NeuN (Millipore) were used for immunochemistry of brain sections.
Novel Object Recognition Test.
The novel object recognition task was conducted in an isolated arena (50 cm long × 50 cm wide × 40 cm high). Five-week-old Fmr1-KO mice and WT littermates were used in this study. The investigator was blind to the genotype. For three consecutive days before the testing day, animals were allowed to explore the empty arena in which the task was to be performed for 10 min. On the fourth day (training and familiarization session) the mouse was placed in the center of the arena between and equidistant from two identical objects and was allowed to explore freely for 10 min. The time the mouse spent exploring each object was recorded. The mouse was then placed in a holding cage for 24 h. The next day (test session), one of the objects was replaced with a novel object. The mouse was placed in the arena for an additional 10 min, and the time spent exploring each object was recorded. Mice that did not spend a minimum of 10 s investigating one or both objects were excluded from the study. The preference index was calculated by dividing the time (in seconds) spent exploring the novel object by the total time spent exploring the two objects. A preference index >0.5 indicates an ability to discriminate the novel object from the familiar object. Exploration was defined as orienting the nose toward the object and approaching the object to a distance <2 cm between the nose and the object. Resting, grooming, or sitting on the object was not considered as exploration.
Statistical Analysis.
Data are presented as means ± SEM. The Kolmogorov–Smirnov test was used to analyze normal distribution of data. The Student’s t test (unpaired, two-tailed) was used for two-group comparisons, and two-way ANOVA followed by a Tukey’s test was used for experiments involving more than two groups. Unless otherwise indicated, n = the number of animals used in the analysis. Statistical significance was defined as P < 0.05. The statistics are listed in tables in SI Appendix.
Supplementary Material
Acknowledgments
We thank Analytical Imaging Facility and Neuro Core (Albert Einstein College of Medicine) for expert advice/assistance in microscopy, and all the members of the R.S.Z. laboratories for their constructive comments. This work was supported by NIH Grant MH-092877 and a generous grant from the F. M. Kirby Foundation (to R.S.Z.). R.S.Z. is the F. M. Kirby Professor in Neural Repair and Protection. M.V.L.B. is supported by NIH Grants NS-095029 and NS-108695 (Co-Principal Investigator Prof. Jun Chen of University Pittsburgh). M.V.L.B. is the Sylvia and Robert S. Olnick Professor of Neuroscience. J.Y. is the recipient of a FRAXA Research Foundation fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808247115/-/DCSupplemental.
References
- 1.Bhakar AL, Dölen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: Causes, diagnosis, mechanisms, and therapeutics. J Clin Invest. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrahams BS, Geschwind DH. Advances in autism genetics: On the threshold of a new neurobiology. Nat Rev Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCary LM, Roberts JE. Early identification of autism in fragile X syndrome: A review. J Intellect Disabil Res. 2013;57:803–814. doi: 10.1111/j.1365-2788.2012.01609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Penagarikano O, Mulle JG, Warren ST. The pathophysiology of fragile X syndrome. Annu Rev Genomics Hum Genet. 2007;8:109–129. doi: 10.1146/annurev.genom.8.080706.092249. [DOI] [PubMed] [Google Scholar]
- 6.Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 2015;16:595–605. doi: 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrie SC, Brems H, Legius E, Bagni C. Cognitive dysfunctions in intellectual disabilities: The contributions of the Ras-MAPK and PI3K-AKT-mTOR pathways. Annu Rev Genomics Hum Genet. 2017;18:115–142. doi: 10.1146/annurev-genom-091416-035332. [DOI] [PubMed] [Google Scholar]
- 8.Kelleher RJ, 3rd, Bear MF. The autistic neuron: Troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Darnell JC, Klann E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross C, et al. Increased expression of the PI3K enhancer PIKE mediates deficits in synaptic plasticity and behavior in fragile X syndrome. Cell Rep. 2015;11:727–736. doi: 10.1016/j.celrep.2015.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 12.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huber KM, Klann E, Costa-Mattioli M, Zukin RS. Dysregulation of mammalian target of rapamycin signaling in mouse models of autism. J Neurosci. 2015;35:13836–13842. doi: 10.1523/JNEUROSCI.2656-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma A, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoeffer CA, et al. Altered mTOR signaling and enhanced CYFIP2 expression levels in subjects with fragile X syndrome. Genes Brain Behav. 2012;11:332–341. doi: 10.1111/j.1601-183X.2012.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris H, Rubinsztein DC. Control of autophagy as a therapy for neurodegenerative disease. Nat Rev Neurol. 2011;8:108–117. doi: 10.1038/nrneurol.2011.200. [DOI] [PubMed] [Google Scholar]
- 18.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoeffer CA, Klann E. mTOR signaling: At the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa-Mattioli M, Monteggia LM. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat Neurosci. 2013;16:1537–1543. doi: 10.1038/nn.3546. [DOI] [PubMed] [Google Scholar]
- 25.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Maday S, Holzbaur EL. Compartment-specific regulation of autophagy in primary neurons. J Neurosci. 2016;36:5933–5945. doi: 10.1523/JNEUROSCI.4401-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharya A, et al. Genetic removal of p70 S6 kinase 1 corrects molecular, synaptic, and behavioral phenotypes in fragile X syndrome mice. Neuron. 2012;76:325–337. doi: 10.1016/j.neuron.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levenga J, et al. Subregion-specific dendritic spine abnormalities in the hippocampus of Fmr1 KO mice. Neurobiol Learn Mem. 2011;95:467–472. doi: 10.1016/j.nlm.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 29.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loos B, du Toit A, Hofmeyr JH. Defining and measuring autophagosome flux—Concept and reality. Autophagy. 2014;10:2087–2096. doi: 10.4161/15548627.2014.973338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Y, et al. Endolysosomal deficits augment mitochondria pathology in spinal motor neurons of asymptomatic fALS mice. Neuron. 2015;87:355–370. doi: 10.1016/j.neuron.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: The roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 36.Busquets-Garcia A, et al. Targeting the endocannabinoid system in the treatment of fragile X syndrome. Nat Med. 2013;19:603–607. doi: 10.1038/nm.3127. [DOI] [PubMed] [Google Scholar]
- 37.Pyronneau A, et al. Aberrant Rac1-cofilin signaling mediates defects in dendritic spines, synaptic function, and sensory perception in fragile X syndrome. Sci Signal. 2017;10:eaan0852. doi: 10.1126/scisignal.aan0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santini E, et al. Exaggerated translation causes synaptic and behavioural aberrations associated with autism. Nature. 2013;493:411–415. doi: 10.1038/nature11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, et al. Neuronal mTORC1 is required for maintaining the nonreactive state of astrocytes. J Biol Chem. 2017;292:100–111. doi: 10.1074/jbc.M116.744482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Y, et al. Regulation of morphine-induced synaptic alterations: Role of oxidative stress, ER stress, and autophagy. J Cell Biol. 2016;215:245–258. doi: 10.1083/jcb.201605065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang G, et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Q, Cai D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IκB kinase β (IKKβ)/NF-κB pathway. J Biol Chem. 2011;286:32324–32332. doi: 10.1074/jbc.M111.254417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh R, et al. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfeiffer BE, et al. Fragile X mental retardation protein is required for synapse elimination by the activity-dependent transcription factor MEF2. Neuron. 2010;66:191–197. doi: 10.1016/j.neuron.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lüscher C, Huber KM. Group 1 mGluR-dependent synaptic long-term depression: Mechanisms and implications for circuitry and disease. Neuron. 2010;65:445–459. doi: 10.1016/j.neuron.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis–Dependent long-term depression in hippocampal area CA1. J Neurophysiol. 2001;86:321–325. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- 48.Fitzjohn SM, et al. A characterisation of long-term depression induced by metabotropic glutamate receptor activation in the rat hippocampus in vitro. J Physiol. 2001;537:421–430. doi: 10.1111/j.1469-7793.2001.00421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Kazdoba TM, Leach PT, Silverman JL, Crawley JN. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis Res. 2014;3:118–133. doi: 10.5582/irdr.2014.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shehata M, Matsumura H, Okubo-Suzuki R, Ohkawa N, Inokuchi K. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci. 2012;32:10413–10422. doi: 10.1523/JNEUROSCI.4533-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai NP, et al. Multiple autism-linked genes mediate synapse elimination via proteasomal degradation of a synaptic scaffold PSD-95. Cell. 2012;151:1581–1594. doi: 10.1016/j.cell.2012.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chowdhury S, et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park S, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jakkamsetti V, et al. Experience-induced Arc/Arg3.1 primes CA1 pyramidal neurons for metabotropic glutamate receptor-dependent long-term synaptic depression. Neuron. 2013;80:72–79. doi: 10.1016/j.neuron.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gkogkas CG, et al. Pharmacogenetic inhibition of eIF4E-dependent Mmp9 mRNA translation reverses fragile X syndrome-like phenotypes. Cell Rep. 2014;9:1742–1755. doi: 10.1016/j.celrep.2014.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osterweil EK, et al. Lovastatin corrects excess protein synthesis and prevents epileptogenesis in a mouse model of fragile X syndrome. Neuron. 2013;77:243–250. doi: 10.1016/j.neuron.2012.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sawicka K, Pyronneau A, Chao M, Bennett MV, Zukin RS. Elevated ERK/p90 ribosomal S6 kinase activity underlies audiogenic seizure susceptibility in fragile X mice. Proc Natl Acad Sci USA. 2016;113:E6290–E6297. doi: 10.1073/pnas.1610812113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Udagawa T, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gretzmeier C, et al. Degradation of protein translation machinery by amino acid starvation-induced macroautophagy. Autophagy. 2017;13:1064–1075. doi: 10.1080/15548627.2016.1274485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kelly SP, Bedwell DM. Both the autophagy and proteasomal pathways facilitate the Ubp3p-dependent depletion of a subset of translation and RNA turnover factors during nitrogen starvation in Saccharomyces cerevisiae. RNA. 2015;21:898–910. doi: 10.1261/rna.045211.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sawicka K, Zukin RS. Dysregulation of mTOR signaling in neuropsychiatric disorders: Therapeutic implications. Neuropsychopharmacology. 2012;37:305–306. doi: 10.1038/npp.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spencer CM, et al. Exaggerated behavioral phenotypes in Fmr1/Fxr2 double knockout mice reveal a functional genetic interaction between fragile X-related proteins. Hum Mol Genet. 2006;15:1984–1994. doi: 10.1093/hmg/ddl121. [DOI] [PubMed] [Google Scholar]
- 64.Sancak Y, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Noh KM, et al. Repressor element-1 silencing transcription factor (REST)-dependent epigenetic remodeling is critical to ischemia-induced neuronal death. Proc Natl Acad Sci USA. 2012;109:E962–E971. doi: 10.1073/pnas.1121568109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyawaki T, et al. The endogenous inhibitor of Akt, CTMP, is critical to ischemia-induced neuronal death. Nat Neurosci. 2009;12:618–626. doi: 10.1038/nn.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Udagawa T, et al. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaneko N, Hwang JY, Gertner M, Pontarelli F, Zukin RS. Casein kinase 1 suppresses activation of REST in insulted hippocampal neurons and halts ischemia-induced neuronal death. J Neurosci. 2014;34:6030–6039. doi: 10.1523/JNEUROSCI.4045-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodenas-Ruano A, Chávez AE, Cossio MJ, Castillo PE, Zukin RS. REST-dependent epigenetic remodeling promotes the developmental switch in synaptic NMDA receptors. Nat Neurosci. 2012;15:1382–1390. doi: 10.1038/nn.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.