Significance
To optimize photosynthetic performance and minimize photooxidative damage, photosynthetic organisms evolved to efficiently balance light energy absorption and electron transport with cellular energy requirements under constantly changing light conditions. The regulation of linear electron flow (LEF) and cyclic electron flow (CEF) contributes to this fine-tuning. Here we present a model of the formation and structural molecular organization of a CEF-performing photosystem I (PSI)–light harvesting complex I (LHCI)–cytochrome (cyt) b6f supercomplex from the green alga Chlamydomonas reinhardtii. Such a structural arrangement could modulate the distinct operation of LEF and CEF to optimize light energy utilization, despite the same individual structural units contributing to these two different functional modes.
Keywords: cyclic electron flow, supercomplex, photosystem I, cytochrome b6f, Chlamydomonas reinhardtii
Abstract
Photosynthetic linear electron flow (LEF) produces ATP and NADPH, while cyclic electron flow (CEF) exclusively drives photophosphorylation to supply extra ATP. The fine-tuning of linear and cyclic electron transport levels allows photosynthetic organisms to balance light energy absorption with cellular energy requirements under constantly changing light conditions. As LEF and CEF share many electron transfer components, a key question is how the same individual structural units contribute to these two different functional modes. Here, we report the structural identification of a photosystem I (PSI)–light harvesting complex I (LHCI)–cytochrome (cyt) b6f supercomplex isolated from the unicellular alga Chlamydomonas reinhardtii under anaerobic conditions, which induces CEF. This provides strong evidence for the model that enhanced CEF is induced by the formation of CEF supercomplexes, when stromal electron carriers are reduced, to generate additional ATP. The additional identification of PSI–LHCI–LHCII complexes is consistent with recent findings that both CEF enhancement and state transitions are triggered by similar conditions, but can occur independently from each other. Single molecule fluorescence correlation spectroscopy indicates a physical association between cyt b6f and fluorescent chlorophyll containing PSI–LHCI supercomplexes. Single particle analysis identified top-view projections of the corresponding PSI–LHCI–cyt b6f supercomplex. Based on molecular modeling and mass spectrometry analyses, we propose a model in which dissociation of LHCA2 and LHCA9 from PSI supports the formation of this CEF supercomplex. This is supported by the finding that a Δlhca2 knockout mutant has constitutively enhanced CEF.
Photosynthesis captures solar energy and stores it in the form of chemical energy, which is essential to support life on Earth. Photosynthetic electron transport operates in two modes: linear (LEF) and cyclic electron flow (CEF). LEF yields ATP and NADPH, while CEF exclusively drives ATP production (1). Fine-tuning LEF and CEF maintains the ATP/NADPH equilibrium and efficient carbon assimilation (2, 3). CEF also plays an important role in photoprotection (4, 5) as it maintains the necessary ΔpH across the thylakoid membrane to allow energy-dependent nonphotochemical quenching and to control the rate limiting step of LEF (6). The dynamic tuning between LEF and CEF is therefore essential for efficient photosynthesis.
LEF involves in-series activity of photosystem II (PSII), cytochrome (cyt) b6f, and photosystem I (PSI), while CEF involves only PSI and cyt b6f. During CEF, electrons released by PSI are reinjected into the photosynthetic electron transport chain at the plastoquinone (PQ) pool or at the stromal side of the cyt b6f complex. The fact that LEF and CEF share many electron transfer components (e.g., PSI and cyt b6f) raises the question of how these membrane protein complexes can contribute to both functional modes. Extensive biochemical and biophysical analyses using the green alga Chlamydomonas reinhardtii suggest that efficient CEF depends on the formation of a CEF supercomplex consisting of PSI, cyt b6f, and subunits ferredoxin–NADP–oxidoreductase (FNR), Proton Gradient Regulation-Like 1 (PGRL1), Anaerobic Response 1 (ANR1), and Calcium Sensor (CAS). The CEF supercomplex is proposed to enhance CEF over LEF when stromal electron carriers are reduced (excess NADPH) and ATP is limiting (7–10). However, structural evidence for this supercomplex in C. reinhardtii is lacking, probably due to its putative dynamic nature.
Here, under CEF-inducing anaerobic conditions, a sucrose density gradient (SDG) fraction with CEF activity (7, 8) was isolated from C. reinhardtii, and a PSI–light harvesting complex I (LHCI)–cyt b6f-containing CEF supercomplex within it, was structurally characterized. The physical association between PSI–LHCI and cyt b6f was supported using single molecule fluorescence (SMF) correlation spectroscopy. Immunoblot and mass spectrometry (MS) analyses also clearly identified PSI, LHCI, cyt b6f as well as FNR, PGRL1, ANR1, and CAS. Their structural organization was characterized using crosslinking, MS, and single particle analysis (SPA). In Chlamydomonas PSI–LHCI, the LHCA2 and LHCA9 subunits are located at its PSAG/H side (11) similar to the recently resolved PSI structure of a red alga (12). Our CEF data suggest a dynamic dissociation/association model, in which LHCA2 and LHCA9 dissociate from PSI–LHCI enabling CEF supercomplex formation.
Results
Identification of a PSI–LHCI–cyt b6f Supercomplex.
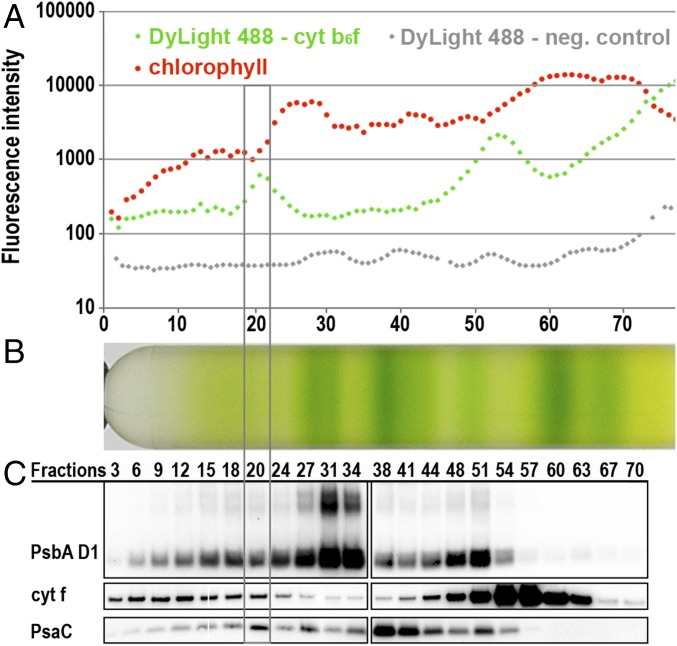
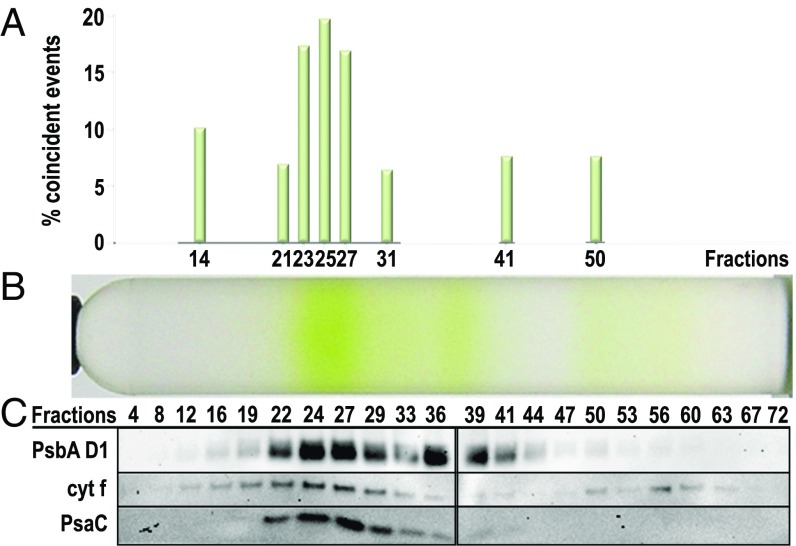
CEF supercomplexes of C. reinhardtii were isolated from anaerobically cultured cells (7, 8). Isolated thylakoid membranes were solubilized with n-dodecyl α-D-maltoside (α-DDM) and fractionated using SDG centrifugation (7, 8). Immunoblot analysis identified a high molecular weight SDG fraction containing the major CEF supercomplex components PSI and cyt b6f (Fig. 1 B and C). Previous work had demonstrated CEF activities in the same SDG fractions, but it remained possible that this was due to (i) colocalization of cyt b6f and PSI in small residual membrane patches (since α-DDM is a mild detergent) or (ii) comigration of separate cyt b6f and PSI supercomplexes on the SDG (e.g., due to the presence of other molecular partners such as LHCII trimers, ATPase dimers, or NDH). We therefore used SMF correlation spectroscopy (SI Appendix, Figs. S1–S3) to demonstrate a single molecule, physical interaction between cyt b6f and PSI–LHCI complexes (Fig. 2 and SI Appendix, Fig. S3; see SI Appendix for more details). Total fluorescence profiles across the SDG fractions showed that the locations of cyt f (labeled with green DyLight 488), as well as PSI and PSII in the SDG correspond well with increases in green fluorescence (Fig. 1 A and C) and red chlorophyll fluorescence signals (Fig. 1 A and B), respectively. The fraction 19–22 pool was repurified (Fig. 1 and SI Appendix, SI Materials and Methods) via a subsequent SDG (Fig. 2B) and measured by SMF correlation spectroscopy to discriminate between single (red or green) vs. coincident (red and green) fluorescent events (SI Appendix, Fig. S2 B and C) corresponding to discrete PSI–LHCI and cyt b6f molecules vs. associated PSI–LHCI–cyt b6f supercomplexes (13, 14). The frequency of SMF coincident events (Fig. 2A) in the numbered fractions of the repurified complex gradient (Fig. 2B) clearly revealed that DyLight-tagged cyt b6f is physically associated with a chlorophyll fluorescent complex in subfractions 23–27, consistent with the location of cyt f and PsaC (Fig. 2C). Coincident events were substantially higher than in the control fractions measured and exceeded the number of coincident peaks of very high molecular weight fractions (e.g., fraction 14), in which broad SMF peaks were occasionally visible due to the presence of small membrane patches (SI Appendix, Fig. S3B). Importantly, these membrane patches contributing to false positive coincident events were not visible in fractions 23–27, reinforcing the conclusion that the observed coincidence is due to genuine CEF supercomplexes. Although an interaction of cyt f with PSII cannot be excluded based on these measurements alone, functional interaction between PSI–LHCI and cyt b6f has been reported (7, 8, 10), while a PSII–cyt b6f supercomplex is unprecedented. Furthermore, immunoblotting and electron microscopy (EM) confirmed that the concentration of PSI in these fractions was approximately 10 times that of PSII (see below). Collectively these results provide evidence for a physical association between PSI–LHCI and cyt b6f essential for the formation of CEF supercomplexes in solution.
Fig. 1.
Identification of the CEF supercomplex peak fraction in SDG by SMF spectroscopy and Western blot analysis. (A) Fluorescence intensity screening of each SDG fraction (log scale) confirms good correlation of red chlorophyll fluorescence with the location of the photosynthetic complexes in the SDG (B) and reveals two main peaks of green fluorescent DyLight 488–trisNTA labeled cyt f. As control for the green fluorescent signal, a SDG of an unlabeled cyt f His-Tag strain was screened (displayed in gray). (B) SDG of anaerobic α-DDM solubilized cyt f His-Tag thylakoids separated into 77 fractions. (C) Immunoblot detection of cyt f, the PSI subunit PSAC, and the PSII subunit PsbA D1 show that the PSAC signal peaks with the higher molecular weight green fluorescent peak signal of the DyLight 488–trisNTA labeled cyt f at fraction 20.
Fig. 2.
Cytochrome b6f is physically associated with chlorophyll fluorescent proteins in the CEF supercomplex sucrose density region revealed by SMF coincidence analysis. (A) Coincident events, i.e., the simultaneous bursts of green and red fluorescence, indicative of the physical association of DyLight 488-labeled cyt f and chlorophyll fluorescent proteins, are most abundant in the CEF supercomplex region of the SDG. The frequency of coincident events relative to total fluorescent events recorded over a period of 60 s is plotted for selected fractions over the corresponding SDG. A false positive rate of 5% was applied to exclude the possible random excitation of two single fluorescent proteins as experimentally examined previously (13). (B) Pooled CEF supercomplex fractions from five SDGs of a cyt f His6-tag strain were concentrated, labeled with DyLight 488–trisNTA, and ultracentrifuged on a subsequent SDG to enrich for potential CEF supercomplexes. (C) Immunoblot detection of cyt f, PSAC, and Psba D1 over the SDG fractions confirms that the highest frequency of coincident events correlates with the localization of these proteins in the high molecular weight CEF supercomplex region.
Structural Characterization of the CEF Supercomplex.
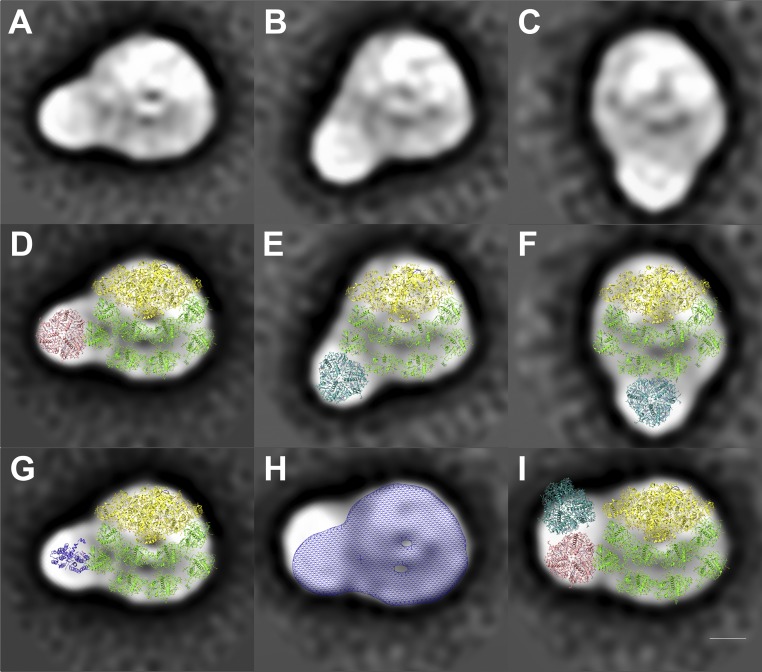
Initial single particle analysis revealed heterogeneity within the PSI–LHCI–cyt b6f supercomplex SDG fraction (SI Appendix, Fig. S4). Consequently, subsequent EM preparations employed an additional gentle affinity purification step using a His-tagged PSI–LHCI (PSAA His-Tag) construct (15). To stabilize this supercomplex, chemical protein crosslinking was performed during the solubilization step before affinity purification (Materials and Methods). This enabled the identification of putative CEF supercomplexes by transmission electron microscopy (TEM) SPA. A total number of 2,708 micrographs of negatively stained particles were recorded and yielded a total of 526,519 projection images of protein complexes (SI Appendix, Fig. S4B). Using SPA with Relion (16), these particles were classified into 350 classes (SI Appendix, Fig. S4C). Parallel liquid chromatography (LC)-MS/MS analysis was conducted (SI Appendix, Fig. S8) and it identified contaminants including mitochondrial NADH dehydrogenase (79,709 particles), mitochondrial ATPase (26,704 particles), and PSII–LHCII complexes (14,664 particles), and the corresponding SPA classes were identified (SI Appendix, Fig. S5) and removed from the dataset, yielding 160,819 potential PSI–LHCI-containing particles (SI Appendix, Fig. S6A). A subset of 52,316 particles were identified as top-view projections of single PSI–LHCI complexes (50 classes, see SI Appendix, Fig. S6B), as opposed to 12,179 top-view projections that contained additional density adjacent to the PSI–LHCI complex (SI Appendix, Fig. S6C), potentially consisting of cyt b6f or LHCs. Next, tilt views and PSI–LHCI complexes lacking well-connected additional densities were eliminated from this dataset, yielding 1,139 particles. These were classified into 16 classes (SI Appendix, Fig. S7) and analyzed (Fig. 3) to confirm the ability to distinguish between PSI–LHCI–cyt b6f supercomplexes (Fig. 4) and PSI–LHCI–LHCII supercomplexes (Fig. 3 A–F, see Discussion).
Fig. 3.
Structural characterization of potential PSI–LHCI–LHCII supercomplexes from C. reinhardtii by single particle electron microscopy. A–C show three of the projections from SI Appendix, Fig. S6, which have densities additional to those of the PSI–LHCI supercomplex. (D–F) These projection maps have been overlaid with the densities of the PSI core complex [yellow (17), Protein Data Bank (PDB) 4Y28] and the LHCA proteins (green, PDB 4Y28). A LHCII trimer [magenta or cyan (42), PDB 1RWT] can be seen to fit well into these three densities. (G) Modeling a cyt b6f monomer [purple (19), PDB 1Q90] into the additional density next to PSI–LHCI supercomplex (A) shows a poor fit. (H) Overlay of the large PSI supercomplex projection map from Fig. 4A and a meshed density map of A. (I) Poor fit of two LHCII trimers (42) into the projection map from Fig. 4A next to the PSI–LHCI complex. This suggests that the additional densities in A–C are likely LHCII trimers, while the new density in H and Fig. 4A best fits a cyt b6f dimer. (Scale bar: 5 nm.)
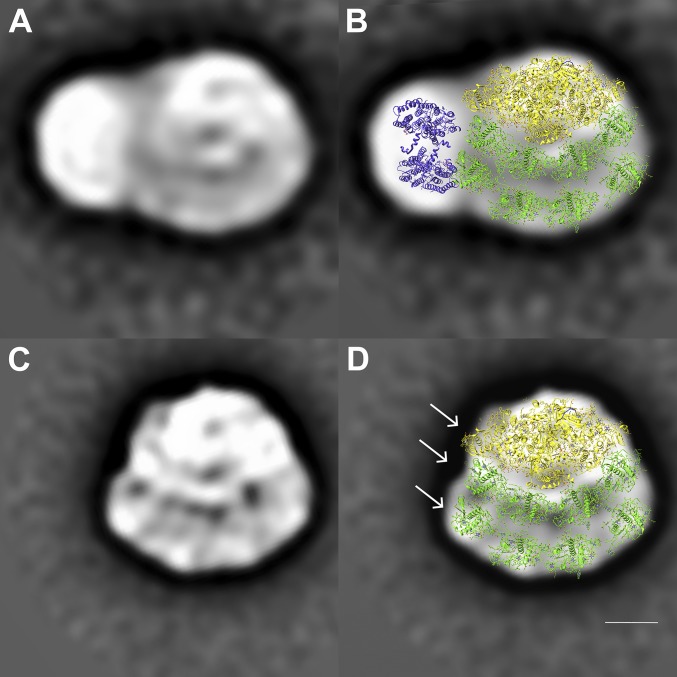
Fig. 4.
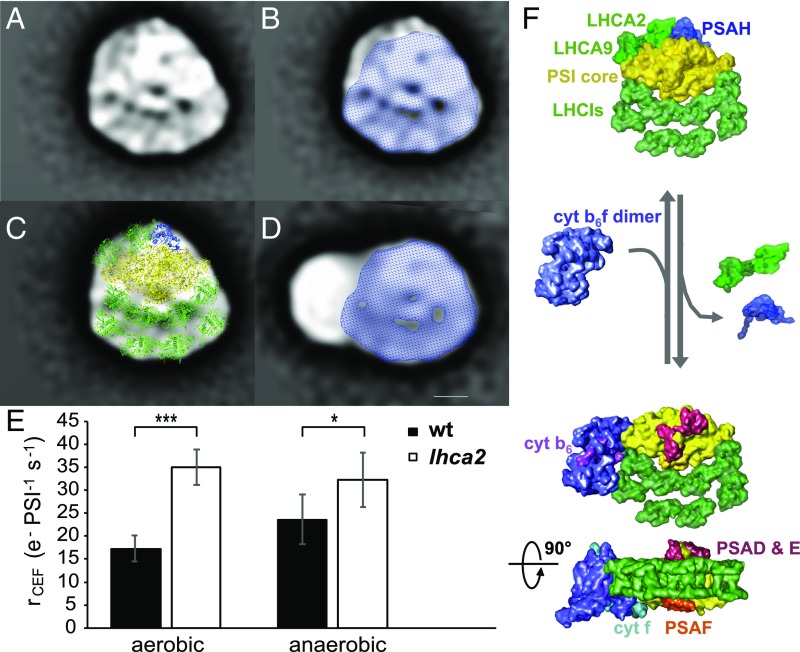
Structural characterization of a PSI–LHCI–cyt b6f supercomplex from C. reinhardtii by single particle transmission electron microscopy. (A) Averaged TEM projection map of a large supercomplex consisting of PSI and LHCI with an additional particle at its PSAG LHCA1 side (sum of 132 particles, representing 50% of classes in SI Appendix, Fig. S7). (B) Structural assignment of this supercomplex based on fitting with the crystal structures of the PSI–LHCI complex (17) and the cyt b6f complex (19) [Protein Data Bank (PDB) accession nos. 4Y28 and 1Q90, respectively]. The PSI core complex is shown in yellow with LHCA proteins highlighted in green. The cyt b6f complex is shown in purple. (C) Averaged projection map of a PSI–LHCI complex missing PSAH at its core. (D) Structural assignment of the PSI–LHCI complex similar to B. Eight LHCA proteins were modeled into the double-layered LHCI belt according to ref. 11. Arrows indicate the position of PSAG (yellow) and two LHCA1 subunits (green). (Scale bar: 5 nm.)
A projection map of a PSI–LHCI-containing supercomplex representing 50% of classes of SI Appendix, Fig. S7 (SI Appendix), is depicted in Fig. 4A. The molecular model overlay of PSI–LHCI (Fig. 4B, yellow and green, respectively) clearly identifies the PSI–LHCI supercomplex (17, 18) and an additional density adjacent to its PSAG–LHCA1 side that can accommodate a cyt b6f dimer (Fig. 4B, purple, ref. 19; see also SI Appendix, Fig. S9). Corresponding PSI–LHCI supercomplex controls are provided in Fig. 4 C and D. Based on this, the first top view of the dynamic CEF supercomplex of the green alga C. reinhardtii containing PSI, LHCI, and cyt b6f is proposed (Fig. 4B).
Fig. 5 provides more detailed structural insights into factors proposed to control the formation of this CEF supercomplex. The PSI dataset (SI Appendix, Fig. S6B) included a larger PSI–LHCI supercomplex (Fig. 5A) than is shown in Fig. 4C (see Fig. 5B, blue shading) highlighting the dynamic nature of these supercomplexes. The additional (Fig. 5B, unshaded) density on the PSAG side is attributed to LHCA2, LHCA9, and PSAH (Fig. 5C) based on refs. 11 and 18. These subunits appear to have dissociated in many isolated PSI–LHCI particles (Fig. 4 C and D) and are also absent in the CEF supercomplex (Fig. 4B). This suggests that PSAG and LHCA1 form an interface with the cyt b6f dimer (Fig. 5 C and D).
Fig. 5.
Dissociation of LHCA2 and LHCA9 from the PSI–LHCI complex favors the association of PSI–LHCI–cyt b6f supercomplexes and enhances cyclic electron flow. (A) Averaged TEM projection map of a PSI–LHCI complex with an additional density at its core. (B) Overlay of the PSI–LHCI complex from A with Fig. 4C. (C) Structural assignment of the PSI–LHCI complex from A based on fitting with the crystal structures of the PSI–LHCI complex (17). The additional densities compared with the smaller PSI–LHCI complex (B) were modeled with PSAH (blue) and two additional LHCA proteins (green) at the PSI core according to ref. 11. (D) Overlay of the CEF supercomplex projection map from Fig. 4A with the PSI–LHCI complex from Fig. 4C. (Scale bar: 5 nm.) (E) Cyclic electron transfer rates of a Δlhca2 mutant compared with wild-type levels in aerobic and anaerobic conditions. Rates were measured in steady state upon a transition from darkness to light with ∼130 μE m−2⋅s−1 light intensity. To exclude contribution of PSII to the electron transfer, cultures were treated with 40 μM DCMU. Anaerobic conditions were achieved by addition of 100 mM glucose and 2 mg⋅mL−1 glucose oxidase. To alleviate PSI acceptor side limitation upon transition to anaerobiosis, anaerobic samples were kept in the darkness for 40 min and continuously illuminated for 2 min before the rate measurements (n = 6 biological replicates ± SD). Statistical analysis: one-way ANOVA followed by a Tukey test for pairwise comparison of the means (***P < 0.001; *P < 0.05). (F) Structural model of CEF supercomplex formation upon dissociation of LHCA2, LHCA9, and PSAH from the PSI–LHCI complex. The PSI core is shown in yellow, LHCI proteins in bright green, cyt b6f in purple. Plastocyanin (cyt f in cyan, PSAF in orange) and ferredoxin (PSAD/E in red and cyt b6 in pink)-binding regions are indicated. All high-resolution components have been filtered to 20 Å to avoid overinterpretation.
A Proposed Role for LHCA2 and LHCA9 in CEF Supercomplex Formation.
The PSI–LHCI supercomplex projection map (Fig. 5A) fits well with the newly proposed model of Chlamydomonas PSI–LHCI (11). According to crosslinking and interaction studies, Ozawa et al. (11) concluded that LHCA2 and LHCA9 are not included in the two LHCI layers (Fig. 5C, Bottom, green), but are located at the other side of the PSI core (Fig. 5C, Top, green) and associate with PSAB and PSAH. This subunit assignment fits the additional PSI density identified here (Fig. 5 B and C). This model therefore differs from the one presented in ref. 18. Importantly, quantitative MS analysis (SI Appendix, Fig. S8) confirmed that LHCA2 and LHCA9 together with PSAH are most easily lost during purification (SI Appendix, Fig. S8A), consistent with Fig. 5. Strikingly, a Δlhca2 knockout mutant showed enhanced cyclic electron flow compared with wild-type (WT) levels under anaerobic conditions; even under aerobic conditions, CEF rates in Δlhca2 were already as high as for WT in anaerobic conditions (Fig. 5E). This suggests that the dissociation of LHCA2 and LHCA9 from the PSI–LHCI supercomplex is important for the assembly of the CEF supercomplex and the promotion of CEF (Fig. 5F).
Discussion
Structure of the Proposed CEF Supercomplex.
In 2010, Iwai et al. (7) presented biochemical evidence for the existence of a CEF supercomplex in the green alga Chlamydomonas. Here, this biochemical evidence is explained in structural terms. First, SMF spectroscopy supported the existence of discrete detergent solubilized supercomplexes containing both cyt b6f and PSI–LHCI in solution (Figs. 1 and 2 and SI Appendix, Fig. S2); using MS, the CEF supercomplex preparation was shown to contain FNR, PGRL1, ANR1, and CAS (SI Appendix, Fig. S8) which are important for CEF (8, 9). Second, having identified the presence of intact PSI–LHCI–cyt b6f supercomplexes (SI Appendix, Fig. S2), over 500,000 TEM projection images of molecules in this preparation were analyzed by SPA (SI Appendix, Figs. S4–S7). This analysis yielded a top-view projection map of the proposed CEF supercomplex that is able to accommodate the PSI core (Fig. 4B, yellow), its eight LHCI antenna proteins (Fig. 4B, green), and a cyt b6f dimer (Fig. 4B, purple). Third, these TEM data combined with functional data from LHCA2 knockouts yielded a model, which incorporates all of the above information and implies a role for LHCA2 and LHCA9 in the CEF supercomplex assembly process (Fig. 5 E and F).
The assembly of the CEF supercomplex when stromal electron carriers are reduced to produce extra ATP (20, 21) places the PSI–LHCI supercomplex and the cyt b6f dimer in close proximity to favor CEF over LEF. As electron transport via plastocyanin and ferredoxin is thought to be diffusion limited (22), the regulation of the distances by structural alignment of the PSI–LHCI supercomplex with cyt b6f could control electron transfer kinetics, as seen via regulation of thylakoid stacking in vascular plants (23).
CEF Supercomplex Assembly and the Promotion of Cyclic Electron Flow.
A surprising finding from the structural data (Fig. 5) was the absence of densities corresponding to LHCA2, LHCA9, and PSAH in the CEF supercomplex (Fig. 5 D vs. B). This led us to examine the functional consequence of LHCA2 knockout on CEF. Strikingly, the Δlhca2 knockout mutant (11) showed constitutively high CEF rates under aerobic conditions, that exceeded WT rates in anaerobic conditions (Fig. 5E). This suggests potential roles for LHCA2 and LHCA9, whose association to the PSI core is unstable in the absence of LHCA2 (11), during the CEF supercomplex assembly process: LHCA2 and LHCA9 could therefore block the CEF supercomplex formation, and their removal could expose a binding site for cyt b6f required to drive CEF supercomplex assembly. This suggests a role for LHCs above and beyond their light harvesting and energy dissipation functions. Notably, LHCA6 is important for NADP(H) dehydrogenase–PSI supercomplex formation in Arabidopsis (24, 25), independently showing the role of a LHCA polypeptide in supercomplex formation. Closer analysis of a series of identified PSI–LHCI supercomplexes with additional putative bound LHCII trimers shows that other LHC complexes could potentially competitively block cyt b6f dimer binding (e.g., Fig. 3H, see below).
State Transitions–CEF.
Both state transitions and CEF enhancement are triggered by similar conditions, for example, the redox state of the chloroplast, although these processes can occur independently of one another (8, 10). Hence, the presence of different PSI–LHCI-containing supercomplexes under anaerobic conditions is expected. To eliminate the possibility that the proposed CEF supercomplex could represent a PSI–LHCI–LHCII complex, we modeled two LHCII trimers into this density; these fitted poorly (Fig. 3I). While the focus of this paper is on the CEF supercomplex, it is of note that we also identified three PSI–LHCI–LHCII complexes (Fig. 3) with an LHCII trimer-like density localized at the LHCI belt side of the complex. These complexes represented the other 50% of classes in SI Appendix, Fig. S7. MS data confirmed that LHCII proteins were less depleted by PSI His-Tag purification than PSII core subunits (SI Appendix, Fig. S8F), indicating that additional LHCIIs might indeed be attached to PSI–LHCI. The supramolecular organization of these PSI–LHCI–LHCII supercomplexes differs substantially from other C. reinhardtii PSI–LHCI–LHCII supercomplexes already described, in which two LHCII trimers and one LHCII monomer were attached to the PSAH PSAL side of PSI opposite the LHCI ring (26). In contrast, in the PSI–LHCI–LHCII complexes identified here, an LHCII trimer localized at previously unreported positions at the outer LHCI belt side (Fig. 3 D–F), which are similar to a recent structure described in Arabidopsis thaliana where an LHCII trimer was associated with PSI at the side of LHCA2 and LHCA3 (27).
The density assigned to the cyt b6f dimer at the PSAG–LHCA1 side of PSI–LHCI in the newly identified CEF supercomplex (Fig. 5) is located similarly to that observed in TEM images of detergent solubilized PSI–LHCI–cyt b6f complexes from A. thaliana (27). In the Chlamydomonas CEF supercomplex, the cyt b6f dimer interacts via its long side (Figs. 4B and 5D); in contrast in A. thaliana, the cyt b6f dimer is reported to interact with PSI–LHCI via its short side (27). Since Chlamydomonas PSI is larger than plant PSI due to higher numbers of LHCI subunits forming a second LHCI belt and the potentially algae-specific positions of LHCA2 and LHCA9 with respect to the PSI core, differences may exist in regulation of CEF assembly formation in plants compared with Chlamydomonas. The binding of the cyt b6f dimer to the PSI–LHCI supercomplex positions the plastocyanin binding sites of cyt f (28, 29) and PSAF (30) close to one another, making the distance for electron transfer of plastocyanin between cyt b6f and PSI–LHCI at the luminal side relatively short. However, electron transfer partitioning between LEF and CEF is most likely regulated at the level of PQ reduction (31) and therefore located toward the stromal side. Cyt b6f dimer associated with its long side to PSI–LHCI would bring the more centrally located components involved in the stromal electron transfer, such as cyt b6 and Rieske subunits as well as the PSAA/PSAB reaction center and ferredoxin binding sites PSAC, PSAD, and PSAE (32, 33) into much closer proximity compared with a short side-bound cyt b6f, with cyt f and PSAF still in close proximity.
Building on these advances, the next challenge is to obtain a physically stable and pure CEF supercomplex for future atomic resolution structure determination to identify the position of additional small proteins in the CEF supercomplex like FNR, PGRL1, ANR1, and CAS (7–9). These subunits were identified in the CEF supercomplex SDG fraction (SI Appendix, Fig. S8C) and are expected to be attached to large supercomplexes (7–9) to enable them to migrate to this high molecular density. We attribute the low proportion of intact complexes on negative stain TEM grids to the labile nature of the supercomplex. A better understanding of the role of LHCA2 and LHCA9 in CEF assembly could lead to the production of physically stable supercomplexes, which would greatly assist in solving its atomic structure.
Materials and Methods
Strains and Culture Conditions.
The C. reinhardtii strains cyt f–His-Tag (19, 34), PSAA–His-Tag (15), a Δlhca2 (35), and CC-4533 (WT) were used in this study. Cells were grown in tris acetate phosphate (TAP) media (36) (22 °C, 50 µM photons m−2⋅s−1, 120 rpm shaking). For thylakoid isolation, cells were grown to 3–4 × 106 cells mL−1 and harvested by centrifugation (4,600 × g, 5 min, 25 °C), resuspending to 2 × 108 cells mL−1 in H1 buffer (25 mM Hepes-KOH pH 7.5, 5 mM MgCl2, 0.3 M sucrose). Anaerobic conditions were induced using 2 mg⋅mL−1 glucose oxidase (Aspergillus niger) and 50 units⋅mL−1 catalase (bovine liver, Sigma-Aldrich) with 100 mM glucose in the dark for 60 min (10).
Thylakoid and Photosynthetic Complex Isolation and DyLight 488–Tris–NTA Labeling of Photosynthetic Complexes.
Thylakoids and photosynthetic complexes were isolated according to refs. 37 and 38; see also SI Appendix, SI Materials and Methods. Cyt f–His-Tag was labeled with DyLight 488– Tris–NTA before solubilization and SDG-based purification (SI Appendix, SI Materials and Methods).
Single Molecule Fluorescence Measurements.
Single molecule spectroscopy (SI Appendix, Fig. S2) was performed based on refs. 13 and 14 and SI Appendix, SI Materials and Methods. Two excitation lasers (488 nm and 561 nm) focused in solution using a 40×/1.2 N.A. water immersion objective (Zeiss) simultaneously excite green (here: the DyLight 488-labeled cyt f protein, see SI Appendix, SI Materials and Methods for more details) and red (here: chlorophyll-containing proteins) fluorophores. Fluorescence was collected and separated using a 565-nm dichroic mirror; signal from DyLight 488-labeled cyt f protein was passed through a 525/20-nm band pass filter, while chlorophyll fluorescence was filtered by a 580-nm long pass filter. The fluorescence of the two channels was recorded simultaneously in 1-ms time bins for 60 s. For single-molecule coincidence detection, the coincidence ratio was calculated as in ref. 13.
Immunoblot Analysis.
A fixed volume (20 μL) of representative fractions across the SDG gradient was analyzed by 4–12% Bis-Tris SDS/PAGE (Invitrogen). Proteins were transferred to PVDF Millipore membrane (Merck) with the XCell Blot II Module (Invitrogen) and blocked with skimmed milk. The membrane was incubated with antibodies against PSAC (1:1,000; Agrisera), Psba D1 (1:10,000; Agrisera), and cyt f (1:5,000; Agrisera), and anti-rabbit IgG (HRP, 1:2,500; Sigma Aldrich) as the secondary antibody. Signal detection was performed with ECL (Amersham GE Healthcare).
Chemical Crosslinking.
To crosslink proteins with disuccinimidyl suberate (DSS) (Thermo Fisher), PSAA His-Tag thylakoids were resuspended in Hepes buffer (50 M Hepes-KOH pH 8, 5 mM MgCl2) and freshly dissolved DSS (in DMSO, 20 mg/mL stock concentration) was added to the sample (final concentration, 0.15 mg/mL) before solubilization of proteins. The crosslinking reaction was performed for 30 min at room temperature in the dark with occasional inversion and stopped by adding Tris buffer (final concentration, 15 mM pH 7.5, 1 mM EDTA).
Purification of His-Tagged Proteins.
His-tagged proteins were purified by immobilized metal ion chromatography according to ref. 15 with the following modifications. PSI–LHCI–cyt b6f containing fractions from six SDGs were pooled (∼4 mL) and loaded onto a 1-mL HiTrap HP column (GE Healthcare) preequilibrated with 5 mM tricine-KOH pH 8, 0.02% α-DDM, 10 mM NaCl, 5 mM MgSO4, 0.5 M sucrose, 2 mM imidazole. The column was washed with two washing buffers, with increasing the imidazole concentrations (10 mM to 20 mM) and decreasing the sucrose concentration (0.5 M to 0 M, 10 mL each). Elution was performed by increasing the imidazole concentration to 200 mM (4 elution fractions, 500 µL each).
Mass Spectrometry.
For quantitative mass spectrometric analysis, the PSAA His-Tag strain was isotopically labeled with 14N and 15N. The pooled 15N labeled PSI–LHCI–cyt b6f fractions (before His-Tag purification) were mixed 1:1 with the His-Tag purified eluate of the 14N labeled PSI–LHCI–cyt b6f sample. Duplicates were performed with a label swap. Samples were digested with trypsin in a 0.5-mL Amicon Ultra ultrafiltration device (30-kDa cutoff; Millipore) (39) with minor modifications. Mass spectrometry was performed according to ref. 38. Identification and quantification of peptide spectrum matches were conducted in the framework of Ursgal (40) and using pyQms (41). See SI Appendix, SI Materials and Methods for details.
Transmission Electron Microscopy.
A total of 5 µL of the PSAA–His-Tag purified SDG fraction was applied to glow-discharged 400-mesh copper TEM grids coated with a thin continuous film of evaporated carbon and complexes stained with 2% uranyl acetate (wt/vol). Single particles were imaged on a Tecnai 12 TEM operated at 120 kV (FEI Company) connected to a Direct Electron LC-1100 lens-coupled 4k × 4k CCD camera (nominal magnification of 67,000×, 2× pixel binning, 4.34 Å pixel size at specimen level). An initial 2D class average was calculated from manually picked particles to enable automated particle selection using RELION (16). A total of 526,519 particles were collected from 2,708 digital micrographs (examples shown in SI Appendix, Fig. S4 A and B, circular mask of 380 Å). Single particle images were analyzed with RELION (16) (see SI Appendix, SI Materials and Methods for details).
Spectroscopic Measurements.
P700 absorption and electrochromic shift signal measurements were performed with a LED pump-probe JTS-10 spectrophotometer (BioLogic) as described previously (10). Single turnover measurements used a dye laser emitting at 640 nm, pumped by the second harmonic of a Minilite II Nd:YAG laser (Continuum). C. reinhardtii cells were harvested and resuspended to a 20 µg⋅mL−1 chlorophyll concentration [20 mM Hepes, pH 7.2, 10% Ficoll (wt/vol)], and incubated (20 min, dark, constant shaking to avoid anaerobiosis). Anaerobic conditions were reached as described above. To eliminate contribution of linear electron flow, the PSII inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU, 40 µM) was added. Electron flow rates were determined as the product of kox [P700red] (10). More details about experimental procedures are listed in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Hawat for help with preparation of MS/MS samples and H. Nüsse and U. Keller for their kind permission and assistance to glow discharge grids at the EM facility of the Institute for Medical and Biophysics (University of Münster, Germany). B.H. acknowledges support from Australian Research Council Grants DP130100346 and DP160101018. M.H. acknowledges support from Deutsche Forschungsgemeinschaft (DFG) Grant HI 739/13-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809973115/-/DCSupplemental.
References
- 1.Arnon DI, Allen MB, Whatley FR. Photosynthesis by isolated chloroplasts. Nature. 1954;174:394–396. doi: 10.1038/174394a0. [DOI] [PubMed] [Google Scholar]
- 2.Bassham JA, Benson AA, Calvin M. The path of carbon in photosynthesis. J Biol Chem. 1950;185:781–787. [PubMed] [Google Scholar]
- 3.Calvin M, Benson AA. The path of carbon in photosynthesis. Science. 1948;107:476–480. doi: 10.1126/science.107.2784.476. [DOI] [PubMed] [Google Scholar]
- 4.Allen JF. Cyclic, pseudocyclic and noncyclic photophosphorylation: New links in the chain. Trends Plant Sci. 2003;8:15–19. doi: 10.1016/s1360-1385(02)00006-7. [DOI] [PubMed] [Google Scholar]
- 5.Munekage Y, et al. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature. 2004;429:579–582. doi: 10.1038/nature02598. [DOI] [PubMed] [Google Scholar]
- 6.Finazzi G, Rappaport F. In vivo characterization of the electrochemical proton gradient generated in darkness in green algae and its kinetic effects on cytochrome b6f turnover. Biochemistry. 1998;37:9999–10005. doi: 10.1021/bi980320j. [DOI] [PubMed] [Google Scholar]
- 7.Iwai M, et al. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature. 2010;464:1210–1213. doi: 10.1038/nature08885. [DOI] [PubMed] [Google Scholar]
- 8.Terashima M, et al. Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc Natl Acad Sci USA. 2012;109:17717–17722. doi: 10.1073/pnas.1207118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petroutsos D, et al. The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell. 2011;23:2950–2963. doi: 10.1105/tpc.111.087973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi H, Clowez S, Wollman F-A, Vallon O, Rappaport F. Cyclic electron flow is redox-controlled but independent of state transition. Nat Commun. 2013;4:1954. doi: 10.1038/ncomms2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozawa S-I, et al. Configuration of ten light-harvesting chlorophyll a/b complex I subunits in Chlamydomonas reinhardtii photosystem I. Plant Physiol. doi: 10.1104/pp.18.00749. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pi X, et al. Unique organization of photosystem I-light-harvesting supercomplex revealed by cryo-EM from a red alga. Proc Natl Acad Sci USA. 2018;115:4423–4428. doi: 10.1073/pnas.1722482115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gambin Y, et al. Single-molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae. eLife. 2013;3:e01434. doi: 10.7554/eLife.01434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sierecki E, et al. Nanomolar oligomerization and selective co-aggregation of α-synuclein pathogenic mutants revealed by single-molecule fluorescence. Sci Rep. 2016;6:37630. doi: 10.1038/srep37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulis G, Narasimhulu KV, Fox LN, Redding KE. Purification of His6-tagged photosystem I from Chlamydomonas reinhardtii. Photosynth Res. 2008;96:51–60. doi: 10.1007/s11120-007-9283-9. [DOI] [PubMed] [Google Scholar]
- 16.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazor Y, Borovikova A, Nelson N. The structure of plant photosystem I super-complex at 2.8 Å resolution. Elife. 2015;4:e07433. doi: 10.7554/eLife.07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drop B, et al. Photosystem I of Chlamydomonas reinhardtii contains nine light-harvesting complexes (Lhca) located on one side of the core. J Biol Chem. 2011;286:44878–44887. doi: 10.1074/jbc.M111.301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stroebel D, Choquet Y, Popot J-L, Picot D. An atypical haem in the cytochrome b(6)f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 20.Lucker B, Kramer DM. Regulation of cyclic electron flow in Chlamydomonas reinhardtii under fluctuating carbon availability. Photosynth Res. 2013;117:449–459. doi: 10.1007/s11120-013-9932-0. [DOI] [PubMed] [Google Scholar]
- 21.Alric J. Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii: (II) Involvement of the PGR5–PGRL1 pathway under anaerobic conditions. Biochim Biophys Acta. 2014;1837:825–834. doi: 10.1016/j.bbabio.2014.01.024. [DOI] [PubMed] [Google Scholar]
- 22.Kirchhoff H. Diffusion of molecules and macromolecules in thylakoid membranes. Biochim Biophys Acta. 2014;1837:495–502. doi: 10.1016/j.bbabio.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 23.Wood WHJ, et al. Dynamic thylakoid stacking regulates the balance between linear and cyclic photosynthetic electron transfer. Nat Plants. 2018;4:116–127. doi: 10.1038/s41477-017-0092-7. [DOI] [PubMed] [Google Scholar]
- 24.Peng L, Fukao Y, Fujiwara M, Takami T, Shikanai T. Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell. 2009;21:3623–3640. doi: 10.1105/tpc.109.068791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouřil R, et al. Structural characterization of a plant photosystem I and NAD(P)H dehydrogenase supercomplex. Plant J. 2014;77:568–576. doi: 10.1111/tpj.12402. [DOI] [PubMed] [Google Scholar]
- 26.Drop B, Yadav KNS, Boekema EJ, Croce R. Consequences of state transitions on the structural and functional organization of photosystem I in the green alga Chlamydomonas reinhardtii. Plant J. 2014;78:181–191. doi: 10.1111/tpj.12459. [DOI] [PubMed] [Google Scholar]
- 27.Yadav KNS, et al. Supercomplexes of plant photosystem I with cytochrome b6f, light-harvesting complex II and NDH. Biochim Biophys Acta. 2017;1858:12–20. doi: 10.1016/j.bbabio.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Soriano GM, Ponamarev MV, Tae GS, Cramer WA. Effect of the interdomain basic region of cytochrome f on its redox reactions in vivo. Biochemistry. 1996;35:14590–14598. doi: 10.1021/bi9616211. [DOI] [PubMed] [Google Scholar]
- 29.Illerhaus J, et al. Dynamic interaction of plastocyanin with the cytochrome bf complex. J Biol Chem. 2000;275:17590–17595. doi: 10.1074/jbc.275.23.17590. [DOI] [PubMed] [Google Scholar]
- 30.Hippler M, et al. The plastocyanin binding domain of photosystem I. EMBO J. 1996;15:6374–6384. [PMC free article] [PubMed] [Google Scholar]
- 31.Dumas L, Chazaux M, Peltier G, Johnson X, Alric J. Cytochrome b 6 f function and localization, phosphorylation state of thylakoid membrane proteins and consequences on cyclic electron flow. Photosynth Res. 2016;129:307–320. doi: 10.1007/s11120-016-0298-y. [DOI] [PubMed] [Google Scholar]
- 32.Sétif P. Ferredoxin and flavodoxin reduction by photosystem I. Biochim Biophys Acta. 2001;1507:161–179. doi: 10.1016/s0005-2728(01)00205-5. [DOI] [PubMed] [Google Scholar]
- 33.Sétif P, Fischer N, Lagoutte B, Bottin H, Rochaix JD. The ferredoxin docking site of photosystem I. Biochim Biophys Acta. 2002;1555:204–209. doi: 10.1016/s0005-2728(02)00279-7. [DOI] [PubMed] [Google Scholar]
- 34.Choquet Y, Zito F, Wostrikoff K, Wollman F. Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell. 2003;15:1443–1454. doi: 10.1105/tpc.011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, et al. An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell. 2016;28:367–387. doi: 10.1105/tpc.15.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorman DS, Levine RP. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hippler M, Drepper F, Farah J, Rochaix JD. Fast electron transfer from cytochrome c6 and plastocyanin to photosystem I of Chlamydomonas reinhardtii requires PsaF. Biochemistry. 1997;36:6343–6349. doi: 10.1021/bi970082c. [DOI] [PubMed] [Google Scholar]
- 38.Bergner SV, et al. State transition7-dependent phosphorylation is modulated by changing environmental conditions, and its absence triggers remodeling of photosynthetic protein complexes. Plant Physiol. 2015;168:615–634. doi: 10.1104/pp.15.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 40.Kremer LPM, Leufken J, Oyunchimeg P, Schulze S, Fufezan C. Ursgal, universal Python module combining common bottom-up proteomics tools for large-scale analysis. J Proteome Res. 2016;15:788–794. doi: 10.1021/acs.jproteome.5b00860. [DOI] [PubMed] [Google Scholar]
- 41.Leufken J, et al. pyQms enables universal and accurate quantification of mass spectrometry data. Mol Cell Proteomics. 2017;16:1736–1745. doi: 10.1074/mcp.M117.068007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, et al. Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.