Significance
Exploring the action of G-protein–coupled receptors (GPCRs) is a problem of major fundamental and practical importance. Hence, it is crucial to understand the free-energy landscape of the activation of a GPCR. However, at present it is practically impossible to explore this issue by all-atom simulations. The current work uses a coarse-grained (CG) model to explore the nature of the relevant landscape and the resulting GPCR activation. We explore the energetic determinants of the allosteric mechanism of GPCR activation, shedding light on the structure/activity relationship of the system. This advance can serve as a basis for further progress in understanding GPCRs and related systems. Furthermore, it demonstrates the utility of CG approaches in describing and analyzing large and complex biological systems.
Keywords: GPCR, β2AR, signal transduction, G protein
Abstract
G-protein–coupled receptors (GPCRs) are a large group of membrane-bound receptor proteins that are involved in a plethora of diverse processes (e.g., vision, hormone response). In mammals, and particularly in humans, GPCRs are involved in many signal transduction pathways and, as such, are heavily studied for their immense pharmaceutical potential. Indeed, a large fraction of drugs target various GPCRs, and drug-development is often aimed at GPCRs. Therefore, understanding the activation of GPCRs is a challenge of major importance both from fundamental and practical considerations. And yet, despite the remarkable progress in structural understanding, we still do not have a translation of the structural information to an energy-based picture. Here we use coarse-grained (CG) modeling to chart the free-energy landscape of the activation process of the β-2 adrenergic receptor (β2AR) as a representative GPCR. The landscape provides the needed tool for analyzing the processes that lead to activation of the receptor upon binding of the ligand (adrenaline) while limiting constitutive activation. Our results pave the way to better understand the biological mechanisms of action of the β2AR and GPCRs, from a physical chemistry point of view rather than simply by observing the receptor’s behavior physiologically.
G-protein–coupled receptors (GPCRs) are responsible for the majority of cellular responses to a plethora of different cues, ranging from hormones and neurotransmitters to external agents like light and smell. As such, they are at the entry point of countless processes, particularly in complex multicellular organisms (1). Therefore, GPCRs are important in understanding molecular determinants of human diseases, emphasized by considering that 40–50% of pharmaceutical drugs target them (2, 3), and more novel therapeutics await discovery as our knowledge of these versatile receptors grows.
GPCR functionality is highly diverse because a single receptor molecule can be activated to elucidate different physiological responses mediated by different ligands or G-protein partners. The extracellular domain of GPCRs can bind to agonists, neutral agonists, reverse agonists, or antagonists, and translate this information to the cytoplasmic domain by activating, neutrally perturbing, or deactivating the action of heterotrimeric G proteins, which leads to different downstream signaling events (4, 5). Additional layers of complexity come in the form of the (mostly unexplored) potential oligomerization of the receptors, the allosteric modulators that increase or decrease the affinity of the receptor to ligands [where some allosteric agents affect different orthosteric targets differently (6)] as well as from the activation of G-protein independent pathways (e.g., β-arrestin–mediated) of signal transduction (7).
The spectacular advances in structural and biochemical studies of the relevant complexes of GPCRs with G proteins or other signaling molecules (8–10) give hope for advances in the detailed understanding of the activation process and the involvement of all partners. Specifically, the human β2 adrenoreceptor (β2AR)-Gs complex (11, 12) provides a very promising starting point for modeling studies. Indeed, molecular dynamics (MD) simulation studies of GPCRs have provided interesting insights on the ligand binding and allosteric activation processes (13, 14). However, despite having a detailed structural understanding of GPCR-mediated allosteric activation, we still do not have a computational description of the actual free energies of the different conformations involved in this process. Thus, one of the most important features that has not been explored by computational studies is the nature of the relevant free-energy landscape. The usefulness of having a reasonable free-energy landscape for the action of biological systems has been demonstrated in many previous works by us (15, 16) and other groups (e.g., refs. 17 and 18). The potential of applying energy landscape considerations to GPCRs has been discussed recently by Deupi and Kobilka (19) and exploiting such a concept by realistic free-energy calculations should be of great importance.
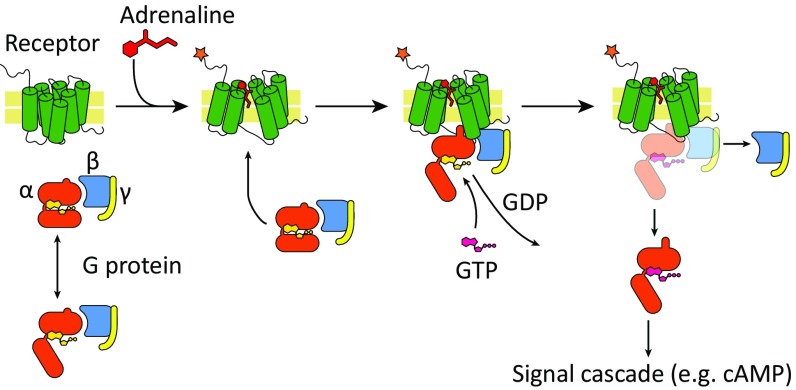
The general mode of activation of GPCRs is now well understood on a phenomenological level, as is illustrated in Fig. 1. As depicted in the figure, the activation process starts upon binding of an agonist, which shifts the allosteric equilibrium to the active form of the receptor, opening a cavity on its cytoplasmic face. Whether a G protein is prebound to the receptor or binds as a consequence of the activation, the next step is a change in the conformation of the G protein (entailing separation of the two major domains of the α-subunit, and penetration of a helical segment, termed α5, into the cavity of the activated receptor) which facilitates an exchange of GDP with GTP [the rate-limiting step in G-protein activation (20)]. The G protein with the bound GTP dissociates subsequently into the α-subunit and βγ-heterodimer, which elicit downstream signal transduction within the cell (21). Despite the above general information, it is important to move to a more detailed description in terms of the relevant free-energy landscape.
Fig. 1.
Schematics of the GPCR-activation pathway. The receptor is activated upon binding of agonist (adrenaline in the case of the β2AR), while the G protein is bound to GDP (studies showing conversion between the conformations of free G protein are mentioned in the main text). G protein binds to the activated receptor, undergoes the conformational changes required to facilitate exchange of GDP with GTP. The G protein is bound to GTP and then dissociates into the α-subunit and βγ-heterodimer, which elicit downstream signal transduction within the cell (e.g., increase or decrease of cAMP through activation or inhibition of adenylyl cyclase, respectively).
Considering the overwhelming complexity of GPCRs, it seems to us that the most effective current way of exploring the activation landscape is to start with coarse-grained (CG) modeling, that can produce an approximated landscape and would allow one to explore key issues. Thus, it may be very useful to construct an initial energy landscape for the activation of β2AR, using our electrostatics-based CG model (22, 23) and related approaches (24). Our approaches have been effective in exploring problems similar to β2AR activation, including conformational landscapes of molecular machines [e.g., ATPase (15) and myosin (16)]. Our approach also allowed some advances in exploring the action of exchange factors (e.g., CDC25) on Ras (25), where we made some progress using mutation experiments even without direct structural information about CDC25.
In this work, we began by looking at the very core components of the signal transduction pathway, including in our modeling the receptor and the G protein (considering the agonist implicitly). We successfully calculated an energy landscape that agrees with experimental results, in terms of the conformational selection and subsequent events (GDP-release and signal transduction). This work also forms a foundation for future studies, which should be fruitful as more components are added and a complete model for all known partners in the GPCR-signaling system—including oligomerization, allosteric modulators, and so forth—are constructed.
The present study is also used as a deductive tool, clarifying the importance of the landscape concept and a warning from the use of the “soft” implication of the presumed importance of dynamical effects in GPCR and related systems. Because, in fact, the protein motions are simply determined by the free-energy landscape, rather than determining the biological action.
Results and Discussion
Building Structural Models for Limiting Cases.
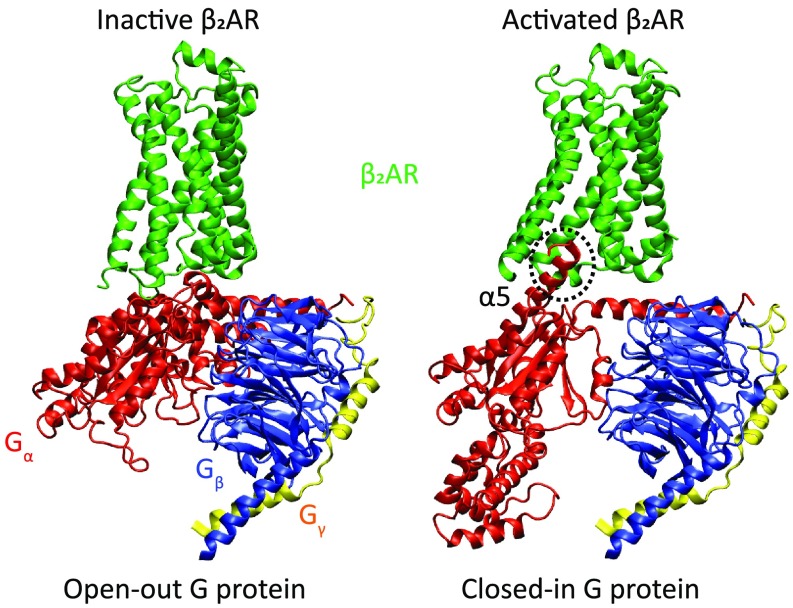
The structural models for this study were based on existing crystal structures of the β2AR and G protein. The structure of the activated state, bound to open G protein (Bos taurus heterotrimer Gs) was based on PDB ID code 3SN6 (11), whereas the structure of the inactive receptor was based on PDB ID code 2RH1 (26). The α-subunit of Gs (abbreviated Gα herein) is composed of two main domains: the Ras-like domain (RD) and the α-helical domain (AHD) (SI Appendix, Fig. S1), and the two domains undergo a very large separation upon binding to an activated GPCR (11, 27). The “open” conformation was taken from PDB ID code 3SN6, whereas the closed conformation was based on PDB ID code 1GP2 (28). The missing loops were constructed based on existing segments from the available structures, as mentioned above, by comparing to the simulation results of Dror et al. (29) and de novo design. The different assemblies were constructed by aligning the various subunits to existing complexes, with minor modifications and atomistic MD simulations to avoid clashes. Finally, the C terminus of the α-subunit (often referred to as α5) was modeled in two conformations, termed “in” and “out” herein [standing for in the activated receptor cavity and out of the receptor cavity; described as disorder-to-order for out-to-in elsewhere (30)], based on the original conformation where α5 is bound to the receptor (i.e., PDB ID code 3SN6) and the structure in PDB ID code 5JS8 (31) (where α5 is free, and consequently partially disordered), respectively (see Fig. 2 for representative configurations). All assemblies were energy-minimized and simulated inside a membrane grid and embedded in a water sphere. Finally, the all-atom protein structures were converted to our CG model (SI Appendix, Fig. S2), and the total energy of the system was computed as previously described (22, 23). The relative free energies of all of the states and corresponding transitions are presented in Fig. 3.
Fig. 2.
Protein structures of representative assemblies (the selection criterion was visual clarity) used in this study (see main text and Materials and Methods for details). The receptor is shown in green, and the G protein α-, β-, and γ-subunits are shown in red, blue, and yellow, respectively. The G-protein α-subunit α5 helix mentioned in the text is labeled and its C terminus, which undergoes a disorder-to-order transition, is marked with a dashed circle (where it is ordered, or “in”). Note that the specific complex in PDB ID code 3SN6 is the activated receptor bound to open-in G protein, which is not shown here.
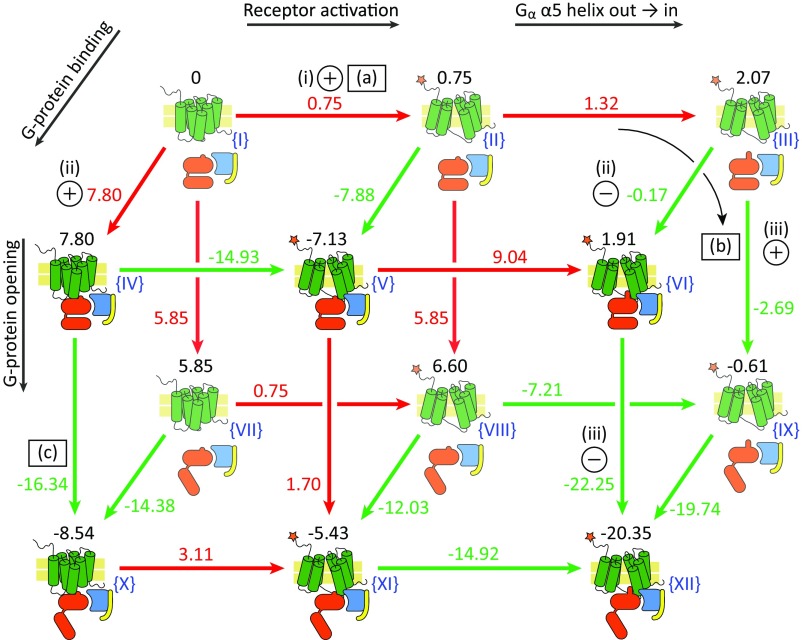
Fig. 3.
The CG energies of all conceivable configurations of the GPCR–G-protein system explored in this work. The total relative energy of each configuration is marked above the cartoon (in kilocalories per mole) and Roman numbers in blue are used to label each configuration. The receptor is shown as green cylinders, with an orange star highlighting the activated conformation. The G-protein α-, β-, and γ-subunits are shown in red, blue, and yellow, respectively. The arrows are shown in red and green for an endergonic or exergonic reaction, respectively. The circles with “+” or “–” in them indicate conditions (for endergonic and exergonic, respectively) that were used to validate the model, as mentioned in Exploring the Free-Energy Landscape. Discrepancies between published results and our calculations are marked with the letters (a), (b), and (c) in squares, and are discussed in Exploring the Free-Energy Landscape. See SI Appendix, Fig. S1 for the relationship between the structures and the cartoons drawn here. The reaction coordinates are described with black arrows; see Exploring the Free-Energy Landscape for more details.
Experimental studies of rhodopsin (a class A GPCR like the β2AR) suggest that the free receptor samples all of the functional conformations and that there are no novel conformations that are being formed upon G-protein binding (32). It is reasonable to assume that a similar case is true for the β2AR, and thus the conformations found in the structures above probably cover the relevant conformational space, with and without the bound G-protein partners.
The binding free energy of the agonist was taken as −8.2 kcal/mol based on the experimental affinity of adrenaline (33), assuming for simplicity that the agonist binds the active conformation with a much higher affinity than the inactive conformation (in general, binding energies for agonists and drugs can be carefully calculated using computational methods) (e.g., ref. 34). The energetics of the GDP and GTP binding was estimated by the protein dipole Langevin dipole semimacroscopic version (PDLD/S) with its linear response approximation version (the PDLD/S-LRA model) (24, 35), which proved successful in many cases (e.g., refs. 36 and 37).
Although studies showed that the receptor can assume several distinct active conformational states (38, 39), we focus here on only one active conformation of the receptor, because this work deals only with the canonical G-protein activation pathway.
Exploring the Free-Energy Landscape.
In studies of biological landscapes, it is often most instructive to look at the “end-points” or “asymptotic points” of the overall landscape (this provides the limits for many possible mechanistic options). In the β2AR system, the most functionally dominant states are: (i) inactive receptor, unbound to agonist (regardless of G-protein state); and (ii) active receptor, bound to an agonist, and bound to a G-protein trimer. Then state (ii) contains two substates: closed (bound to GDP) and “open-in” (ready to release GDP). These limits provide the guide for exploring the asymptotic states (Fig. 3). A dominant feature in the conformational changes of the G-protein α-subunit is the position of the C-terminal α5 helix. Here we use the term “in” for the case when α5 has moved into the binding pocket of the receptor and is more helical (or the same conformation in the absence of a receptor) or “out” for the case when it is outside the binding pocket and somewhat disordered (see SI Appendix, Fig. S1 for more details). It is worth noting that the movement of α5 entails motion in the connected β6 structural element, and the joint movement of the α5-β6 region was experimentally shown to be involved in GDP release (40, 41).
Fig. 3 presents the CG free energies calculated for the end-points, exploring four coordinates: (i) receptor conformations, using active and inactive states; (ii) Gα α5 state, using out and in (disordered and ordered, respectively) conformations; (iii) G-protein α-subunit RD and AHD relative orientation, using the closed and open conformations as the two end-points; and (iv) whether the G protein is bound to the receptor or not. Fig. 3 presents coordinates (i) and (ii) sequentially on the horizontal axis, coordinate (iii) on the vertical axis, and coordinate (iv) as the “depth” axis. The coordinates are also labeled on Fig. 3. In addition to the end-point energies, the ΔG differences between the configurations are calculated and shown in Fig. 3 (on the appropriate arrows). Hence, Fig. 3 allowed us to trace how the different states interconvert, and what was the change in free energy. We wish to emphasize that Fig. 3 presents energies calculated in the absence of agonist, and the landscape in the presence of the agonist (a parametric addition to the energy; see above) is depicted in Figs. 4 and 5.
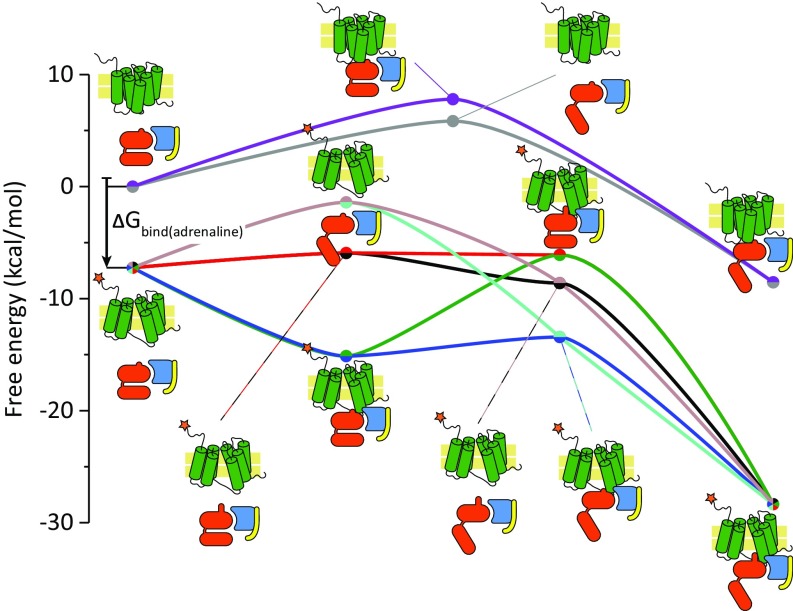
Fig. 4.
Cross-sections of the CG energy landscape. The energies of the receptor and G-protein states, following several linear paths, are depicted by colored lines accompanied by cartoons adjacent to each point (shown as circles). The vertical black arrow denotes the difference in energy associated with binding of the adrenaline agonist. The purple and gray curves (top) denote inactive receptor. The top of the black arrow denotes the energy of the agonist-free activated receptor; the bottom of the arrow and all lines starting at that point denote the agonist-bound receptor states.
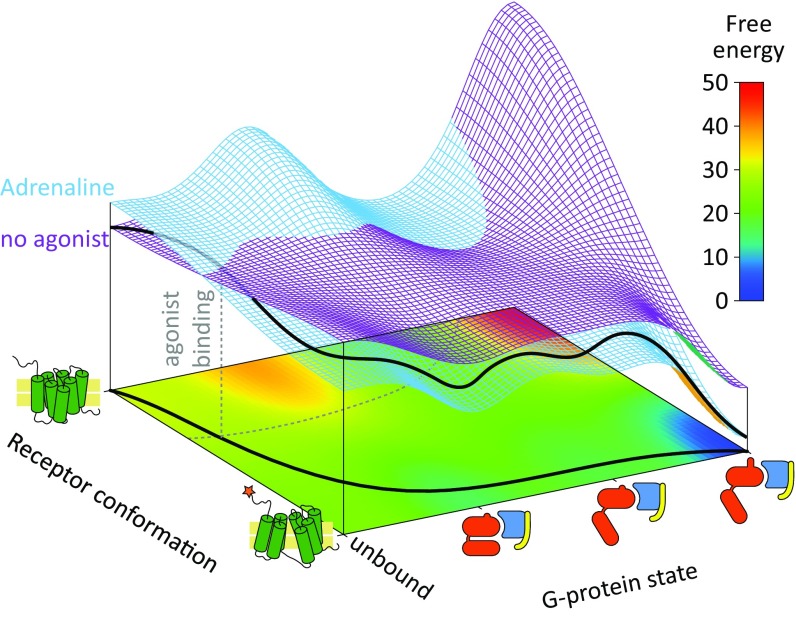
Fig. 5.
Three-dimensional semiquantitative energy landscapes for the activation of the GPCR and G protein system. The horizontal axes represent the conformation of the receptor and the state of the G protein, and the vertical z axis indicates the associated free energy in kilocalories per mole. Two landscapes are presented, for the free energies with the agonist adrenaline and without it (in blue and purple, respectively). A 2D projection map of the free energy (with the agonist) is also presented as a heatmap, in the lower part of the chart (in the xy plane). The G-protein state axis is constructed as a linear sequential coordinate to reduce the number of dimensions. A probable path is depicted by black lines, on the 3D free energy landscape and the projection map, and the intersection between the two landscapes is marked by dashed gray lines.
In terms of agreement with experimental findings, the results in Fig. 3 show a significant promise. The most important conditions that are needed to satisfy for a minimal working model are as follows. (i) The receptor with no bound agonist should remain mostly in the inactive conformation, and equilibrium should shift to the active conformation when the agonist binds. (ii) The G protein should bind the activated receptor but not its inactive conformation. And (iii) The G protein should bind GDP tightly, but then release GDP when bound to an activated receptor (open-in). In terms of these three conditions [shown as pluses (+) and minuses (–) in Fig. 3], our model performs well. We report that the receptor is more stable when it is in the inactive conformation (states I → II in Fig. 3), and the binding energy of the agonist (−8.2 kcal/mol) (33) would shift this equilibrium to the active form. Then, when the receptor is active, our calculations show that binding of the G protein becomes favorable. We also show that the closed conformation of the G protein (closed-out, binding GDP tightly) is converted to the open and ready-to-release GDP conformation (open-in), upon binding to the activated receptor (II → V → XII). We do note, however, that the model and resulting energies are not perfect. Specifically: (a) we report an energy difference between the inactive and active receptor that is surprisingly small (I → II; ∼1 kcal/mol), which indicates that β2AR exhibits high basal activity. This result is discussed below. (b) The transition of Gα from closed-out to open-in is marginally spontaneous (II → IX ΔG is approximately −1 kcal/mol) when the G protein is free, whereas it is only expected to be energetically favorable when the G protein is bound to the receptor, for which our model predicts ΔG is approximately −13 kcal/mol (V → XII). And (c) the G protein can open when bound to the inactive receptor (IV → X; however, the transition from out to in is not sterically possible when the receptor is inactive, and thus release of GDP is unlikely). These discrepancies (marked as squares in Fig. 3) might be a result of inaccuracies in the construction and assembly of the models, at the fine-tuning level, as well as performance gaps of the CG force field. Regardless, the overall picture presented is in line with experimental observations and provides much value. Finally, some indications that our calculations might correspond to actual observations have been reported, as discussed further below.
To further clarify the consequences of our calculations, we present in Fig. 4 1D traces constructing a qualitative energy landscape based on the trends of the asymptotic points shown in Fig. 3 and discussed above. This figure can help in following the change in free energy along the main conformational coordinate changes. Briefly, one can see that the receptor is activated upon binding of the agonist (vertical black arrow in Fig. 4). Also, it is shown that when the receptor is inactive, the conformational changes in the G protein harbor intermediates that are high in energy, making GDP release less likely. This trend is not observed in the activated receptor, where most G-protein intermediates are lower in energy than their precedent state. Of course, it would be useful to obtain the activation barrier for conformational changes by running CG simulations, but this is challenging because of the complexity of the system and is left to subsequent studies. Interestingly, however, we explored the simpler task of moving from the active to the inactive receptor (without the G protein) and obtained an approximate barrier of 3 kcal/mol (using a normal-mode–based method that is described in SI Appendix; see SI Appendix, Fig. S3) that is reasonable and in line with previously reported computational estimates (42).
To reiterate, examining the energy landscapes shown in Figs. 3 and 4 we see that the inactive receptor might promote binding to an open G-protein trimer (albeit with a higher energy intermediate), whereas the activated receptor promotes opening of the bound G protein with no high-energy intermediates, and also allows the movement of helix α5, which is a prerequisite for release of GDP (43). Thus, our computed energies conform with the notion that only interactions with the activated receptor will result in release of GDP from the G protein (44), and hence in signal transduction; regardless of unproductive conformational changes occurring in the G protein, as reported in ref. 45.
To conceptualize our analysis and to clarify the allosteric activation mechanism we also describe the energy landscapes for the system with and without the ligand as a function of the receptor conformation and the states of the G protein. The corresponding description (Fig. 5) illustrates the effect of the ligand in stabilizing the active state, where the landscape guides the system to a configuration that forces the G protein to open and to exchange the GDP by GTP. The difference in energy between the active and inactive receptor is too small for the surfaces to be visibly separated and is also suggestive of high basal activity [see discrepancy (a) above, discussing the small energy difference], and we therefore shifted the calculated ΔG from ∼1 kcal/mol to 4 kcal/mol in Fig. 5 (for visual purposes). At first glance, the calculated energy gap of ∼1 kcal/mol [discrepancy (a) above] appears to be underestimated; however and interestingly, previous studies report that β2AR exhibits high basal activity indeed (46, 47). Furthermore, single-molecule FRET experiments indicate that as much as 20–30% of the receptor population is found in the active state even at the absence of agonist (48, 49). Moreover, recent experimental data lend additional support to our original calculations, indicating that the receptor bound to an agonist is not necessarily the lowest point in energy surface (50). In this case, the activated receptor becomes predominant only after binding the G protein as well. This finding is consistent with several works (e.g., refs. 12, 48, and 51). Thus, our calculated ΔG results are actually encouraging.
Our finding of a fairly low energy difference between the open and closed states of the G protein suggests that even without activation, the G protein might populate both conformations. This is supported by other studies (31, 45). The open state, however, becomes more dominant as it binds the active receptor, as seen in our energy landscape and in the cited experiments. Furthermore, it is important to note that when the conformation is open-out it is still not prone to release GDP as in the open-in conformation.
Although the most straightforward model follows an increase in receptor–G-protein coupling upon agonist binding (e.g., refs. 52 and 53), other studies indicate that GPCRs exist in cells in the form of preformed complexes with their appropriate G-protein partners (54, 55). The precoupling concept brings an interesting perspective that can be considered using our energy landscape, if we reassess the chain of events starting from the precoupled states rather than the dissociated ones (as in Figs. 3–5). Ultimately, the end result and practically all of the discussion above remain the same even with this modification of the analysis. However, a functional precoupling cannot properly explain the amplification of GPCR activation (43).
The model’s usefulness can be expanded by adding more complexity, such as binding of antagonists, binding of different G-protein partners, phosphorylation of the receptor, and binding of β-arrestin, as well as taking into account the effect of oligomerization or allosteric modulators, and their effect on the structure (56) and thus the energy landscape. Note in this respect that a very recent breakthrough study solved the structure of the μ-opioid receptor in complex with a Gi protein (57); our approach can help pave the way to understanding how GPCRs select their partners in a specific way.
Activation of the G Protein
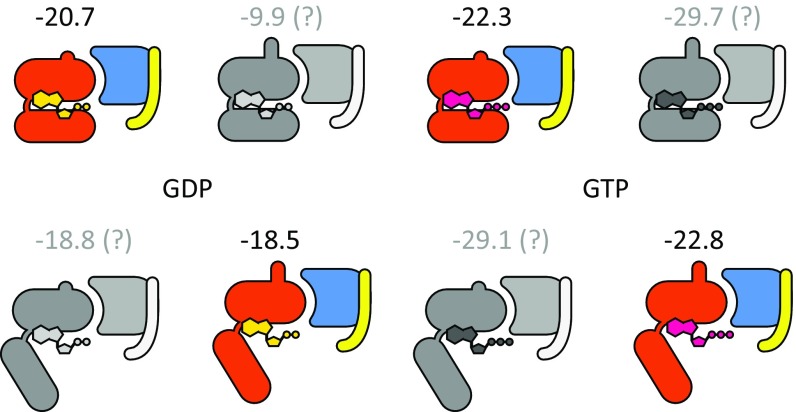
The activation of the G protein (composed of two domains, one of which is the Ras-like domain) is a central mode in the signal transduction of GPCRs and thus it is important to probe its molecular origin. Here we can go back to a related early study (25) of the activation of the exchange factor CDC25 that exchanges GDP for GTP in the Ras cycle. In that case we had demonstrated that the exchange activity is based on an electrostatic control. Thus, we explored if the G protein in the GPCR system is activated in a similar way. To examine this issue, we performed PDLD/S-LRA binding calculations for GDP and GTP to the G protein. The results, presented in Fig. 6 in color, consider only the direct X-ray–derived conformations (i.e., 3SN6 for open-in and 5JS8 for closed-out). The other conformations are presented for comparison in Fig. 6 in grayscale, but will not be considered in the discussion. As seen in Fig. 6, the transition of the G protein from closed to open-in reduces the binding affinity of GDP (from −20.7 to −18.5 kcal/mol) but has little effect on the affinity of GTP (−22.3 to −22.8), so with the larger concentration of GTP [∼10 GTP/GDP concentration ratio (58)] the exchange would be energetically favorable. We note here that a thorough investigation would require considering the kinetic barriers as well. Thus, further detailed studies of the binding of the GTP and GDP are left for subsequent works.
Fig. 6.
PDLD/S-LRA binding energies (in kilocalories per mole) of GDP and GTP to the Gα subunit in the different G-protein states. The colored states are based directly on X-ray structures whereas the grayscale states were modeled using different templates for each feature (the RS-AHD interdomain orientation and the α5 conformation). In the discussion we considered only the X-ray–based models. See SI Appendix, Fig. S1 for a match between the structures and the cartoons drawn here.
Concluding Remarks
Gaining a quantitative or semiquantitative description of the landscape of GPCRs can provide a crucial help in rational design of drugs that control signal transduction processes. This challenge is addressed here in a CG study of the activation of the β2AR. The resulting landscape reproduces key trends in the energetics of the asymptotic states inferred from previous experimental and computational studies, and thus provides us with computer-generated energetic rankings for different protein conformations in the GPCR–G-protein complex. Such results and their changes with different GPCR activators or inhibitors should provide a powerful way of understanding the action of GPCR.
Several works have emphasized the role of dynamics in the activation process either specifically or by implications that can be understood as emphasizing the key importance of dynamical effects (e.g., ref. 59). Here, as is the case in related proposals about the role of dynamics in enzyme catalysis (for review, see ref. 60), we find such implications to be problematic and irrelevant to the functional issue. That is, of course, the change of the landscape can lead to larger flexibility and faster motions, but as long as the relevant barriers are more than a few kilocalories per mole the energetics determine the time of passing the barriers and of moving between different configurations. Thus, the activation involves changes in energetics, and any change of dynamical behavior is the result of the change in energetics and not the reason for it.
Other works in the field discuss thoroughly the concept of allosteric effects within GPCRs (61, 62): the process in which binding of a substrate in one site elicits an effect in another remote site (for example, binding of an agonist to the pocket within the TM domain increases the affinity of the receptor to the G protein at its cytoplasmic face). Such effects are often attributed to “conformational changes,” and we would argue that the allosteric effects are ultimately associated with structurally induced electrostatic effects. In this respect, our model has reviewed the overall energies of the end-states and hasn’t thoroughly addressed the internal changes and their modulation by electrostatic interactions within the protein, to account for the allosteric effect observed. Such considerations are far from trivial and should be addressed in further studies.
Another aspect that is not addressed in the present study is biased agonism, and the related hypothesis that the receptor has more than two conformations (61). These conformations or subconformations are responsible, in part, to the preferential binding of one partner over another (e.g., G protein over β-arrestin, allowing bias to take place). Here we focused on the two conformations that are most relevant for G-protein activation, which are the two conformations with currently solved structures. Subconformations that might be less stable and therefore hard to attain structural information for can be investigated using computational means and can be explored in future studies.
Finally, we wish to point out that our model, and especially the outlook of Fig. 3, is strongly related to and also expands the long-standing ternary complex model (62, 63). The energetic considerations of the ternary complex model should be valid in our model, because they all rely on the same basic principles of thermodynamics.
The present work is a general exploration study that demonstrates the power of model-based CG strategy in charting the landscape of GCPRs. As demonstrated above, the results of our approach are not yet fully quantitative. However, the ability to examine the qualitative nature of the landscape can be a major tool in guiding experimental and theoretical exploration of GCPRs and related systems. Obviously, subsequent studies can explore the barriers between states, the difference between agonists and antagonists, and can also explore the nature of binding to different protein partners (e.g., Gi protein or β-arrestins).
Materials and Methods
The structural models were constructed using a combination of VMD plugins (64), the CHARMM-GUI server (65), and MODELER (66). The PDB IDs used were 3SN6 (11) for active β2AR–G-protein complex and 2RH1 (26) for inactive β2AR. Gs was modeled based on PBD ID codes 3SN6 and 1GP2 (28) for open and closed states, respectively, and modeling of the α5 segment of Gα, which interacts with the receptor, was modeled based on PBD ID codes 3SN6 or 5JS8 (31) for ordered and disordered conformation, respectively. All assemblies were constructed making sure that their amino acid sequences matched exactly (mutating or deleting extraneous residues if needed), to ensure that the only difference between the systems are the coordinates of the atoms. This step is important to make the energy comparison as precise as possible. Finally, all assemblies were embedded in a grid of particles emulating a membrane bilayer, and surrounded by a water sphere using the MOLARIS software package (67). The energy of the systems was minimized using the steepest descent algorithm, and a short relaxation run of 20 ps was performed to locally relax the proteins and remove possible steric clashes. See the SI Appendix for more details.
In calculating the asymptotic CG energies, we selected 10 snapshots from each system, and the proteins were simplified to the CG model of MOLARIS-XG (22, 23). The protonation states of all titratable residues were determined using a Monte Carlo simulation (22), and finally the total energy of the system was calculated as previously described, using model 1 in ref. 23. The energies of all of the snapshots were averaged for each assembly. See the SI Appendix for more details.
The energy profile of the conformational transition, going from the inactive to the active β2AR conformation, was calculated using an underdevelopment method, which calculates the normal modes of the protein, and performs a Monte Carlo simulation to sample the transition. See the SI Appendix for more details.
Supplementary Material
Acknowledgments
We thank the University of Southern California High Performance Computing and Communication Center for computational resources. This work was supported by National Science Foundation Grant MCB 1707167, National Institute of Health Grant R01-AI055926, and the Bridge Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810316115/-/DCSupplemental.
References
- 1.Alberts B. Molecular Biology of the Cell. 6th Ed. Garland Science, Taylor and Francis Group; New York: 2015. p. 984. [Google Scholar]
- 2.Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR drug discovery: New agents, targets and indications. Nat Rev Drug Discov. 2017;16:829–842. doi: 10.1038/nrd.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sriram K, Insel PA. G protein-coupled receptors as targets for approved drugs: How many targets and how many drugs? Mol Pharmacol. 2018;93:251–258. doi: 10.1124/mol.117.111062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reginald RH, Garrett CM. 2007. Biochemistry (Thompson Group, Belmont, CA) 3rd Ed, pp xxxiii, 1059.
- 5.Rasmussen SG, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Roth BL, Irwin JJ, Shoichet BK. Discovery of new GPCR ligands to illuminate new biology. Nat Chem Biol. 2017;13:1143–1151. doi: 10.1038/nchembio.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuss C, Gerber U. G-protein-independent signaling by G-protein-coupled receptors. Trends Neurosci. 2000;23:469–475. doi: 10.1016/s0166-2236(00)01643-x. [DOI] [PubMed] [Google Scholar]
- 8.Jastrzebska B, et al. Rhodopsin-transducin heteropentamer: Three-dimensional structure and biochemical characterization. J Struct Biol. 2011;176:387–394. doi: 10.1016/j.jsb.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson MA, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manglik A, et al. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dror RO, et al. Activation mechanism of the β2-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohlhoff KJ, et al. Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat Chem. 2014;6:15–21. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Astumian RD, Mukherjee S, Warshel A. The physics and physical chemistry of molecular machines. ChemPhysChem. 2016;17:1719–1741. doi: 10.1002/cphc.201600184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alhadeff R, Warshel A. Reexamining the origin of the directionality of myosin V. Proc Natl Acad Sci USA. 2017;114:10426–10431. doi: 10.1073/pnas.1711214114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin JC, Hyeon C, Thirumalai D. Sequence-dependent folding landscapes of adenine riboswitch aptamers. Phys Chem Chem Phys. 2014;16:6376–6382. doi: 10.1039/c3cp53932f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Zheng W, Papoian GA, Wolynes PG. Exploring the free energy landscape of nucleosomes. J Am Chem Soc. 2016;138:8126–8133. doi: 10.1021/jacs.6b02893. [DOI] [PubMed] [Google Scholar]
- 19.Deupi X, Kobilka BK. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (Bethesda) 2010;25:293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higashijima T, Ferguson KM, Sternweis PC, Smigel MD, Gilman AG. Effects of Mg2+ and the beta gamma-subunit complex on the interactions of guanine nucleotides with G proteins. J Biol Chem. 1987;262:762–766. [PubMed] [Google Scholar]
- 21.Lefkowitz RJ. A brief history of G-protein coupled receptors (Nobel Lecture) Angew Chem Int Ed Engl. 2013;52:6366–6378. doi: 10.1002/anie.201301924. [DOI] [PubMed] [Google Scholar]
- 22.Vicatos S, Rychkova A, Mukherjee S, Warshel A. An effective coarse-grained model for biological simulations: Recent refinements and validations. Proteins. 2014;82:1168–1185. doi: 10.1002/prot.24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vorobyov I, Kim I, Chu ZT, Warshel A. Refining the treatment of membrane proteins by coarse-grained models. Proteins. 2016;84:92–117. doi: 10.1002/prot.24958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warshel A, Sharma PK, Kato M, Parson WW. Modeling electrostatic effects in proteins. Biochim Biophys Acta. 2006;1764:1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Muegge I, Schweins T, Langen R, Warshel A. Electrostatic control of GTP and GDP binding in the oncoprotein p21ras. Structure. 1996;4:475–489. doi: 10.1016/s0969-2126(96)00052-4. [DOI] [PubMed] [Google Scholar]
- 26.Cherezov V, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westfield GH, et al. Structural flexibility of the G alpha s alpha-helical domain in the beta2-adrenoceptor Gs complex. Proc Natl Acad Sci USA. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wall MA, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 29.Dror RO, et al. Signal transduction. Structural basis for nucleotide exchange in heterotrimeric G proteins. Science. 2015;348:1361–1365. doi: 10.1126/science.aaa5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flock T, et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goricanec D, et al. Conformational dynamics of a G-protein α subunit is tightly regulated by nucleotide binding. Proc Natl Acad Sci USA. 2016;113:E3629–E3638. doi: 10.1073/pnas.1604125113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Eps N, et al. Gi- and Gs-coupled GPCRs show different modes of G-protein binding. Proc Natl Acad Sci USA. 2018;115:2383–2388. doi: 10.1073/pnas.1721896115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manglik A, Kobilka B. The role of protein dynamics in GPCR function: Insights from the β2AR and rhodopsin. Curr Opin Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vasile S, et al. Characterization of ligand binding to GPCRs through computational methods. Methods Mol Biol. 2018;1705:23–44. doi: 10.1007/978-1-4939-7465-8_2. [DOI] [PubMed] [Google Scholar]
- 35.Warshel A, Russell ST. Calculations of electrostatic interactions in biological systems and in solutions. Q Rev Biophys. 1984;17:283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- 36.Sham YY, Chu ZT, Tao H, Warshel A. Examining methods for calculations of binding free energies: LRA, LIE, PDLD-LRA, and PDLD/S-LRA calculations of ligands binding to an HIV protease. Proteins. 2000;39:393–407. [PubMed] [Google Scholar]
- 37.Alhadeff R, Warshel A. Simulating the function of sodium/proton antiporters. Proc Natl Acad Sci USA. 2015;112:12378–12383. doi: 10.1073/pnas.1516881112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenakin T, Christopoulos A. Signalling bias in new drug discovery: Detection, quantification and therapeutic impact. Nat Rev Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 39.Shonberg J, et al. Biased agonism at G protein-coupled receptors: The promise and the challenges—A medicinal chemistry perspective. Med Res Rev. 2014;34:1286–1330. doi: 10.1002/med.21318. [DOI] [PubMed] [Google Scholar]
- 40.Thomas TC, Schmidt CJ, Neer EJ. G-protein alpha o subunit: Mutation of conserved cysteines identifies a subunit contact surface and alters GDP affinity. Proc Natl Acad Sci USA. 1993;90:10295–10299. doi: 10.1073/pnas.90.21.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iiri T, Herzmark P, Nakamoto JM, van Dop C, Bourne HR. Rapid GDP release from Gs alpha in patients with gain and loss of endocrine function. Nature. 1994;371:164–168. doi: 10.1038/371164a0. [DOI] [PubMed] [Google Scholar]
- 42.Provasi D, Artacho MC, Negri A, Mobarec JC, Filizola M. Ligand-induced modulation of the free-energy landscape of G protein-coupled receptors explored by adaptive biasing techniques. PLOS Comput Biol. 2011;7:e1002193. doi: 10.1371/journal.pcbi.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oldham WM, Hamm HE. Heterotrimeric G protein activation by G-protein-coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. doi: 10.1038/nrm2299. [DOI] [PubMed] [Google Scholar]
- 44.Hamm HE. The many faces of G protein signaling. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y, Zhang L, Zhang XC, Zhao Y. Structural dynamics of Giα protein revealed by single molecule FRET. Biochem Biophys Res Commun. 2017;491:603–608. doi: 10.1016/j.bbrc.2017.07.156. [DOI] [PubMed] [Google Scholar]
- 46.Milligan G. Constitutive activity and inverse agonists of G protein-coupled receptors: A current perspective. Mol Pharmacol. 2003;64:1271–1276. doi: 10.1124/mol.64.6.1271. [DOI] [PubMed] [Google Scholar]
- 47.Bond RA, et al. Physiological effects of inverse agonists in transgenic mice with myocardial overexpression of the beta 2-adrenoceptor. Nature. 1995;374:272–276. doi: 10.1038/374272a0. [DOI] [PubMed] [Google Scholar]
- 48.Lamichhane R, et al. Single-molecule view of basal activity and activation mechanisms of the G protein-coupled receptor β2AR. Proc Natl Acad Sci USA. 2015;112:14254–14259. doi: 10.1073/pnas.1519626112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calebiro D, Sungkaworn T. Single-molecule imaging of GPCR interactions. Trends Pharmacol Sci. 2018;39:109–122. doi: 10.1016/j.tips.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 50.Manglik A, Kruse AC. Structural basis for G protein-coupled receptor activation. Biochemistry. 2017;56:5628–5634. doi: 10.1021/acs.biochem.7b00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregorio GG, et al. Single-molecule analysis of ligand efficacy in β2AR-G-protein activation. Nature. 2017;547:68–73. doi: 10.1038/nature22354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hein P, Frank M, Hoffmann C, Lohse MJ, Bünemann M. Dynamics of receptor/G protein coupling in living cells. EMBO J. 2005;24:4106–4114. doi: 10.1038/sj.emboj.7600870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hein P, et al. Gs activation is time-limiting in initiating receptor-mediated signaling. J Biol Chem. 2006;281:33345–33351. doi: 10.1074/jbc.M606713200. [DOI] [PubMed] [Google Scholar]
- 54.Audet N, et al. Bioluminescence resonance energy transfer assays reveal ligand-specific conformational changes within preformed signaling complexes containing delta-opioid receptors and heterotrimeric G proteins. J Biol Chem. 2008;283:15078–15088. doi: 10.1074/jbc.M707941200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cevheroğlu O, Becker JM, Son CD. GPCR-Gα protein precoupling: Interaction between Ste2p, a yeast GPCR, and Gpa1p, its Gα protein, is formed before ligand binding via the Ste2p C-terminal domain and the Gpa1p N-terminal domain. Biochim Biophys Acta. 2017;1859:2435–2446. doi: 10.1016/j.bbamem.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 56.Liu X, et al. Mechanism of intracellular allosteric β2AR antagonist revealed by X-ray crystal structure. Nature. 2017;548:480–484. doi: 10.1038/nature23652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koehl A, et al. Structure of the µ-opioid receptor-Gi protein complex. Nature. 2018;558:547–552. doi: 10.1038/s41586-018-0219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 59.Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR dynamics: Structures in motion. Chem Rev. 2017;117:139–155. doi: 10.1021/acs.chemrev.6b00177. [DOI] [PubMed] [Google Scholar]
- 60.Warshel A, Bora RP. Perspective: Defining and quantifying the role of dynamics in enzyme catalysis. J Chem Phys. 2016;144:180901. doi: 10.1063/1.4947037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith JS, Lefkowitz RJ, Rajagopal S. Biased signalling: From simple switches to allosteric microprocessors. Nat Rev Drug Discov. 2018;17:243–260. doi: 10.1038/nrd.2017.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mahoney JP, Sunahara RK. Mechanistic insights into GPCR-G protein interactions. Curr Opin Struct Biol. 2016;41:247–254. doi: 10.1016/j.sbi.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Lean A, Stadel JM, Lefkowitz RJ. A ternary complex model explains the agonist-specific binding properties of the adenylate cyclase-coupled beta-adrenergic receptor. J Biol Chem. 1980;255:7108–7117. [PubMed] [Google Scholar]
- 64.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 65.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 66.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2017;1654:39–54. doi: 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
- 67.Lee FS, Chu ZT, Warshel A. Microscopic and semimicroscopic calculations of electrostatic energies in proteins by the POLARIS and ENZYMIX programs. J Comput Chem. 1993;14:161–185. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.