Significance
Engineered mouse models of acute leukemia are critical to understanding the biological mechanisms by which a primary oncogene induces disease. While PAX5 fusion proteins are considered primary oncogenic events in B-ALL, their role in leukemia development is ill-known due to the lack of animal models. This report provides a novel and accurate in vivo model for B-ALL induced by PAX5-ELN fusion protein that establishes a preleukemic phase and recapitulates the key features of human disease, including acquired mutations in genes of the JAK/STAT and RAS/MAPK pathways. This study is of general interest, as it allows a better understanding of the biological mechanism by which an oncoprotein perturbs normal B-cell development and leads to pathological B-ALL.
Keywords: B-cell acute lymphoblastic leukemia, engineered mouse models, PAX5 fusion proteins, leukemia initiation, oncogenic transformation
Abstract
PAX5 is a well-known haploinsufficient tumor suppressor gene in human B-cell precursor acute lymphoblastic leukemia (B-ALL) and is involved in various chromosomal translocations that fuse a part of PAX5 with other partners. However, the role of PAX5 fusion proteins in B-ALL initiation and transformation is ill-known. We previously reported a new recurrent t(7;9)(q11;p13) chromosomal translocation in human B-ALL that juxtaposed PAX5 to the coding sequence of elastin (ELN). To study the function of the resulting PAX5-ELN fusion protein in B-ALL development, we generated a knockin mouse model in which the PAX5-ELN transgene is expressed specifically in B cells. PAX5-ELN–expressing mice efficiently developed B-ALL with an incidence of 80%. Leukemic transformation was associated with recurrent secondary mutations on Ptpn11, Kras, Pax5, and Jak3 genes affecting key signaling pathways required for cell proliferation. Our functional studies demonstrate that PAX5-ELN affected B-cell development in vitro and in vivo featuring an aberrant expansion of the pro-B cell compartment at the preleukemic stage. Finally, our molecular and computational approaches identified PAX5-ELN–regulated gene candidates that establish the molecular bases of the preleukemic state to drive B-ALL initiation. Hence, our study provides a new in vivo model of human B-ALL and strongly implicates PAX5 fusion proteins as potent oncoproteins in leukemia development.
B-cell precursor acute lymphoblastic leukemia (B-ALL) is the most common pediatric cancer. B-ALL is characterized by a blockade of B-cell differentiation combined with an uncontrolled proliferation of blastic cells. Current chemotherapy is efficient at inducing long-term remission in childhood B-ALL, but the most common cause of treatment failure remains relapse that occurs in 15 to 20% of patients (1). The prognosis is even worse in adult B-ALL, as only 30% of adults achieve long-term disease-free survival (2).
B-cell development is initiated by the entry of hematopoietic progenitors into the B-cell lineage transcription program and the concomitant sequential rearrangement of Ig genes through V(D)J recombination, ultimately leading to the generation of immunocompetent plasma cells. B-cell development can be dissected into pre-pro-B, pro-B, pre-B, immature B, and mature B-cell populations corresponding to different stages of differentiation (3). PAX5 is critical from early stages of B-cell development up to mature B cells (4). B-cell differentiation is completely blocked at the pro-B stage in Pax5 knockout mice, revealing its importance for early B lymphogenesis (5). Indeed, PAX5 plays a critical role in B-cell lineage commitment by activating the transcription of B cell-specific genes such as CD19 and BLK and suppressing alternative lineage choices (6–8).
PAX5 is the main target of genetic alterations in B-ALL. Heterozygous deletions and loss-of-function mutations of PAX5 are found in more than one-third of human B-ALL (9–11). These alterations result in loss of PAX5 expression and impairment of DNA-binding activity and/or transcriptional activity of PAX5. PAX5 is also rearranged in 2.6% of pediatric B-ALL cases, being fused to various fusion partners (9, 12–14). PAX5 translocations have been associated with a blockade of B-cell differentiation, as illustrated by PAX5-ETV6 and PAX5-FOXP1, which fuse the PAX5 paired domain to ETV6 and FOXP1 transcription factors, respectively (15).
We previously reported the molecular characterization of a new chromosomal t(7;9)(q11;p13) translocation in two cases of adult B-ALL. This translocation juxtaposed the 5′ region of PAX5 and almost the entire sequence of elastin (ELN) (13). The resulting PAX5-ELN fusion protein had conserved the nuclear localization sequence (NLS) and the DNA-binding paired-box domain of PAX5, and could therefore act as a constitutive repressor of the residual wild-type PAX5. Indeed, similar to PAX5-ETV6 and PAX5-FOXP1, transient expression of PAX5-ELN displayed a dominant-negative effect on PAX5 by down-regulating the transcription of PAX5 target genes (11, 13, 16, 17). However, the generality of the dominant-negative effect was recently questioned in vivo (15).
A detailed and dynamic analysis of the PAX5-ELN effect on B-ALL initiation and transformation requires the availability of a mouse model that allows a B-cell restricted expression of PAX5-ELN. Here, we demonstrate, by using a knockin (KI) mouse line, that PAX5-ELN is an initiating oncogenic event that perturbs normal B-cell development by aberrantly expanding pro-B cells at the earliest steps of leukemogenesis. Nonetheless, complete B-ALL transformation induced by PAX5-ELN involved the acquisition of additional mutations and the deregulated expression of multiple genes. Our study provides novel insights into the functional role of PAX5-ELN as a potent oncoprotein in B-ALL development, and should help to elucidate the molecular mechanisms that underlie B-ALL initiation and transformation.
Results
Constitutive Expression of Human PAX5-ELN Leads to B-ALL Development.
To investigate the B-leukemic potential of PAX5-ELN, we developed a KI mouse model in which the cDNA encoding the human PAX5-ELN fusion protein was expressed in B-cell progenitors. This was achieved by inserting the human cDNA at the IgH locus under the control of a VH promoter (PVH) and the endogenous Eμ enhancer whose activity is triggered early in B-cell development (18). To avoid transcriptional readthrough from upstream promoters at different developmental stages, a pause/polyadenylation site (19) was added upstream of the ectopic PVH promoter (Fig. 1A and SI Appendix, Fig. S1). Unless otherwise indicated, the experiments have been performed on heterozygous mice.
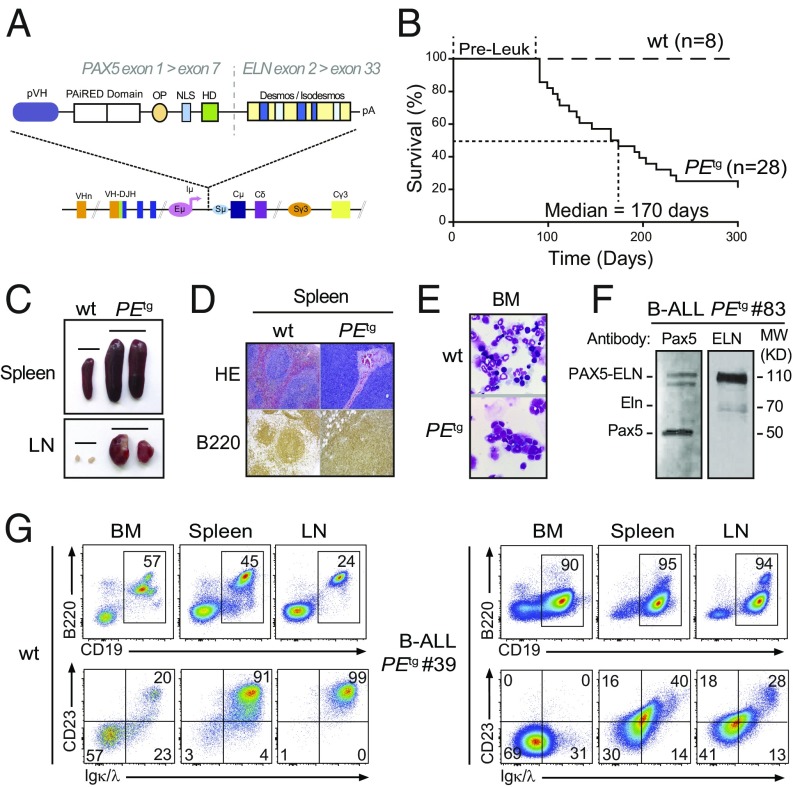
Fig. 1.
Human PAX5-ELN expression induces efficient B-ALL development. (A) Generation of knockin mouse model expressing PAX5-ELN fusion protein. The sequence encoding the human PAX5-ELN fusion protein was inserted downstream of Eμ enhancer. Desmos, Desmosine; HD, homeodomain; OP, octapeptide; pVH, VH gene promoter. (B) Kaplan–Meier curves of the time to leukemia for cohorts of PEtg mice (n = 28). WT mice (n = 8) were used as controls. Pre-Leuk, preleukemic time. (C–E) PAX5-ELN induces B-ALL development characterized by leukemic cell invasion in the bone marrow, spleen, and lymph nodes. (C) Pictures of spleens (Upper) and LNs (Lower) from WT and leukemic PEtg mice are shown. (D) Staining with hematoxylin and eosin (HE) and immunohistochemistry of B220 are shown of spleens from WT and leukemic PEtg mice. (E) Pictures of May-Grünwald–Giemsa–stained cytospin of BM cells from WT and leukemic PEtg mice. (F) Protein extract of leukemic cells from a B-ALL PEtg mouse (no. 83) was subjected to immunoblotting with anti–N-terminal Pax5 (N19) antibody for the detection of PAX5-ELN and endogenous Pax5 and with anti-ELN antibody for the detection of PAX5-ELN and endogenous Eln. (G) Total cells from the BM, spleen, and LNs of WT (Left) and leukemic PEtg (Right) mouse (no. 39) were immunophenotyped using the B220, CD19, CD23, and Igκ/λ markers.
Mutant mice that constitutively expressed PAX5-ELN (hereafter PEtg mice) efficiently developed leukemia with a penetrance of 80% at 300 d (Fig. 1B). Leukemic development was associated with a splenomegaly (Fig. 1C, Upper) characterized by a massive infiltration of B220+ cells and a dramatic perturbation of the spleen architecture (Fig. 1D). Moreover, PEtg mice developed a lymphadenopathy (Fig. 1C, Lower) and exhibited blast cells in the bone marrow (BM) (Fig. 1E). Importantly, although the expression of PAX5-ELN was driven by IgH regulatory sequences, immunoblot analysis of protein extracts with a PAX5 paired domain-specific antibody revealed that the abundance of PAX5-ELN was not higher than that of endogenous PAX5 (Fig. 1F). This observation indicates that the reported effects on PEtg mice cannot be ascribed to high expression levels of PAX5-ELN fusion protein. Immunophenotypic characterization showed that BM, spleen, and lymph nodes (LNs) of leukemic PEtg mice contained an aberrant proportion of CD19+ B cells associated with an aberrant and variable expression of CD23 and Igκ/λ markers (Fig. 1G and SI Appendix, Fig. S2).
Clonal Transformation and Collaborating Events with PAX5-ELN Oncoprotein.
The IgH variable region can broadly be divided into the VH domain, including the distal VHJ558 and the proximal VH7183 gene families, and the DHJH domain, comprising a dozen DH segments followed by four JH segments (Fig. 2A, Upper) (20). Assembly of the IgH variable region involves two recombination steps: first DH to JH, followed by VH to DHJH. To determine the rearrangement status of the IgH locus in leukemic cells, we performed a qPCR-based V(D)J recombination assay (21) on genomic DNA purified from blasts of five independent B-ALL mice (Fig. 2A, Upper). The data reveal prominent DHJH and VHDHJH rearrangements, involving both proximal and distal VH segments, in all B-ALL samples (Fig. 2A, Lower). These results indicate that PAX5-ELN fusion protein induces the clonal transformation of a B-cell progenitor that has already rearranged its IgH locus, and support the notion that PAX5-ELN acts as a potent B-ALL oncoprotein.
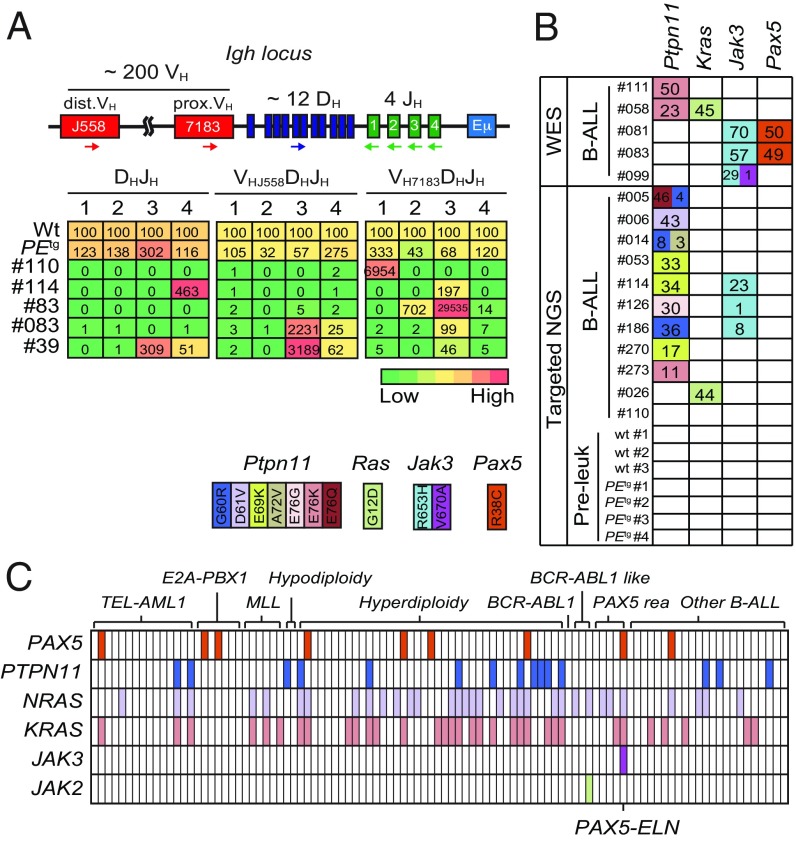
Fig. 2.
Clonal selection in PAX5-ELN–induced B-ALL. (A) Schematic of the mouse IgH locus (Upper). The locus is composed of ∼200 VH (red boxes), ∼12 DH (blue), and 4 JH (green) segments that undergo recombination in B-cell precursors to produce a functional VHDHJH unit. Genomic DNAs were prepared from purified PEtg pro-B cells and five leukemic PEtg mice and subjected to quantitative PCR to quantify DHJH and VHDHJH rearrangements using primers that bind the indicated gene segments. dVH, distal VH; pVH, proximal VH. PCR of the HS5 element downstream of the 3′ regulatory region was performed for normalization of DNA input. PCR was performed in triplicate. Quantification of DHJH and VHDHJH rearrangements is represented as a heat map (Lower). DNA from purified WT pro-B cells was used as a control and normalization (100% of the signal) for each gene rearrangement ranked in each column. (B) Recurrent mutations in B-ALL blasts induced by PAX5-ELN. Whole-exome sequencing (WES) was performed on BM cells from five leukemic PEtg mice. Mutations found on Ptpn11, Kras, Pax5, and Jak3 genes were verified and screened for recurrence by targeted next-generation sequencing (NGS) on BM cells from 11 leukemic, 3 WT, and 4 preleukemic 30-d-old PEtg mice. Each row represents a leukemia sample, and each column represents a genetic alteration. Colors indicate the position of the mutation, and numbers represent the variant allele frequency of each mutation. (C) Mutations on PTPN11, KRAS, NRAS, JAK3, JAK2, and PAX5 genes were screened for recurrence on 101 B-ALL patient samples. Each column represents a B-ALL sample classified according to the indicated oncogenic subtypes, and each row represents a genetic alteration. Colored boxes indicate the presence of a mutation, listed in Dataset S1.
Our observations on B-ALL transformation delay (>90 d) (Fig. 1B) led us to suspect potential acquisition of secondary mutations. To further identify these additional genetic alterations that potentially cooperate with PAX5-ELN, we performed whole murine exome sequencing in five PEtg leukemias and identified recurrent somatic mutations in Ptpn11, Kras, Jak3, and Pax5 genes. These mutations were also found after specific sequencing of these loci in an additional 11 PEtg leukemias (Fig. 2B). These findings strengthen the notion that aberrant activation of JAK/STAT and/or RAS/MAPK signaling pathways is required for an overt B-cell leukemia transformation in our model.
To address the critical question of the recurrence of these somatic mutations in human B-ALL, we performed the targeted sequencing of several exons of PAX5, PTPN11, NRAS, KRAS, JAK3, and JAK2 genes in a cohort of 101 pediatric B-ALL patients (Fig. 2C and Dataset S1). Importantly, we detected recurrent mutations in PAX5 (9/101), PTPN11 (15/101), NRAS (33/101), and KRAS (33/101) genes in all of the different B-ALL oncogenic subtypes (Fig. 2C). Interestingly, the patient cohort contained five PAX5-rearranged B-ALL patients, including one with the PAX5-ELN translocation that exhibited, just as in our mouse line, PAX5, NRAS, KRAS, and JAK3 mutations (Fig. 2C and Dataset S1). Together, the above data establish that our PAX5-ELN mouse model recapitulates the multistep pathogenesis of human B-ALL.
PAX5-ELN Oncoprotein Perturbs Early B-Cell Progenitor Cell Differentiation at Preleukemic Stage.
The survival curve of PEtg mice revealed a leukemia onset starting at 90 d after birth (Fig. 1B). This leukemia development latency allowed us to explore the effect of PAX5-ELN oncoprotein on the early steps of leukemogenesis. Cytological examination of the preleukemic PEtg mice confirmed the absence of blasts in the BM (SI Appendix, Fig. S3A) and any obvious perturbation of the main cell lineages (SI Appendix, Fig. S3B). Moreover, the absence of detection of Ptpn11, Kras, Pax5, and Jak3 mutations in the BM of preleukemic PEtg mice (Fig. 2B) allowed us to precisely address the role of PAX5-ELN in leukemia initiation, before the onset of clonal transformation induced by the acquisition of additional cooperating oncogenic events.
To identify the earliest cell types that are affected by PAX5-ELN oncoprotein, we analyzed B-cell progenitors (SI Appendix, Fig. S3C) in the BM of PEtg mice during early and late preleukemic stages, namely at 30 and 90 d after birth, respectively. We observed that PAX5-ELN significantly induced a three- to fourfold expansion of the pro-B population at 30 and 90 d, whereas pre-pro-B and pre-B populations were not affected (Fig. 3 A and B). Interestingly, 30-d-old PEtg pro-B cells exhibited a polyclonal profile for DHJH and VHDJH gene rearrangements that was roughly comparable to their WT counterparts (Fig. 2A), indicating that PAX5-ELN–induced pro-B cell expansion was not associated with a clonal selection at the preleukemic stage. In addition, this pro-B cell expansion was associated with a reduction of the immature and circulating B-cell populations in the BM (Fig. 3 A and B), indicating that PAX5-ELN partially blocked B-cell differentiation at the preleukemic stage. Accordingly, we observed a moderate but significant reduction of spleen size in preleukemic PEtg mice compared with age-matched WT mice (SI Appendix, Fig. S4A), suggesting that PAX5-ELN retained B-cell loading in peripheral lymphoid organs. Indeed, while the overall distribution of B-cell populations was unaffected by PAX5-ELN in the spleen (SI Appendix, Fig. S4B), the absolute number of immature and mature splenic B cells was significantly reduced in preleukemic PEtg mice (SI Appendix, Fig. S4C). The above results indicate that PAX5-ELN oncoprotein expands preleukemic pro-B cells at the expense of subsequent stages of maturation in vivo. Finally, we confirmed that PAX5-ELN altered the intrinsic capacity of differentiation of pro-B cells using in vitro functional approaches, and had a restricted expression and effect on the B-cell lineage (SI Appendix, Text and Figs. S5 and S6).
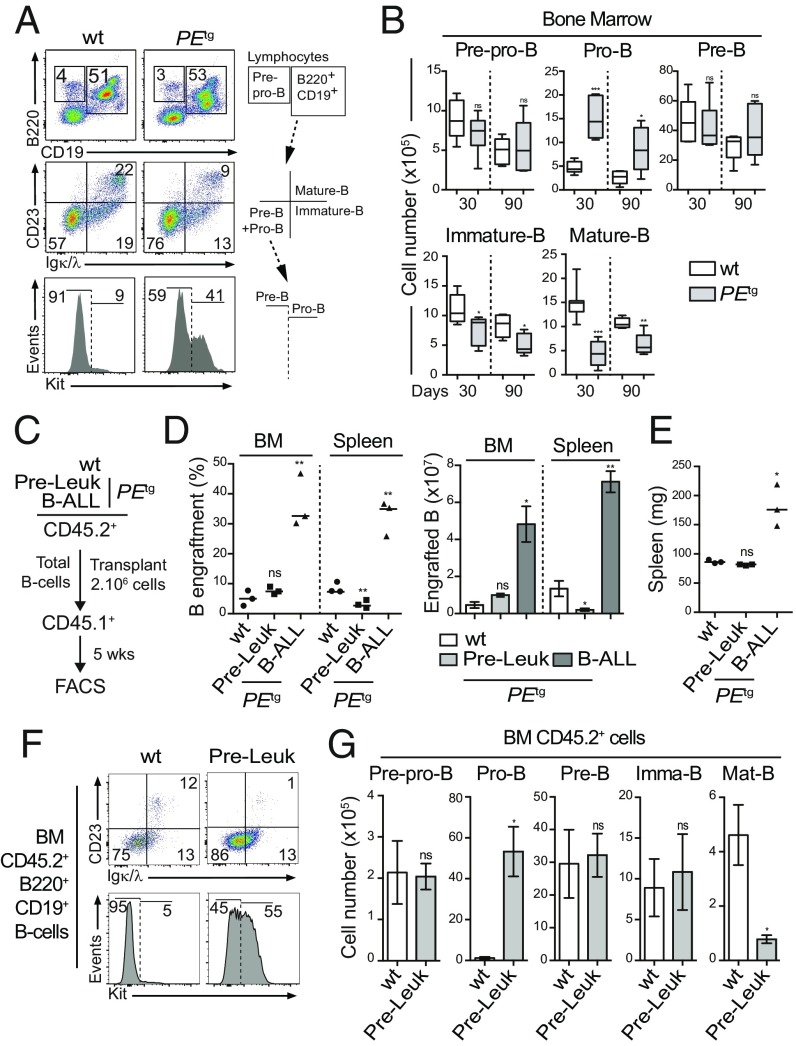
Fig. 3.
Pro-B cells are expanded by PAX5-ELN at a preleukemic phase. (A and B) Preleukemic PEtg pro-B cells exhibit an aberrant expansion potential in vivo. Immunophenotyping of BM cells from 30-d-old WT and PEtg mice was performed using B220, CD19, CD23, Igκ/λ, and Kit markers (A, Left), and a gating strategy was applied to discriminate B-cell subpopulations (A, Right and SI Appendix, Fig. S2). Absolute numbers of pre-pro-B, pro-B, pre-B, immature B, and mature B cells from the BM of 30- and 90-d-old WT and PEtg mice were calculated (B; n = 5 to 8 mice per condition). (C–E) Preleukemic and leukemic PEtg B cells exhibit different engraftment potentials. Total B cells from the BM of preleukemic and B-ALL PEtg mice (CD45.2+) were purified and i.v. transplanted in recipient mice (CD45.1+, n = 3, 2.106 cells per mouse) pretreated with 30 mg/kg busulfan (C). The same number of total B cells from the BM of WT mice was transplanted in parallel as control (C; n = 3). The proportion (D, Left) and absolute numbers (D, Right) of donor-derived (CD45.2+) B cells in the recipient BM were calculated 5 wk after transplantation. Spleen weights of engrafted mice were compared (E). (F and G) Preleukemic PEtg pro-B cells expand after transplantation. Immunophenotyping of engrafted B cells (CD45.2+B220+CD19+) in the BM was performed (F), and absolute numbers of donor-derived (CD45.2+) preleukemic pre-pro-B, pre-B, pro-B, immature B, and mature B cells in the recipient BM were calculated (G). Error bars represent standard deviations, ***P < 0.0005, **P < 0.005, and *P < 0.05; ns, nonsignificant.
PAX5-ELN Oncoprotein Confers an Aberrant Expansion Potential to Preleukemic Pro-B Cells.
We first evaluated the effect of PAX5-ELN on B-cell progenitor turnover using a total BM transplantation assay. The data strongly suggested that PAX5-ELN induced an aberrant expansion potential to preleukemic pro-B/pre-B progenitors before B-ALL transformation (SI Appendix, Text and Fig. S7 A–D). To strengthen this notion, we compared the reconstitution potential of WT, preleukemic, and leukemic PEtg B cells. Equal numbers of total B cells purified from the BM of WT, preleukemic, and leukemic PEtg mice were transplanted into congenic mice (Fig. 3C). Five weeks after transplantation, engraftment efficiencies of WT and preleukemic PEtg B cells were similarly low (Fig. 3D), and recipient mice did not exhibit splenomegaly (Fig. 3E). Interestingly, the analysis of engrafted B-cell compartments in the BM of recipient mice revealed an aberrant proportion of PEtg donor-derived pro-B cells specifically (65 ± 15-fold expansion) (Fig. 3 F and G and SI Appendix, Fig. S7E). This striking expansion was not associated with the acquisition of Ptpn11, Kras, Pax5, or Jak3 mutations (SI Appendix, Fig. S7F). In addition, consistent with our results in steady-state conditions, we observed a moderate but significant diminution of the engraftment efficiency of preleukemic PEtg B cells in the spleen compared with WT controls (Fig. 3D). This was due to a reduction of the number of donor-derived immature and mature B cells (SI Appendix, Fig. S7 G and H).
Thus, our transplantation experiments with preleukemic B cells provide additional evidence to support that PAX5-ELN induces an aberrant self-renewal activity to normal pro-B cells before clonal and malignant transformation. On the other hand, recipient mice transplanted with clonal leukemic PEtg cells exhibited a high level of engraftment in the BM (Fig. 3D) associated with an important blast invasion in the spleen (Fig. 3 D and E), and with the presence of Ptpn11 mutations (SI Appendix, Fig. S7F), characteristic of the spreading of a clonal population.
Combined, our functional approaches establish a sharp difference in the reconstitution potential between clonal and nonclonal B-cell populations in our model. Furthermore, they indicate that the pro-B cell compartment is abnormally expanded by PAX5-ELN oncoprotein at the preleukemic stage, and therefore represents the likely cellular target of leukemia initiation.
Gene Regulation by PAX5-ELN in Pro-B Cells.
Based on transient transfection assays, PAX5 fusion proteins including PAX5-ELN, PAX5-ETV6, and PAX5-FOXP1 were thought to act as dominant-negatives by affecting the transcriptional activity of WT PAX5 (11, 13, 16, 17). However, recent evidence in vivo revealed that PAX5-ETV6 or PAX5-FOXP1 fusion proteins marginally modified the expression of PAX5 target genes (15, 22). To address this hypothesis in our in vivo model, we first compared the gene expression profiles of purified pro-B cells from preleukemic PEtg mice and age-matched WT mice (SI Appendix, Fig. S8A). Transcriptome analysis identified 145 up-regulated and 49 down-regulated genes with an expression difference of more than 1.5-fold and an adjusted P value of <0.05 in PAX5-ELN–expressing pro-B cells (Fig. 4A and Dataset S2). In parallel, we established a list of ex vivo PAX5-modified genes by comparing the gene expression profiles of Pax5−/− embryonic liver (E17.5) pro-B cells retrovirally transduced with either MIE or MIE-PAX5. This strategy led to the identification of 174 PAX5–up-regulated and 438 PAX5-repressed genes (Fig. 4A and Dataset S3). Interestingly, the gene set enrichment analysis of a well-established list of in vivo PAX5-regulated genes, arising from in vivo gene expression profiles of murine Pax5+/+ and Pax5−/− pro-B cells (22), with our new ex vivo PAX5 gene signature demonstrates a global similarity between the two approaches (SI Appendix, Fig. S8B). We determined the overlap of our ex vivo PAX5-activated genes with PAX5-ELN–repressed genes. This analysis revealed that only one gene was activated by PAX5 and repressed by PAX5-ELN. Conversely, only 14 of the PAX5-repressed genes were activated by PAX5-ELN in pro-B cells (Fig. 4B and Dataset S4A). Closely similar results were observed when we used the well-established list of in vivo PAX5-regulated genes (22) (SI Appendix, Fig. S8C and Dataset S4B), further validating our approach. In addition, we observed by qPCR that ectopic expression of PAX5-ELN in preleukemic pro-B cells was not associated with the down-regulation of endogenous Pax5 and its two common target genes CD19 and CD79a (SI Appendix, Fig. S8D). Finally, we confirmed that the majority of the genes modified by PAX5-ETV6 and PAX5-FOXP1 in pro-B cells (15) did not overlap with our PAX5-regulated genes (SI Appendix, Fig. S8 E and F and Dataset S4 C and D). Together, similar to PAX5-ETV6 and PAX5-FOXP1 (15), our results indicate that, in vivo, PAX5-ELN does not generally antagonize the normal functions of PAX5 in preleukemic pro-B cells.
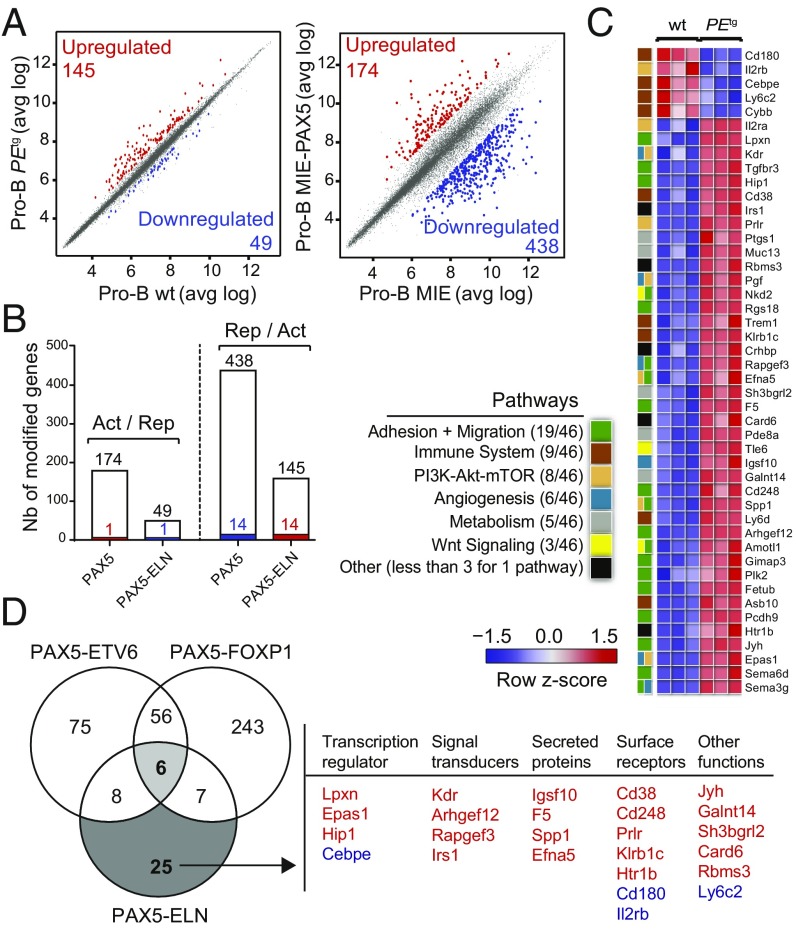
Fig. 4.
(A) Scatter plot of gene expression differences between in vivo purified WT and PEtg preleukemic pro-B cells (Left) and between ex vivo E17.5 fetal liver Pax5−/− pro-B cells transduced with either MIE-PAX5 or MIE retroviral vectors (Right) based on three independent microarray experiments. The normalized expression data of individual coding genes (indicated by dots) were plotted as the average log ratio (avg log). Up- and down-regulated genes with an expression difference of >1.5- and >3-fold, and an adjusted P value of <0.05, are colored in red or blue, respectively. (B) Absence of a general dominant-negative effect of PAX5-ELN on ex vivo PAX5-regulated genes in pro-B cells. Comparison of PAX5-activated and PAX5-ELN–repressed genes (Left) and of PAX5-repressed and PAX5-ELN–activated genes (Right) in preleukemic pro-B cells. Overlap indicates that one gene was activated by PAX5 and repressed by PAX5-ELN and 14 genes of the PAX5-repressed genes were activated by PAX5-ELN as represented by colored bars. (C) Heat map displaying the differential expression of PAX5-ELN–activated (red) and –repressed (blue) genes in WT (n = 3) and PEtg (n = 3) pro-B cells. The 46 PAX5-ELN–modified genes were selected on the basis of an expression difference of >2-fold (P < 0.05) and for encoding a protein implicated in one of the indicated pathways. The expression value of each gene is visualized according to the indicated scale. The pathway annotation is shown (Left). (D) Venn diagram indicating the overlap between PAX5-ETV6–, PAX5-FOXP1– (15), and PAX5-ELN–modified genes in preleukemic pro-B cells, selected for an expression difference of >3-, 3-, and 2-fold, respectively (Left). PAX5-ELN–modified genes that were included neither in PAX5-ETV6 nor in PAX5-FOXP1 signatures are listed according to their biological functions as transcriptional regulators, signal transducers, secreting proteins, and surface receptors. PAX5-ELN–activated and –repressed genes are indicated in red and blue, respectively.
To gain insight into the molecular regulation of PAX5-ELN in the early steps of B-ALL development, we focused on the genes presenting a twofold change of expression when comparing preleukemic PEtg with WT pro-B cells. Forty-one activated and five repressed PAX5-ELN genes were identified in preleukemic PEtg pro-B cells (Fig. 4C and Dataset S2). To identify shared molecular programs induced by PAX5-ETV6, PAX5-FOXP1, and PAX5-ELN, we overlapped the lists of their targets in preleukemic pro-B cells (15) (Fig. 4D). Notably, we found only six common genes, including genes encoding for a signal transducer (Gimap3), a secreted protein (Sema3g), and surface receptors (Il2rα and Trem1) (SI Appendix, Fig. S9A). In contrast, we identified 25 PAX5-ELN–modified genes, which were modified neither by PAX5-ETV6 nor by PAX5-FOXP1 (Fig. 4D). Interestingly, these genes encoded four signal transducers, four secreted proteins, and seven surface receptors but also the four transcriptional regulators Lpxn, Epas1, Hip1, and Cebpe (Fig. 4D).
The above results suggest that PAX5-ELN predominantly regulates an independent molecular program in preleukemic pro-B cells. To provide additional support to this notion, we categorized PAX5-ELN–modified genes according to pathways and identified activated (act) and repressed (rep) genes coding for proteins involved in the following processes: adhesion and migration (19 act), immune system (5 act, 4 rep), PI3K-mTOR signaling (7 act, 1 rep), angiogenesis (6 act), metabolism (5 act), and Wnt signaling (3 act) (Fig. 4C). Most of these pathways are strongly deregulated in PAX5-ETV6–induced B-ALL cells compared with their preleukemic counterparts (15), suggesting that PAX5-ELN activates at the preleukemic stage a molecular program required for B-ALL development in PAX5-ETV6tg mice. Interestingly, we identified six genes (Epas1, Arhgef12, Fetub, Rapgef3, Galnt14, and Cd248) activated by PAX5-ELN in preleukemic pro-B cells that were shown to be specifically activated in B-ALL PAX5-ETV6 cells (SI Appendix, Fig. S9B).
In conclusion, we have identified regulated PAX5-ELN genes in preleukemic pro-B cells with important functions in several signaling pathways and gene candidates that establish the molecular bases to drive B-ALL development.
Discussion
Genome-wide profiling identified the PAX5 gene as the most frequent target of somatic mutation in human B-ALL (11). Nonetheless, while PAX5 acts as a main tumor suppressor gene in B-ALL, B-cell development is normal in heterozygous Pax5+/− mice (5, 23), unless primed by chemical or retroviral mutagenesis (24), suggesting oncogenic cooperation for B-ALL transformation. Indeed, the tumor suppressor functions of Pax5 in B leukemogenesis were revealed under constitutive activation of STAT5, JAK1, and JAK3 (24, 25). PAX5 is also involved in reciprocal translocations leading to the fusion of its N-terminal domain with the C-terminal sequence of a second transcription factor such as ETV6 and FOXP1 (11, 26). In contrast with heterozygous PAX5 deletions, which are considered to have a secondary role during late stages of leukemogenesis, it is generally assumed that PAX5 fusion proteins act as primary oncogenic events altering normal B-cell development in the early steps of the disease (14). A wide diversity of fusion partners has been described (9, 14). Remarkably, they all conserve the N-terminal DNA-binding region of PAX5 and the NLS but lack the potent C-terminal transcriptional regulatory domains, suggesting that the PAX5 part of the chimeric protein is required for leukemogenesis.
The loss of PAX5 transcriptional domains in the chimeric proteins led to the notion that the fusion proteins might act as constitutive repressors, as shown in various reporter assays (11, 13, 16, 27). Consequently, it was inferred that B-ALL might arise as a result of an antagonizing effect of the fusion proteins on PAX5 activity. The situation may be more complex, as illustrated by the recent finding that PAX5-ETV6 and PAX5-FOXP1 do not actually modify the expression of PAX5 target genes but rather implicate pre-B cell receptor, migration, and adhesion signaling pathways (15). PAX5-ELN rearrangement is unusual because it does not involve a transcription factor but an extracellular matrix protein (13). Furthermore, PAX5-ELN is one of the chimeric proteins that conserves the largest part of PAX5. Although PAX5-ELN blocked the transcription of PAX5 target genes in transactivation assays (13), our results indicate that PAX5-ELN was able to repress only one of the PAX5-activated genes and to activate a small subset of the PAX5-repressed genes, strongly suggesting that PAX5-ELN does not generally antagonize the normal function of PAX5 in preleukemic pro-B cells. Thus, in line with PAX5-ETV6 and PAX5-FOXP1 models (15), our in vivo data do not support the generality of a dominant-negative mechanism for PAX5 fusion proteins.
Several hypotheses could be put forward to account for this discrepancy. One possible explanation could be merely quantitative. In heterozygous mice, thus mimicking the situation in patients, the concentration of the fusion protein may be below the threshold required to exert a dominant-negative effect, whereas this threshold may be readily achieved in transient reporter assays. As mentioned above, expression of a small subset of PAX5-regulated genes was impacted by PAX5-ELN, suggesting that there are instances in vivo where the dominant-negative effect is exerted by the oncoprotein. Alternatively, though not mutually exclusively, fusion proteins may be subjected to different regulatory mechanisms (half-life, posttranslational modifications, interacting partners, etc.) in vivo from the time lapse of transient expression. In this context, it should be noted that although the expression of PAX5-ELN cDNA was driven by the powerful IgH regulatory elements, PAX5-ELN levels were not higher than endogenous PAX5 levels, excluding an inappropriate ectopic expression of the fusion protein in our model. Importantly, although the insertion of Pax5 into the IgH locus led to T lymphomas (28), the weak expression of PAX5-ELN in thymocytes did not perturb T-cell development, underlying the restricted effect of PAX5-ELN expression on the B-cell lineage.
Interestingly, while PAX5-ETV6 and PAX5-FOXP1 arrested lymphopoiesis at the pro-B/pre-B cell transition, they did not induce leukemia on their own in the corresponding mutant mice (15), suggesting the requirement of cooperating mutations for B-ALL transformation. Specifically, in contrast to the PAX5-ETV6 mouse model that required oncogenic cooperation (i.e., loss of the Cdkn2a/b tumor suppressor locus) for leukemia development (15), PAX5-ELN efficiently induced B-ALL in mice. In this context, most deregulated pathways in preleukemic PAX5-ELN pro-B cells were also strongly deregulated in PAX5-ETV6–induced B-ALL cells. Further analysis revealed that PAX5-ELN predominantly regulated an independent molecular program in preleukemic pro-B cells, in which activation of six genes, also found activated in B-ALL PAX5-ETV6 cells (15), may explain the stronger potential of PAX5-ELN to induce B-ALL compared with PAX5-ETV6.
B cell-specific, constitutive expression of PAX5-ELN led to clonal transformation associated with the acquisition of additional mutations in key components of the JAK/STAT and RAS/MAPK pathways. These pathways represent two common targets of somatic mutations in other oncogene-induced B-ALL mice (29, 30) and in human B-ALL (11), including PAX5-ELN B-ALL cases (31, 32). Our analysis of a B-ALL patient cohort revealed the presence of PAX5, PTPN11, NRAS, KRAS, and JAK3 mutations within different B-ALL oncogenic subtypes including PAX5-rearranged leukemias and a PAX5-ELN B-ALL case.
This study clearly demonstrated the effect of PAX5-ELN on leukemia initiation at the preleukemic stage, before malignant transformation. The partial blockade of differentiation induced by PAX5-ELN was associated with an aberrant expansion of pro-B cells, which was also revealed by transplantation assays. Importantly, this proliferative advantage was not associated with an obvious clonal selection and therefore suggests that PAX5-ELN confers self-renewal properties to pro-B cells, a situation that is reminiscent of the E2A-PBX1 mouse model (30). This observation supports the view that aberrant self-renewal activity of lymphoid progenitors induced by a primary oncogene can be an initiating event in leukemia development. This establishes a preleukemic stage setting genetic instability and accumulated genetic alterations that cooperate to lead to fully transformed B-ALL (33). Emerging evidence suggests that primary genetic alteration can convert normal committed progenitors into preleukemic stem cells by reprogramming aberrant self-renewal properties both in lymphoid and myeloid lineages (33–35). Importantly, recent findings proposed that long-lasting preleukemic stem cells were hidden within the bulk of leukemic cells, and served as a reservoir for disease relapse (36, 37). An important challenge is to identify and target these leukemic initiating cells that represent an extremely rare subpopulation in patients. In this respect, the availability of the PAX5-ELN mouse model should be valuable. Additionally, by recapitulating the different steps of the human disease, the model represents a major opportunity to develop new therapeutic strategies on both B-ALL initiation and transformation, and a robust and reproducible tool for preclinical studies of drug screening and development.
Experimental Procedures
Generation of the PAX5-ELN knockin mouse model, FACS analysis, Ig rearrangement assay, whole-exome sequencing and specific resequencing, transplantation assay, microarray experiment, retroviral transduction, coculture of pro-B cells, RT-PCR, Western blot, and statistical analysis are described in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge the cytometry and cell-sorting facility of the CRCT (INSERM U1037) and the IPBS animal facility for technical assistance. We are grateful to Manon Farcé and Laetitia Ligat from the facilities of the CRCT for assistance with flow cytometry and microscopy, respectively. We thank the Anexplo/Genotoul platforms for technical assistance (UMS006 and histology department). This research was supported by Institut National du Cancer (INCa) R11190BB-INCA 2011-131-PAX5, ARC Grant SFI20101201888, and FEDER Grant CITTIL. G.C., N.R., C.C., and N.S.N.H. were supported by INCa Grant R11190BB-INCA 2011-131-PAX5. The team is supported by association Laurette FUGAIN, Ligue nationale contre le cancer, “111 des arts,” Association Capucine, Société Française des Cancers de l’Enfant, and la région Occitanie. Work in the A.A.K. lab is supported by INCa, ANR, and Fondation ARC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721678115/-/DCSupplemental.
References
- 1.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: An update. J Clin Oncol. 2011;29:551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 3.Hardy RR, et al. B-cell commitment, development and selection. Immunol Rev. 2000;175:23–32. [PubMed] [Google Scholar]
- 4.Adams B, et al. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 5.Urbánek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 6.Souabni A, Cobaleda C, Schebesta M, Busslinger M. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 2002;17:781–793. doi: 10.1016/s1074-7613(02)00472-7. [DOI] [PubMed] [Google Scholar]
- 7.Nera KP, et al. Loss of Pax5 promotes plasma cell differentiation. Immunity. 2006;24:283–293. doi: 10.1016/j.immuni.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Nutt SL, Heavey B, Rolink AG, Busslinger M. Pillars article: Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999. 401: 556–562. J Immunol. 2015;195:766–772. [PubMed] [Google Scholar]
- 9.Nebral K, et al. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:134–143. doi: 10.1038/leu.2008.306. [DOI] [PubMed] [Google Scholar]
- 10.Familiades J, et al. PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploinsufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: A GRAALL study. Leukemia. 2009;23:1989–1998. doi: 10.1038/leu.2009.135. [DOI] [PubMed] [Google Scholar]
- 11.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 12.Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: A master regulator of B cell development and leukemogenesis. Adv Immunol. 2011;111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2. [DOI] [PubMed] [Google Scholar]
- 13.Bousquet M, et al. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood. 2007;109:3417–3423. doi: 10.1182/blood-2006-05-025221. [DOI] [PubMed] [Google Scholar]
- 14.Coyaud E, et al. Wide diversity of PAX5 alterations in B-ALL: A Groupe Francophone de Cytogenetique Hematologique study. Blood. 2010;115:3089–3097. doi: 10.1182/blood-2009-07-234229. [DOI] [PubMed] [Google Scholar]
- 15.Smeenk L, et al. Molecular role of the PAX5-ETV6 oncoprotein in promoting B-cell acute lymphoblastic leukemia. EMBO J. 2017;36:718–735. doi: 10.15252/embj.201695495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurahashi S, et al. PAX5-PML acts as a dual dominant-negative form of both PAX5 and PML. Oncogene. 2011;30:1822–1830. doi: 10.1038/onc.2010.554. [DOI] [PubMed] [Google Scholar]
- 17.Kawamata N, Pennella MA, Woo JL, Berk AJ, Koeffler HP. Dominant-negative mechanism of leukemogenic PAX5 fusions. Oncogene. 2012;31:966–977. doi: 10.1038/onc.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp DJ, Harris AW, Cory S, Adams JM. Expression of the immunoglobulin C mu gene in mouse T and B lymphoid and myeloid cell lines. Proc Natl Acad Sci USA. 1980;77:2876–2880. doi: 10.1073/pnas.77.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haddad D, et al. Sense transcription through the S region is essential for immunoglobulin class switch recombination. EMBO J. 2011;30:1608–1620. doi: 10.1038/emboj.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 21.Braikia FZ, Chemin G, Moutahir M, Khamlichi AA. Quantification of V(D)J recombination by real-time quantitative PCR. Immunol Lett. 2014;162:119–123. doi: 10.1016/j.imlet.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Revilla-I-Domingo R, et al. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–3146. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 24.Dang J, et al. PAX5 is a tumor suppressor in mouse mutagenesis models of acute lymphoblastic leukemia. Blood. 2015;125:3609–3617. doi: 10.1182/blood-2015-02-626127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heltemes-Harris LM, et al. Ebf1 or Pax5 haploinsufficiency synergizes with STAT5 activation to initiate acute lymphoblastic leukemia. J Exp Med. 2011;208:1135–1149. doi: 10.1084/jem.20101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cazzaniga G, et al. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001;61:4666–4670. [PubMed] [Google Scholar]
- 27.Fazio G, et al. PAX5/ETV6 alters the gene expression profile of precursor B cells with opposite dominant effect on endogenous PAX5. Leukemia. 2013;27:992–995. doi: 10.1038/leu.2012.281. [DOI] [PubMed] [Google Scholar]
- 28.Souabni A, Jochum W, Busslinger M. Oncogenic role of Pax5 in the T-lymphoid lineage upon ectopic expression from the immunoglobulin heavy-chain locus. Blood. 2007;109:281–289. doi: 10.1182/blood-2006-03-009670. [DOI] [PubMed] [Google Scholar]
- 29.van der Weyden L, et al. Somatic drivers of B-ALL in a model of ETV6-RUNX1; Pax5(+/−) leukemia. BMC Cancer. 2015;15:585. doi: 10.1186/s12885-015-1586-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duque-Afonso J, et al. Comparative genomics reveals multistep pathogenesis of E2A-PBX1 acute lymphoblastic leukemia. J Clin Invest. 2015;125:3667–3680. doi: 10.1172/JCI81158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullighan CG, et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc Natl Acad Sci USA. 2009;106:9414–9418. doi: 10.1073/pnas.0811761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denk D, Bradtke J, König M, Strehl S. PAX5 fusion genes in t(7;9)(q11.2;p13) leukemia: A case report and review of the literature. Mol Cytogenet. 2014;7:13. doi: 10.1186/1755-8166-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong D, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–339. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 34.Gerby B, et al. SCL, LMO1 and Notch1 reprogram thymocytes into self-renewing cells. PLoS Genet. 2014;10:e1004768. doi: 10.1371/journal.pgen.1004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krivtsov AV, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- 36.Shlush LI, et al. HALT Pan-Leukemia Gene Panel Consortium Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333, and erratum (2014) 508:420. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.