Significance
Clathrin-mediated endocytosis (CME) determines the internalization of receptors and their downstream signaling. We discovered that CME is differentially regulated by specific signaling kinases in cancer cells. In particular, ERK1/2-mediated phosphorylation of the FCH/F-BAR and double SH3 domains-containing protein 2 (FCHSD2) regulates CME and the trafficking and signaling activities of EGF receptors. This reciprocal interaction negatively regulates cancer proliferation and migration. The expression level of FCHSD2 is positively correlated with higher lung cancer patient survival rates. This study identifies signaling pathways that impinge on the endocytic machinery and reveals a molecular nexus for crosstalk between intracellular signaling and CME. Cancer cells specifically adapt this crosstalk as a determinant for tumor progression, which has implications for novel therapeutics against cancers.
Keywords: epidermal growth factor receptor, signal transduction, nonsmall cell lung cancer, Nervous Wreck
Abstract
Clathrin-mediated endocytosis (CME) regulates the uptake of cell-surface receptors as well as their downstream signaling activities. We recently reported that signaling can reciprocally regulate CME in cancer cells and that this crosstalk can contribute to cancer progression. To further explore the nature and extent of the crosstalk between signaling and CME in cancer cell biology, we analyzed a panel of oncogenic signaling kinase inhibitors for their effects on CME across a panel of normal and cancerous cells. Inhibition of several kinases selectively affected CME in cancer cells, including inhibition of ERK1/2, which selectively inhibited CME by decreasing the rate of clathrin-coated pit (CCP) initiation. We identified an ERK1/2 substrate, the FCH/F-BAR and SH3 domain-containing protein FCHSD2, as being essential for the ERK1/2-dependent effects on CME and CCP initiation. Our data suggest that ERK1/2 phosphorylation activates FCHSD2 and regulates EGF receptor (EGFR) endocytic trafficking as well as downstream signaling activities. Loss of FCHSD2 activity in nonsmall cell lung cancer (NSCLC) cells leads to increased cell-surface expression and altered signaling downstream of EGFR, resulting in enhanced cell proliferation and migration. The expression level of FCHSD2 is positively correlated with higher NSCLC patient survival rates, suggesting that FCHSD2 can negatively affect cancer progression. These findings provide insight into the mechanisms and consequences of the reciprocal regulation of signaling and CME in cancer cells.
Cancer progression involves tumor cell proliferation and metastasis driven, in part, by altered intracellular signaling downstream of plasma membrane (PM) receptors (1, 2). In tumor cells, signaling activities can be altered by dysregulated endocytic trafficking that increases receptor recycling and decreases lysosomal targeting and degradation (3–5). Hence, a link between endocytosis and cancer behavior has been suggested (3, 6–8). Clathrin-mediated endocytosis (CME) is the major endocytic pathway that determines the rates of internalization of PM receptors, regulates their cell-surface expression, and controls their downstream signaling activities (9, 10). Despite the well-known fact that CME can regulate intracellular signaling pathways (11), whether and how signaling can feed back to regulate CME during cancer cell progression has been less studied.
CME occurs when clathrin-coated pits (CCPs) assemble on the cell surface, concentrate cargo molecules (i.e., receptors and their bound ligands), invaginate, and undergo membrane fission catalyzed by the GTPase dynamin to release clathrin-coated vesicles into the cytosol. Once considered a constitutive process, recent studies have demonstrated that CME can be dynamically regulated in response to cargo and downstream signaling pathways (12–15). For example, in nonsmall cell lung cancer (NSCLC) cells, dynamin-1 (Dyn1), the neuron-enriched isoform, is frequently up-regulated and can be activated downstream of a protein kinase B (Akt)/GSK3β signaling cascade, resulting in increased rates of CCP initiation and dysregulated CCP maturation (15). Moreover, the clathrin light chain b isoform (CLCb), which functions primarily to regulate clathrin assembly and is neuronally enriched (16, 17), is also selectively up-regulated in NSCLC cells (18). CLCb-dependent alterations in CME lead to Dyn1 activation via increased Akt/GSK3β phosphorylation associated with APPL1-positive signaling endosomes (18). The crosstalk between signaling and CME contributes to abnormal trafficking of EGF receptors (EGFRs) and altered downstream signaling, leading to enhanced cancer cell migration and metastasis (18). Accordingly, it is important to further understand the nature and extent of interactions between CME and intracellular signaling in cancer cell biology.
To address this issue, we analyzed a panel of oncogenic signaling kinase inhibitors for their differential effects on CME in several human noncancerous and cancer cells. We identified ERK1/2 activity as being selectively required for rapid CME and CCP initiation in cancer cells and FCH/F-BAR and double SH3 domains-containing protein 2 [FCHSD2; Nervous Wreck (Nwk) in Drosophila] as the downstream kinase substrate responsible for these effects. Our study provides insight into the mechanisms and consequences of the reciprocal regulation of signaling and CME in cancer cells.
Results
Oncogenic Signaling Differentially Affects CME in Cancer Cells.
Recent studies have shown that the crosstalk between intracellular signaling and CME can affect cancer progression and metastasis (8, 18). Thus, it is important to determine the extent of this crosstalk as well as the signaling pathways that impinge on the regulation of CME in cancer cells. To this end, we prioritized a set of kinases and GTPases that are often implicated in cancer-relevant signaling pathways and assessed the effects of their validated inhibitors (19–23) on transferrin receptor (TfnR) endocytosis, a canonical marker for the quantification of CME (24, 25). Initially, we chose noncancerous ARPE-19 cells, which have been routinely used to study TfnR endocytosis (24), and compared these with H1299 NSCLC cells, which exhibit crosstalk between signaling and CME (15, 18).
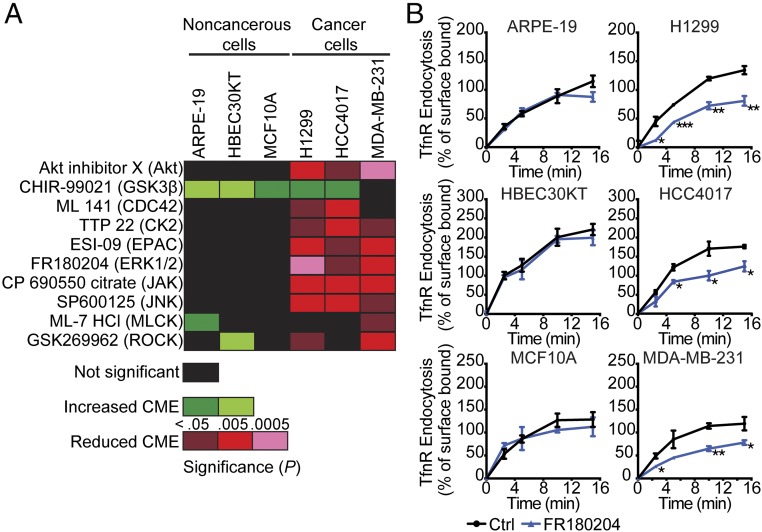
Strikingly, 10 of the 21 inhibitors we screened differentially affected CME in H1299 vs. ARPE cells (SI Appendix, Fig. S1). This included inhibition of Akt, which we previously showed functions through an Akt/GSK3β signaling cascade to activate Dyn1 and enhance endocytosis in H1299 and A549 NSCLC cells (15, 18, 26). To further validate these findings, we extended the functional comparison of these 10 inhibitors to more closely matched cancerous and noncancer cell lines. We used another NSCLC cell line, HCC4017, together with the syngeneic nontumorigenic HBEC cell line, HBEC30KT, derived from the same patient (27). In addition, we compared the breast cancer cell line MDA-MB-231 with normal human mammary epithelial cells, MCF10A (28). Consistent with our initial screen, inhibition of a subset of these kinases significantly reduced CME activity in most cancer cell lines without affecting CME in their noncancerous counterparts (Fig. 1A). These findings reveal the unexpected differential regulation of CME in cancer cells by distinct intracellular signaling pathways.
Fig. 1.
Differential effects of kinase inhibitors on CME in cancer cells. (A) Heatmap illustrating results from a systematic analysis of the effects of pharmacological kinase inhibitors on TfnR endocytosis in human noncancerous and cancer cell lines. (B) Absolute rates of endocytosis of TfnR were measured in different pairs of human noncancerous and cancer cells without or with treatment with the ERK1/2 inhibitor FR180204 (10 μM). The percentage of internalized TfnR was calculated relative to the initial surface TfnR. Data represent mean ± SEM (n = 3). Two-tailed Student’s t tests were used to assess statistical significance for comparison with controls (Ctrl). *P < 0.05, **P < 0.005, ***P < 0.0005.
Inhibiting some of these kinases selectively enhanced CME in noncancerous vs. cancer cells. GSK3β enhanced rates of CME in most cell lines tested, presumably through activation of Dyn1 (15, 26). Interestingly, inhibition of myosin light chain kinase (MLCK) and Rho kinase (ROCK) accelerated endocytosis in a subset of noncancer cell lines while inhibiting CME in a subset of cancer cells. As these kinases control the actin cytoskeleton, these results could reflect known cell-type differences in the role of actin in cancer vs. noncancer cells (29, 30). While it will be important to investigate how each of these signaling pathways impinges on CME, we chose to focus our further studies on ERK signaling, as the EGFR/Ras/ERK1/2 pathway is a major driver of, and therapeutic target for, multiple cancers (31–34). Inhibition of ERK1/2 selectively inhibits CME in all cancer cell lines tested without affecting CME in several noncancerous cell lines (Fig. 1B). All subsequent studies were performed in ARPE-19 and H1299 cells and in the syngeneic pair HCC4017 and HBEC30KT. In addition, given that the absolute rates of TfnR uptake differ between cell lines (Fig. 1B), data were normalized to the percent of TfnR uptake in treated cells relative to their untreated controls.
ERK1/2 Specifically Regulates CME in Cancer Cells.
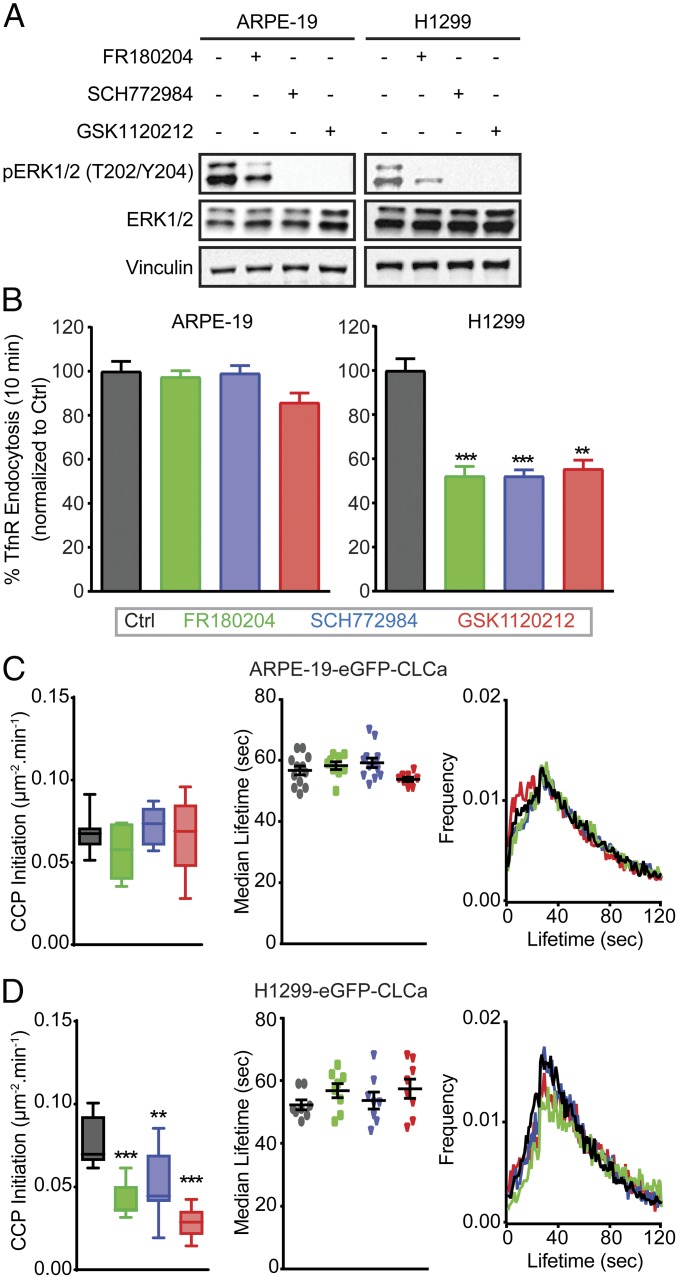
ERK1/2 signaling, downstream of EGFR and Ras is highly activated in many cancers. To confirm that ERK1/2 activity is selectively involved in the regulation of CME in cancer cells, we used a second ERK1/2 inhibitor (SCH772984) as well as a well-characterized inhibitor of the essential upstream kinase, mitogen-activated protein kinase kinase (MEK, GSK1120212). As expected, these ERK1/2-signaling inhibitors efficiently decreased ERK1/2 phosphorylation (Fig. 2A and SI Appendix, Fig. S2A). Importantly, they also specifically inhibited CME both in H1229 cells relative to ARPE-19 cells (Fig. 2B) and in HCC4017 cells relative to HBEC30KT cells (SI Appendix, Fig. S2B).
Fig. 2.
ERK1/2 selectively affects CME activities in cancer cells. (A) Representative Western blots used to measure the efficiencies of kinase inhibitors in reducing ERK1/2 phosphorylation in control, ERK1/2 inhibitor (FR180204 and SCH772984; 10 μM)-treated or MEK1/2 inhibitor (GSK1120212; 10 μM)-treated ARPE-19 and H1299 cells. See SI Appendix, Fig. S2A for quantitation. (B) Endocytosis of TfnR was measured in ARPE-19 and H1299 cells untreated (Ctrl) or incubated with the indicated inhibitors, as described above. All data were normalized to the percentage of TfnR internalized after 10 min in control cells and represent mean ± SEM (n = 3). See Fig. 1B for examples of absolute kinetics of TfnR uptake, which vary among cell lines. (C and D) Rates of CCP initiation (Left), median lifetimes (Middle) and average lifetime distribution (Right) of all bona fide CCPs in control and inhibitor-treated ARPE-19 cells (C) and H1299 cells (D). Data in C and D were obtained from at least 15 cells per condition (>10,000 CCPs per condition). The box plots represent median, 25th, and 75th percentiles, and error bars indicate the outermost data points. Two-tailed Student’s t tests were used to assess statistical significance for comparison with controls. **P < 0.005, ***P < 0.0005.
CME is a multistep process that involves CCP initiation, stabilization, maturation, and finally fission. To determine which step(s) of CME was affected upon ERK1/2 inhibition, we monitored CCP dynamics using total internal reflection fluorescence microscopy (TIR-FM) (15, 18) in ARPE-19 and H1299 cells stably expressing EGFP-clathrin light chain a (EGFP-CLCa) and in HCC4017 cells stably expressing SNAP-CLCa. CCPs were detected and tracked with high sensitivity and in a comprehensive and unbiased manner using cmeAnalysis (35) to measure initiation rates and lifetimes. As expected, CCP dynamics were unchanged in the ARPE-19 cells upon inhibitor treatment (Fig. 2C). However, inhibition of ERK1/2 signaling dramatically reduced the rate of CCP initiation without significantly altering the lifetime distribution or median lifetimes of CCPs in both H1299 cells (Fig. 2D) and HCC4017 cancer cells (SI Appendix, Fig. S2C). These data indicate that ERK1/2 signaling differentially regulates the first key step of CME (i.e., CCP initiation) (36) in cancer cells. As SCH772984 was the most potent ERK inhibitor, it was used in all further studies.
FCHSD2 Regulates CME in Cancer Cells.
ERK1/2 kinases are ubiquitously expressed serine/threonine kinases that regulate diverse biological processes (37–40) by phosphorylating multiple substrates (41). We screened the literature for ERK1/2 substrates and identified FCHSD2 (42), the human ortholog of Nwk in Drosophila, which has been implicated in membrane remodeling (43) and the regulation of growth factor signaling (44, 45) at the synapse. FCHSD2 was of interest because it shares an N-terminal F-BAR domain with FCH domain-only (FCHo) proteins known to play a role in CCP initiation (46, 47) and has two C-terminal SH3 domains shown to interact with components of the actin and CME machinery (48).
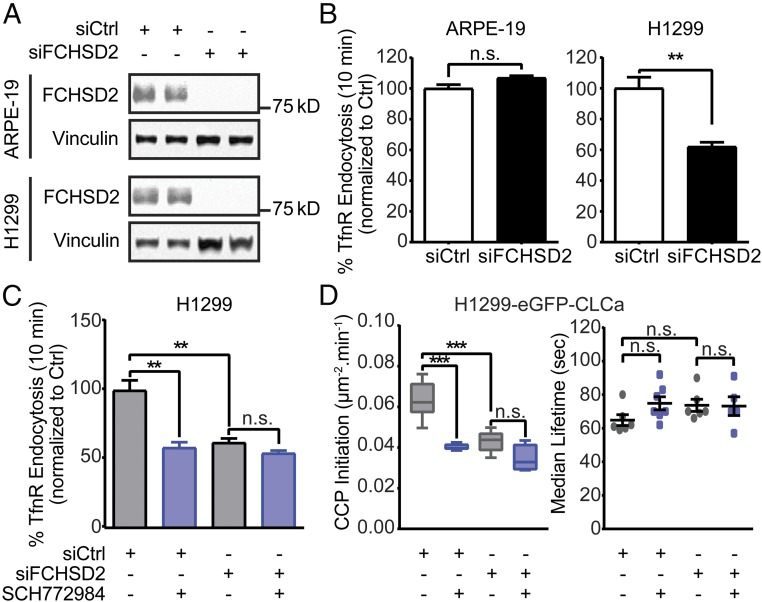
To test its potential role in CME, we first investigated the effects of efficient siRNA-mediated knockdown of FCHSD2 (Fig. 3A and SI Appendix, Fig. S3A) in noncancerous and cancer cells. Like ERK1/2 inhibition, depletion of FCHSD2 did not significantly alter CME activities in nontumorigenic ARPE-19 or HBEC30KT cells but potently inhibited CME in H1299 and HCC4017 NSCLC cells (Fig. 3B and SI Appendix, Fig. S3B). We confirmed that FCHSD2 is expressed in all these cell lines, albeit at different levels that do not correlate with their sensitivity to FCHSD2 knockdown (i.e., H1299 > HBEK30KT > HCC4017 ∼ ARPE) (SI Appendix, Fig. S4). Importantly, FCHSD2-depleted cancer cells were no longer sensitive to ERK1/2 inhibition (Fig. 3C and SI Appendix, Fig. S3C), even though they remained sensitive to SP600125 (SI Appendix, Fig. S3C), a JNK inhibitor that also specifically affects CME in cancer cells (Fig. 1A). These data suggest that FCHSD2 might be the CME-relevant downstream target of ERK. Consistent with this, like ERK inhibition, knockdown of FCHSD2 specifically inhibited the rate of CCP initiation without affecting CCP lifetimes (Fig. 3D). The reduced rate of CCP initiation in FCHSD2-depleted cells was now insensitive to ERK1/2 inhibition. Together, these data demonstrate that FCHSD2 plays a role in CME, especially in CCP initiation, and suggest that it functions as a relevant downstream effector of ERK1/2 kinases.
Fig. 3.
FCHSD2 is specifically enlisted for CME in cancer cells. (A) Representative Western blots and the quantification of FCHSD2 knockdown efficiency in control and FCHSD2 siRNA-treated ARPE-19 and H1299 cells. (B) Endocytosis of TfnR was measured in control and FCHSD2 siRNA-treated ARPE-19 and H1299 cells. All data were normalized to control as in Fig. 2. (C) Endocytosis of TfnR was measured, as described in B in control and FCHSD2 siRNA-treated H1299 cells without or with treatment with the ERK1/2 inhibitor SCH772984 (10 μM). Data in A–C represent mean ± SEM (n = 3). (D) Rates of initiation (no. CCPs⋅μm2⋅min) (Left) and median lifetimes (Right) of bona fide CCPs in control and FCHSD2 siRNA-treated H1299 cells treated or not treated with the ERK1/2 inhibitor SCH772984 (10 μM). Data were obtained from at least 15 cells per condition (>10,000 CCPs per condition). The box plots represent the median, 25th, and 75th percentiles, and error bars indicate the outermost data points. Two-tailed Student’s t tests were used to assess statistical significance for the indicated dataset. **P < 0.005, ***P < 0.0005; n.s., not significant.
ERK1/2 Directly Regulates FCHSD2-Mediated CME in Cancer Cells.
The phosphorylation of FCHSD2 on S681 within a canonical ERK phosphorylation motif (PXSP) was identified in a large-scale screen for ERK2 substrates using an analog-sensitive ERK2 mutant (42). We first confirmed EGF-dependent phosphorylation of this site in serum-starved and EGF-treated HCC4017 cells by phospho-proteomic analysis (SI Appendix, Table S1).
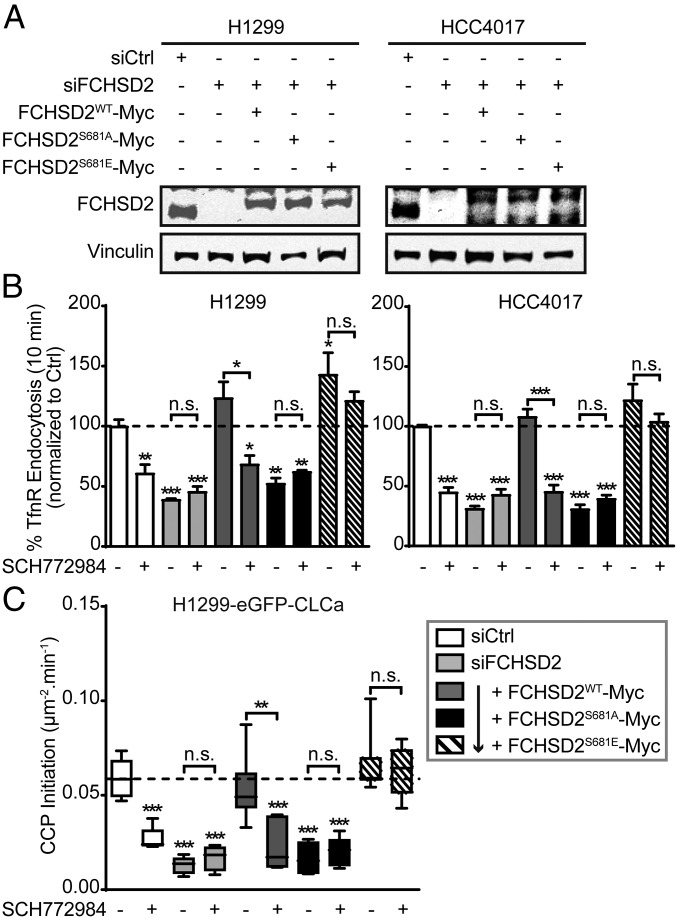
To test whether FCHSD2 was indeed the ERK1/2 substrate responsible for cancer cell-specific regulation of CME, we performed FCHSD2 siRNA-mediated knockdown in H1299 and HCC4017 cells followed by reconstitution with wild-type FCHSD2 (FCHSD2WT-Myc), a mutant that cannot be phosphorylated by ERK1/2 (FCHSD2S681A-Myc), or a phosphomimetic mutant (FCHSD2S681E-Myc) at nearly endogenous levels (Fig. 4A). As expected, reconstitution of FCHSD2 siRNA-depleted H1299 or HCC4017 cells with FCHSD2WT-Myc fully rescued CME activity and restored sensitivity to ERK1/2 inhibition (Fig. 4B, dark gray bars). In contrast, reconstitution with nonphosphorylatable FCHSD2S681A-Myc failed to rescue CME or restore sensitivity to ERK1/2 inhibition (Fig. 4B, black bars). Reconstitution with FCHSD2S681E-Myc also rescued CME in siRNA-treated cells. However, in contrast to cells expressing WT FCHSD2, CME in cells reconstituted with the phosphomimetic FCHSD2 mutant was now resistant to ERK1/2 inhibition (Fig. 4B, hatched bars). Together these data provide strong evidence that phosphorylation of FCHSD2 on S681 is indeed required for the ERK1/2-dependent effects on CME in cancer cells.
Fig. 4.
ERK1/2-mediated phosphorylation of FCHSD2 is required for CME in cancer cells. (A) Representative Western blots of FCHSD2 expression in controls, FCHSD2 knockdown (KD) cells, or FCHSD2–KD cells reconstituted with FCHSD2WT-Myc, FCHSD2S681A-Myc, or FCHSD2S681E-Myc. (B) Endocytosis of TfnR was measured in controls, FCHSD2–KD cells, or FCHSD2–KD cells reconstituted with FCHSD2WT-Myc, FCHSD2S681A-Myc, or FCHSD2S681E-Myc, in the absence or presence of the ERK1/2 inhibitor SCH772984 (10 μM). All data were normalized to control and represent mean ± SEM (n = 3). (C) Rates of initiation (no. CCPs⋅μm2⋅min) of bona fide CCPs in the cells as described in B. Data were obtained from at least 15 cells per condition (>10,000 CCPs per condition); the box plots represent median, 25th, and 75th percentiles, and error bars represent the outermost data points. Two-tailed Student’s t tests were used to assess statistical significance for comparison with siCtrl without SCH772984 treatment and for the indicated dataset. *P < 0.05, **P < 0.005, ***P < 0.0005; n.s., not significant.
Reconstitution of FCHSD2-depleted cells with FCHSD2WT-Myc, but not with the FCHSD2S681A-Myc mutant, restored ERK1/2-dependent rates of CCP initiation (Fig. 4C, dark gray and black bars). The fully restored rate of CCP initiation in siRNA-treated cells reconstituted with FCHSD2S681E-Myc was no longer sensitive to ERK1/2 inhibition (Fig. 4C, hatched bars). These results establish that the effects of ERK1/2 inhibition on CME and CCP initiation are mediated by FCHSD2 activity through phosphorylation of S681.
ERK1/2 Activity Is Required for FCHSD2 Recruitment to the PM and CCPs.
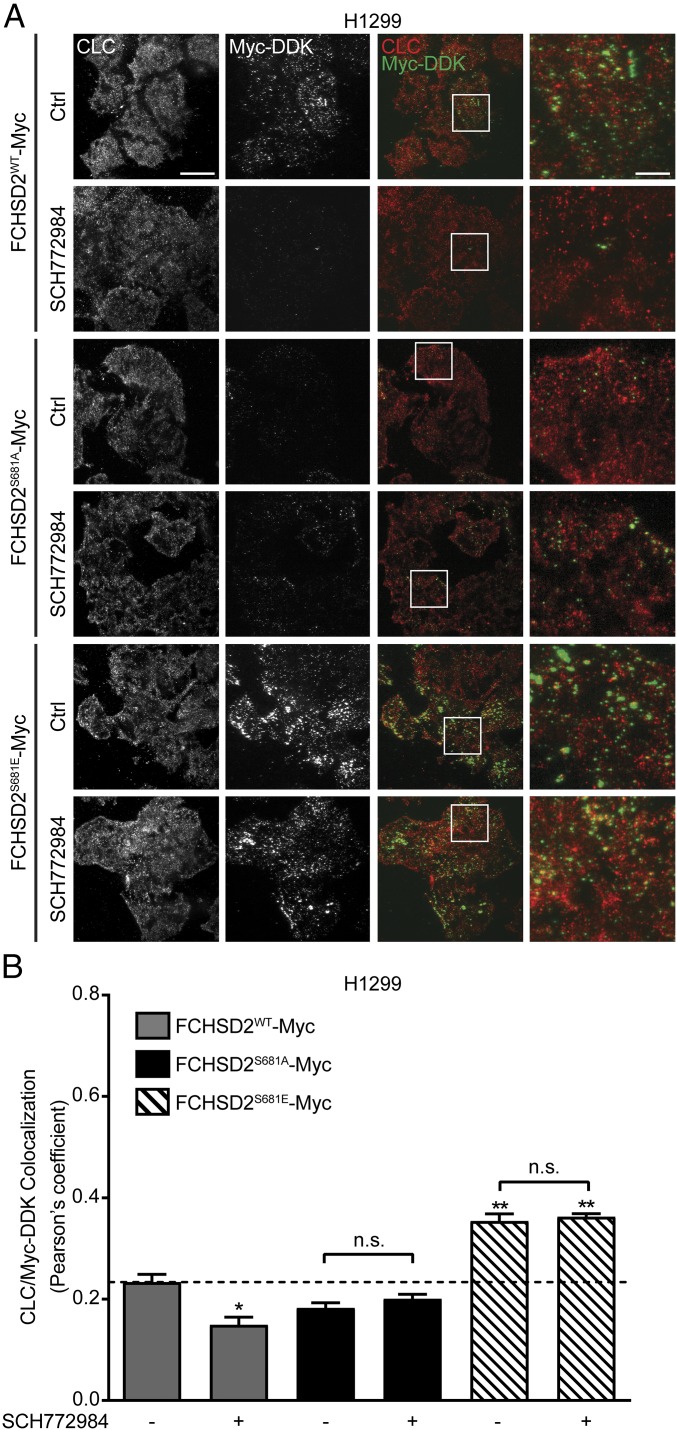
Nwk has been shown to regulate actin assembly through activation of neural Wiskott–Aldrich syndrome protein (N-WASP) (49), and studies in Drosophila have suggested it regulates sorting in early endosomal compartments (45). Thus, the effects on CCP initiation might be indirect. To test this, we examined the subcellular localization of FCHSD2. While the commercially available antibodies could be validated by siRNA knockdown for Western blotting, immunofluorescent staining was nonspecific (SI Appendix, Fig. S5A). In addition, C-terminally GFP-tagged FCHSD2 formed aggregates in the perinuclear region of the cell (SI Appendix, Fig. S5A) that were not seen with myc-tagged protein (SI Appendix, Fig. S5C). Given that the myc-tagged protein was fully functional (Fig. 4), we confirmed the specificity of Myc-DDK epitope antibodies by Western blotting and immunofluorescence (SI Appendix, Fig. S5 B and C) and used this antibody for colocalization studies in H1299 and HCC4017 cells expressing FCHSD2WT-Myc, FCHSD2S681A-Myc, or FCHSD2S681E-Myc (Fig. 5 and SI Appendix, Fig. S6).
Fig. 5.
FCHSD2 recruitment to the PM and CCPs is regulated by ERK1/2 activity. (A) Representative TIR-FM images of CLC and Myc-DDK immunofluorescence staining in FCHSD2WT-Myc–, FCHSD2S681A-Myc–, and FCHSD2S681E-Myc–transduced H1299 cells in the absence or presence of the ERK1/2 inhibitor SCH772984 (10 μM). (Scale bars: 25 μm in overviews, 6.25 μm in the magnified views at far right.) (B) Colocalization of CLC and Myc-DDK immunofluorescence staining in the cells as described in A. Data were obtained from n = 3 independent experiments with at least 40 cells in total per condition and represent the mean ± SEM. Two-tailed Student’s t tests were used to assess statistical significance for comparison with FCHSD2WT-Myc without SCH772984 treatment and for the indicated dataset. *P < 0.05, **P < 0.005; n.s., not significant.
Total internal reflection fluorescence (TIRF) imaging of H1299 (Fig. 5A) and HCC4017 (SI Appendix, Fig. S6A) cells revealed that FCHSD2WT-Myc could be detected on the PM in punctate structures partially colocalizing with clathrin (Fig. 5B and SI Appendix, Fig. S6B). Recruitment of FCHSD2 to the PM was regulated by ERK1/2 activity. Thus, upon inhibition of ERK1/2, the recruitment of FCHSD2WT-Myc to the PM was significantly reduced (Fig. 5A), as was its colocalization with clathrin (Fig. 5B). Moreover, the nonphosphorylatable FCHSD2S681A-Myc was inefficiently recruited to the PM, whereas FCHSD2S681E-Myc exhibited increased PM recruitment. Both mutants were now resistant to ERK1/2 inhibition. Although the colocalization of FCHSD2S681E-Myc with clathrin was significantly increased relative to WT cells, the degree of colocalization remained at <40% in H1299 cells (Fig. 5B) and ∼50% in HCC4017 cells (SI Appendix, Fig. S6B). This partial colocalization might indicate an only transient association of FCHSD2 with CCPs. In any case, these data suggest a phosphorylation-dependent recruitment of FCHSD2 to the PM in a position to regulate CME.
FCHSD2 Regulates the Endocytic Trafficking and Surface Expression of EGFR.
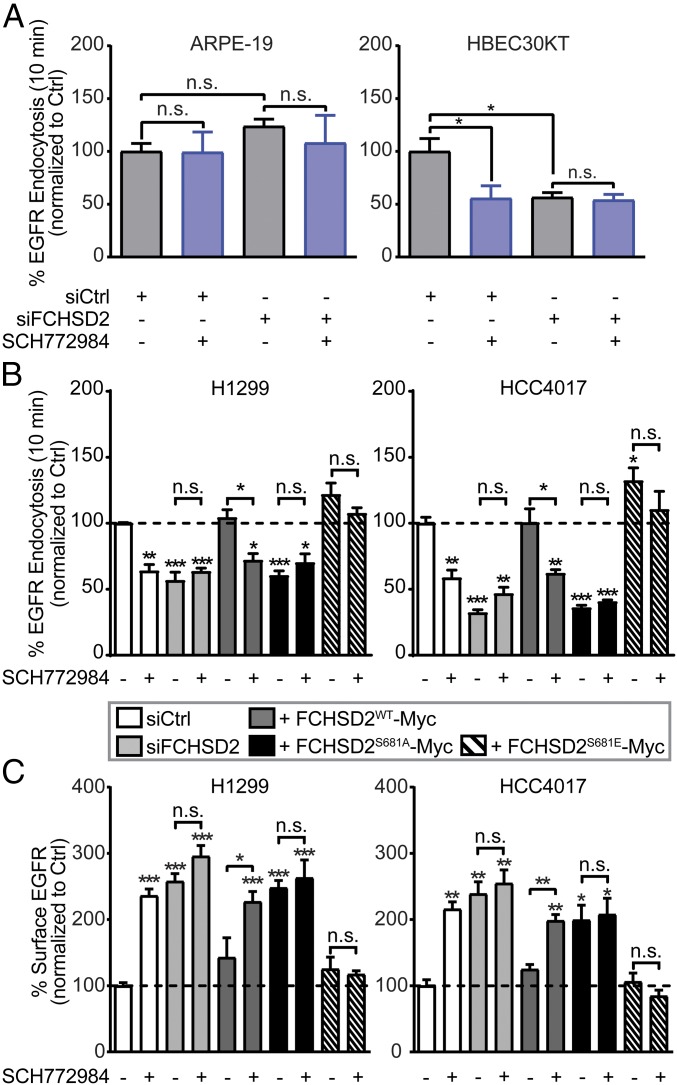
Many properties of progressive cancer cells are driven by altered signaling downstream of membrane receptors (50). EGFR is the predominant oncogenic receptor in NSCLC, and its downstream signaling can be changed by alterations in endocytic trafficking (51, 52). Given that the crosstalk between ERK1/2 signaling and FCHSD2 functions to regulate CME, we next examined whether FCHSD2 might be selectively required for EGFR endocytosis even in noncancerous cells. Consistent with TfnR endocytosis, neither ERK1/2 inhibition nor siRNA-mediated knockdown of FCHSD2 significantly affected EGFR endocytosis in nontumorigenic ARPE-19 cells (Fig. 6A). However, in HBEC30KT cells, EGFR uptake was more sensitive to ERK1/2 inhibition and FCHSD2 knockdown than was TfnR uptake (compare Fig. 6A with SI Appendix, Figs. S2B and S3B).
Fig. 6.
FCHSD2 mediates EGFR trafficking in cancer cells. (A) Endocytosis of EGFR was measured in control and FCHSD2 siRNA-treated ARPE-19 and HBEC30KT cells not treated or treated with the ERK1/2 inhibitor SCH772984 (10 μM). (B) Endocytosis of EGFR was measured in the control cells, FCHSD2–KD cells, or FCHSD2–KD cells reconstituted with FCHSD2WT-Myc, FCHSD2S681A-Myc, or FCHSD2S681E-Myc in the absence or presence of the ERK1/2 inhibitor SCH772984 (10 μM). (C) Surface levels of EGFR in the cells as described in B. All data were normalized to control and represent mean ± SEM (n = 3). Two-tailed Student’s t tests were used to assess statistical significance for comparison with siCtrl without SCH772984 treatment and for the indicated dataset. *P < 0.05, **P < 0.005, ***P < 0.0005; n.s., not significant.
As expected, ERK1/2 inhibition reduced EGFR endocytosis in H1299 and HCC4017 NSCLC cells (Fig. 6B), resulting in an approximately twofold increase in the level of surface EGFR (Fig. 6C, white bars). Knockdown of FCHSD2 similarly resulted in a significant decrease in the rate of EGFR endocytosis and an increase in surface EGFR, and these effects were insensitive to ERK1/2 inhibition (Fig. 6 B and C, light gray bars). Reconstitution of FCHSD2-depleted cells with FCHSD2WT-Myc restored EGFR endocytosis rates and EGFR surface levels as well as sensitivity to ERK1/2 inhibition (Fig. 6 B and C, dark gray bars). In contrast, FCHSD2S681A-Myc failed to restore EGFR trafficking or ERK1/2 sensitivity (Fig. 6 B and C, black bars), but reconstitution with FCHSD2S681E-Myc increased rates of EGFR endocytosis and reduced the level of surface EGFR relative to siFCHSD2-treated cells, both effects were resistant to ERK1/2 inhibition (Fig. 6 B and C, hatched bars).
FCHSD2 Negatively Regulates EGFR Signaling, Proliferation, and Migration.
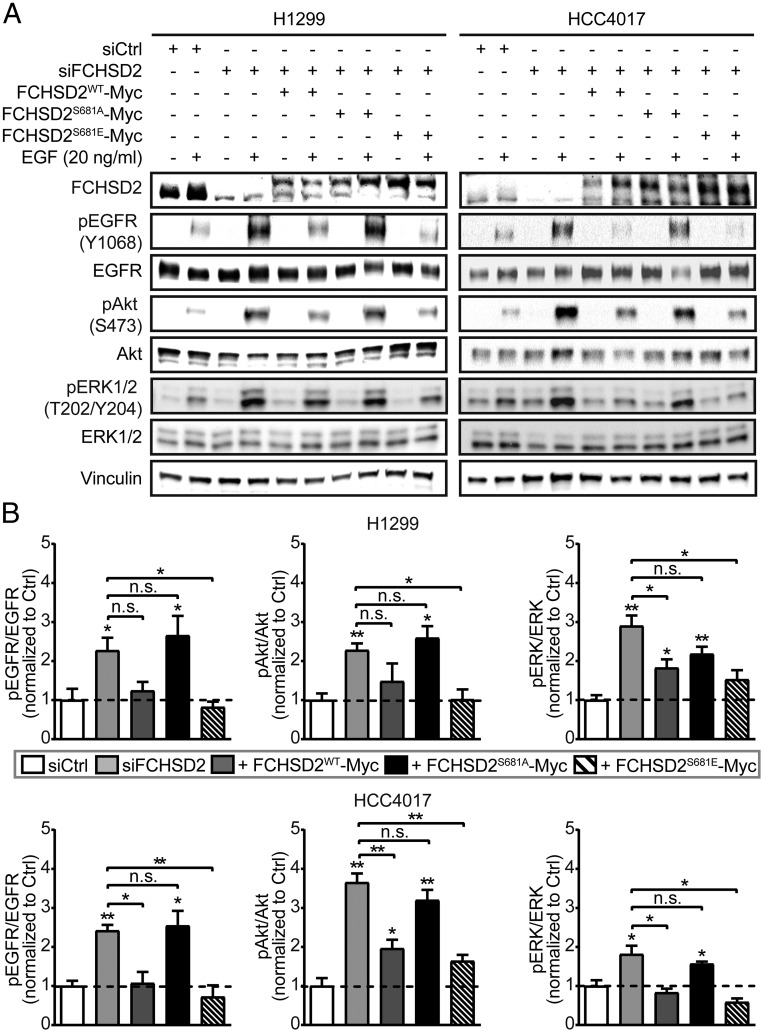
We next measured the downstream consequences of these changes in EGFR endocytosis and surface expression on EGFR signaling and cell behavior. Depletion of FCHSD2 resulted in dramatic increases in several EGF-dependent downstream signaling events, including tyrosine phosphorylation of the EGFR and activation of Akt and ERK1/2 in both H1299 and HCC4017 cells (Fig. 7 and Dataset S1). Reintroducing FCHSD2WT-Myc, but not FCHSD2S681A-Myc, again suppressed these signaling activities (Fig. 7 and Dataset S1). Expression of FCHSD2S681E-Myc also reduced signaling activities to control levels (Fig. 7 and Dataset S1). These findings suggest that the crosstalk between ERK1/2 kinases and FCHSD2 contributes to the endocytic trafficking of EGFRs and reciprocally regulates its downstream signaling activities.
Fig. 7.
FCHSD2 negatively regulates EGFR signaling in cancer cells. (A) Representative Western blots of EGFR signaling activities upon EGF stimulation (20 ng/mL) for 10 min after serum starvation for 16 h in the control cells, FCHSD2–KD cells, and FCHSD2–KD cells reconstituted with FCHSD2WT-Myc, FCHSD2S681A-Myc, or FCHSD2S681E-Myc. (B) Quantification of pEGFR/EGFR, pAkt/Akt, and pERK/ERK intensity ratios in the cells as described in A. All data were normalized to control and represent mean ± SEM (n = 3). Two-tailed Student’s t tests were used to assess statistical significance for comparison with siCtrl and for the indicated dataset. *P < 0.05, **P < 0.005; n.s., not significant.
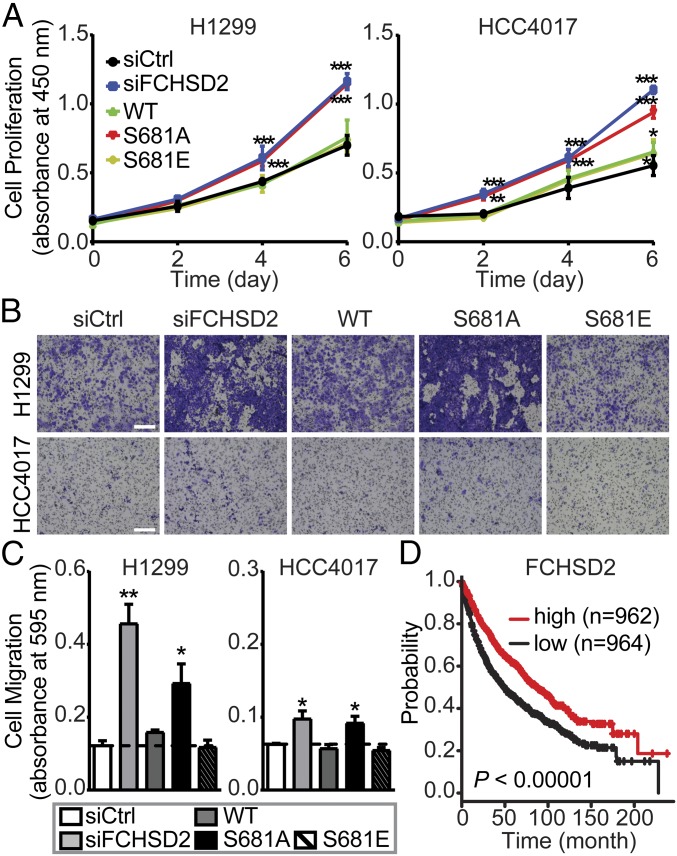
EGFR signaling affects both cancer cell proliferation and migration (52). Therefore, we examined the effects of FCHSD2-dependent alterations on the endocytic trafficking of EGFR and its signaling activities on these cancer cell behaviors. Loss of FCHSD2 increased the rates of proliferation (Fig. 8A) in the NSCLC cells. These effects were reversed upon reconstitution with FCHSD2WT-Myc or FCHSD2S681E-Myc but less so by FCHSD2S681A-Myc (Fig. 8A).
Fig. 8.
FCHSD2 functions as a negative regulator for cancer progression. (A) Cell proliferation abilities were measured in the control, FCHSD2–KD cells, and FCHSD2–KD cells reconstituted with FCHSD2WT-Myc, FCHSD2S681A-Myc, or FCHSD2S681E-Myc. ***P < 0.0005. (B) Representative images of cell migration in the cells as described in A. (Scale bars, 250 μm.) (C) Quantification of migrating cells in B. Data in A and C represent mean ± SEM (n = 3). Two-tailed Student’s t tests were used to assess statistical significance for comparison with siCtrl. *P < 0.05, **P < 0.005. (D) Kaplan–Meier survival analysis of NSCLC patients was performed in FCHSD2 high- and low-expression cohorts.
Similarly, EGF-dependent migration through Transwell filters was enhanced by siRNA knockdown of FCHSD2 (Fig. 8 B and C). Reconstitution with either FCHSD2WT-Myc or FCHSD2S681E-Myc returned migration rates to control levels, whereas reconstitution with FCHSD2S681A-Myc less potently suppressed the migration abilities (Fig. 8 B and C), especially in the more rapidly migrating H1299 cells.
Finally, we examined the relationship between FCHSD2 expression and survival by mining clinical data and found that NSCLC patients with relatively high FCHSD2 expression had significantly better survival rates than those in the low-expression group (Fig. 8D). Together, these data suggest that FCHSD2 functions as a negative regulator for cancer aggressiveness.
Discussion
The reciprocal crosstalk between signaling and CME in cancer cells contributes to abnormal trafficking of surface receptors and altered downstream signaling (3, 7, 18). These findings have led to the hypothesis that CME can be “adapted” by cancer cells to support enhanced tumor cell proliferation and metastasis (8). Here we explored the scope of this crosstalk and found that CME can be differentially regulated in cancer cells by several oncogenic signaling kinases. These studies reveal a more extensive crosstalk between signaling and CME than previously appreciated. Further studies are needed to dissect the direct or indirect roles and relevant substrates for the many signaling pathways that selectively impinge on CME in cancer cells.
We focused on the effects of ERK, given its importance as a major driver and therapeutic target for multiple cancers (31–34). Our data suggest that ERK1/2 inhibition decreased the rates of CME of TfnR and EGFR in cancer cells by specifically inhibiting CCP initiation, the critical first step in CME. ERK1/2 activity accelerated CME and CCP initiation rates through phosphorylation and activation of FCHSD2. When activated, FCHSD2 negatively regulates EGFR signaling and cell proliferation and migration in NSCLC cells. This study, together with our previous findings (15, 18), further supports the concept of adaptive CME and reveals a another mechanism for the reciprocal regulation of signaling and CME in cancer cells.
The domain structure of FCHSD2 and its known protein interactions (SI Appendix, Fig. S7) are suggestive of a role in CME (53, 54). Indeed, FCHSD2 is a member of the mild curvature-generating F-BAR family of proteins, whose prototypical members, FCHo1/2, have been shown to function in the early stages of CCP initiation and stabilization (46, 48). A recent study, published while this manuscript was in preparation, confirmed a role for FCHSD2 in CME (55). As these researchers used HeLa cells as a model, their data are consistent with ours showing activation of FCHSD2 in cancer cells. However, based on their analysis of CCP dynamics, these authors concluded that FCHSD2 is required to activate actin and facilitate later stages of CCP maturation: They did not measure the rates of CCP initiation. While we did not detect an effect of FCHSD2 knockdown on CCP maturation, the differences here may lie in how CCP dynamics were analyzed. We employed an unbiased, comprehensive analysis of all CCPs (35), whereas the previous study analyzed only a subset of CCPs corresponding to the “brightest spots” and then “manually selected for events showing fluorescence profiles compatible with typical endocytic events” (55). It is possible that the maturation of the brightest and thus largest CCPs may be selectively dependent on actin, as has been shown (56), and that FCHSD2 may have a dual role in CME. Further studies are needed to resolve differences between these two reports and to elucidate the mechanism(s) by which FCHSD2 regulates CME.
Many F-BAR proteins contain SH3 domains (48, 57), and FCHSD2 encodes two that interact with other components of the endocytic machinery, including dynamin, N-WASP, and intersectin1/2 (ITSN1/2) (44, 58). The above-mentioned study (55) reported that FCHSD2-Venus was recruited to CCPs through interactions with intersectin and speculated that FCHSD2 functioned to regulate N-WASP–dependent actin assembly during intermediate stages of CME. Which of these partners, if any, is required for FCHSD2 function in CCP initiation remains to be determined.
Much more is known about Nwk, the Drosophila ortholog of FCHSD2, which has been studied exclusively in the context of endocytic trafficking and bone morphogenic protein (BMP) signaling at neuromuscular junctions (44, 45, 58). In addition to its interactions with N-WASP, shibire, and Dap160 (SI Appendix, Fig. S7) (44, 58), Nwk physically and functionally interacts with the endosomal sorting factor SNX16 to regulate sorting of BMP along the endosomal pathway (45, 58). Whether FCHSD2 also functions in endosomal sorting in mammalian cells remains to be determined. Interestingly, just as FCHSD2 appears to negatively regulate EGFR signaling and cell proliferation, Nwk is known to negatively regulate BMP signaling as well as synaptic growth (44, 58).
Nwk displays intramolecular, autoinhibitory interactions between its F-BAR and C-terminal SH3b domain (SI Appendix, Fig. S7) that inhibit both membrane binding of the F-BAR domain and protein interactions with the SH3a domain. Release of autoinhibitory SH3b domain interactions is required for activation of Nwk (49, 59) and FCHSD2 (55). We found that ERK1/2-mediated phosphorylation of FCHSD2 on S681 is also critical for its activity. This phosphorylation site, which resides in the C-terminal region distal to SH3b, is not conserved in Nwk (SI Appendix, Fig. S7) and may reflect an additional level of regulation that renders mammalian FCHSD2 activity responsive to extracellular signaling. We speculate that phosphorylation of S681 either directly disrupts the autoinhibitory interactions of the adjacent SH3b domain or indirectly increases the binding affinity of SH3b domain ligands to relieve the autoinhibition. Alternatively, phosphorylation may alter FCHSD2 interactions with other proteins independently of its own activation. Clearly, further investigation is needed to test these hypotheses regarding the regulation of FCHSD2.
Our results, together with those of Almeida-Souza et al. (55), extend FCHSD2 function outside the nervous system and to earlier stages of the endocytic pathway. Strikingly, FCHSD2 becomes the third component of the neuronal endocytic machinery, along with CLCb and Dyn1 (60), that allows differential regulation of CME in cancer cells. Endocytic trafficking plays a critical role in regulating and maintaining signaling at the synapse. Whether adaptive endocytosis can result from co-opting other components of the synaptic machinery to alter endocytic trafficking and signaling properties in cancer cells remains to be determined.
Hyperactivity of EGFR signaling at the cell surface is a common feature among different types of cancers and is widely considered advantageous for tumor progression (61). Why then would ERK1/2 activation downstream of EGFR activate EGFR endocytosis and suppress EGFR proliferative and migratory signaling? We propose several, not mutually exclusive, possibilities for the functional significance of this potentially negative feedback loop. One possibility relates to the well-established spatiotemporal regulation of EGFR signaling (62, 63) and differences in downstream signaling triggered by PM-associated vs. endosome-associated EGFR (64–68). Thus, ERK1/2 and FCSDH2-dependent increases in EGFR uptake and its redistribution to endosomal compartments could alter signaling pathways and downstream cellular responses in ways beneficial to the metastatic cancer cell. A second possibility relates to the documented saturability of EGFR sorting within endosomal compartments (69). Thus, the increased efficiency of EGFR uptake might be needed to ensure saturation of downstream endosomal sorting machinery leading to increased rates of recycling and sustained EGFR signaling. Alternatively, recent evidence has suggested that a subset of EGFR signaling might occur specifically within CCPs (70). Thus, ERK- and FCHSD2-dependent increases in CCP initiation might function to increase the number of these signaling platforms that trigger a subset of downstream signaling responses. Finally, it is important to note that both FCHSD2 and its Drosophila homolog Nwk function as negative regulators of signaling (this study and refs. 44, 45, 58). Thus, FCHSD2 activity may play a role in buffering EGFR signaling within the tumor environment where local concentrations of EGF can be high compared with those seen by metastatic cells (71). In this scenario, FCHSD2’s specific up-regulation in cancer cells could reflect a compensatory mechanism to counteract the detrimental effects of hyperactive EGFR signaling.
Finally, it is also possible that the effects of FCHSD2 on signaling vary depending on the signaling receptor. For example, c-Met is believed to signal primarily from endosomal compartments (72); thus increased endocytosis might enhance this receptor’s downstream signaling. Indeed, while we have found that high levels of FCHSD2 expression correlate with better survival rates in human NSCLC patients, others have reported that high levels of FCHSD2 correlate with poor prognosis in acute myeloid leukemia patients (73). Thus, it will be important to examine the effects of FCHSD2 activity on signaling downstream of other cell-surface receptors.
Overall, our study reveals extensive crosstalk between signaling and CME that is often specific to cancer cells. Based on our findings reported here and elsewhere (74, 75), we suggest that cancer cells can adapt this crosstalk as a determinant for tumor progression. These cancer cell-specific adaptations of the endocytic pathway may provide opportunities of the development of novel therapeutic strategies against cancer.
Materials and Methods
Cell Culture and Reagents.
ARPE-19 cells (from ATCC) were cultivated in DMEM/F12 (Thermo Fisher Scientific) supplemented with 10% (vol/vol) FCS (HyClone). H1299 and HCC4017 NSCLC cells [from John Minna, University of Texas Southwestern Medical Center (UTSWMC), Dallas] were grown in RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% (vol/vol) FCS. HBEC30KT, nontransformed human bronchial epithelial cells (from John Minna) were cultivated in complete keratinocyte-SFM medium (Thermo Fisher Scientific). MCF10A cells (from ATCC) were grown in complete mammary epithelial cell growth medium (Lonza). MDA-MB-231 breast adenocarcinoma cells (from R. Brekken, UTSWMC, Dallas) were grown in DMEM containing high-glucose medium (Thermo Fisher Scientific) supplemented with 10% (vol/vol) FCS. The following inhibitors and their final concentrations, shown in parenthesis, were used in this study: Akt inhibitor X (10 μM), the CaMK-II inhibitor KN93 (3.7 μM), the CDC42 inhibitor ML 141 (20 μM), the CDK inhibitor Aminopurvalanol A (200 nM), the EPAC inhibitor ESI-09 (32 μM), the ERK inhibitor FR180204 (10 μM), the GSK3β inhibitor CHIR-99021 (10 μM), and Src inhibitor I (440 nM) were from MilliporeSigma. The B-Raf inhibitor SB590885 (20 nM), the ERK inhibitor SCH772984 (10 μM), the JAK inhibitor CP 690550 citrate (10 nM), and the JNK inhibitor SP600125 (900 nM) were from Selleck Chemicals. The CK2 inhibitor TTP 22 (1 μM), the MLCK inhibitor ML-7 HCl (3 μM), the mTOR inhibitor rapamycin (10 nM), the p38 inhibitor SB239063 (450 nM), and the PKA inhibitor KT-5720 (600 nM) were from Santa Cruz. The MEK inhibitor GSK1120212 (10 μM) and the PI3K inhibitor BKM120 (1 μM) were from MedChemExpress. The PDK1 inhibitor GSK2334470 (100 nM), the PTEN inhibitor VO-OHpic (460 nM), and the ROCK inhibitor GSK269962 (40 nM) were from Tocris Bioscience. The PKC inhibitor Gö-6983 (600 nM) was from Cayman Chemical. Protease and phosphatase inhibitor mixtures were from Roche. All other chemicals were reagent grade and were purchased from Sigma.
Endocytosis Assays.
TfnR and EGFR endocytosis assays were performed exactly as described (18) using 4 µg/mL anti-TfnR mAb (HTR-D65) or 20 ng/mL biotinylated-EGF (Thermo Fisher Scientific) (conditions under which EGF uptake is clathrin dependent), respectively, and using cells grown overnight in gelatin-coated 96-well plates and plated at a density of 3 × 104 cells per well. For TfnR and EGFR endocytosis assays using inhibitors, cells were preincubated in the absence (i.e., control) or presence of the indicated inhibitors at the concentrations described above for 30 min at 37 °C before internalization assays were performed in the continued absence or presence of the respective inhibitors.
Western Blotting.
Cells were washed three times with PBS and were harvested and resuspended in 150–200 μL of reducing Laemmli sample buffer. The cell lysate was boiled for 10 min, and 50 μg of cell lysate was loaded onto an SDS gel. After transfer to a nitrocellulose membrane, membranes were blocked with either 5% powdered milk or 5% BSA and were probed with antibodies against the following proteins: pERK1/2 T202/Y204 (no. 4370S; Cell Signaling), ERK1/2 (no. 4695S; Cell Signaling), Myc-DDK tag (no. TA50011-100; OriGene), pEGFR Y1068 (no. 3777S; Cell Signaling), EGFR (no. 4267S; Cell Signaling), pAkt S473 (no. 4060L; Cell Signaling), Akt (no. 9272S; Cell Signaling), and vinculin (no.V9131; MilliporeSigma), which was used as internal control, according to the manufacturers’ instructions. For FCHSD2 we used no. PA5-58432 (Thermo Fisher Scientific), which detected an ∼85-kDa band that was specifically knocked down by siFCHSD2 treatment at 1:500 dilution. Note that the manufacturer’s data show a 100-kDa band, which we also detected, but this band did not respond to siRNA knockdown. The calculated molecular mass for the 740-aa variant of FCHSD2 (accession number O94868, UniProtKB) is ∼84 kDa. HRP-conjugated secondary antibodies (no. G21234 and no. G21040; Thermo Fisher Scientific) were used according to the manufacturer’s instructions.
Quantitative analysis was performed by using ImageJ software (NIH). For EGF-dependent signaling, the H1299 and HCC4017 cells (5 × 105 cells) were seeded in six-well plates containing RPMI 1640 with 10% FCS. Eight hours after seeding, cells were washed three times with PBS and starved in RPMI 1640 without FCS for 16 h. The cells then were left untreated or treated with 20 ng/mL of EGF for 10 min. After the EGF treatment, cells were washed three times with PBS and were harvested and resuspended in 150–200 μL of reducing Laemmli sample buffer, and the cell lysates were subjected to Western blotting as described above.
TIR-FM and Image Data Analysis.
TIR-FM was performed as previously described (13). Briefly, ARPE-19 and H1299 cells stably expressing eGFP-CLCa and HCC4017 cells expressing SNAP-CLCa bound to SNAP-Cell TMR-Star (New England BioLabs) were imaged using a 100× 1.49 NA Apo TIRF objective (Nikon) mounted on a Ti-Eclipse inverted microscope with a Perfect Focus System (Nikon). TIR-FM illumination was achieved using the Diskovery Platform (Andor Technology). During imaging, cells were maintained at 37 °C in complete culture medium. Time-lapse image sequences were acquired at a penetration depth of 80 nm and a frame rate of 1 Hz using a sCMOS camera with 6.5-μm pixel size (pco.edge). For TIR-FM using inhibitors, cells were preincubated in the absence (i.e., control) or presence of the indicated inhibitors described above for 30 min at 37 °C, followed by imaging in the absence or presence of the respective inhibitors. CCP lifetime distribution and initiation density analyses were carried out in Matlab (MathWorks), using cmeAnalysisPackage (35).
Mutagenesis of FCHSD2 and Generation of Phosphomutants.
The FCHSD2 (NM_014824) human-tagged ORF clone lentiviral construct was purchased from OriGene (no. RC221241L3). The FCHSD2 Myc-tagged phosphomutants (S681A and S681E) were generated by standard site-directed mutagenesis (76). The primer sequences for the mutagenesis of S681A and S681E are 5′-CTGTACTTTCCCCGGGCTCCTTCAGCAAACG-3′ and 5′-GCTCCCTGTACTTTCCCCGGGAGCCTTCAGCAAACGAAAAAAG-3′, respectively. All primers were purchased from Integrated DNA Technologies.
siRNA Transfection and FCHSD2 Constructs Transduction.
Cells were treated with the siRNA pool targeting FCHSD2 (no. E-021240-00-0010; Dharmacon) using RNAiMAX (Thermo Fisher Scientific) to silence the endogenous protein. Briefly, 50 nM of the indicated siRNA pool and 6.5 μL of RNAiMAX reagent were added in 1 mL of OptiMEM (Thermo Fisher Scientific) in each well of a six-well plate and were incubated for 20 min at room temperature. Cells were resuspended in 1 mL of culture medium, seeded in each well of a six-well plate containing the mixed siRNA–lipid complex, and incubated for 48 h, followed by experiments. The AllStars Negative siRNA nontargeting sequence was purchased from Qiagen (no.SI03650318).
FCHSD2WT-Myc, FCHSD2S681A-Myc, or FCHSD2S681E-Myc fusion proteins or C-terminally GFP-tagged wild-type FCHSD2 lentiviral construct (no. RC221214L2; OriGene) transduction in H1299 or HCC4017 cells was performed using lentiviral vectors. The lentiviral vectors and lentiviral packing plasmids [pSPAX2 (Gag/Pol) and pMD2G] were cotransfected into HEK293T cells using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions for virus production. The virus-conditioned medium was used to infect target cells in the presence of 10 μg/mL polybrene (MilliporeSigma). Cells were collected 48 h postinfection for experiments.
Immunofluorescence Staining and Colocalization Analysis.
Cells were washed three times with PBS, fixed with 4% (wt/vol) paraformaldehyde (Electron Microscopy Sciences) for 30 min at 37 °C, and permeabilized using 0.05% saponin (wt/vol) (MilliporeSigma) for 10 min. Cells were blocked with 5% BSA and probed with antibodies against FCHSD2 (no. PA5-58432; Thermo Fisher Scientific) and Myc-DDK tag (no. TA50011-100; OriGene) according to the manufacturers’ instructions. The rabbit polyclonal CLC antibody used in this study (1:150 dilution) was produced in our laboratory using KLH-conjugated synthetic peptide between 27 and 45 amino acids from the N-terminal region of human CLC. Alexa Fluor-conjugated secondary antibodies (no. A-31571 and no. A-11036; Thermo Fisher Scientific) were used according to the manufacturer’s instructions. TIR-FM was performed as described above. Colocalization (Pearson’s coefficient) analysis was performed by using ImageJ software (NIH).
Cell Proliferation.
Cells were seeded on 24-well plates at a density of 2 × 104 cells per well on day 0. On day 2 cell numbers were determined, and trypsinized cells were reseeded at the initial density. This was repeated on day 4 and day 6. The cell proliferation ability was measured by using the CCK-8 Counting Kit (Dojindo Molecular Technologies) according to the manufacturer’s instructions. Briefly, cells were incubated with culture medium containing the CCK-8 solution for 1 h at 37 °C, and then the absorbance was read at 450 nm.
Cell Migration.
Tumor cell migration ability was evaluated in 6.5-mm Transwells with 8.0-µm pore polyester membrane inserts (no. 3464; Corning) in 24-well plates. Cells (1 × 105) were resuspended in 300 μL of RPMI 1640 without FCS and were seeded on each Transwell insert, and each well of the 24-well plate was filled with 700 μL of complete culture medium. After incubation for 16 h at 37 °C, the cells remaining on the upper surfaces of the membrane were removed by wet cotton swabs. Cells that had migrated to the lower surfaces of the membrane were fixed with ice-cold methanol (Pharmco) for 10 min and were stained with 0.5% crystal violet (MilliporeSigma) for 15 min. After imaging, cells were washed three times with ddH2O and were incubated in 1% SDS solution (MilliporeSigma) for 5 min at room temperature; then the number of cells was quantified by the absorbance at 595 nm.
Analysis of Kaplan–Meier Survival Data.
NSCLC patient survival data were downloaded from the Kaplan–Meier plotter database (77). Analysis of NSCLC patients was performed in FCHSD2 high- and low-expression cohorts. P value was calculated by log rank test (77).
Supplementary Material
Acknowledgments
We thank Richard Carr III for initiating this direction of research while he was a postdoctoral fellow in the S.L.S. laboratory; members of the S.L.S. laboratory for critically reading the manuscript; Heather Grossman, Kim Reed, and Marcel Mettlen for technical assistance in plasmid preparation and microscopy, respectively; and Andrew Lemoff and the University of Texas Southwestern Proteomics Core Facility for help with mass spectrometry and proteomic analysis. The work was supported by NIH Grants GM73165 (to S.L.S. and Gaudenz Danuser, Principal Investigator on Grant GM73165) and MH61345 (to S.L.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810209115/-/DCSupplemental.
References
- 1.Bacac M, Stamenkovic I. Metastatic cancer cell. Annu Rev Pathol. 2008;3:221–247. doi: 10.1146/annurev.pathmechdis.3.121806.151523. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Mellman I, Yarden Y. Endocytosis and cancer. Cold Spring Harb Perspect Biol. 2013;5:a016949. doi: 10.1101/cshperspect.a016949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sigismund S, et al. Endocytosis and signaling: Cell logistics shape the eukaryotic cell plan. Physiol Rev. 2012;92:273–366. doi: 10.1152/physrev.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paul NR, Jacquemet G, Caswell PT. Endocytic trafficking of integrins in cell migration. Curr Biol. 2015;25:R1092–R1105. doi: 10.1016/j.cub.2015.09.049. [DOI] [PubMed] [Google Scholar]
- 6.Lanzetti L, Di Fiore PP. Endocytosis and cancer: An ‘insider’ network with dangerous liaisons. Traffic. 2008;9:2011–2021. doi: 10.1111/j.1600-0854.2008.00816.x. [DOI] [PubMed] [Google Scholar]
- 7.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: An emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 8.Schmid SL. Reciprocal regulation of signaling and endocytosis: Implications for the evolving cancer cell. J Cell Biol. 2017;216:2623–2632. doi: 10.1083/jcb.201705017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 10.Gonnord P, Blouin CM, Lamaze C. Membrane trafficking and signaling: Two sides of the same coin. Semin Cell Dev Biol. 2012;23:154–164. doi: 10.1016/j.semcdb.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Sorkin A, von Zastrow M. Endocytosis and signalling: Intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Fiore PP, von Zastrow M. Endocytosis, signaling, and beyond. Cold Spring Harb Perspect Biol. 2014;6:a016865. doi: 10.1101/cshperspect.a016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loerke D, et al. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009;7:e57. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puthenveedu MA, von Zastrow M. Cargo regulates clathrin-coated pit dynamics. Cell. 2006;127:113–124. doi: 10.1016/j.cell.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 15.Reis CR, et al. Crosstalk between Akt/GSK3β signaling and dynamin-1 regulates clathrin-mediated endocytosis. EMBO J. 2015;34:2132–2146. doi: 10.15252/embj.201591518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acton SL, Brodsky FM. Predominance of clathrin light chain LCb correlates with the presence of a regulated secretory pathway. J Cell Biol. 1990;111:1419–1426. doi: 10.1083/jcb.111.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brodsky FM, et al. Clathrin light chains: Arrays of protein motifs that regulate coated-vesicle dynamics. Trends Biochem Sci. 1991;16:208–213. doi: 10.1016/0968-0004(91)90087-c. [DOI] [PubMed] [Google Scholar]
- 18.Chen PH, et al. Crosstalk between CLCb/Dyn1-mediated adaptive clathrin-mediated endocytosis and epidermal growth factor receptor signaling increases metastasis. Dev Cell. 2017;40:278–288.e5. doi: 10.1016/j.devcel.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe H, et al. Discovery of a highly potent and selective MEK inhibitor: GSK1120212 (JTP-74057 DMSO solvate) ACS Med Chem Lett. 2011;2:320–324. doi: 10.1021/ml200004g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins JM, et al. Comprehensive characterization of the published kinase inhibitor set. Nat Biotechnol. 2016;34:95–103. doi: 10.1038/nbt.3374. [DOI] [PubMed] [Google Scholar]
- 21.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 22.Ohori M, et al. Identification of a selective ERK inhibitor and structural determination of the inhibitor-ERK2 complex. Biochem Biophys Res Commun. 2005;336:357–363. doi: 10.1016/j.bbrc.2005.08.082. [DOI] [PubMed] [Google Scholar]
- 23.Wilkes EH, Terfve C, Gribben JG, Saez-Rodriguez J, Cutillas PR. Empirical inference of circuitry and plasticity in a kinase signaling network. Proc Natl Acad Sci USA. 2015;112:7719–7724. doi: 10.1073/pnas.1423344112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elkin SR, et al. A systematic analysis reveals heterogeneous changes in the endocytic activities of cancer cells. Cancer Res. 2015;75:4640–4650. doi: 10.1158/0008-5472.CAN-15-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srinivasan S, et al. A noncanonical role for dynamin-1 in regulating early stages of clathrin-mediated endocytosis in non-neuronal cells. PLoS Biol. 2018;16:e2005377. doi: 10.1371/journal.pbio.2005377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramirez RD, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–9034. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 28.Qu Y, et al. Evaluation of MCF10A as a reliable model for normal human mammary epithelial cells. PLoS One. 2015;10:e0131285. doi: 10.1371/journal.pone.0131285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujimoto LM, Roth R, Heuser JE, Schmid SL. Actin assembly plays a variable, but not obligatory role in receptor-mediated endocytosis in mammalian cells. Traffic. 2000;1:161–171. doi: 10.1034/j.1600-0854.2000.010208.x. [DOI] [PubMed] [Google Scholar]
- 30.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 31.Efferth T. Signal transduction pathways of the epidermal growth factor receptor in colorectal cancer and their inhibition by small molecules. Curr Med Chem. 2012;19:5735–5744. doi: 10.2174/092986712803988884. [DOI] [PubMed] [Google Scholar]
- 32.Fitzgerald TL, et al. Roles of EGFR and KRAS and their downstream signaling pathways in pancreatic cancer and pancreatic cancer stem cells. Adv Biol Regul. 2015;59:65–81. doi: 10.1016/j.jbior.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Lo HW. Targeting Ras-RAF-ERK and its interactive pathways as a novel therapy for malignant gliomas. Curr Cancer Drug Targets. 2010;10:840–848. doi: 10.2174/156800910793357970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montagut C, Settleman J. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 2009;283:125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 35.Aguet F, Antonescu CN, Mettlen M, Schmid SL, Danuser G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev Cell. 2013;26:279–291. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrlich M, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 37.Aouadi M, Binetruy B, Caron L, Le Marchand-Brustel Y, Bost F. Role of MAPKs in development and differentiation: Lessons from knockout mice. Biochimie. 2006;88:1091–1098. doi: 10.1016/j.biochi.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Bost F, Aouadi M, Caron L, Binétruy B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie. 2005;87:51–56. doi: 10.1016/j.biochi.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 39.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 40.Govindarajan A, Kelleher RJ, Tonegawa S. A clustered plasticity model of long-term memory engrams. Nat Rev Neurosci. 2006;7:575–583. doi: 10.1038/nrn1937. [DOI] [PubMed] [Google Scholar]
- 41.Yoon S, Seger R. The extracellular signal-regulated kinase: Multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 42.Carlson SM, et al. Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. Sci Signal. 2011;4:rs11. doi: 10.1126/scisignal.2002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becalska AN, et al. Formation of membrane ridges and scallops by the F-BAR protein Nervous Wreck. Mol Biol Cell. 2013;24:2406–2418. doi: 10.1091/mbc.E13-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connor-Giles KM, Ho LL, Ganetzky B. Nervous wreck interacts with thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron. 2008;58:507–518. doi: 10.1016/j.neuron.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodal AA, et al. A presynaptic endosomal trafficking pathway controls synaptic growth signaling. J Cell Biol. 2011;193:201–217. doi: 10.1083/jcb.201009052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henne WM, et al. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma L, et al. Transient Fcho1/2⋅Eps15/R⋅AP-2 nanoclusters prime the AP-2 clathrin adaptor for cargo binding. Dev Cell. 2016;37:428–443. doi: 10.1016/j.devcel.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Y, et al. Endocytosis and membrane receptor internalization: Implication of F-BAR protein carom. Front Biosci. 2017;22:1439–1457. doi: 10.2741/4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stanishneva-Konovalova TB, et al. Coordinated autoinhibition of F-BAR domain membrane binding and WASp activation by Nervous Wreck. Proc Natl Acad Sci USA. 2016;113:E5552–E5561. doi: 10.1073/pnas.1524412113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sangwan V, Park M. Receptor tyrosine kinases: Role in cancer progression. Curr Oncol. 2006;13:191–193. [PMC free article] [PubMed] [Google Scholar]
- 51.Shtiegman K, et al. Defective ubiquitinylation of EGFR mutants of lung cancer confers prolonged signaling. Oncogene. 2007;26:6968–6978. doi: 10.1038/sj.onc.1210503. [DOI] [PubMed] [Google Scholar]
- 52.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu S, et al. Analysis for Carom complex, signaling and function by database mining. Front Biosci. 2016;21:856–872. doi: 10.2741/4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Itoh T, et al. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Almeida-Souza L, et al. A flat BAR protein promotes actin polymerizatioin at the base of clathrin coated pits. Cell. 2018;174:325–337.e14. doi: 10.1016/j.cell.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cureton DK, Massol RH, Whelan SP, Kirchhausen T. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 2010;6:e1001127. doi: 10.1371/journal.ppat.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu S, Xiong X, Zhao X, Yang X, Wang H. F-BAR family proteins, emerging regulators for cell membrane dynamic changes-from structure to human diseases. J Hematol Oncol. 2015;8:47. doi: 10.1186/s13045-015-0144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodal AA, Motola-Barnes RN, Littleton JT. Nervous wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J Neurosci. 2008;28:8316–8325. doi: 10.1523/JNEUROSCI.2304-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelley CF, et al. Membrane charge directs the outcome of F-BAR domain lipid binding and autoregulation. Cell Rep. 2015;13:2597–2609. doi: 10.1016/j.celrep.2015.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–1321. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reis CR, Chen PH, Bendris N, Schmid SL. TRAIL-death receptor endocytosis and apoptosis are selectively regulated by dynamin-1 activation. Proc Natl Acad Sci USA. 2017;114:504–509. doi: 10.1073/pnas.1615072114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jutten B, Rouschop KM. EGFR signaling and autophagy dependence for growth, survival, and therapy resistance. Cell Cycle. 2014;13:42–51. doi: 10.4161/cc.27518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ray M, Salgia R, Vokes EE. The role of EGFR inhibition in the treatment of non-small cell lung cancer. Oncologist. 2009;14:1116–1130. doi: 10.1634/theoncologist.2009-0054. [DOI] [PubMed] [Google Scholar]
- 64.Galperin E, Sorkin A. Endosomal targeting of MEK2 requires RAF, MEK kinase activity and clathrin-dependent endocytosis. Traffic. 2008;9:1776–1790. doi: 10.1111/j.1600-0854.2008.00788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorkin A. Internalization of the epidermal growth factor receptor: Role in signalling. Biochem Soc Trans. 2001;29:480–484. doi: 10.1042/bst0290480. [DOI] [PubMed] [Google Scholar]
- 66.Vieira AV, Lamaze C, Schmid SL. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 67.Villaseñor R, Nonaka H, Del Conte-Zerial P, Kalaidzidis Y, Zerial M. Regulation of EGFR signal transduction by analogue-to-digital conversion in endosomes. eLife. 2015;4:e06156. doi: 10.7554/eLife.06156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, You L, Xie J, Pan H, Han W. The roles of subcellularly located EGFR in autophagy. Cell Signal. 2017;35:223–230. doi: 10.1016/j.cellsig.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 69.French AR, Sudlow GP, Wiley HS, Lauffenburger DA. Postendocytic trafficking of epidermal growth factor-receptor complexes is mediated through saturable and specific endosomal interactions. J Biol Chem. 1994;269:15749–15755. [PubMed] [Google Scholar]
- 70.Wei Y, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154:1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sigismund S, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci USA. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joffre C, et al. A direct role for Met endocytosis in tumorigenesis. Nat Cell Biol. 2011;13:827–837. doi: 10.1038/ncb2257. [DOI] [PubMed] [Google Scholar]
- 73.Han Y, et al. FCHSD2 predicts response to chemotherapy in acute myeloid leukemia patients. Leuk Res. 2012;36:1339–1346. doi: 10.1016/j.leukres.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 74.Corcelle E, et al. Disruption of autophagy at the maturation step by the carcinogen lindane is associated with the sustained mitogen-activated protein kinase/extracellular signal-regulated kinase activity. Cancer Res. 2006;66:6861–6870. doi: 10.1158/0008-5472.CAN-05-3557. [DOI] [PubMed] [Google Scholar]
- 75.Wang J, et al. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J Biol Chem. 2009;284:21412–21424. doi: 10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reikofski J, Tao BY. Polymerase chain reaction (PCR) techniques for site-directed mutagenesis. Biotechnol Adv. 1992;10:535–547. doi: 10.1016/0734-9750(92)91451-j. [DOI] [PubMed] [Google Scholar]
- 77.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.