Significance
Exosomes are tiny vesicles secreted by cells that play an important role in cell-to-cell communication. The RNA, DNA, and proteins in exosomes are reported to be critical biomarkers for precision diagnostics and therapeutics. Exosome analysis thus offers an approach to learn health conditions without tissue biopsy. Clinical application and fundamental study of exosomes require analytical tools capable of physical and biochemical characterization. The challenge in developing such tools lies in that conventional optical microscopy is not suitable for exosome analysis due to its limited sensitivity. In this paper, we report a plasmonic microscopy capable of imaging single exosomes and quantitative analysis of physical (size) and biochemical (interaction with antibody) information.
Keywords: extracellular vesicle, plasmonic microscopy, label-free, molecular interaction, nanoparticle
Abstract
Exosomes play an important role in numerous cellular processes. Fundamental study and practical use of exosomes are significantly constrained by the lack of analytical tools capable of physical and biochemical characterization. In this paper, we present an optical approach capable of imaging single exosomes in a label-free manner, using interferometric plasmonic microscopy. We demonstrate monitoring of the real-time adsorption of exosomes onto a chemically modified Au surface, calculating the image intensity, and determining the size distribution. The sizing capability enables us to quantitatively measure the membrane fusion activity between exosomes and liposomes. We also report the recording of the dynamic interaction between exosomes and antibodies at the single-exosome level, and the tracking of hit-stay-run behavior of exosomes on an antibody-coated surface. We anticipate that the proposed method will contribute to clinical exosome analysis and to the exploration of fundamental issues such as the exosome–antibody binding kinetics.
Exosomes are tiny membrane vesicles (30–150 nm in diameter), which carry rich molecular information about the parent cells (1). They play an important role in numerous cellular processes, including cell–cell communication, angiogenesis, immune response, and cancer metastasis (2–6). Despite their importance, practical use of exosomes is constrained by the lack of analytical tools capable of their physical and molecular characterization (7–9). Conventional analytical methods, such as transmission electron microscopy (TEM) and ELISAs, often require large amounts of samples and extensive sample-preparation processes (10). New techniques, including nanoparticle tracking analysis (NTA) (11, 12), tunable resistive pulse sensing (13, 14), flow cytometry (FC) (15, 16), and surface plasmon resonance (SPR) (17, 18) are powerful, but have their own limitations in throughput, sensitivity, or multiplexing capability (8, 9). Fundamental knowledge about how exosomes interact with cells to alter the physiology is difficult to discover, since conventional optical microscopy is not able to visualize exosomes, and complex labeling is required (19, 20).
Light scattering is the basis of multiple technologies for exosome analysis including NTA and FC. However, limitation in sensitivity hinders the dynamic imaging of single exosomes. Given the fact that scattered light intensity by a nanoobject scales with the sixth power of object diameter, and that there are many additional spurious scattering sources, such as out-of-focus objects in the medium and surface inhomogeneity, the challenge of detecting pure light scattering from single exosomes becomes evident. Plasmonic microscopy is superior in exosome analysis because it images only the samples within 200 nm of the surface, and thus rejects noise from out-of-focus sources in the medium. Resonant detection enables the measurement of the binding kinetics between exosomes and antibodies (17, 18), but heterogeneity information is lost due to the lack of single-exosome analysis capability. SPR microscopy (SPRM) with a high-numerical-aperture (N.A.) objective has been established in the last two decades as a unique tool for imaging biological samples, including cells (21), viruses (22), and DNA molecules (23). However, direct imaging of single exosomes has not been realized due to limited sensitivity.
In this paper, we report an optical method for imaging and size analysis of exosomes with interferometric plasmonic microscopy (iPM). iPM is similar to SPRM, but with higher sensitivity and spatial resolution achieved through optimized detection methodologies and image-processing techniques. The development of SPRM toward iPM for the detection, imaging, and quantitative analysis of single exosomes is a nontrivial task because the expected signal, which is about a hundredth of the detectable signal in current state-of-the-art SPRM, is much smaller than the intensity fluctuations originating from illumination and other spurious scattering sources.
Results and Discussion
Principle of iPM Detection.
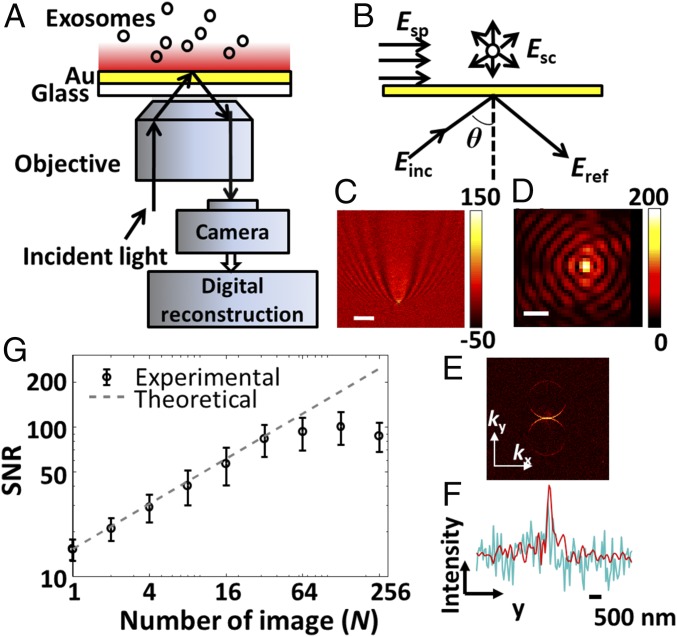
iPM was built in the Kreschmenn configuration (Fig. 1A). A laser light at a wavelength of 637 nm was collimated and illuminated a Au chip at a highly inclined incident angle θ, stimulating surface plasmons (SPs) on the Au surface. Reflective light, together with the light scattered by the object, was then collected and imaged onto a complementary-metal-oxide-semiconductor (CMOS) camera. The intrinsic principle of iPM is thus common-path interferometry (Fig. 1B) (24). The recorded iPM image results from the interference (Eq. 1) between the reflective light (Eref) and the SPs scattered by the object at near-field (Esc), which back-propagates onto the far-field detector. The interferometric scattering signal by an object () manifests itself as a small variation over a large background (). This background image was recorded before adsorption, and subtracted from images after adsorption to reveal the small intensity variation induced by objects (SI Appendix, Fig. S1). We note that iPM signals scale with D3 (D denotes diameter of the object), compared with D6 in pure scattering detection ().
| [1] |
SPs penetrate into the medium within only 200 nm above the metal surface, and thus only objects within this range will scatter the SPs and induce an intensity change in the iPM images. The recorded image of an object (Fig. 1C) appears as a unique pattern, causing difficulties in quantitative analysis and lowering spatial resolution. Back-propagation of the scattered SPs into far-field emission is directional, and the scattered light will only emit from the glass slide within a small range of angles. This effect is obvious if we examine the recorded image in k space by taking the 2D fast Fourier transform (2D-FFT) of the intensity image (Fig. 1E). By simulating the reflected light as a planar wave, we adopted a digital image-processing algorithm (SI Appendix, Fig. S1) (25) to reconstruct the iPM image of an object as a diffraction-limited spot (Fig. 1D), with a spatial resolution of ∼260 nm (Fig. 1F). Since the scattered iPM signals are confined in the two ring areas, the spatial filter in the image-reconstruction algorithm also rejects the noise (Fig. 1F).
Fig. 1.
iPM system. (A) Schematic of iPM. (B) Interferometric scattering model of iPM. (C) Images of a 100-nm silica nanoparticle without (scale bar: 3 µm) and (D) with (scale bar: 300 nm) image-reconstruction process. (E) k-space image by taking 2D-FFT of C. (F) Longitudinal intensity profile across the particle in C (cyanine) and D (red). (G) SNR in iPM detection of 100-nm silica nanoparticles with the running-average algorithm, and the theoretical shot-noise limitation (dashed line).
Noise Limitation in iPM Detection.
In a well-established iPM experiment, the shot-noise-induced fluctuation of the intensity is the dominating noise source due to the large background in the image. The signal-to-noise ratio (SNR) of iPM (SI Appendix, Fig. S2) was thus improved by a running-average algorithm in the temporal domain (SI Appendix, Fig. S1), which scaled with N1/2 (N denotes number of images averaged) in a range up to n = 32 (Fig. 1G). The running-average strategy requires a high recording rate with enough photon flux on the CMOS detector, which is not suitable for pure scattering detection but is easily fulfilled in iPM. In practical iPM experiments, the incident angle was fixed at the angle at which 10% of the incident light was reflected (SI Appendix, Fig. S3), and the recording frame rate was 320 frames per second (fps) at 512 × 512 pixels with a 33.28 × 33.28-μm2 full field-of-view area. The running average was carried out with n = 32 images.
Calibrating iPM with Silica Nanoparticles.
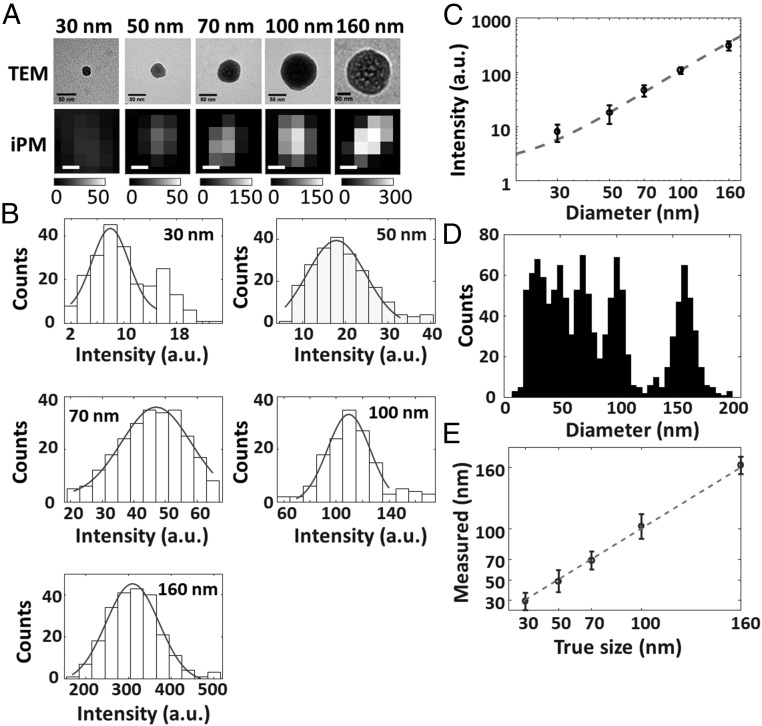
As a scattering-based detection technique, the iPM image intensity is intrinsically dependent on the size and refractive index of the objects. For exosome sizing, a calibration curve of iPM image intensity versus size is a prerequisite. Silica nanoparticles are preferred for calibration purposes due to them having a similar refractive index as exosomes. The diameters of standardized silica nanoparticles used for calibration were 30, 50, 70, 100, and 160 nm to cover the size range of exosomes. Typical TEM images and corresponding iPM images of silica nanoparticles are shown in Fig. 2A for comparison. While iPM did not spatially resolve the nanoparticle in as much detail as TEM due to the diffraction limit, the correlation between iPM intensity and particle size is clear. To quantify this correlation, the iPM intensity of each silica nanoparticle was calculated as the difference between the peak and mean background intensity (SI Appendix, Fig. S2). The statistical histograms of iPM intensity were then constructed from hundreds of recorded silica nanoparticles (Fig. 2B), and each was fitted with a Gaussian function to determine the corresponding intensity values (mean ± SD, n > 150). The second peak as indicated by the arrow in the histogram of 30-nm silica nanoparticles was attributed to the dimers. The calibration curve was then plotted as the iPM intensity versus the diameter of silica nanoparticles (Fig. 2C), and fitted the numerical model well (SI Appendix, Fig. S4).
Fig. 2.
iPM calibration with silica nanoparticles. (A) TEM (scale bar: 50 nm) and iPM images (scale bar: 100 nm) of standardized silica nanoparticles. (B) Recorded iPM intensity distribution of silica nanoparticles. (C) Calibration curve of iPM intensity vs. diameter fitted with the numerical model. (D) Size distribution of silica nanoparticles in a mixture as determined by iPM. (E) Comparison of nanoparticle size in the mixture determined by iPM and the true labeled value.
One key feature required for exosome analysis is the capability of composite sample analysis, because precise isolation of exosomes is challenging and cumbersome (1). We recorded the iPM images of a mixture containing silica nanoparticles with mean diameters of 30, 50, 70, 100, and 160 nm, calculated the iPM intensity of each individual particle bond to the Au surface, and determined the size distribution from hundreds of particles with the calibration curve (Fig. 2D). Five peaks were observed at 27.8 ± 8.9, 46.9 ± 10.2, 68.4 ± 9.6, 104.3 ± 11.3, and 166.1 ± 8.3 nm, showing good correspondence with the true value (Fig. 2E). The results thus suggest a sizing error of ∼10 nm in iPM detection.
Real-Time Monitoring and Analysis of Exosome Adsorption Process.
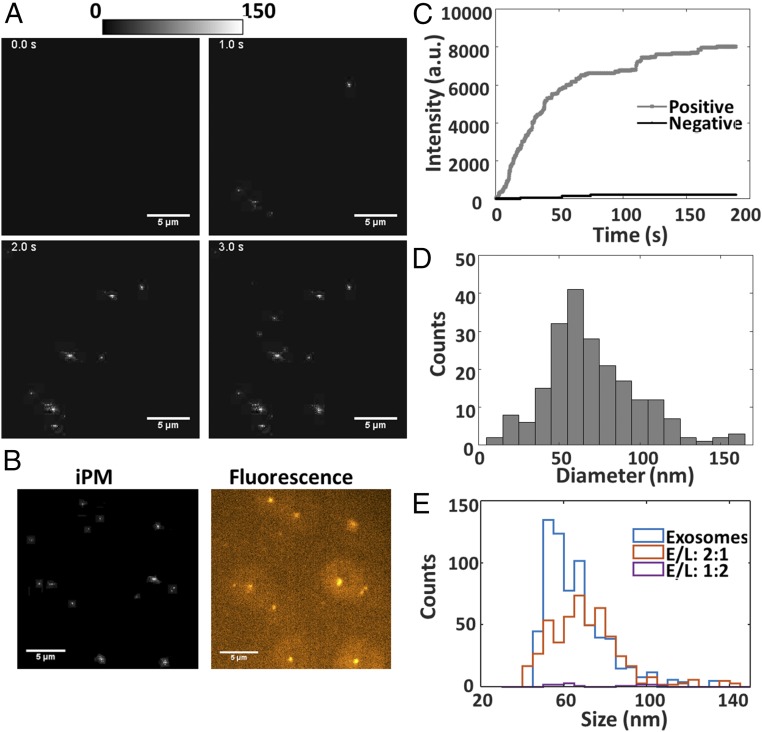
Exosomes were isolated and purified from the medium after culturing A549 cell lines for 72 h using differential centrifugation. Before iPM recording, we first examined the samples with TEM to confirm exosome isolation (SI Appendix, Fig. S5), and determined the concentration to be ∼109/mL with NTA. The adsorption of exosomes is expected to be dependent on the surface charge of the Au surface, due to the negative charge on exosomes membrane surface (SI Appendix, Fig. S6). We thus recorded the real-time adsorption of exosomes (Movie S1) onto Au surfaces modified with HS-PEG-NH2 (10,000 Da), and show typical time-lapse snapshots in Fig. 3A. Individual exosomes were easily distinguishable as bright spots. As a label-free technology, iPM lacks the specificity in identifying adsorbed objects. However, iPM is easily integrated with fluorescent microscopy. We labeled the lipid membrane of exosomes with DiIC18(3) dye, and recorded the fluorescent image, which corresponds well with the iPM images (Fig. 3B).
Fig. 3.
iPM detection of exosomes. (A) Snapshots of exosome adsorption onto a positive-charge-modified Au surface. (B) iPM and fluorescent images of exosomes. (C) Real-time binding curve of exosomes onto positive- or negative-charge-modified Au surface. (D) Size distribution of exosomes determined by iPM. (E) Size distribution of exosomes and exosome-liposome complex absorbed on positive-charge-modified Au surface, prepared at different exosome/liposome ratios (E/L). (Scale bar: 5 μm.)
A real-time binding curve was plotted by summing the iPM intensity of each individual exosome frame-by-frame (Fig. 2C). Compared with conventional plasmonic assays that measure the light intensity induced by a large number of exosomes, iPM offers single-exosome sensitivity. As expected, binding events were observed only on the Au surface modified with positive charge, but few observed on a negatively charged surface (Fig. 3C). The sizes of hundreds of exosomes were determined with the calibration curve in Fig. 2C, as shown in Fig. 3D. A continuous distribution of the exosome diameter between 30 and 150 nm was observed, with a peak value at ∼62 nm. The size distribution is significantly lower than the NTA results (SI Appendix, Fig. S7), which gives a peak value at ∼120 nm. This is reasonable, because NTA measures the hydrodynamic diameter instead of the true particle size.
Quantitative Study of Membrane Fusion Between Exosomes and Liposomes.
Exosomes have been recognized as natural drug delivery cargo (26), and membrane fusion between exosomes and liposomes has emerged as a potential approach for better design of drug-delivery systems (27). We performed the quantitative study of the fusion activity between exosomes and cationic liposomes. First, we recorded the liposome adsorption onto a negative-charge-modified Au surface and determined the size distribution of liposomes to be ∼64 nm (SI Appendix, Fig. S8). This value is also much smaller than that obtained by NTA analysis (SI Appendix, Fig. S7), but closer to the size observed in TEM images (SI Appendix, Fig. S4). Few liposomes were detected on the positive-charge-modified Au surface due to the positively charged liposome membrane surface (SI Appendix, Fig. S6). We then prepared the mixture of exosomes and liposomes at dose ratios of 2:1 and 1:2, incubated under 37 °C for 30 min to allow the membrane fusion, and performed the iPM analysis on a positive-charge-modified gold surface to obtain the size distribution of particles (Fig. 3E and SI Appendix). For pure exosomes, we identified more than 600 individual exosomes in 5 min, showing normal size distribution with a mean diameter of ∼56 nm. For the mixture of exosomes and liposomes at a ratio of 2:1, less than 400 particles were detected, and the mean diameter increased to ∼75 nm. The increase of mean diameter was also confirmed by the NTA analysis (SI Appendix, Fig. S7). Meanwhile, for the mixture of exosomes and liposomes at a ratio of 1:2, only a few particles (less than 20) were detected. We attribute the differences to the net surface charge of the exosomes/liposomes complex after membrane fusion (SI Appendix, Fig. S8C).
The Hit-Stay-Run Behavior of Exosomes on Antibody-Coated Surfaces.
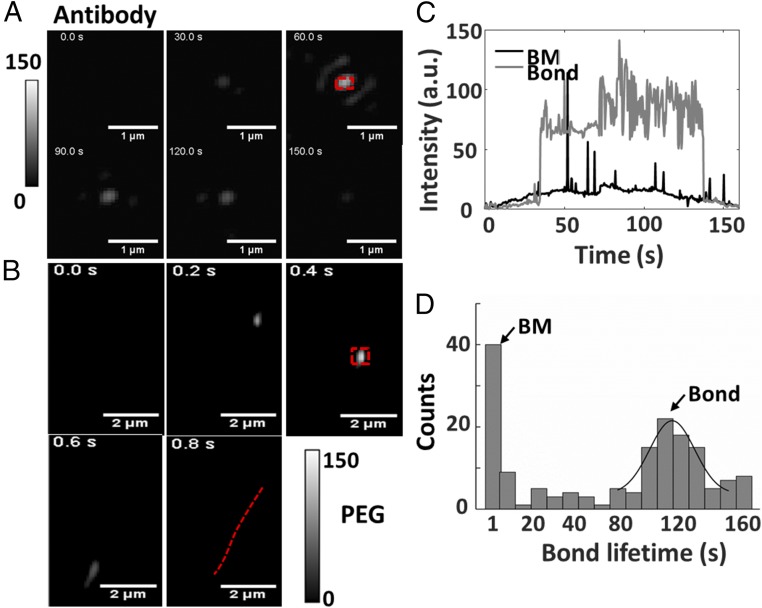
Measuring the adsorption of exosomes onto an antibody-coated surface enables the molecular profiling of exosomes with plasmonic sensors (17, 18). We examined the adsorption of exosomes onto a CD 63 antibody-coated Au surface as an example to explore the origin of the plasmonic signal at the single-exosome level. The behavior of exosomes differs greatly from that on a positive-charge-modified Au surface described above or a (1-Mercapto-11-undecyl)hexa(ethylene glycol) (PEG)-coated surface. On a PEG-coated surface, exosome nanoparticles were imaged to appear, move, and leave immediately, which is due to Brownian motion (BM) (Fig. 4B). This is reasonable because a PEG coating is well known to block nonspecific binding of exosomes (17). On an antibody-coated surface, an intermediate hit-stay-run behavior was observed. Most exosomes bonded to the antibody, stayed for a while, and left afterward (Fig. 4A and Movie S2), while some exosomes underwent BM similar to that on a PEG-coated surface. We attribute the distinct hit-stay-run behavior on the antibody-coated surface to the relatively weaker binding force on antibodies compared with that on the charged surface.
Fig. 4.
iPM imaging of exosome–antibody interaction. (A) Snapshots of exosomes bonded to CD 63 antibody-coated and (B) PEG-coated gold surface; the red dashed line shows the BM trajectory. (C) Real-time iPM signals of an exosome bonded to antibodies or that underwent BM. (D) Distribution of exosome–antibody bond lifetime measured on a CD 63-coated Au surface.
To quantitatively study the binding dynamics between exosomes and antibodies, real-time intensity profiles of the regions indicated in Fig. 4 A and B were plotted in Fig. 4C. Each peak on the BM profile represented a transient event. Owing to the evanescent wave nature of SPs, iPM intensity is quite sensitive to vertical direction, and thus a large fluctuation induced by small motion in the vertical direction was observed on the line profile of bond exosomes. We then determined the bond lifetime of each exosome on the surface as the duration of the plateau and the spikes on the line profiles. With further development, the bond lifetime may serve as a potential indicator for estimating the binding-energy landscape between exosomes and antibodies (28). A continuous distribution from less than 1 s to more than 160 s was obtained, with two obvious peaks (Fig. 4D). The peak at less than 1 s corresponds to BM, and the peak at ∼110 s describes the typical lifetime of the bond between an exosome and the CD63 antibodies. The relatively long bond lifetime was due to the multiple binding sites on the exosome surface to the antibodies.
Real-Time Tracking of Exosome Motion.
The imaging capability of iPM enables real-time tracking of individual exosomes, which is important for understanding the intra- and intercellular trafficking process of exosomes. The iPM image of exosome nanoparticles shows a normal distribution, and the central location can be accurately determined by fitting with a 2D Gaussian model (Fig. 5A). Fig. 5 B and C shows typical locations of an exosome bond to positive-charge-modified and antibody-coated Au surfaces in 1 min, respectively, and typical BM trajectories of exosome on a PEG-coated surface were shown in Fig. 5D. To quantify the difference, the mean velocity of exosome motion was calculated as the total distance traveled versus time of the trajectory (mean ± SD, n = 52, Fig. 5E) The difference in velocity was due to the external binding force exerted on exosomes from antibodies or the charge on Au surface, which confines the motion of exosomes.
Fig. 5.
Tracking and localization of single exosomes with iPM. (A) The 2D iPM intensity profile of an exosome, and its localization by 2D Gaussian fitting. (Scale bar: 300 nm.) (B) Localization of an exosome bond to a positive-charge-modified, and antibody-coated (C) Au surface. (D) BM trajectories of four exosomes on a PEG-coated Au surface. (E) mean velocity of exosomes on positive-charge-modified (Au), antibody-coated (Antibody), and PEG-coated (PEG) Au surfaces.
We anticipate that, with further development, the proposed iPM technology could be useful for exploring exosomes as a cancer biomarker for diagnostics and for evaluating tumor response to therapy in translational settings. Although the exosome samples in the current study were isolated from cell-line models after ultracentrifugation, direct analysis of clinical samples after simple filtration on plasmonic platforms is believed to be reasonable (17). While iPM offers exceptional sensitivity in dynamic analysis of single exosomes, the throughput for screening multiple clinical samples is yet to be optimized. Integration with microfluidic fluid control, micropatterning of antibodies, and full automation will be the key to pushing iPM toward successful translation into clinical applications (17, 18).
Conclusions
We have demonstrated iPM as an efficient analytical approach for visualizing and analyzing single exosomes. We optimize iPM sensitivity and spatial resolution to image single-exosome nanoparticles as small as 30 nm. The capability of iPM in sizing and tracking single exosomes enables the quantitative study of membrane fusion between exosomes and liposomes, as well as the exosome–antibody interaction. We discovered that exosomes’ behavior occurred in a unique hit-stay-run pattern on an antibody-coated surface, which offers a potential approach for estimating the binding force. With further development and validation, iPM could help explore fundamental questions about exosome-mediated intercellular communication and tumor microenvironments, and also be useful for identifying exosomes as cancer biomarkers for diagnostics and for exploring exosomes as targeted delivery systems. Technical modifications, including the use of fast cameras to improve temporal resolution (29) for analyzing rapid exosome-mediated processes, and the integration with fluorescence microscopy for multiplexing (30) will further extend the application of iPM.
Methods
iPM System.
The iPM system was built on an inverted microscope (Nikon Eclipse Ti-S) using a high-N.A. 100× oil immersion objective (N.A. = 1.49). The iPM chips were BK7 (VWR Corporation) glass coverslips coated with 2 nm of chromium and then 47 nm of Au by evaporation. Incident light from a 637-nm fiber-pigtailed laser (OBIS 637 nm LX FP 100 mW) was refocused and collimated, and illuminated the Au chip in the Kreschmenn configuration. A CMOS camera (Prime TM; Photometrics) was used with the system for image recording. Images were acquired at 512 × 512 pixels (full field of view: 33.28 × 33.28 µm2) and a frame rate of 320 fps. A motorized XY stage (Ludl Electronic Products, Ltd.) was incorporated on the microscope to translate the sensor chip.
iPM Image Processing.
Images recorded by the CMOS camera were running-averaged every 32 frames before digital reconstruction to minimize shot noise. Then the first image was subtracted from each image to remove background noise. The image reconstruction was performed in MATLAB (MathWorks) with self-developed codes. The wave vector of SPs was determined by calculating the center and radius of the ring in k space. Deconvolution was done in k space using the point-spread function obtained experimentally, by aligning 30 individual images of 100-nm silica nanoparticles to the maximum intensity point, and averaging after alignment.
Silica Nanoparticle Calibration.
Silica nanoparticles were purchased from MikroNano. The silica nanoparticles were diluted with PBS at 1:1,000 vol/vol, and ultrasonicated for 30 min for redispersion. For iPM calibration, 20 µL of nanoparticle solution was injected into the channel, and images were recorded immediately for 5 min. iPM intensity is determined as the mean intensity of a 3 × 3-pixel area around the maximum intensity pixel, after subtraction of mean background intensity.
Cell Culture and Exosomes Isolation.
Human lung cancer A549 cells (American Type Culture Collection, ATCC) were maintained in Dulbeco’s modified essential medium (HyClone; GE Healthcare) culture medium supplemented with exosome-depleted 10% FBS (Invitrogen, Thermo Fisher Scientific) and 1% (vol/vol) penicillin-streptomycin (Invitrogen, Thermo Fisher Scientific). Cancer cells were incubated in a humidified incubator (Thermo Fisher Scientific) at 37 °C with 5% CO2 for 72 h. We collected the cell culture medium by centrifugation at 4,000 × g for 10 min to remove the cells. The supernatant was further centrifuged at 10,000 × g for 10 min to remove cell debris. After filtering using a 0.22-μm membrane filter (Millipore), the supernatant was further ultracentrifuged at 100,000 × g for 90 min at 4 °C to retain the precipitated pellets of extracellular vesicles (EVs). The EV pellets were washed with 30 mL PBS once, and precipitated by a second ultracentrifugation at 100,000 × g for 90 min at 4 °C, and then supernatant was discarded. The purified EV was redispersed in 1× PBS solution (pH = 7.4) for use. Bio-TEM images were obtained by 120-kV TEM (Tecnai G2 SpritBiotwin). Size distribution was analyzed using a NanoSight NS300 instrument (Malvern Instruments Ltd.). The zeta potential was tested using Zetasizer ZSP (Malvern Instruments Ltd.).
Fluorescence Imaging of Exosomes.
To label the lipid membrane of exosomes, DiIC18(3) (AT Bioquest) was diluted with PBS to a concentration of 5 mM, and mixed with exosome samples at a ratio between DiIC18(3) molecules and exosomes of ∼100:1. A laser source at 561 nm (300 mW; Olympus Optical System) coupled with optical fiber was used for fluorescence excitation. To minimize out-of-focus fluorescence from medium, the excitation light was introduced at a highly inclined angle around 53° through the total internal reflection fluorescence module. Emitted fluorescence signals were collected with a 405-/488-/561-/640-nm filter set (Chroma). The exposure time of the CMOS camera was set to 5 s for optimized contrast. Pseudocolor was added to the fluorescent image with Image J software.
Surface Modification.
The Au chip was rinsed with deionized water and ethanol, and then blown dry with nitrogen gas. The chip was then further cleaned with hydrogen flame and immediately submerged into 10 mM HS-PEG-NH2 (Mr 10,000 Da; Nanocs) water/ethanol (1:1) solution for positive-charge modification, or 10 mM 3-Mercaptopropionic acid (Sigma-Aldrich) water/ethanol (1:1) solution for negative-charge modification, or 1 mM (1-Mercapto-11-undecyl)hexa(ethylene glycol) (Sigma-Aldrich) for PEG coating. After overnight incubation at room temperature, the chip was rinsed with deionized water and ethanol, and then blown dry with nitrogen gas.
Antibody Coating.
Mouse monoclonal CD63 antibody was purchased from NOVUS. The monoclonal antibody was bound onto the Au surface using a modified N-ethyl-N’-(3-(dimethylamino)propyl)carbodiimide (EDC) and N-hydroxysuccinimide (NHS) reaction (31). The Au surface was treated by trace oxygen plasma for 5 min, and then immediately immersed in 0.1 mg/mL mercapto acetic acid solution for carboxylation for 20 min. The chip was rinsed with PBS buffer, and then activated in 1 mL molar equivalent of NHS and EDC Mes buffer (pH = 5.5) solution for 30 min at 37 °C. Following the activation of carboxyl groups, the chip was incubated in 4 μg/mL of the Monoclonal CD63 antibody PBS buffer solution at 37 °C in the dark overnight. The chip was rinsed twice with PBS buffer to remove unbound antibody before use.
Preparation of Cationic Liposome Particles.
Cationic liposomes were prepared according to a method described elsewhere (32). N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium chloride (DOTAP) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were purchased from Lipids, GmbH. In brief, the dichloromethane mixture solution of 1 mg DOTAP and 1.2 mg DOPE was removed from the solvent, formed the lipid film with the help of rotary evaporation for 6–8 h at 30 °C, and the film was hydrated with PBS (1×, pH = 7.4) buffer with ultrasonication for 30 s. The resulting suspension was incubated at 37 °C for 1 h and extruded through a polycarbonate membrane with pore sizes of 100 nm (Whatman) 30 times.
Supplementary Material
Acknowledgments
The authors thank the Shanghai Pujiang Program (Grant 18PJ1406400), the National Natural Science Foundation of China (Grant 31771088), and the National Key Research and Development Program of China (Grants 2017YFC0107603 and 2017ZX10203205-006-002) for financial support.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Matlab codes for iPM image processing have been deposited on GitHub and are available at https://github.com/laoyxiaoy/iPM.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804548115/-/DCSupplemental.
References
- 1.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30:3.22.1–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 2.Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 3.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 4.Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 6.Tkach M, Théry C. Communication by extracellular vesicles: Where we are and where we need to go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 7.van der Pol E, et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12:1182–1192. doi: 10.1111/jth.12602. [DOI] [PubMed] [Google Scholar]
- 8.Chia BS, Low YP, Wang Q, Li P, Gao Z. Advances in exosome quantification techniques. TrAC Trends Anal Chem. 2017;86:93–106. [Google Scholar]
- 9.Koritzinsky EH, Street JM, Star RA, Yuen PST. Quantification of exosomes. J Cell Physiol. 2017;232:1587–1590. doi: 10.1002/jcp.25387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 11.Gardiner C, Ferreira YJ, Dragovic RA, Redman CWG, Sargent IL. Extracellular vesicle sizing and enumeration by nanoparticle tracking analysis. J Extracell Vesicles. 2013;2:19671. doi: 10.3402/jev.v2i0.19671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filipe V, Hawe A, Jiskoot W. Critical evaluation of nanoparticle tracking analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayley H, Martin CR. Resistive-pulse sensing-from microbes to molecules. Chem Rev. 2000;100:2575–2594. doi: 10.1021/cr980099g. [DOI] [PubMed] [Google Scholar]
- 14.Vogel R, et al. Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor. Anal Chem. 2011;83:3499–3506. doi: 10.1021/ac200195n. [DOI] [PubMed] [Google Scholar]
- 15.Orozco AF, Lewis DE. Flow cytometric analysis of circulating microparticles in plasma. Cytometry A. 2010;77:502–514. doi: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian Y, et al. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12:671–680. doi: 10.1021/acsnano.7b07782. [DOI] [PubMed] [Google Scholar]
- 17.Im H, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol. 2014;32:490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu L, et al. Label-free quantitative detection of tumor-derived exosomes through surface plasmon resonance imaging. Anal Chem. 2014;86:8857–8864. doi: 10.1021/ac5023056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zong S, et al. Single molecule localization imaging of exosomes using blinking silicon quantum dots. Nanotechnology. 2017;29:065705. doi: 10.1088/1361-6528/aaa375. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, et al. Visualization and intracellular dynamic tracking of exosomes and exosomal miRNAs using single molecule localization microscopy. Nanoscale. 2018;10:5154–5162. doi: 10.1039/c7nr08800k. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, et al. Label-free measuring and mapping of binding kinetics of membrane proteins in single living cells. Nat Chem. 2012;4:846–853. doi: 10.1038/nchem.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang S, et al. Label-free imaging, detection, and mass measurement of single viruses by surface plasmon resonance. Proc Natl Acad Sci USA. 2010;107:16028–16032. doi: 10.1073/pnas.1005264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu H, Shan X, Wang S, Chen H, Tao N. Plasmonic imaging and detection of single DNA molecules. ACS Nano. 2014;8:3427–3433. doi: 10.1021/nn4062885. [DOI] [PubMed] [Google Scholar]
- 24.Yu H, Shan X, Wang S, Chen H, Tao N. Molecular scale origin of surface plasmon resonance biosensors. Anal Chem. 2014;86:8992–8997. doi: 10.1021/ac501363z. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Shan X, Wang S, Tao N. Achieving high spatial resolution surface plasmon resonance microscopy with image reconstruction. Anal Chem. 2017;89:2704–2707. doi: 10.1021/acs.analchem.6b05049. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, et al. Cell-derived exosomes as promising carriers for drug delivery and targeted therapy. Curr Cancer Drug Targets. 2017;18:347–354. doi: 10.2174/1568009617666170710120311. [DOI] [PubMed] [Google Scholar]
- 27.Sato YT, et al. Engineering hybrid exosomes by membrane fusion with liposomes. Sci Rep. 2016;6:21933. doi: 10.1038/srep21933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans E. Probing the relation between force–lifetime–and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 29.Yu H, et al. Tracking fast cellular membrane dynamics with sub-nm accuracy in the normal direction. Nanoscale. 2018;10:5133–5139. doi: 10.1039/c7nr09483c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J, et al. A signal-amplifiable biochip quantifies extracellular vesicle-associated RNAs for early cancer detection. Nat Commun. 2017;8:1683. doi: 10.1038/s41467-017-01942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sam S, et al. Semiquantitative study of the EDC/NHS activation of acid terminal groups at modified porous silicon surfaces. Langmuir. 2010;26:809–814. doi: 10.1021/la902220a. [DOI] [PubMed] [Google Scholar]
- 32.Gao X, Huang L. Cationic liposome-mediated gene transfer. Gene Ther. 1995;2:710–722. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.