Significance
Neurogenetic studies demonstrate that copy number variations (CNVs) in the ASTN2 gene occur in patients with neurodevelopmental disorders (NDDs), including autism spectrum. Here, we show that ASTN2 associates with recycling and degradative vesicles in cerebellar neurons, and binds to and promotes the endocytic trafficking and degradation of synaptic proteins. Overexpression of ASTN2 in neurons increases synaptic activity and reduces the levels of ASTN2 binding partners, an effect dependent on its FNIII domain, which is recurrently perturbed by CNVs in patients with NDDs. These findings suggest that ASTN2 is a key regulator of dynamic trafficking of synaptic proteins and lend support to the idea that aberrant regulation of protein homeostasis in neurons is a contributing cause of complex NDDs.
Keywords: autism spectrum disorder, protein degradation, synapse, cerebellum, protein trafficking
Abstract
Surface protein dynamics dictate synaptic connectivity and function in neuronal circuits. ASTN2, a gene disrupted by copy number variations (CNVs) in neurodevelopmental disorders, including autism spectrum, was previously shown to regulate the surface expression of ASTN1 in glial-guided neuronal migration. Here, we demonstrate that ASTN2 binds to and regulates the surface expression of multiple synaptic proteins in postmigratory neurons by endocytosis, resulting in modulation of synaptic activity. In cerebellar Purkinje cells (PCs), by immunogold electron microscopy, ASTN2 localizes primarily to endocytic and autophagocytic vesicles in the cell soma and in subsets of dendritic spines. Overexpression of ASTN2 in PCs, but not of ASTN2 lacking the FNIII domain, recurrently disrupted by CNVs in patients, including in a family presented here, increases inhibitory and excitatory postsynaptic activity and reduces levels of ASTN2 binding partners. Our data suggest a fundamental role for ASTN2 in dynamic regulation of surface proteins by endocytic trafficking and protein degradation.
ASTN2 is a large vertebrate-specific transmembrane protein, expressed in the developing and adult brain primarily, and with the highest levels detected in the cerebellum, but also at lower levels in the cortex, the olfactory bulb, and the dentate gyrus of the hippocampus (1). Previously, we showed that ASTN2 interacts with ASTN1, a surface membrane protein that regulates glial-guided neuronal migration (1–4). Recently, copy number variants (CNVs) of ASTN2, both deletions and duplications (SI Appendix, Fig. S1), were identified in patients with neurodevelopmental disorders (NDDs), including autism spectrum disorder (ASD), schizophrenia, attention-deficit/hyperactivity disorder (ADHD), bipolar disease, intellectual disability (ID), and global developmental delay (5–9). In particular, ASTN2 CNVs mainly affected the MAC/Perforin (MACPF) and FNIII encoding regions of the gene and were identified as a significant risk factor for ASD in males in a study of 89,985 subjects (10).
Despite shared protein homology, ASTN2, but not ASTN1, is highly expressed in the adult cerebellum long after completion of neuronal migration, which is suggestive of key additional roles unrelated to migration. While the cerebellum has traditionally been associated with motor control, recent evidence has suggested nonmotor functions, including language, visuospatial memory, attention, and emotion (11–13). In particular, loss of cerebellar Purkinje cells (PCs) is one of the most consistent findings in postmortem studies in patients with ASD (14). Moreover, specific targeting of cerebellar neurons in mouse models of ASD-associated genes leads to impaired cerebellar learning (15) and social behaviors (16). The mechanism of action of ASTN2 in postmigratory neurons and how it may contribute to the pathophysiology of NDDs is currently unknown.
Here, we describe a family with a paternally inherited intragenic ASTN2 duplication and NDDs, including ASD and, most notably, learning difficulty and speech and language delay. By immunogold electron microscopy (EM), we show that ASTN2 localizes primarily to endocytic and autophagocytic vesicles in PC soma and to subsets of dendritic spines. By immunoprecipitation (IP)/mass spectrometry (MS), we identify ASTN2 binding partners, including C1q, Neuroligins, ROCK2, and SLC12a5 (KCC2), and show that ASTN2 removes surface proteins by endocytosis. Further, ASTN2 is found in a subset of vesicles along the entire endosomal pathway and links to the endosomal trafficking machinery via binding to the adaptor protein AP-2 and the vacuolar protein-sorting–associated protein 36 (VPS36). Importantly, consistent with a role in regulating the surface expression of key synaptic proteins, while conditional overexpression of ASTN2 in PCs increases synaptic strength, ASTN2 with deletion of the FNIII domain, the region recurrently disrupted by CNVs in patients, including the family presented here, is inefficient at changing synaptic activity. At the molecular level, overexpression of ASTN2 results in reduced protein levels of its synaptic binding partners. Our study identifies ASTN2 as a molecule that modulates the composition of the surface membrane proteome. We propose that the intragenic ASTN2 CNVs in patients result in misregulation of surface protein turnover, which is crucial for normal synaptic activity.
Results
Paternally Inherited ASTN2 CNVs in a Family with ASD, ID, and Speech and Language Delay.
SNP array genetic testing of a child presented at 19 months of age identified a 171-kb duplication at 9q33.1, affecting exons 17–20 of ASTN2 (Dataset S1). The CNV was present in the father and in three of five children, indicating a paternally inherited heterozygous duplication. The children displayed a range of NDDs (Dataset S1), including ID and ASD. Two features in particular stood out in the affected children, namely, learning difficulty and speech and language delay regardless of other diagnoses.
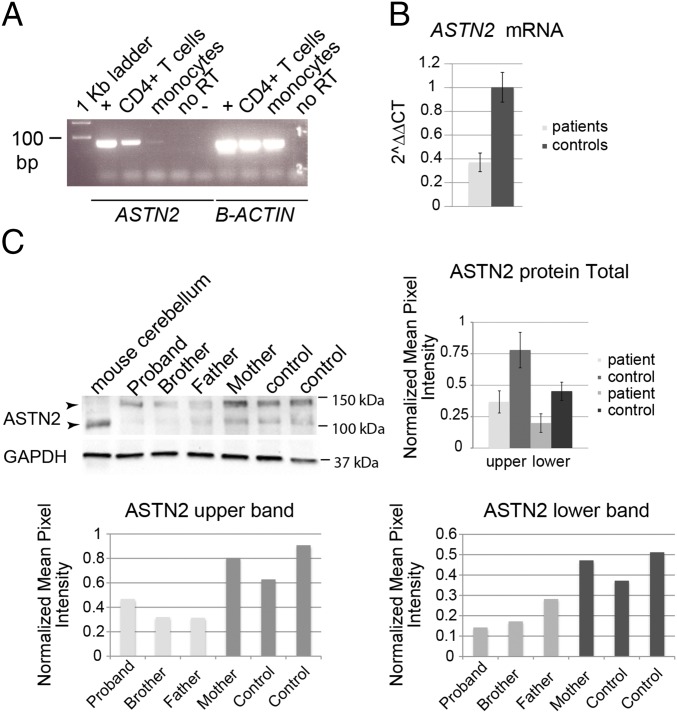
To investigate how the duplication of exons 17–20, which code for part of the MACPF domain and part of the FNIII domain of ASTN2, affects ASTN2 expression, we obtained peripheral blood mononuclear cells (PBMCs) from this patient family, where ASTN2 expression was detected in the CD4+ T cell fraction (Fig. 1A). The duplication was predicted to result in either an mRNA encoding an intact MACPF domain but a truncated FNIII domain due to the creation of a frameshift stop codon (termed JDUP; SI Appendix, Fig. S2) or nonsense-mediated decay of the mRNA. In CD4+ T cells isolated from two of the three boys with the ASTN2 CNV as well as the father, ASTN2 was reduced by ∼30–50% compared with controls, including the mother (Fig. 1B). We detected two protein bands, one of which is absent in the mouse, both of which were ∼50% lower in patients compared with controls, by Western blot (Fig. 1C). These results suggest that the duplication causes loss of half of the ASTN2 transcripts and protein due to a frameshift stop codon in one allele and subsequent mRNA decay. While the mRNA quantification (Fig. 1B) suggests that the majority of the duplicated mRNA undergoes nonsense-mediated decay, we cannot exclude the possibility that low levels of the truncated protein (termed JDUP) is expressed in patients, as the antibody used does not recognize JDUP (verified with a deletion construct).
Fig. 1.
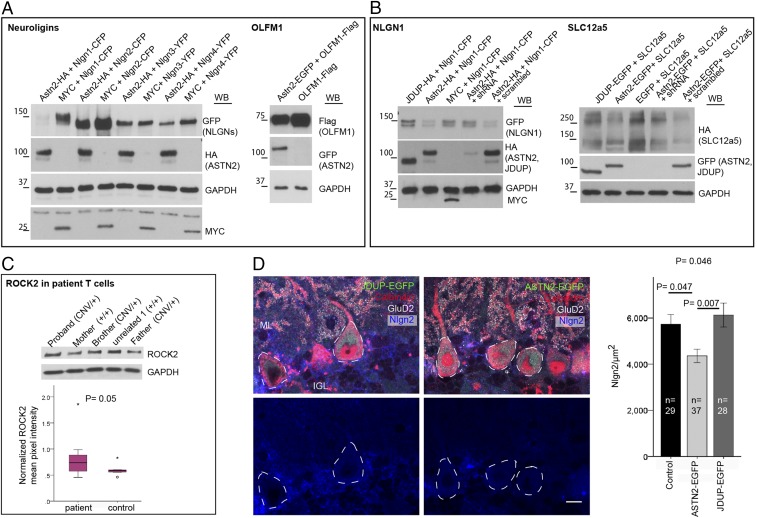
ASTN2 expression in patients. (A) Expression of ASTN2 detected in human CD4+ T cells but not in monocytes. Positive (human fibroblasts), negative (no template), and no RT controls are indicated. (B) ASTN2 mRNA levels, expressed as 2ΔΔCT (cycle time by quantitative RT-PCR) in relation to GUSB (endogenous control). Protein levels in ASTN2 CNV patient T cells versus controls (C, Left) are quantified in the graph (C, Right). (C, Bottom) Quantifications of individual ASTN2 bands [upper (Left) and lower (Right) bands] in relation to GAPDH are shown. N = 3 patients and 3 controls. Bars show mean ± 1 SD.
In the DECIPHER database (https://decipher.sanger.ac.uk), which currently contains clinical and genetic information on 11,887 patients with NDD, ID is reported in 11 of 18 (61%) patients with ASTN2 CNVs (both deletions and duplications), which is a slightly higher rate of occurrence than in the overall NDD population [6,735 of 11,887 patients (57%)]. Speech and language delay was reported in five of 18 patients (28%) with ASTN2 CNVs, which is also above the rate observed in the overall NDD population [2,505 of 11887 patients (21%)]. In relation to other genes that are highly associated with either ID or ASD/ID, patients with ASTN2 CNVs fell above the median for ID among the investigated genes (above six of eight ASD-associated genes and within the range of the ID genes) and on the median among these genes for speech and language delay (SI Appendix, Fig. S1B). Thus, ID is the single most commonly occurring feature in patients with ASTN2 CNVs (both deletions and duplications), followed by speech and language delay, including in patients diagnosed with ASD.
ASTN2 Protein Localization in the Brain.
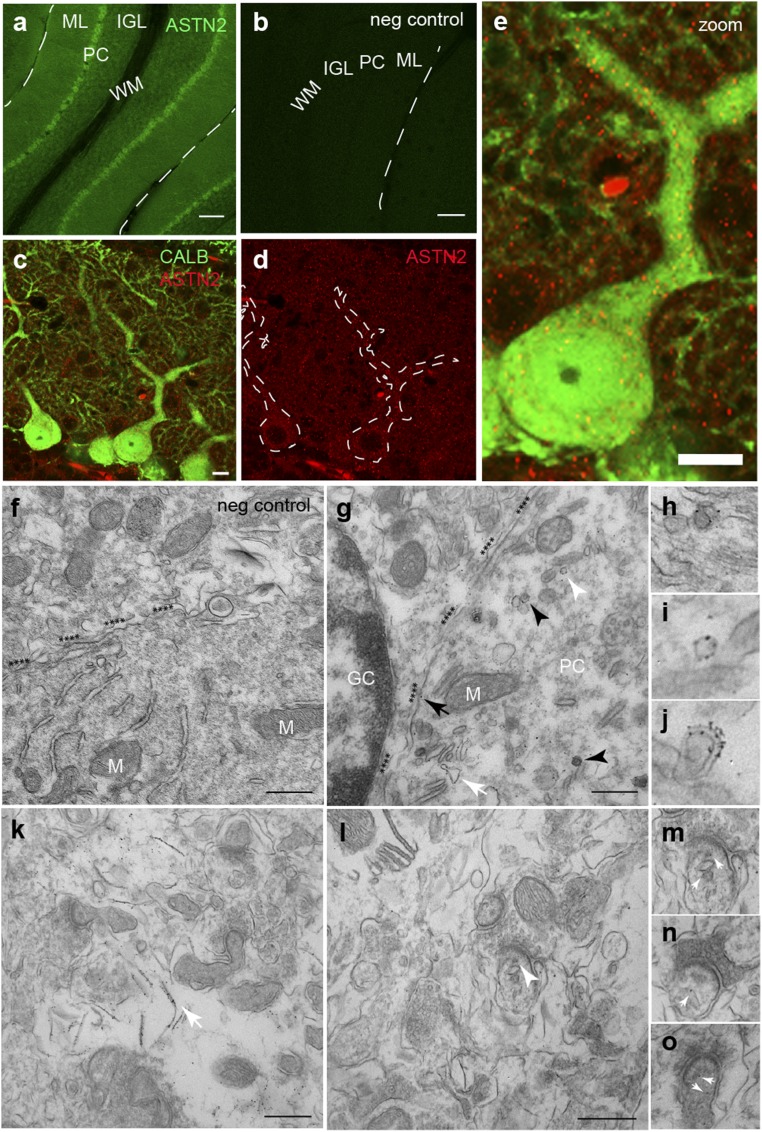
To investigate the function of ASTN2, we first analyzed its subcellular localization in the mouse cerebellum, which is the strongest site of expression in the brain (1). Immunohistochemistry (antibody validation is discussed in ref. 1 and illustrated in SI Appendix, Fig. S3 A and B) in the postnatal mouse cerebellum [postnatal day 15 (P15) and P28] showed ASTN2 in granule cells (GCs), in the molecular layer, and at higher levels in PCs (Fig. 2A). In PCs, punctate labeling was detected in the PC body, in the dendritic stalk, and in dendrites (Fig. 2 C–E). Immuno-EM labeling revealed that ASTN2 localized to membranes in the endoplasmic reticulum (ER) and in small, round trafficking vesicles near the ER, the Golgi, and the plasma membrane of PCs (Fig. 2G). ASTN2 also localized to endocytic vesicles (Fig. 2 G–I) and autophagosomes (Fig. 2 G and J). A subset of dendritic spines, mostly in proximal regions of PCs, was positive for ASTN2. In labeled spines, ASTN2 localized to membranes near, but not directly at, the postsynaptic density (Fig. 2 L–O). Colabeling of ASTN2 with recycling (Rab4), early (Rab5), and late (Rab7) endosomal markers revealed that a small subset of ASTN2 puncta localizes to all these fractions of the endocytic pathway in the cerebellum (SI Appendix, Fig. S4). The punctate expression pattern of ASTN2 and its localization to membranes of endocytic and autophagocytic vesicles suggests involvement in trafficking, and its presence proximal to synapses in postmigratory neurons raised the possibility that ASTN2 is involved in synaptic function.
Fig. 2.
ASTN2 subcellular protein localization in the cerebellum. Sagittal sections of cerebellum labeled with antibodies against ASTN2 (green) (A) and against ASTN2 (red) and Calbindin (green) (C–E) at P15. (B) Negative (neg) control (no primary antibody). Dotted lines in D outline PC bodies and primary dendrites. (E) Zoomed-in view of one PC. (F–O) Immunogold EM labeling of ASTN2 at P28. (F) Negative control (no primary). (G) ASTN2 labeling in a PC soma associated with the plasma membrane (highlighted by asterisks and black arrow), membranes of the ER (white arrow), trafficking vesicles (white arrowhead and I), and autophagosomes (black arrowheads and J). (H) High-power image showing ASTN2 labeling associated with an endocytic vesicle at the plasma membrane. (K) PC dendrite in the ML with ASTN2 labeling on membranous structures within the dendrite (arrow). Synapses in this image are negative for ASTN2. (L) PC dendritic area with positive labeling in a spine (white arrowhead). (M–O) Higher magnification examples of PC dendritic spines showing ASTN2 labeling (arrows). IGL, internal granule layer; M, mitochondria; ML, molecular layer; WM, white matter. (Scale bars: A and B, 100 μm; C–E, 10 μm; F, G, K, and L, 0.5 μm.)
ASTN2 Binds to and Reduces the Surface Expression of Synaptic Proteins by Endocytosis.
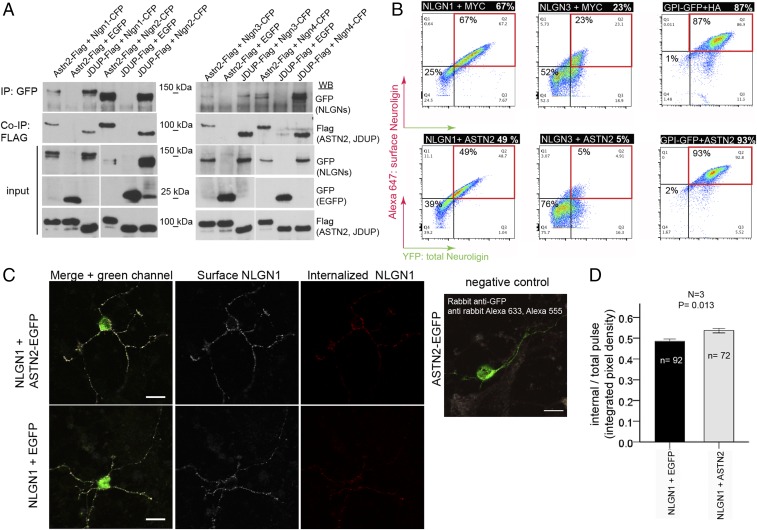
To investigate whether ASTN2 has a synaptic role, we first examined if it binds to key adhesion proteins known to regulate PC synaptic function. Co-IP experiments revealed that ASTN2 interacts with members of the Neuroligin family, as does the truncated JDUP version (Fig. 3A). By Western blot, NLGN1/2 interacted more strongly with ASTN2 than with JDUP, while NLGN3/4 interacted more strongly with JDUP than with ASTN2. Thus, ASTN2 binds to Neuroligins, and the presence of the FNIII domain differentially impacts the affinity of ASTN2 for different binding partners.
Fig. 3.
ASTN2 regulation of Neuroligin surface expression by protein–protein binding and endocytosis. (A) Western blots showing co-IP of ASTN2 and JDUP with Neuroligins (Nlgn) 1–4 in HEK 293T cells. (B) Live immunolabeling of surface Neuroligin (NLGN) expression (Alexa-647, red quadrants) in HEK 293T cells analyzed by flow cytometry in cells coexpressing NLGN1-HA-YFP or NLGN3-YFP with either a MYC control vector (Top) or ASTN2-HA (Bottom). (Right) Surface glycosylphosphatidylinositol (GPI)-anchored EGFP is unaltered by ASTN2. (C, Left) Pulse–chase labeling of NLGN1-HA-YFP coexpressed with either EGFP or ASTN2-EGFP in GCs showing surface (white) and internalized (red) NLGN1 labeling after a 20-min chase. (C, Right) Negative control, showing that the EGFP from ASTN2-EGFP (or EGFP) is not detected on the surface. (Scale bars: 10 μm.) (D) Quantification of the pulse–chase expressed as integrated pixel density (sum of all pixel intensities per area minus the background) of the internal labeling divided by the integrated pixel density of the total pulse (red + white). The graph shows mean ± 1 SEM. N, number of experiments; n, total number of cells analyzed. The P value was calculated by analysis of covariance (Methods). WB, Western blot.
To investigate whether ASTN2 regulates the surface expression of Neuroligins, we quantified the surface expression of NLGN1-YFP by live immunolabeling and flow cytometry in the presence and absence of ASTN2 in HEK 293T cells. We found a reduction in surface NLGN1 in cells cotransfected with ASTN2 compared with cells without (Fig. 3B, 49% versus 67%). This reduction was even more marked for NLGN3 in the presence of ASTN2 (Fig. 3B, 23% versus 5%). However, ASTN2 did not reduce the surface expression of glycosylphosphatidylinositol-anchored surface EGFP (Fig. 3B). Hence ASTN2 specifically removes Neuroligins from the surface of HEK cells due to protein–protein interaction.
We then examined whether ASTN2 expression also reduced surface NLGN1 in neurons. For these experiments, we chose to target GCs, which are far more numerous than PCs; also express ASTN2; and, together with PCs, are the main neuronal subtype in the cerebellum. We used overexpression to disrupt the stoichiometry of ASTN2 protein complexes, as knockdown of ASTN2 protein was not possible in neurons (SI Appendix, Fig. S3) possibly due to the extremely long half-life of ASTNs in the brain (17). As seen in HEK 293T cells, GCs grown in culture for 14 days and cotransfected with Nlgn1 and Astn2 had reduced surface expression of NLGN1 compared with controls by live immunolabeling of surface NLGN1-YFP (SI Appendix, Fig. S5). To investigate whether the reduction in surface expression is due to endocytosis versus potential changes in surface insertion of NLGN1 upon expression from plasmids, we carried out pulse–chase labeling of surface NLGN1-YFP. GCs that coexpressed NLGN1 and ASTN2 had higher levels of internalized NLGN1 after a 20-min chase than GCs that expressed NLGN1 and a control plasmid (Fig. 3 C and D). Thus, ASTN2 interacts with several key synaptic adhesion proteins and can reduce their surface expression in neurons and HEK cells by endocytosis.
ASTN2 Binds a Number of Proteins Suggestive of Trafficking of Multiple Protein Complexes in Neurons.
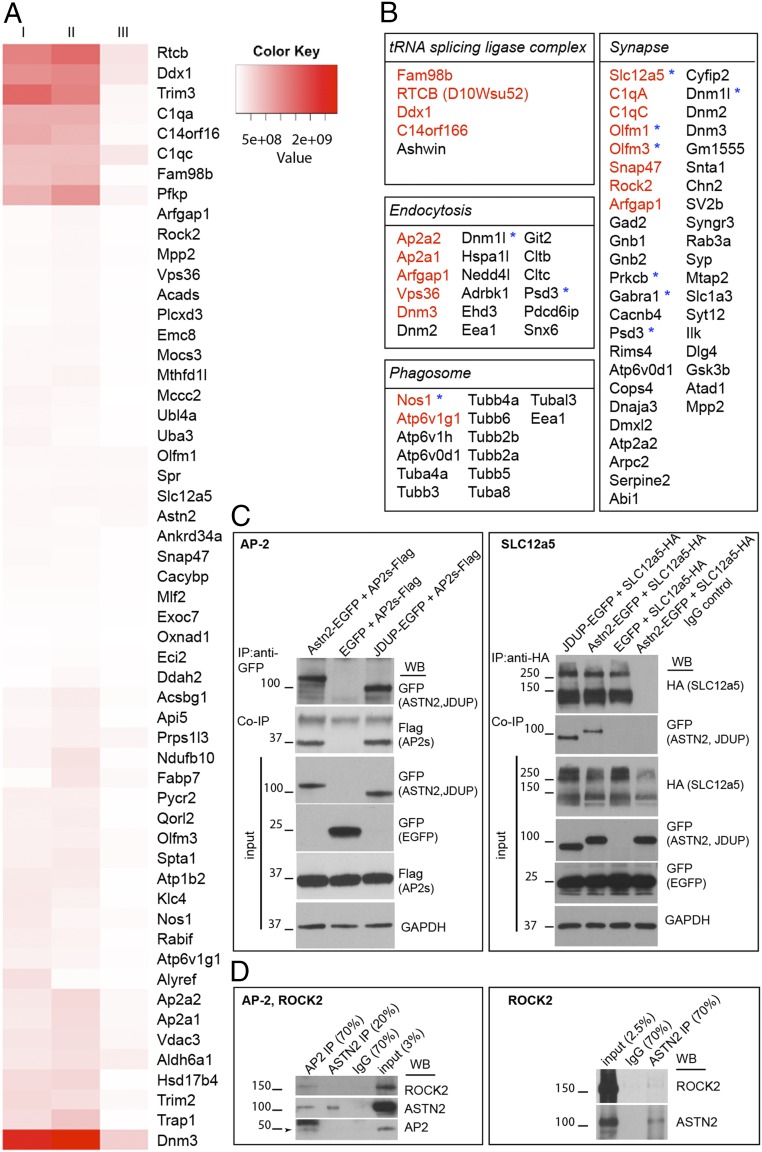
To identify additional ASTN2 binding partners in an unbiased manner, we carried out IP of ASTN2 from the P22–P28 mouse cerebellum, followed by MS analysis. An initial round of experiments with duplicate samples was followed by a second experiment with more stringent washes and the inclusion of a further negative control in which the ASTN2 antibody was affinity-removed from the antisera (SI Appendix, Fig. S6A). The combined experiments identified 466 proteins enriched in the ASTN2 IP compared with IgG or the depleted ASTN2 sera samples (Dataset S2). Further refinement of the list to only include proteins with at least three peptide hits that were ≥1.5-fold enriched in the ASTN2 IP versus the IgG or depleted anti-ASTN2 sera yielded 57 proteins (Fig. 4A). We identified AP-2, an adaptor protein in Clathrin-mediated endocytosis from the plasma membrane, and VPS36, found on sorting endosomes. We also identified multiple proteins involved in synaptic form and function, such as C1q, which has been shown to mediate synaptic pruning (18); OLFM1/3, which form complexes with AMPA receptors (19) and were recently identified as ASD candidates (20); ROCK2, a Rho-kinase that regulates spine morphology and synaptic activity through regulation of the cytoskeleton (21); and SLC12a5 (KCC2), a potassium/chloride cotransporter that regulates the intracellular chloride ion gradient as well as dendritic spine morphogenesis (22, 23) and is also implicated in ASD (24–26). Thus, ASTN2 interacts with multiple proteins that regulate synaptic activity. We confirmed these interactions by co-IP and Western blot in HEK 293T cells cotransfected with AP-2, SLC12a5, C1q, or OLFM1 and either ASTN2 or JDUP or EGFP/MYC (control) constructs (Fig. 4C and SI Appendix, Fig. S6B). Furthermore, we detected co-IP of ASTN2/ROCK2/AP-2 (Fig. 4D) and ASTN2/NGLN2/AP-2 (SI Appendix, Fig. S6C) in vivo. Functional enrichment analysis of the proteins identified categories (Fig. 4B) such as phagosome, endocytosis, synapse, microtubule-associated processes, and tRNA splicing ligase complex (not investigated further here). A number of the interacting proteins identified are, like ASTN2, implicated in ASD pathogenesis (Fig. 4B, asterisks). Taken together, our IP/MS experiments show that ASTN2 binds to proteins involved in vesicle trafficking and synaptic function, including synaptic pruning proteins, ion transporters, accessory proteins to ligand-gated ion channels, and proteins involved in cytoskeletal rearrangements, suggesting that ASTN2 possibly promotes the trafficking of multiple surface proteins.
Fig. 4.
Protein interactors of ASTN2 identified by IP plus LC-MS/MS. (A) Heat map of the top 57 candidate interacting proteins enriched in ASTN2 IPs in three experiments from P22–P28 cerebellar lysates. The intensity of the map is based on the MS intensity spectra values (Dataset S2). IP I and IP II are biological and technical replicates. IP III is a third biological replicate, which was washed more stringently and performed separately (Methods). (B) Lists of identified proteins by functional enrichment. Proteins belonging to the list of top hits (in A) are shown in red. ASD-associated proteins found by cross-referencing Dataset S2 to the SFARI Gene human ASD-gene list (https://gene.sfari.org) are marked with blue asterisks. Western blots show co-IPs of AP-2 (sigma fragment, Ap2s) and SLC12a5 with ASTN2 or JDUP in HEK 293T cells (C) and of AP-2, ROCK2, and ASTN2 in cerebellar lysates at P22 (D). The protein ladder is shown in kilodaltons. In the SLC12a5 blot, GFP appears in all samples due to the existence of an internal ribosome entry site-EGFP in the SLC12a5-HA construct.
ASTN2, but Not the FNIII Truncation, Induces Degradation of Surface Proteins.
In our flow cytometry analyses (Fig. 3B), where HEK 293T cells transfected with NLGN1/3-YFP, with and without ASTN2, were analyzed for surface and internal YFP expression, an increase in the percentage of cells that did not express NLGN1 or NLGN3 in the presence of ASTN2 was observed (bottom left quadrants of graphs), as opposed to an increase in cells that expressed NLGN1/3 internally but not on the cell surface (bottom right quadrants of graphs), suggesting that ASTN2 not only internalizes proteins but also induces degradation. Indeed, we detected reduced expression of the identified synaptic binding partners (Fig. 5 A and B, NLGN1-4, SLC12a5, and OLFM1) but no change in the levels of the adaptor protein AP-2 or GAPDH (Figs. 4C and 5 A and B) upon ASTN2 overexpression compared with controls in HEK 293T cells by Western blot. Importantly, while coexpression of NLGN1 or SLC12a5 with ASTN2 resulted in reduced levels of both, this reduction was much less marked upon coexpression with JDUP (Fig. 5B). Thus, coexpression of ASTN2, but not the JDUP truncation, markedly reduced protein levels. Moreover, shRNA-mediated knockdown of ASTN2 resulted in similar levels of NLGN1 and SLC12a5 to cells without ASTN2 protein or cells with JDUP, suggesting that the reduction in the levels of ASTN2 binding partners only occurs in the presence of intact ASTN2. Together, our data suggest that ASTN2 promotes the internalization and degradation of surface proteins.
Fig. 5.
ASTN2 reduces the levels of interacting proteins. (A) Western blots showing reduced protein levels of NLGN1–NLGN4 and OLFM1 in HEK 293T cells in the presence of ASTN2 compared with MYC (control) or OLFM1 alone. (B) Western blots showing reduced expression of NLGN1 and SLC12a5 in HEK 293T cells in the presence of ASTN2 or ASTN2 coexpressed with a scrambled plasmid, but less so in the presence of JDUP or MYC or when ASTN2 is knocked down with shRNA. GAPDH was used as an internal control for protein loading. (C) Representative Western blot showing ROCK2 levels in ASTN2 CNV patient T cells. The controls consisted of the mother and unrelated healthy subjects. Quantification of ASTN2 normalized to GAPDH in four technical replicates of three patients and three controls is shown in the box plot. (D) Conditional expression of ASTN2-EGFP and JDUP-EGFP (green) in sagittal sections of PCP2-Cre+ cerebella labeled with antibodies against Calbindin (red), NLGN2 (blue), and GluD2 (white). Quantification of NLGN2 levels (corrected integrated pixel density) in PC somas is shown (outlined by dashed lines) upon ASTN2 versus JDUP overexpression or control (PCP2-Cre−/− mice injected with conditional ASTN2-EGFP virus). (Scale bar: 10 μm.) The graph shows mean ± 1 SEM. n, total number of cells analyzed from three mice per condition. The P value at the top was determined by analysis of covariance, and those closer to bars were determined by post hoc tests between groups.
To further examine the idea that ASTN2 promotes protein degradation, we searched the top 57 protein hits identified by MS to see if any membrane proteins identified were also found in CD4+ patient T cells, where ASTN2 levels are reduced. Among the 57 hits, only ROCK2 is expressed in neurons as well as in T cells. Although like ASTN2, ROCK2 levels were variable among patients, they were, on average, higher in patients compared with controls (Fig. 5C). The expression level of the lower ASTN2 band (Fig. 1C, Bottom Right graph) inversely correlated with ROCK2 levels among patients. Together, our data suggest that ASTN2 plays a fundamental role in modulating the dynamic localization and degradation of several protein complexes in multiple cell types.
ASTN2 Modulates Synaptic Activity.
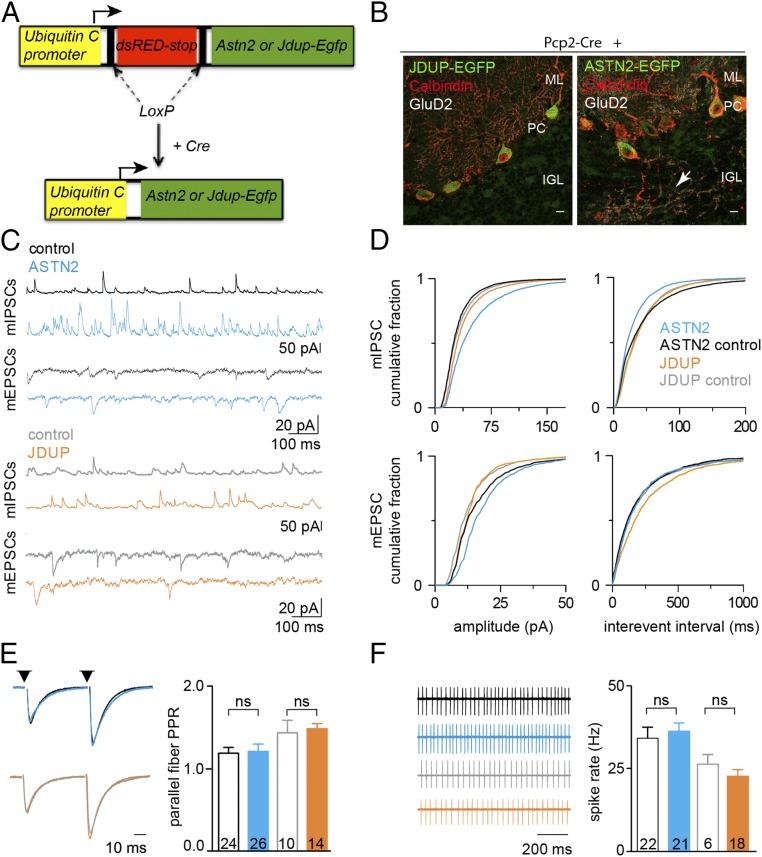
To investigate whether manipulation of ASTN2 levels impacts synaptic function in the cerebellum, we generated conditional lentiviruses expressing either the full-length EGFP-tagged ASTN2 (pFU-cASTN2-EGFP) or a truncated version (pFU-cJDUP-EGFP) lacking the FNIII domain. Overexpression approaches have generally been reported to be more sensitive in revealing roles for adhesion molecules (27) and proteins in multimeric complexes during synaptogenesis, due to the disruption of the stoichiometry of complexes and unmasking of functions otherwise compensated for by homologous proteins in loss-of-function approaches (28). Viruses were stereotactically injected in vivo into Pcp2-Cre+ cerebella at P0–P2 (Fig. 6A) to target PCs. EGFP expression, restricted by Pcp2-Cre to PCs only, was observed 3–4 wk after viral injection (Fig. 6B), allowing analysis of synaptic activity approximately at the stage when the IP/MS and the immuno-EM analyses were carried out (P22–P28). Interestingly, ASTN2-EGFP expression, but not JDUP-EGFP expression, resulted in mislocalization of some PCs to ectopic locations within the internal GC layer and in the white matter. The ectopic PCs had developed dendritic trees despite being mislocalized (Fig. 6B and SI Appendix, Fig. S7).
Fig. 6.
Effect of ASTN2 overexpression on synaptic activity of PCs. (A) Schematic of conditional lentiviral vectors. Expression of ASTN2-EGFP or JDUP-EGFP is driven by the Ubiquitin C promoter in the presence of Cre. (B) Sagittal sections showing JDUP-EGFP and ASTN2-EGFP (green) expression in PCs marked by Calbindin (red) and GluD2 (white) 3–4 wk after injection into Pcp2-Cre+ mice. The arrow indicates an ectopic PC and its dendritic tree in the IGL of an ASTN2-EGFP–injected mouse. (Scale bars: 10 μm.) (C) mIPSCs (Top) and mEPSCs (Bottom) postsynaptic currents in controls (PCP2-Cre−/−; black, n = 21 cells) and ASTN2-expressing (PCP2-Cre+; blue, n = 14 cells) PCs and in control (PCP2-Cre−/−; gray, n = 10 cells) and JDUP-expressing (PCP2-Cre+; orange, n = 12 cells) PCs. (D) Cumulative histograms of the amplitude (Left) and frequency (Right) of miniature events in control (black or gray) and ASTN2-expressing (blue) or JDUP-expressing (orange) PCs. Distributions were compared using the Mann–Whitney U test and were found to differ significantly between ASTN2 and controls in all measurements (P < 0.0001), except for mEPSC frequency, which was the same between control and ASTN2 but significantly different between control and JDUP (P < 0.0001). (E, Left) Evoked parallel fiber EPSCs [membrane potential (Vm) ∼ −75 mV, 50-ms interstimulus interval, arrowheads]. (E, Right) Summary graph of paired-pulse ratios (PPR) (mean ± 1 SEM). (F, Left) Cell-attached recordings of spontaneous spiking. (F, Right) Summary graph (mean ± 1 SEM) of spontaneous firing rates. The total numbers of cells recorded from five to seven animals per condition are shown. IGL, internal granule layer; ML, molecular layer; ns, not significant.
To test the properties of intrinsic excitability and synaptic transmission onto PCs that expressed either ASTN2-EGFP or JDUP-EGFP, we performed whole-cell electrophysiological recordings in acute brain slices from injected Pcp2-Cre+ animals 3–4 wk after viral injections (P21–P35). Control recordings were performed on EGFP-negative PCs from Pcp2-Cre−/− littermates injected with the same conditional viruses. Miniature excitatory/inhibitory postsynaptic currents (mEPSCs/mIPSCs) were recorded to assess nonevoked, quantal synaptic input, primarily from the parallel fibers (mEPSCs) and the inhibitory stellate and basket cells (mIPSCs). In PCs expressing ASTN2-EGFP, we found a significant increase in both mIPSC amplitude [maximum difference (Δmax) = 25.2%] and frequency (Δmax = 13.8%) and an increase in mEPSC amplitude (Δmax = 21.5%) in the same PCs. There was no change in the frequency of mEPSCs (Δmax = 5.9%; Fig. 6 C and D). In PCs expressing JDUP-EGFP, there was a much less marked increase in mIPSC amplitudes (Δmax = 11.4%; Fig. 6 C and D) and no change in frequency (Δmax = 2.3%). In addition, we observed a less marked increase in mEPSC amplitudes (Δmax = 11.9%) but a significant decrease in mEPSC frequency (Δmax = 13.5%). These results indicate changes in the synaptic strength of PCs, with the strongest effect on mIPSCs upon ASTN2-EGFP expression.
We also tested evoked excitation from parallel fibers and found that the paired-pulse ratio was unchanged, suggesting no difference in presynaptic release probability (Fig. 6E). In addition, we did not observe any differences in the spontaneous spiking of either ASTN2-EGFP– or JDUP-EGFP–expressing cells as measured by noninvasive cell-attached recordings (Fig. 6F). These results suggest that ASTN2 overexpression increases synaptic strength primarily by altering the postsynaptic response to neurotransmitters, rather than the intrinsic excitability of PCs or presynaptic release dynamics. Importantly, we did not observe the same degree of changes with expression of JDUP-EGFP as we did with ASTN2-EGFP. Finally, comparison of NLGN2 expression, which is the most highly expressed of the Neuroligins in the cerebellum (29), in the soma of targeted PCs revealed a significant decrease in ASTN2-EGFP PCs compared with JDUP-EGFP or control PCs from Pcp2-Cre−/− cerebella injected with the same conditional viruses (Fig. 5D), further corroborating our earlier findings that ASTN2 overexpression induces degradation of synaptic binding partners.
Discussion
Using immunogold EM, biochemical, electrophysiological, and functional assays, we demonstrate a role for ASTN2 in controlling protein trafficking and homeostasis in synaptic function. We detect ASTN2 in dendritic spines of neurons and in trafficking vesicles, and identify binding to several synaptic and trafficking proteins. Consistent with this interpretation, ASTN2 binds the Clathrin adapter AP-2. In postmitotic PCs, the sole output neuron of the cerebellar cortex and a cell type with documented loss in patients with ASD (14), overexpression of ASTN2 increased synaptic strength and decreased protein levels of synaptic binding partners. Our analyses suggest a role for ASTN2 in controlling surface membrane protein dynamics and underscore the contribution of impairment in protein trafficking to NDDs.
The results reported here compared the effect of ASTN2 overexpression with that of a truncated form lacking the FNIII domain. These experiments were informative in showing that removal of the FNIII domain interfered with the ability of ASTN2 to promote protein degradation, but not its ability to interact with binding partners. Overexpression in cell types that normally express ASTN2 provided a powerful means to study the consequences of disrupting the stoichiometry of ASTN2 in its native protein complexes. It should be noted that knockdown of ASTN2 protein was achieved in HEK cells, but not in neurons (SI Appendix, Fig. S3), possibly due to the formation of protein complexes in neurons, which promote the perdurance of the protein. Indeed, two pulse–chase studies examining protein turnover in the mouse brain found that ASTN1, the homolog of ASTN2, is present even after 1 month following in vivo isotopic labeling (17, 30). It is therefore likely that ASTN2 is also an extremely long-lived protein.
The interpretation that ASTN2 is a long-lived protein is also consistent with the reported stability of ASTN2 protein at pH 4.0 (31), which would allow it to traffic binding partners through the lower pH endosomal compartments of the endosomal/lysosomal system. In line with a trafficking role, the ASTN2 protein sequence (SI Appendix, Fig. S2) contains tyrosine-based sorting signals recognized by the adaptor proteins AP-1–AP-4 and a dileucine-based signal recognized by Golgi-localized gamma-adaptin ear containing ARF-binding (GGA) proteins (localized to endosomes), which are also involved in endosomal trafficking (32), as well as lysosomal sorting signals like those found in lysosomal membrane proteins LAMP1/LAMP2. These signals are recognized not only at the plasma membrane but also at sorting stations, such as the endosomes (32), suggesting that ASTN2 is likely involved in multiple steps of endosomal/lysosomal trafficking and not just at the surface membrane. This is consistent with our EM data, which showed that ASTN2 localizes to membranes and vesicles throughout the cell soma (Fig. 2D), and with our colocalization experiment with endosomal markers, where ASTN2 was found in a small fraction of early as well as late endosomes (SI Appendix, Fig. S4). Furthermore, our MS and biochemical data show that ASTN2 binds various endosomal trafficking and sorting proteins, including AP-2 and VPS36 [part of the endosomal sorting complex required for transport II (ESCRTII) complex], and controls the surface removal and degradation of a number of synaptic proteins. Finally, our EM analysis showed ASTN2 also on autophagosomes. As a large body of work shows that autophagic vesicles fuse with endosomes and lysosomes (33, 34), it will be interesting to further examine the involvement of ASTN2 in the interplay between autophagy and endosomal/lysosomal trafficking. We note that our findings do not exclude a role for ASTN2 in protein degradation pathways other than the endosomal/lysosomal system.
In addition to components of vesicle trafficking, the identification of interactions with multiple proteins involved in synaptic pruning [C1q (18)], AMPA receptor accessory proteins [OLFM1/3 (19)], ion transport [SLC12a5 (22)], and proteins regulating synaptic adhesion and activity [ROCK2 (21) and NLGN1-4 (29)] suggests that ASTN2 may modulate the composition of multiple protein complexes that impact synaptic form and function. In PCs, the sum of these interactions is increased postsynaptic activity upon ASTN2 overexpression. We speculate that the ability of ASTN2 to remove surface proteins also caused the mislocalization of PCs in vivo (SI Appendix, Fig. S7). The mechanisms that keep PCs in place are not fully understood, however.
Of note, a previous proteomic study of synaptosomal fractions prepared from mouse and human brains detected ASTN2 (35), corroborating our finding that ASTN2 is indeed present near synapses. The largest synaptic activity changes observed upon ASTN2 overexpression were increased mIPSC frequency and amplitude. Increases in mini-frequency are commonly associated with increased numbers of synapses, while increases in mini-amplitude often reflect increased numbers of postsynaptic receptors. However, other mechanisms could also contribute, such as alterations in single-channel conductance, receptor desensitization, changes in intracellular ion concentrations due to alterations in plasma membrane ion transporters, or fine-scale structural changes (36). Given the finding that ASTN2 internalizes synaptic and cell surface proteins, we favor the second set of scenarios whereby removal of proteins accessory to channels, such as OLFM1/3 and NLGN2 or the ion transporter SLC12a5, changes ligand-gated, channel-mediated responses. Excessive removal and degradation of accessory proteins could also leave receptors stranded on the cell surface, with their normal activity disrupted. Moreover, reduced levels of SLC12a5 could result in an increase in both mEPSCs and mIPSCs, as reported by a study in the hippocampus of SLC12a5-deficient mice (37). It should be noted that although NLGNs were not identified by IP/MS, which samples the most abundant and stable interactions, two NLGN1/3 peptides were identified with a targeted MS approach. It is possible that other transient interactions were not detected by our method and would need to be investigated in a candidate protein approach. Overall, our data suggest that as the primary role of ASTN2 appears to be in trafficking of an array of synaptic proteins, manipulations of ASTN2 can result in diverse synaptic modifications, including changes in postsynaptic receptor expression and synaptic strength depending on context. Further studies on the effect of loss of ASTN2 await development of a genetic mouse model.
Interestingly, the JDUP truncation did not abolish, but rather altered, the interaction of ASTN2 with its binding partners, increasing its affinity for some (NLGN3/4) while decreasing it for others (NLGN1/2; Fig. 3A). These differences in affinities could underlie the differing effects on mIPSC and mEPSC events induced by ASTN2 versus JDUP (Fig. 6). While ASTN2 clearly interacts with and regulates the availability of a number of proteins, a systems level analysis using advanced live cell imaging combined with single-molecule tracking of multiple proteins in action is needed to understand the combined effects of manipulation of an ASTN2-mediated trafficking pathway.
As reported here and elsewhere, patients with ASTN2 CNVs can manifest a spectrum of NDD phenotypes, even within the same family (Dataset S1). Our analysis of the association of ASTN2 CNVs with specific features commonly reported in such patients revealed a high level of association with ID and delayed speech and language. Based on our findings that an ASTN2-mediated protein trafficking pathway modulates synaptic strength, we propose that ASTN2 CNVs (truncating duplications and deletions) in patients with NDDs cause an accumulation of surface proteins. It appears critical for neurons to respond to inputs and modify their surface proteome in a rapid fashion. Activity-dependent changes in gene expression have been well documented, but more rapid changes in the composition of the synaptic proteome, through protein trafficking and degradation, would indeed offer quicker means to adjust to such inputs. Interestingly, Astn2 levels have been reported to change in response to stress in the CA3 region of the hippocampus in mice (38), suggesting the possibility that ASTN2 may modulate the surface proteome in response to activity in neurons.
ASTN2 is most abundantly expressed in the cerebellum, with far lower levels in the cortex, the olfactory bulb, and the dentate gyrus of the hippocampus. Although the contribution of the cerebellum to ASD, ID, or speech and language development is poorly understood, neuroimaging studies show both functional and neuroanatomical evidence for the critical importance of the cerebellum (39–41). Computational studies mapping the spatiotemporal coexpression of ASD-associated genes, including ASTN2, show that cortical projection neurons (layer 5/6) and the cerebellar cortex are the two most prevalent sites of ASD gene coexpression (42, 43). Furthermore, long-term depression, which is thought to be essential for many forms of cerebellar learning, is altered at parallel fiber/PC synapses in mice with targeted disruptions of several ASD-associated genes (16, 44–47). The present findings suggest that ASTN2 is a key regulator of dynamic trafficking of synaptic proteins in the cerebellum and lend support to the idea that aberrant regulation of protein homeostasis is a contributing cause of complex NDDs such as ASD and ID (48).
Methods
Human Subjects.
Patient clinical information and PBMCs were collected at Kennedy Krieger Institute upon receipt of informed consent from all subjects. This study was approved by the Kennedy Krieger Institute Institutional Review Board and The Rockefeller University Institutional Review Board.
Mice.
C57BL/6J mice (The Jackson Laboratory) were used unless stated otherwise. All procedures were performed according to guidelines approved by The Rockefeller University Institutional Animal Care and Use Committee. Both males and females were used for all studies and were randomly allocated to control and test groups.
RT-PCR and Quantitative RT-PCR.
RT-PCR and quantitative RT-PCR assays were carried out using standard procedures. Details are provided in SI Appendix, Supplementary Materials and Methods.
Primary Cell and Cell Line Culture.
HEK 293T/17 cells (catalog no. Crl-11268; American Type Culture Collection) were grown at 37 °C/5% CO2 in DMEM/F12, 10% heat-inactivated FBS, 4 mM l-glutamine, and 100 U/mL penicillin/streptomycin (all from Gibco). Mixed cerebellar cultures were prepared from P6–P8 pups and cultured in serum-containing medium as previously described (1, 49). Half of the medium was replaced with fresh medium without serum every 3–4 d for the duration of culture. More information on PBMC isolation and culture is provided in SI Appendix, Supplementary Materials and Methods.
Immunohistochemistry/Cytochemistry.
P15 or older mice were fixed by perfusion with 4% paraformaldehyde (PFA) and sectioned sagittally at 50 μm (Leica Vibratome). In vitro-cultured cells (described above) were grown on glass coverslips (no. 1.5 thickness; Fisher Scientific) and fixed for 15 min at room temperature in 4% PFA. Immunohistochemistry was carried out according to standard protocols. Detailed information is provided in SI Appendix, Supplementary Materials and Methods.
Antibodies Used for Immunohistochemistry, IP, and Western Blot.
Information on antibodies used for immunohistochemistry, IP, and Western blot is provided in SI Appendix, Supplementary Materials and Methods.
cDNA/shRNA Constructs.
Information on cDNA/shRNA constructs is provided in SI Appendix, Supplementary Materials and Methods.
Knockdown of ASTN2.
Information on knockdown of ASTN2 is provided in SI Appendix, Supplementary Materials and Methods.
Pre-Embedding Nanogold Immunolabeling and EM.
P28 mice were perfusion-fixed with 4% PFA. Midsagittal vibratome sections (50 μm) were prepared and incubated in blocking solution [3% BSA, 0.1% saponin in 0.1 M sodium cacodylate buffer (pH 7.4)] for 2 h at room temperature, followed by incubation in anti-ASTN2 antibody in blocking solution for 48 h at 4 °C. After washing (four times for 1 h each time in sodium cacodylate buffer), the sections were incubated for 2 h at room temperature with secondary antibody (1:100, Nanogold-IgG anti-rabbit 2003; Nanoprobes), washed four times for 1 h each time in 0.1% saponin in 0.1 M sodium cacodylate buffer (pH 7.4), and then fixed in 2.5% glutaraldehyde (Sigma) overnight at 4 °C. The sections underwent silver enhancement (HQ Silver Enhancement 2012; Nanoprobes) and gold toning using a 0.1% solution of gold chloride (HT1004; Sigma) according to the manufacturer’s description. The tissue was postfixed with 1% osmium tetroxide for 1 h on ice. Sections underwent en bloc staining with 1% uranyl acetate for 30 min, were dehydrated in a graded series of ethanol, were incubated for 10 min in acetone, were infiltrated with an Eponate 12 Embedding Kit (Ted Pella), and were polymerized for 48 h at 60 °C. Ultrathin sections (70 nm) prepared from three to four midsagittal sections per mouse were imaged on a JEOL JEM-100CX microscope at 80 kV and a digital imaging system (XR41-C; Advanced Microscopy Technology Corp.). For negative controls, sections were processed as described but with omission of the primary antibody. A total of 224 images were analyzed.
IP, Intracellular Cross-Linking of Proteins, Depletion of ASTN2 Antisera, and Western Blot.
For in vitro IPs, proteins from transfected HEK 293T cells were extracted in either radioimmunoprecipitation assay (RIPA) buffer (Thermo Scientific) or a customized IP buffer [50 mM Tris (pH 7.4), 200 mM NaCl, 5 mM MgCl2, 5 mM NaF, 1.5% octyl β-d-glucopyranoside (Abcam), 1× protease inhibitor mixture, 10 units of Benzonase Nuclease (Sigma)]. For in vivo IPs, proteins were extracted from P22–P28 cerebella using the customized IP buffer (as described above). Overnight IPs with 1.8-mg (in vivo) or 0.5-mg (in vitro) protein inputs were carried out according to standard protocols using Dynabeads Protein G (Invitrogen) cross-linked with antibodies using Bis-sulfosuccinimidyl suberate (BS3) cross-linking according to the manufacturer’s description (Thermo Scientific). As controls, either normal IgG from the same species as the antibody (Santa Cruz Biotechnology) or “anti-ASTN2 depleted antisera” were used. The anti-ASTN2 depleted antisera were prepared by incubating the anti-ASTN2 antibody with N-terminal biotinylated peptide against which the antibody had been raised (KITCEEKMVSMARNTYGETKGR) in the customized IP buffer. The antibody/peptide mix was then pulled out with Dynabeads MyOne Streptavidin T1 beads (Invitrogen), and the volume containing the antisera depleted of ASTN2 antibody was cross-linked to Dynabeads Protein G for use as a negative control. Depletion of anti-ASTN2 antibody was confirmed by Western blot analysis on cerebellar lysates (SI Appendix, Fig. S6A). Intracellular cross-linking of proteins was carried out before in vivo IPs with disuccinimidyl suberate (Thermo Scientific) according to the manufacturer’s descriptions. Western blots were carried out according to standard protocols using SDS/PAGE gels (Fisher) and Immobilon-P transfer membranes (Millipore). Blots were developed using an ECL Western Blotting Kit (GE Healthcare) or SuperSignal West Pico kit (Thermo Scientific) and exposed to X-ray film (Kodak).
Proteomics.
IPs were prepared as described earlier using 1.8 mg protein from whole cerebellar lysates (P22–P28) with antibody cross-linked beads. Immunoprecipitated proteins were eluted with 8 M urea (GE Healthcare) in 0.1 M ammonium bicarbonate (Sigma) and 10 mM DTT (Sigma). Cysteines were alkylated with iodoacetamide (Sigma). Samples were then diluted below 4 M urea before digesting with LysC (Waco Chemicals) for 6 h, after which urea was diluted below 2 M for overnight trypsin (Promega) digestion. Peptides were desalted by StaGE tips and processed for nano liquid chromatography (LC)-tandem MS (MS/MS) in data-dependent mode (Dionex U3000 coupled to a QExactive or QExactive Plus mass spectrometer; ThermoFisher Scientific). Generated LC-MS/MS data were queried against UniProt’s complete mouse proteome (https://www.uniprot.org/proteomes/UP000000589, downloaded July 2014) concatenated with common contaminants, and peptides were identified and quantified using Proteome Discoverer 1.4 (ThermoScientific) and Mascot 2.5.1 (Matrix Sciences) with fully tryptic restraints (Trypsin/P) and up to three missed cleavages with protein N-terminal acetylation and methionine oxidation as variable modifications and cysteine carbamidomethylation as a stable modification. Peptide matches required 5 ppm accuracy in MS1 and 20 millimass units in MS2, with a 1% false discovery rate filter using Percolator (50). To create the list of 466 identified proteins, all proteins that were only present in IgG samples were filtered out, as were proteins that showed enrichment in the IgG control over the ASTN2 IP in the most stringently washed experiment (III). The most stringent list of 57 proteins (Fig. 4) was created by including only proteins with at least three peptide hits that were ≥1.5-fold enriched in the combined ASTN2 I + II IPs/I + II IgG IPs or in ASTN2 III IP/depleted anti-ASTN2 sera. Functional enrichment analysis was carried out using STRING (v10.0, https://string-db.org/cgi/input.pl).
Flow Cytometry.
Information on flow cytometry is provided in SI Appendix, Supplementary Materials and Methods.
Surface and Pulse–Chase Labeling in Neurons.
At 14 days in vitro (DIV14), mixed cerebellar cultures (prepared as described earlier) were transfected with either “Nlgn1-HA-YFP + Astn2-EGFP” or “Nlgn1-HA-YFP + EGFP” plasmids using Lipofectamine 2000. On DIV17, cells were incubated with anti-GFP (1:500) diluted in culture medium containing 10 mM Hepes (Sigma) at 4 °C for 20 min, followed by two washes with medium plus 10 mM Hepes on ice to prevent endocytosis. For surface-labeling experiments (SI Appendix, Fig. S5), cells were fixed and processed as previously described. For pulse–chase experiments (Fig. 3), cells were incubated in fresh medium for a 20-min chase period at 35 °C/5% CO2. Cells were then incubated with anti-rabbit Alexa-633 (1:300) for 30 min at 4 °C to label all pulsed NLGN1-HA-YFP left on the cell surface, followed by two washes in medium on ice to wash away unbound antibodies. The cells were then fixed and processed as described earlier to detect internalized (anti-rabbit Alexa-555 secondary) and total (mouse anti-HA primary, followed by anti-mouse Alexa-405 secondary) protein. Control experiments were carried out to ensure that surface labeling did not occur with the GFP antibody on cells that expressed EGFP or ASTN2-EGFP only, as neither protein is exposed on the surface membrane, while the YFP tag of NLGN1-HA-YFP is positioned outside the plasma membrane, and hence detected by live labeling.
Imaging.
Information on imaging is provided in SI Appendix, Supplementary Materials and Methods.
Virus Production and in Vivo Viral Injections.
Vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped lentiviruses were produced with the pFU-cASTN2-EGFP and pfU-cJDUP-EGFP plasmids as previously reported (51). Viruses were collected and concentrated 45 h after transfection, and the pH of the media was kept between 7 and 7.3. Neonatal (24–30 h old) first generations from hemizygous PCP2-Cre breeding pairs [B6.Cg-Tg(Pcp2-cre)3555Jdhu/J; The Jackson Laboratory] were cryoanesthetized and injected using a modified protocol of Kim et al. (52), utilizing a 10-μL Hamilton syringe (catalog no. 1701-RN; Hamilton) fitted with a custom 32-gauge needle (no. 4 point style, 12° angle, 9.52-mm length; catalog no 7803-04; Hamilton). The needle was inserted perpendicular to the occipital plate at a depth of ∼2.5 mm, centering the tip in-line with the anterior/posterior axis and between the ears. All procedures were approved by the Duke University Institutional Animal Care and Use Committee and were in compliance with regulations.
Electrophysiology.
Acute sagittal slices (250 μm thick) were prepared from the cerebellar vermis of 3- to 4-wk-old injected mice (PCP2-Cre+) and control littermates (PCP2-Cre−/−). Slices were cut in an ice-cold potassium cutting solution (53) consisting of 130 mM K-gluconate, 15 mM KCl, 0.05 mM EGTA, 20 mM Hepes, and 25 mM glucose (pH 7.4) with KOH, and were transferred to an incubation chamber containing artificial cerebrospinal fluid (aCSF) composed of 125 mM NaCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 2.5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 25 glucose (pH 7.3, 310 Osm). Electrophysiological recordings were performed at 32–33 °C using a MultiClamp 700B Amplifier (Axon Instruments), with signals digitized at 50 kHz and filtered at 10 kHz. All whole-cell recordings were performed using a cesium-based internal solution containing 140 mM Cs-gluconate, 15 mM Hepes, 0.5 mM EGTA, 2 mM tetraethylammonium chloride, 2 mM MgATP, 0.3 mM NaGTP, and 10 mM phosphocreatine-Tris2, 2 mM QX 314-Cl. The pH was adjusted to 7.2 with CsOH. For parallel fiber stimulation experiments, glass monopolar electrodes (2–3 MΩ) were filled with aCSF and current was generated using a stimulus isolation unit (ISO-Flex Stimulus Isolator; A.M.P.I.). Spontaneous miniature synaptic currents were recorded in the presence of tetrodotoxin (0.5 μM; Tocris). IPSCs were recorded at the empirically determined EPSC reversal potential (∼ +10 mV), and EPSCs were recorded at the IPSC reversal potential (∼ −75 mV). Membrane potentials were not corrected for the liquid junction potential. Series resistance was monitored with a −5-mV hyperpolarizing pulse, and only recordings that remained stable over the period of data collection were used. The mIPSCs and mEPSCs were analyzed using MiniAnalysis software (v6.0.3; Synaptosoft, Inc.), using a 1-kHz low-pass Butterworth filter and a detection threshold set to 5× (for IPSCs) or 10× (for EPSCs) higher than baseline noise. So that no individual recording biased our distributions, 400 mIPSCs and 120 mEPSCs from each cell were randomly selected to establish the amplitude and frequency distributions of events across conditions. To measure the paired-pulse ratio, parallel fibers were stimulated at 20 Hz.
Quantification and Statistics.
Observations were replicated in at least three independent experiments (technical replicates). Data represented in graphs are both biological (pooled or individual animals/starting material) and technical (repeated multiple times) replicates, except for Fig. 1C, where individual data points are shown for human samples. Pixel intensities of Western blots and immunolabeling were quantified using ImageJ (NIH). Surface and internal labeling of NLG1-HA-YFP, as well as NLGN2 labeling in PCs in vivo, was quantified as follows: Each cell and its processes, including dendritic spines in the case of GCs and the cell soma only in the case of PCs in vivo, were outlined. The “integrated density” was measured (sum of all pixel intensities per square micrometer). The “mean fluorescence background” of each channel was also measured by selecting an area containing no cells. The “corrected fluorescence” was then calculated per cell as follows: integrated density − (area of selected cell * mean fluorescence intensity of image). For GCs, 20 cells per coverslip and two coverslips per condition were imaged from three independent experiments. The data were plotted as mean ± 1 SEM. The “total” in Fig. 5D represents the sum of the total pulse (internal labeling + surface labeling values). All data were checked for normality with the Shapiro–Wilk test. Outliers, identified in box plots, were removed, and nonnormal data were natural log-transformed to obtain the normal distribution. Specifically, four outliers were removed out of 164 data points in Fig. 5D. In general, data were analyzed by ANOVA, but if a covariate was present (e.g., “area” or “total pulse”), then analysis of covariance was used, taking these covariates into account. Where applicable, P values were calculated assuming equal variances among groups (tested with Levene’s test) and were two-sided unless stated otherwise. Differences between groups when more than two were present were identified by Bonferroni’s post hoc test. The total number of cells per condition (n) analyzed is stated on each bar, and the total number of experiments is given as N. All electrophysiology data were analyzed with the Mann–Whitney U test using GraphPad Prism software and Clampfit (Molecular Devices). The Δmax describes the percentage of maximum difference between each pair of distributions. Significant statistical difference between distributions of mEPSC and mIPSC amplitudes and frequencies is defined by P < 0.01.
Supplementary Material
Acknowledgments
We thank Dr. Inés Ibañez-Tallon (The Rockefeller University) for the pFU-cMVIIA-PE plasmid and for helpful discussions. We are indebted to Drs. Beatriz Antolin-Fontes and Nathalie Blachere (The Rockefeller University); Kunihiro Uryu and Nadine Soplop (Rockefeller University EM Facility); Pablo Ariel and Alison North (Rockefeller University Bio-Imaging); Svetlana Mazel, Selamawit Tadesse, and Stanka Semova (Rockefeller University Flow Cytometry); and Brian Dill (Rockefeller University Proteomics) for expert advice and technical assistance. We thank Drs. Mustafa Sahin (Boston Children’s Hospital), David Solecki (St. Jude Children’s Research Hospital), and Eve Govek (The Rockefeller University) for helpful comments on the manuscript and Yin Fang for technical support. We also thank Leila Jamal and Dr. Denise Batista (Kennedy Krieger Institute) for communicating details of patient CNVs and facilitating sample collection. The Neuroligin constructs were kind gifts from Drs. Ann-Marie Craig (University of British Columbia) and Peter Scheiffele (University of Basel), and the SLC12a5 construct was from Dr. Pavel Uvarov (University of Helsinki). This work was supported by the Eugene W. Chinery 2012 Trust (M.E.H.); the Renate, Hans, and Maria Hofmann Trust (H.B. and M.E.H.), NSF Predoctoral Fellowship DGF 1106401 (to T.R.F.), and National Institute of Neurological Disorders and Stroke Grant R01 NS096289-01A1 (to C.H.). The Rockefeller University Proteomics Resource Center acknowledges funding from the Leona M. and Harry B. Helmsley Charitable Trust. This study makes use of data generated by the DECIPHER community; a full list of centers that contributed to the generation of the data is available from https://decipher.sanger.ac.uk/, and funding was provided by the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809382115/-/DCSupplemental.
References
- 1.Wilson PM, Fryer RH, Fang Y, Hatten ME. Astn2, a novel member of the astrotactin gene family, regulates the trafficking of ASTN1 during glial-guided neuronal migration. J Neurosci. 2010;30:8529–8540. doi: 10.1523/JNEUROSCI.0032-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stitt TN, Hatten ME. Antibodies that recognize astrotactin block granule neuron binding to astroglia. Neuron. 1990;5:639–649. doi: 10.1016/0896-6273(90)90218-5. [DOI] [PubMed] [Google Scholar]
- 3.Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. doi: 10.1242/dev.129.4.965. [DOI] [PubMed] [Google Scholar]
- 4.Fishell G, Hatten ME. Astrotactin provides a receptor system for CNS neuronal migration. Development. 1991;113:755–765. doi: 10.1242/dev.113.3.755. [DOI] [PubMed] [Google Scholar]
- 5.Lesch KP, et al. Molecular genetics of adult ADHD: Converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm (Vienna) 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- 6.Vrijenhoek T, et al. Genetic Risk and Outcome in Psychosis (GROUP) Consortium Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernardini L, et al. High-resolution SNP arrays in mental retardation diagnostics: How much do we gain? Eur J Hum Genet. 2010;18:178–185. doi: 10.1038/ejhg.2009.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lionel AC, et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci Transl Med. 2011;3:95ra75. doi: 10.1126/scitranslmed.3002464. [DOI] [PubMed] [Google Scholar]
- 10.Lionel AC, et al. Disruption of the ASTN2/TRIM32 locus at 9q33.1 is a risk factor in males for autism spectrum disorders, ADHD and other neurodevelopmental phenotypes. Hum Mol Genet. 2014;23:2752–2768. doi: 10.1093/hmg/ddt669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glickstein M. What does the cerebellum really do? Curr Biol. 2007;17:R824–R827. doi: 10.1016/j.cub.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Timmann D, Daum I. Cerebellar contributions to cognitive functions: A progress report after two decades of research. Cerebellum. 2007;6:159–162. doi: 10.1080/14734220701496448. [DOI] [PubMed] [Google Scholar]
- 13.Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annu Rev Neurosci. 2009;32:413–434. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- 14.Fatemi SH, et al. Consensus paper: Pathological role of the cerebellum in autism. Cerebellum. 2012;11:777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kloth AD, et al. Cerebellar associative sensory learning defects in five mouse autism models. eLife. 2015;4:e06085. doi: 10.7554/eLife.06085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai PT, et al. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci USA. 2010;107:14508–14513. doi: 10.1073/pnas.1006551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 19.Schwenk J, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- 20.Krishnan A, et al. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci. 2016;19:1454–1462. doi: 10.1038/nn.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Z, Meng Y, Asrar S, Todorovski Z, Jia Z. A critical role of Rho-kinase ROCK2 in the regulation of spine and synaptic function. Neuropharmacology. 2009;56:81–89. doi: 10.1016/j.neuropharm.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 22.Rivera C, et al. The K+/Cl- co-transporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature. 1999;397:251–255. doi: 10.1038/16697. [DOI] [PubMed] [Google Scholar]
- 23.Li H, et al. KCC2 interacts with the dendritic cytoskeleton to promote spine development. Neuron. 2007;56:1019–1033. doi: 10.1016/j.neuron.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 24.Merner ND, et al. Regulatory domain or CpG site variation in SLC12A5, encoding the chloride transporter KCC2, in human autism and schizophrenia. Front Cell Neurosci. 2015;9:386. doi: 10.3389/fncel.2015.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang X, et al. KCC2 rescues functional deficits in human neurons derived from patients with Rett syndrome. Proc Natl Acad Sci USA. 2016;113:751–756. doi: 10.1073/pnas.1524013113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee A, et al. Jointly reduced inhibition and excitation underlies circuit-wide changes in cortical processing in Rett syndrome. Proc Natl Acad Sci USA. 2016;113:E7287–E7296. doi: 10.1073/pnas.1615330113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prelich G. Gene overexpression: Uses, mechanisms, and interpretation. Genetics. 2012;190:841–854. doi: 10.1534/genetics.111.136911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, et al. Neuroligins sculpt cerebellar Purkinje-cell circuits by differential control of distinct classes of synapses. Neuron. 2015;87:781–796. doi: 10.1016/j.neuron.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heo S, et al. Identification of long-lived synaptic proteins by proteomic analysis of synaptosome protein turnover. Proc Natl Acad Sci USA. 2018;115:E3827–E3836. doi: 10.1073/pnas.1720956115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni T, Harlos K, Gilbert R. Structure of astrotactin-2: A conserved vertebrate-specific and perforin-like membrane protein involved in neuronal development. Open Biol. 2016;6:160053. doi: 10.1098/rsob.160053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 33.Otomo A, Pan L, Hadano S. Dysregulation of the autophagy-endolysosomal system in amyotrophic lateral sclerosis and related motor neuron diseases. Neurol Res Int. 2012;2012:498428. doi: 10.1155/2012/498428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. J Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayés A, et al. Comparative study of human and mouse postsynaptic proteomes finds high compositional conservation and abundance differences for key synaptic proteins. PLoS One. 2012;7:e46683. doi: 10.1371/journal.pone.0046683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walmsley B, Alvarez FJ, Fyffe RE. Diversity of structure and function at mammalian central synapses. Trends Neurosci. 1998;21:81–88. doi: 10.1016/s0166-2236(97)01170-3. [DOI] [PubMed] [Google Scholar]
- 37.Khalilov I, et al. Enhanced synaptic activity and epileptiform events in the embryonic KCC2 deficient hippocampus. Front Cell Neurosci. 2011;5:23. doi: 10.3389/fncel.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marrocco J, et al. A sexually dimorphic pre-stressed translational signature in CA3 pyramidal neurons of BDNF Val66Met mice. Nat Commun. 2017;8:808. doi: 10.1038/s41467-017-01014-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen G, Courchesne E. Differential effects of developmental cerebellar abnormality on cognitive and motor functions in the cerebellum: An fMRI study of autism. Am J Psychiatry. 2003;160:262–273. doi: 10.1176/appi.ajp.160.2.262. [DOI] [PubMed] [Google Scholar]
- 40.Amaral DG, Schumann CM, Nordahl CW. Neuroanatomy of autism. Trends Neurosci. 2008;31:137–145. doi: 10.1016/j.tins.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Stoodley CJ, et al. Altered cerebellar connectivity in autism and cerebellar-mediated rescue of autism-related behaviors in mice. Nat Neurosci. 2017;20:1744–1751. doi: 10.1038/s41593-017-0004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parikshak NN, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koekkoek SK, et al. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in fragile X syndrome. Neuron. 2005;47:339–352. doi: 10.1016/j.neuron.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Baudouin SJ, et al. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 2012;338:128–132. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- 46.Reith RM, et al. Loss of Tsc2 in Purkinje cells is associated with autistic-like behavior in a mouse model of tuberous sclerosis complex. Neurobiol Dis. 2013;51:93–103. doi: 10.1016/j.nbd.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 47.Piochon C, et al. Cerebellar plasticity and motor learning deficits in a copy-number variation mouse model of autism. Nat Commun. 2014;5:5586, and erratum (2015) 6:6014. doi: 10.1038/ncomms6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sztainberg Y, Zoghbi HY. Lessons learned from studying syndromic autism spectrum disorders. Nat Neurosci. 2016;19:1408–1417. doi: 10.1038/nn.4420. [DOI] [PubMed] [Google Scholar]
- 49.Baptista CA, Hatten ME, Blazeski R, Mason CA. Cell-cell interactions influence survival and differentiation of purified Purkinje cells in vitro. Neuron. 1994;12:243–260. doi: 10.1016/0896-6273(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 50.Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 51.Auer S, et al. Silencing neurotransmission with membrane-tethered toxins. Nat Methods. 2010;7:229–236. doi: 10.1038/nmeth.1425. [DOI] [PubMed] [Google Scholar]
- 52.Kim JY, et al. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. Eur J Neurosci. 2013;37:1203–1220. doi: 10.1111/ejn.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dugué GP, Dumoulin A, Triller A, Dieudonné S. Target-dependent use of co-released inhibitory transmitters at central synapses. J Neurosci. 2005;25:6490–6498. doi: 10.1523/JNEUROSCI.1500-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.