Significance
Bacterial pathogens cause plant diseases that threaten the global food supply. To control diseases, it is important to understand how pathogenic bacteria evade plant defense and promote infection. We identify from the phytopathogen Pseudomonas syringae a small-molecule virulence factor—phevamine A. Both the chemical structure and mode of action of phevamine A are different from known bacterial phytotoxins. Phevamine A promotes bacterial growth by suppressing plant immune responses, including both early (the generation of reactive oxygen species) and late (the deposition of cell wall reinforcing callose in leaves and leaf cell death) markers. This work uncovers a widely distributed, small-molecule virulence factor and shows the power of a multidisciplinary approach to identify small molecules important for plant infection.
Keywords: natural products, genome mining, phytopathogen, virulence factor, plant immunity
Abstract
Bacterial plant pathogens cause significant crop damage worldwide. They invade plant cells by producing a variety of virulence factors, including small-molecule toxins and phytohormone mimics. Virulence of the model pathogen Pseudomonas syringae pv. tomato DC3000 (Pto) is regulated in part by the sigma factor HrpL. Our study of the HrpL regulon identified an uncharacterized, three-gene operon in Pto that is controlled by HrpL and related to the Erwinia hrp-associated systemic virulence (hsv) operon. Here, we demonstrate that the hsv operon contributes to the virulence of Pto on Arabidopsis thaliana and suppresses bacteria-induced immune responses. We show that the hsv-encoded enzymes in Pto synthesize a small molecule, phevamine A. This molecule consists of l-phenylalanine, l-valine, and a modified spermidine, and is different from known small molecules produced by phytopathogens. We show that phevamine A suppresses a potentiation effect of spermidine and l-arginine on the reactive oxygen species burst generated upon recognition of bacterial flagellin. The hsv operon is found in the genomes of divergent bacterial genera, including ∼37% of P. syringae genomes, suggesting that phevamine A is a widely distributed virulence factor in phytopathogens. Our work identifies a small-molecule virulence factor and reveals a mechanism by which bacterial pathogens overcome plant defense. This work highlights the power of omics approaches in identifying important small molecules in bacteria–host interactions.
Bacterial small molecules play key roles in pathogen–plant interactions. Pseudomonas syringae, a phylogenetically diverse species of bacteria responsible for many crop diseases, employs a myriad of virulence factors, including secreted protein effectors delivered to host cells by the type III secretion system (T3SS) and small molecules (1, 2). Coronatine, syringomycin, syringopeptin, tabtoxin, and phaseolotoxin are among the small-molecule virulence factors produced by various strains of P. syringae, acting as phytohormone mimics or phytotoxins (1). For example, coronatine disrupts plant immune signaling by mimicking the structure of the plant hormone jasmonic acid-isoleucine (1, 3), and phaseolotoxin contributes to virulence by interfering with the synthesis of l-arginine and polyamines in plants (4, 5). While many protein effectors have been extensively studied in P. syringae, only a limited number of small molecules have been identified. The genomes of P. syringae and other phytopathogens encode potential biosynthetic enzymes (6, 7). Identifying the cryptic small molecules synthesized by these enzymes may reveal new virulence mechanisms that could be targeted to control plant infection.
Many phytopathogenic virulence factors function to suppress plant immune responses (8, 9). The first layer of the plant immune system relies on the recognition of conserved microbe-associated molecular patterns (MAMPs) by host pattern recognition receptors to generate MAMP-triggered immunity (MTI) (10). The MTI signaling cascade includes receptor kinase activation, reactive oxygen species (ROS) production, calcium influx, mitogen-activated protein kinase activation, defense gene activation, and callose deposition at the plant cell wall (SI Appendix, Fig. S1); each of these steps can be targeted by virulence factors (10). The sigma factor HrpL controls the expression of P. syringae virulence factors including the T3SS and its associated effector genes (11). HrpL also regulates genes responsible for the biosynthesis of coronatine in some, but not all, P. syringae strains (12, 13). Our recent study identified a number of uncharacterized HrpL-regulated genes that encode putative biosynthetic enzymes for small molecules (12). We therefore proposed that these HrpL-regulated genes are responsible for synthesizing small molecules that play a role in virulence (12).
In this study, we focus on an HrpL-regulated three-gene biosynthetic operon PSPTO_0873–0875 from P. syringae pv. tomato DC3000 (Pto) (12, 14). This operon shares 66–85% homology with hrp (hypersensitive response and pathogenicity)-associated systemic virulence (hsv) genes from Erwinia amylovora (15). The hsv operon was implicated in the virulence of E. amylovora on apple shoots (16), but the small molecule(s) produced by hsv-encoded enzymes are unknown. Here, we confirm that the Pto hsv operon acts as a P. syringae virulence factor on Arabidopsis thaliana and demonstrate that the hsv operon dampens host immune responses in both A. thaliana and Nicotiana benthamiana. We demonstrate that the enzymes encoded by the hsv operon synthesize a small molecule that is important for virulence. By integrating heterologous expression, metabolomics, and in vitro biosynthesis, we identify a bioactive small molecule, phevamine A, a conjugate of l-phenylalanine, l-valine, and a modified spermidine. We further show that phevamine A suppresses the potentiation of MAMP-induced ROS bursts by spermidine and l-arginine. Thus, phevamine A is a small-molecule virulence factor that promotes bacterial growth and virulence, in part by suppressing plant immune responses.
Results and Discussion
The P. syringae hsv Operon Promotes Virulence and Suppresses Defense Responses.
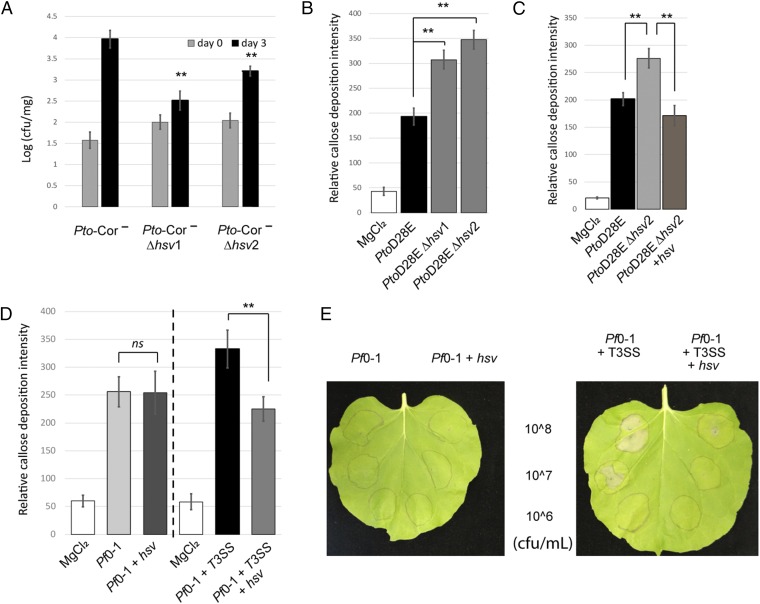
To investigate the function of the hsv operon, we first generated independent clean deletion mutants in Pto. The parental strain, Pto, and the two mutants, Pto_∆hsv1, Pto_∆hsv2 were used to infect A. thaliana Col-0 seedlings by dip inoculation (SI Appendix, Materials and Methods) and the growth of these bacteria was monitored. We observed that the mutants grew slightly less than the Pto strains (SI Appendix, Fig. S2) in two independent experiments, but this reduced growth trend was not statistically significant. This observation is not surprising, as Pto is an aggressive pathogen on A. thaliana and its large suite of virulence factors can act collectively (17). It is therefore rare to observe significant loss of virulence phenotypes when deleting a single candidate virulence factor from this strain. To circumvent this problem, we used a weakly pathogenic Pto derivative deficient in coronatine production (Pto DC3118, hereafter Pto-Cor−) (17) to generate independent hsv mutants Pto-Cor−Δhsv1, and Pto-Cor−Δhsv2. Three days postinfection, significantly less bacteria for Pto-Cor−Δhsv1 and Pto-Cor−Δhsv2 were recovered from the seedlings than Pto-Cor− (Fig. 1A). This result indicates that hsv is required for full virulence in planta.
Fig. 1.
The hsv operon is required for virulence and suppresses defense responses. (A) Pto-Cor−Δhsv mutants display reduced growth on A. thaliana seedlings compared with Pto-Cor−. This experiment was repeated three times with similar results. (B) PtoD28EΔhsv mutants induce higher callose deposition than PtoD28E on 4-wk-old A. thaliana leaves. The experiment is a representative of four independent replicates. (C) Complementation of hsv in PtoD28EΔhsv2 (PtoD28EΔhsv2 + NptII::hsv) reestablishes callose deposition to levels triggered by PtoD28E. This assay is a representative of two independent replicates. (D) The hsv operon suppresses callose deposition when coexpressed in Pf0-1 with T3SS, but not when expressed in Pf0-1. The callose deposition was monitored as in B after infiltration of Pf0-1 or Pf0-1 expressing hsv. This assay was performed twice with similar results. Pf0-1+T3SS and Pf0-1+T3SS-expressing hsv were tested separately with at least three replicate experiments for each strain. (E) The hsv operon in Pf0-1 suppresses T3SS-mediated cell death in N. benthamiana. Pf0-1-derived strains were inoculated at three different concentrations (colony-forming units per milliliter). Pictures were taken 20-h postinfiltration. This assay was repeated four times with similar results. ** indicates t test, P value < 0.01. Error bars represent ±SE. Relative callose intensity represents the number of callose deposits observed per field. cfu/mg, colony-forming units per milligram of plant tissue; ns, not significant.
We next addressed whether the hsv operon had an effect on host callose deposition, a cell wall reinforcing response typically triggered during MTI (18, 19) (SI Appendix, Fig. S1). Several type III secretion effector proteins can inhibit callose deposition (20–24) and thus may obscure phenotypes resulting from hsv deletion. We therefore generated two independent hsv clean deletions in PtoD28E (PtoD28EΔhsv1 and PtoD28EΔhsv2), a strain lacking 28 different type III secretion effectors (25). PtoD28E, PtoD28EΔhsv1, and PtoD28EΔhsv2 were used to infiltrate 4-wk-old leaves of A. thaliana. Both PtoD28EΔhsv alleles induced higher callose deposition than PtoD28E (Fig. 1B); complementation of PtoD28EΔhsv2 with constitutively expressed hsv reduced the level of callose back to that induced by PtoD28E (Fig. 1C). These results demonstrate that hsv suppresses a late marker of plant defense at the cell wall.
Conversely, we also examined whether hsv expression is sufficient to suppress plant callose deposition induced by a nonpathogenic bacterium. The hsv operon was constitutively expressed in Pseudomonas fluorescens Pf0-1 (Pf0-1), a strain that lacks both hsv and a T3SS, but still induces callose deposition (Fig. 1D) (26). Expression of hsv had no effect on the level of Pf0-1-induced callose deposition (Fig. 1D). A Pf0-1 derivative engineered to contain a complete P. syringae T3SS locus (Pf0-1+T3SS) (27) induced more callose than wild-type Pf0-1 (SI Appendix, Fig. S3), suggesting that Pf0-1 T3SS components are recognized by the plant (28). Expression of hsv in the Pf0-1+T3SS strain reduced the level of callose back to the level elicited by wild-type Pf0-1 (Fig. 1D and SI Appendix, Fig. S3). Thus, hsv suppresses the callose deposition induced by Pf0-1 T3SS components. To extend this observation, we evaluated the activities of these P. fluorescens strains on a second plant species, N. benthamiana. Infiltration of N. benthamiana leaves with Pf0-1 or Pf0-1-expressing hsv had no effect; the tissue infiltrated with bacteria was identical to the noninfiltrated tissue (Fig. 1E). In contrast, infiltration with Pf0-1+T3SS elicited a dose-dependent leaf cell death. This response was suppressed by hsv expression (Fig. 1E). Thus, hsv suppresses the cell death caused by a component of the T3SS or the linked harpin gene transferred from P. syringae pv. syringae to Pf0-1 (29, 30).
hsv-Encoded Enzymes Synthesize Three Related Small Molecules.
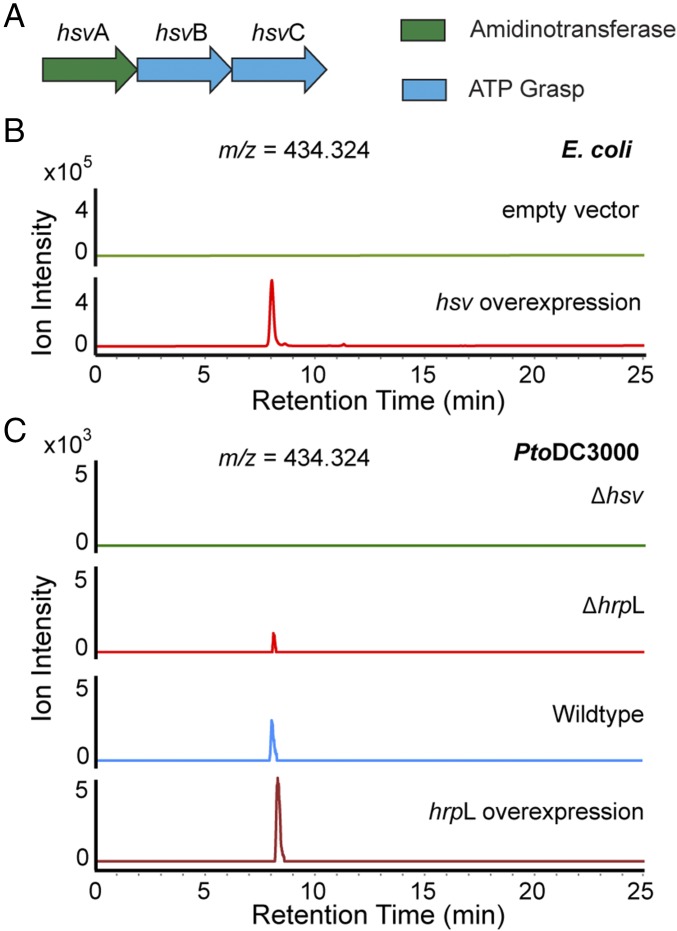
The hsv operon encodes three enzymes, including a putative amidinotransferase, HsvA, and two putative ATP-grasp–type enzymes, HsvB and HsvC (Fig. 2A). To test the hypothesis that these hsv-encoded enzymes synthesize small molecules, we conducted hsv heterologous expression and comparative metabolomics experiments. The hsv cluster was overexpressed in Escherichia coli as a heterologous host to enhance small molecule production. Organic extracts of culture supernatants were analyzed using liquid-chromatography-coupled high-resolution mass spectrometry (LC-HRMS) to generate metabolomic profiles for comparison. Three species with mass-to-charge ratios (m/z) of 287.255, 434.324, and 581.392 were present at high levels in the hsv-expressing E. coli, but were absent in the control E. coli carrying the empty vector (Fig. 2B and SI Appendix, Figs. S4–S6). The species with the m/z of 287.255 and 434.324 were also detected by LC-HRMS in the culture extract of the wild-type Pto, but not in Pto∆hsv (Fig. 2C and SI Appendix, Fig. S5). The production of the metabolite with the m/z of 434.324 is also observed in Pto overexpressing hrpL, but is reduced by hrpL deletion (Fig. 2C), consistent with the regulation of hsv by hrpL (12). This metabolite was detected at the same level in the supernatants of both Pf0-1- and Pf0-1+T3SS-expressing hsv (SI Appendix, Fig. S7), indicating that secretion of the compound is independent of the T3SS.
Fig. 2.
Metabolomics reveals the small molecules synthesized by hsv-encoded enzymes. (A) Graphical representation of the hsv operon. (B) Comparative metabolomics analysis in E. coli. Overexpression of hsv produces a molecule with an m/z of 434.324 (green), which is absent in E. coli harboring the empty vector (red). (C) Comparative metabolomics analysis in Pto. The molecule with the m/z of 434.324 is absent in Δhsv (green) and reduced in ΔhrpL (red), but present in both wild-type Pto (blue) and Pto overexpressing hrpL (maroon). Extracted ion chromatograms for 434.324 are shown in B and C. The slight change in retention time is due to the hydrophilicity of this molecule. Experiments presented in B and C were repeated at least three times.
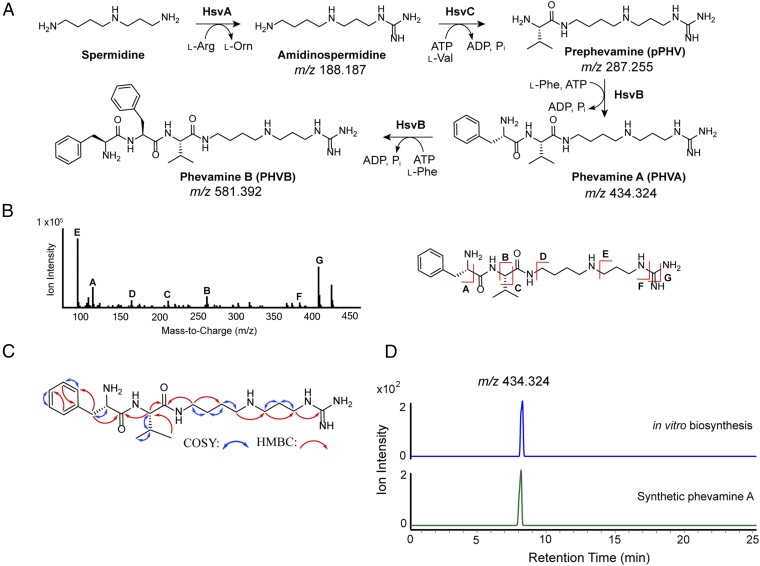
We analyzed the structures of the identified metabolites from E. coli and P. syringae strains expressing the hsv operon by tandem MS, which revealed that these metabolites share similar MS fragments (SI Appendix, Figs. S8 and S9) and likely possess related structures. The mass differences between 287.255 and 434.324, and between 434.324 and 581.392 are both 147.068, corresponding to the mass of a phenylalanine in an amide linkage. Based on these data, we propose that the structures of the compounds with the m/z of 434.324 and 581.392 contain one and two phenylalanines, respectively. We name the identified metabolites phevamine A (m/z 434.324; PHVA), phevamine B (m/z 581.392; PHVB) and prephevamine (m/z 287.255; pPHV).
In Vitro Biosynthesis of Phevamines Reveals Structures.
To characterize the structures of phevamines, we reconstituted the biosynthetic enzymes encoded by the hsv operon in vitro. HsvA shares homology with amidinotransferases that transfer an amidino group from arginine to an amine, suggesting the presence of an amidino group. HsvB and HsvC belong to the ATP-grasp enzyme superfamily consisting of ATP-dependent enzymes that condense carboxylic acids with amines, suggesting the presence of amide linkages in the phevamines and prephevamine (31). HsvA, HsvB, and HsvC were overexpressed in E. coli and purified as recombinant proteins. We reconstituted the activity of HsvA by incubating l-arginine, the donor of the amidino group for amidinotransferases, in the presence of a variety of physiologically relevant amines as acceptors, including l-lysine as well as spermidine and spermine, which are polyamines produced by P. syringae (32, 33). We found that HsvA preferentially modifies spermidine and generates amidinospermidine (Fig. 3A and SI Appendix, Figs. S10 and S11). Subtracting the mass of amidinospermidine from that of prephevamine results in a mass consistent with a valine in an amide linkage. We therefore proposed amidinospermidine is condensed with l-Val as the second biosynthetic step to generate prephevamine (Fig. 3A). Both ATP-grasp enzymes, HsvB and HsvC, were examined for this condensation activity in the presence of amidinospermidine, l-Val, ATP, and Mg2+. Only HsvC catalyzes prephevamine formation (Fig. 3A and SI Appendix, Fig. S10). Data from analyzing product formation by LC-HRMS and measuring phosphate released in the assay suggest that l-phenylalanine is a structural component of the phevamines (SI Appendix, Fig. S12). To validate this proposal, we incubated prephevamine with HsvB and l-Phe, and the formation of phevamine A and phevamine B was indeed observed (Fig. 3A and SI Appendix, Figs. S10 and S13). This result also indicates that HsvB can catalyze two rounds of condensation with l-Phe. Tandem MS analysis of phevamine A suggests that the amidino group is connected to the propylamine end of spermidine (Fig. 3B and SI Appendix, Fig. S9).
Fig. 3.
Structural characterization and biosynthesis of prephevamine and the phevamines. (A) Biosynthetic pathway of prephevamine and the phevamines based on in vitro characterization. (B) Structural analysis of phevamine A produced by E. coli overexpressing hsv using tandem MS. (C) COSY and (1H,13C)-HMBC correlations of in vitro synthesized and purified phevamine A. (D) Extracted ion chromatograms of phevamine A (m/z, 434.324) produced via in vitro enzymatic synthesis (blue) or total chemical synthesis (green).
Having identified that phevamines consist of l-Phe, l-Val, and amidinospermidine, we next determined the connectivity of these components by NMR analysis. Due to high polarity, phevamines were difficult to isolate from bacterial culture extracts. To isolate sufficient materials for NMR, we developed in vitro biosynthetic methods for phevamine A and phevamine B (Methods). From 16 mL of in vitro enzymatic assays, 5.7 mg of phevamine A and 5.5 mg of phevamine B were purified. These compounds were analyzed by 1H, (1H,1H)-COSY, (1H,13C)-HMBC, and (1H,13C)-HSQC NMR experiments (Fig. 3C and SI Appendix, Figs. S14–S21). The NMR data support that the amidino group is linked to the propylamine side of spermidine and that phevamine B contains an additional l-Phe at the N-terminus of phevamine A. Phevamine B was only detected under conditions of heterologous expression and in vitro synthesis, but not in the culture extract of Pto (SI Appendix, Fig. S6), suggesting that phevamine B is unlikely a physiologically relevant molecule. The production of phevamine B may be due to the overexpression of HsvB in E. coli and the high concentration of HsvB and l-Phe added in vitro. Thus, we focused on phevamine A for further structural characterization. To confirm the absolute stereochemistry, we developed a total synthesis method for phevamine A (SI Appendix, Materials and Methods). Synthetic and in vitro isolated phevamine A exhibit identical 1H and 13C NMR signals, and the same LC retention time and MS fragmentation pattern as the species detected in the bacterial culture extract (Fig. 3D and SI Appendix, Figs. S14–S22). These results confirmed the structure assignment for phevamine A and provided materials for biological activity testing of this molecule. In an initial activity test, we examined the ability of phevamine A to bind ferric iron using a chromeazurol S assay and observed no significant binding to ferric iron (SI Appendix, Fig. S23), suggesting phevamine A is unlikely acting as a siderophore.
Comparative Genomics Suggests a Potential Host Target for Phevamines.
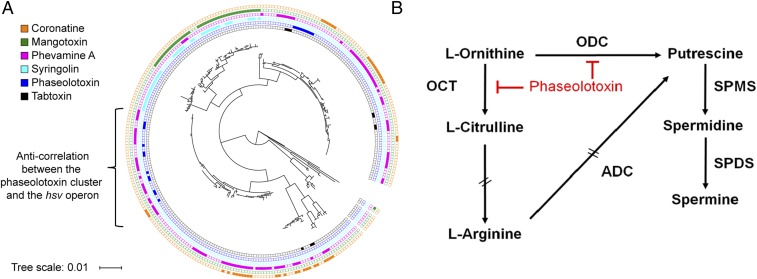
We investigated the phylogenetic distribution of hsv in bacterial genomes using MultiGeneBlast that identifies homologous gene clusters based on sequence similarity and gene synteny (34). We found that hsv is widely distributed across plant-pathogenic bacterial genera including Pseudomonas, Erwinia, and Pantoea. We compared the distribution of the hsv operon to those biosynthetic gene clusters encoding known P. syringae phytotoxins, focusing on coronatine, mangotoxin, syringolin, phaseolotoxin, and tabtoxin (Fig. 4A and Dataset S1) (1, 35). The hsv operon is present in ∼37% of P. syringae (107 out of 292) and is the most widely distributed gene cluster of the six analyzed. The hsv operon rarely cooccurs with any of the aforementioned small-molecule biosynthetic clusters. This anticorrelation is especially prominent between hsv and phaseolotoxin biosynthetic genes in the genomes of otherwise extremely closely related strains of P. syringae pv. actinidiae (36). We previously showed that functionally redundant virulence factors rarely cooccur in the same strain (37). Specifically, the small molecule coronatine and three different type III secretion system protein effectors use four distinct mechanisms to target the same host defense signaling pathway, and only genes responsible for a single mechanism are typically found in any given P. syringae genome (37). Based on the clear anticorrelation of hsv with the phaseolotoxin biosynthetic genes, and the knowledge that phaseolotoxin targets host arginine and polyamine biosynthesis by inhibiting ornithine carbamoyltransferase and ornithine decarboxylase (Fig. 4B) (4, 5, 38), we hypothesized that the action of phevamine A involves polyamines and/or arginine. This hypothesis is consistent with previous implication of polyamine function in plant immune responses (39, 40). Furthermore, our genomic analysis revealed that many P. syringae strains do not contain the biosynthetic gene clusters for phevamine A or the five other phytotoxins described herein (Fig. 4A). These strains could produce different small molecules that might play a role in bacteria–plant interaction.
Fig. 4.
Distribution of the hsv operon and other toxins among P. syringae and graphical representation of polyamine biosynthesis and action of phaseolotoxin. (A) Phylogenetic tree of 292 P. syringae strains and the distribution of coronatine, mangotoxin, phevamine A, syringolin, phaseolotoxin, and tabtoxin biosynthetic genes, from the outer ring to the inner ring. The tree scale represents the nucleotide substitution per site. (B) Phaseolotoxin inhibits the arginine and polyamine pathways by suppressing the activity of ornithine decarboxylase (ODC) and ornithine carbamoyltransferase (OCT). ADC, arginine decarboxylase; SPDS, spermidine synthase; SPMS, spermine synthase. Double dashed arrows represent pathways with intermediates not shown. Reproduced from ref. 60 with permission from American Society for Microbiology.
Polyamines and Arginine Potentiate an Early MTI Response.
We investigated the effect of arginine and polyamines on the MTI response induced by the well-studied MAMP flg22, a short peptide derived from the Pseudomonas flagellin (18). We monitored the ROS burst following recognition of flg22 by the pattern recognition receptor FLS2 (SI Appendix, Fig. S1) (41, 42). The polyamines putrescine, spermidine, and spermine potentiated the flg22-induced ROS burst in N. benthamiana, resulting in an earlier and larger amplitude response (Fig. 5 A and B and SI Appendix, Fig. S24A). The potentiation was dose-dependent (SI Appendix, Fig. S24B). Spermidine alone induced only a slight ROS burst (SI Appendix, Fig. S24C) (40), and the increase of the flg22-induced ROS burst in the presence of spermidine was higher than an additive effect (SI Appendix, Fig. S24C). The potentiation by spermidine was also observed in A. thaliana, but was not observed in the flg22 receptor mutant fls2 (Fig. 5C and SI Appendix, Fig. S25 A and B). Spermidine potentiation was also observed using the elicitor elf18 (SI Appendix, Fig. S25C) (43). Surprisingly, the Arabidopsis Ca2+ burst induced by flg22 was reduced in the presence of spermidine (Fig. 5D and SI Appendix, Figs. S1 and S25D). Arginine had a similar effect on ROS burst in A. thaliana and N. benthamiana, while l-citrulline did not (SI Appendix, Figs. S26 and S27). Thus, spermidine and arginine potentiate MAMP-induced ROS bursts in both plant species, and spermidine modulates the flg22-induced Ca2+ burst, at least in A. thaliana. These results are consistent with previously proposed functions of polyamines in plant defense responses (39, 40), including up-regulation of polyamine biosynthesis in plants following challenge with bacterial pathogens (44–47), subsequent polyamine transport to the apoplast (44), and increased resistance toward Pseudomonas viridiflava observed after exogenous addition of spermine (47).
Fig. 5.
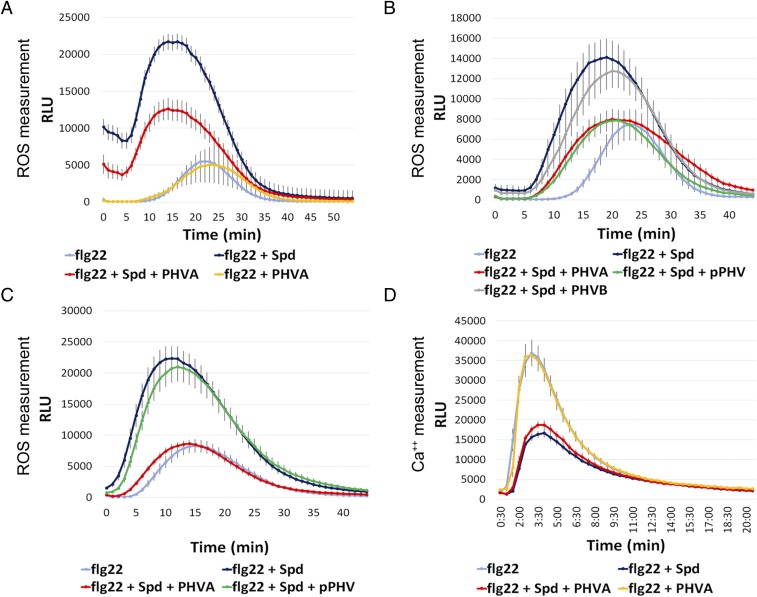
Effect of the phevamines and prephevamine on the spermidine potentiation of the ROS burst and suppression of the calcium burst. (A) Phevamine A suppresses the spermidine potentiation of the flg22-induced ROS burst in N. benthamiana, but does not affect the flg22-induced ROS burst directly. Leaf disks were treated with 50 nM flg22, with or without spermidine at 400 µM, and with or without phevamine A at 400 µM. (B) Phevamine A and prephevamine suppress the spermidine potentiation in N. benthamiana, but phevamine B does not. Leaf disks were treated with flg22 at 10 nM, spermidine, phevamines, and prephevamine were used at 300 µM. (C) In Arabidopsis, phevamine A suppresses the spermidine potentiation, while prephevamine does not. Leaf disks were treated with flg22 at 10 nM, spermidine and phevamines were used at 300 µM. (D) Phevamine A does not affect the flg22-induced Ca2+ burst, or the reduction of this burst by spermidine. Leaf disks were treated with flg22 at 20 nM, spermidine and phevamine A were used at 300 µM. Error bars represent SEs. PHVA, phevamine A; PHVB, phevamine B; pPHV, prephevamine; RLU, relative luminescence units; Spd, spermidine. All experiments presented here were repeated at least three times.
Phevamine A Suppresses Spermidine and Arginine Potentiation of the flg22-Induced ROS Burst.
The anticorrelation of phevamine A- and phaseolotoxin-encoding genes suggested that phevamines might impair the effect of spermidine and arginine on the early MAMP-dependent ROS burst. We therefore tested whether phevamine A could impair the effect of exogenously supplied spermidine and arginine in the MTI-induced ROS burst potentiation assay. Leaf disks of N. benthamiana were challenged with flg22 in the presence of phevamine A, spermidine, or both (Fig. 5A). We observed that phevamine A suppressed the spermidine potentiation of the flg22-induced ROS burst in N. benthamiana, but did not directly affect the flg22 response (Fig. 5A). We noted that prephevamine also suppressed the spermidine potentiation of the flg22-induced ROS burst, but phevamine B did not (Fig. 5B). This result is consistent with our observation that phevamine B is not naturally produced by Pto (SI Appendix, Fig. S6). In contrast, only phevamine A suppressed the spermidine-mediated flg22-induced ROS burst in A. thaliana leaf disks in a similar experimental setup (Fig. 5C). This indicates that the molecular mechanism(s) of the spermidine potentiation might differ slightly between N. benthamiana and A. thaliana.
Spermidine affects both the flg22-induced ROS and Ca2+ bursts (Fig. 5D and SI Appendix, Fig. S24C). We therefore tested the effect of phevamine A on the flg22-induced Ca2+ burst in the presence and absence of spermidine using a transgenic A. thaliana line that expresses the aequorin reporter (48). As noted above, spermidine suppressed the flg22-induced Ca2+ burst, but the addition of phevamine A did not affect this suppression. Phevamine A also had no direct effect on the Ca2+ burst induced by flg22 (Fig. 5D). These data suggest that phevamine A acts downstream of the Ca2+ burst.
Additionally, both phevamine A and prephevamine were tested for their effect on the ROS potentiation mediated by arginine in both N. benthamiana and A. thaliana. Phevamine A, but not prephevamine, could suppress the arginine-mediated potentiation of the flg22-induced ROS burst in N. benthamiana (SI Appendix, Fig. S27). This may imply that the potentiation mediated by spermidine and arginine in N. benthamiana involves slightly divergent molecular mechanism(s), supported by the different potentiation phenotypes for prephevamine. Similarly, only phevamine A and not prephevamine could inhibit the arginine potentiation of the flg22-induced ROS burst in A. thaliana.
Overall, we observed that phevamine A consistently suppressed both the spermidine- and arginine-mediated potentiation of the flg22-induced ROS burst across two divergent plant species. Thus, phevamine A is likely the most physiologically relevant small molecule produced by the hsv operon in Pto.
Conclusions
We identify a bacterial small-molecule virulence factor, phevamine A. The biosynthesis of phevamine A is controlled by the HrpL virulence regulator in P. syringae. Interestingly, phevamine A shares structural similarity with insect polyamine toxins: argiotoxin from the orb-weaver spider, and philanthotoxin from the Egyptian solitary wasp, both of which contain a polyamine and two amino acids, and are powerful neurotoxins that target ion channels (49, 50). The structural similarity suggests related mechanisms, which will be the focus of future studies.
The importance of phevamine A in bacterial virulence has been demonstrated in two different pathogens, E. amylovora (16) and P. syringae in this study. We show that the hsv operon and purified phevamine A can suppress both early and late markers of plant immune responses. Specifically, the hsv operon suppresses T3SS/harpin-induced callose deposition and host cell death. The phylogenetic distribution of the hsv operon suggested that phevamine A is a conserved virulence factor among bacterial phytopathogens. The anticorrelation of hsv and the biosynthetic genes for phaseolotoxin within a closely related clade of P. syrinage pv. actinidiae genomes suggested functional redundancy between phevamine A and phaseolotoxin. We demonstrated that phevamine A impairs plant immune signal potentiation by polyamines and arginine, while it is known that phaseolotoxin targets the synthesis of polyamines and arginine.
Our discovery of phevamine A also provided the opportunity to describe functions for polyamines in the potentiation of the MAMP-induced ROS burst induced by flg22, expanding the role of polyamines as important defense mediators. Our results are consistent with a model (SI Appendix, Fig. S28) in which P. syringae uses either phaseolotoxin or phevamine A to dampen host immune output by altering polyamine biosynthesis or the signaling capacity of polyamines, respectively. We speculate that components of the T3SS and/or harpins may be recognized at the cell surface, acting as elicitors of MTI subject to polyamine potentiation and suppression by phevamine A.
We show that two ATP-grasp enzymes are responsible for assembling phevamine A. These enzymes belong to a large superfamily of enzymes that catalyze amide bond formation by activating carboxylic acids through phosphorylation (51). ATP-grasp–type enzymes are widely used by bacteria for the synthesis of diverse small molecules, including the virulence factors phaseolotoxin, dapdiamide, and mangotoxin (52–54). Studies of these enzymes in a marine proteobacterium and a soil actinomycete have led to the discovery of small molecules with distinct structures but unknown functions (55, 56). Still, most biosynthetic operons containing ATP-grasp–type enzymes have eluded genome-guided discovery. Our study suggests that targeted mining of uncharacterized ATP-grasp enzymes will likely lead to novel small molecules.
Our approach for identifying cryptic small molecules builds on the rich knowledge of pathogen gene expression and integrates comparative genomic analysis, biochemical enzyme reconstitution, and physiological assays. Bacterial genomes harbor many biosynthetic operons of unknown function; therefore, this approach holds potential for identifying many more small molecules essential for bacteria–host interactions.
Methods
Metabolite Extraction for Metabolomics.
E. coli, P. syringae, and P. fluorescens cultures (100 mL) were spun down at 3,500 × g and 4 °C for 10 min. Solvent extractions were performed using an equal volume of supernatant to chloroform-methanol (2:1:1). The top aqueous layer was separated into a round-bottom flask and concentrated under vacuum. The dried-down material was resuspended and transferred to a small glass vial, concentrated under reduced pressure, and stored at −20 °C until MS analysis.
MS and MS/MS Analysis.
Culture extracts, in vitro synthesized phevamines, and chemically synthesized phevamine A, were analyzed by Agilent 6520 accurate-mass quadrupole-time of flight (Q-TOF) LC/MS using a Phenomenex Kinetex 5-µm C18 column (100 Å, 150 mm × 4.60 mm). Mobile phases were water and acetonitrile, each containing 0.1% formic acid. The gradient was held at 2% acetonitrile for 2 min before ramping up to 45% acetonitrile over 17 min at a flow rate of 0.4 mL/min. For each sample, m/z 434.324 was selected at a retention time of 8 ± 5 min and analyzed by MS fragmentation. Targeted MS/MS was conducted using 30-V collision energy, 970.9-ms/s acquisition time, and a 4 m/z isolation width.
Preparative Scale in Vitro Enzymatic Synthesis of Phevamines.
One milliliter, one-pot enzymatic assays for phevamine A were conducted at room temperature for 2 h. Assays contained 5 mM l-arginine, 5 mM spermidine, 1 mM l-valine, 0.5 mM l-phenylalanine, 100 mM Hepes (pH 7.5), 2 mM MgCl2, 1 mM ATP, 20 µM HsvA, 10 µM HsvB, 10 µM HsvC, and water. The reaction was then quenched with 1 mL acetonitrile to precipitate protein overnight at −20 °C. The precipitated proteins were removed by centrifugation. Phevamines were purified from the supernatant using one round of preparative HPLC, and two rounds of analytical HPLC using conditions described in SI Appendix, Materials and Methods.
Plant Material and Bacterial Growth.
A. thaliana was grown in walk-in growth rooms maintained at 21 °C/18 °C (day/night) with a 9-h/15-h (day/night) cycle. N. benthamiana was grown in a walk-in growth room maintained at 26 °C/22 °C with a 12-h/12-h (day/night) cycle and a LGM550 professional LED grow light system (LED Grow Master Global). For maintenance and transformation, Pseudomonas strains were grown in King’s B media at 28 °C. For infiltration or dipping in planta, Pseudomonas strains were grown in liquid culture overnight with the appropriate antibiotics, then plated on a Petri dish and incubated overnight before resuspension in 10 mM MgCl2.
Bacterial Growth Assay.
Pto, Pto∆hsv, Pto-Cor−, and Pto-Cor−∆hsv cells were resuspended in 10 mM MgCl2 to a final concentration of 2 × 105 cfu/mL. Twenty-day-old A. thaliana Col-0 seedlings were dipped in bacterial solutions supplemented with Silwet L-77 (Momentive), and growth was assessed at day 0 and 3-d postinfection as described by Tornero and Dangl (57). Eight samples containing three seedlings were collected for each treatment.
Callose Deposition Measurement.
Three- to 5-wk-old Arabidopsis plants were infiltrated with Pseudomonas strains at an OD600 of 0.2 in 10 mM MgCl2 and collected after about 20 h. To visualize callose deposition, leaves were stained with aniline blue (58). The tissue was cleared and dehydrated with 96% ethanol overnight at 37 °C. Cleared leaves were washed with distilled water and then stained in 0.01% aniline blue in 150 mM K2HPO4 (pH 9.5) for 4 h at room temperature. Stained samples were washed and mounted in distilled water and examined by epifluorescence (LEICA M205 FA) with 100× magnification. Images were taken at the region below the infiltrated zone of each leaf. Counting of accumulated callose foci was carried out using ImageJ (NIH). For each treatment, 10–20 leaves were processed.
Phylogenetic Analysis.
The phylogenetic history of P. syringae was inferred from Yang et al. (37). Protein sequences of toxin biosynthetic genes listed in Dataset S1 were used as basic local alignment search tool (BLAST) queries to search for homologous protein sequences in the 292 P. syringae genomes [available as of January 2017, Pathosystems Resource Integration Center (PATRIC)]. A hit with over 80% protein sequence identity was considered positive. The tree was visualized using interactive tree of life (iTOL) (59).
ROS Burst Measurement.
Leaf disks from 4-wk-old A. thaliana Col-0, or N. benthamiana were placed into a 96-well plate with 100 µl of water in each well. Twelve leaf disks were used per treatment. After overnight incubation for Arabidopsis leaf disks, or 20 h for N. benthamiana leaf disks, each sample was treated with 100 µl of reaction mix, including 17 mg/mL of luminol (Sigma), 10 mg/mL of horseradish peroxidase (Sigma), distilled water, flg22 at concentrations ranging from 5 to 100 nM, l-arginine, and polyamines at concentrations typically ranging from 200 to 800 µM. Phevamines and prephevamine were used at the same concentration as l-arginine (Sigma) or spermidine. Luminescence was measured immediately with 0.5-s integration and 1-min interval over 45 min using a SpectraMax L (Molecular Devices). For each experiment, 8–12 leaf-disks per condition were monitored. The flg22 peptide (QRLSTGSRINSAKDDAAGLQIA) was synthesized by Genscript (41).
Supplementary Material
Acknowledgments
We thank Dr. Albert Bowers for helpful discussions of the manuscript, Dr. Gary Pielak for the naming of the phevamines, Dr. Jillian Tyrrell for cloning the hsv operon into the pLIC-His vector, Dr. Jake Malone for sharing the pBBR5 plasmid, Dr. Jim Jorgenson and Katherine Simpson for helpful discussion of isolation of phevamines, and Kevin Santa Maria for assistance with MultiGeneBlast. This work is supported by the Rita Allen Foundation and the David and Lucile Packard Foundation (B.L.), National Institutes of Health (R00 GM099904 to B.L., 5T32 GM008500 to J.A.B., and R01 GM112739-01 to F.C.S.), the National Science Foundation (IOS-1257373 to J.L.D.), and the Howard Hughes Medical Institute. J.L.D. is an Investigator of the Howard Hughes Medical Institute, and F.C.S. is a Faculty Scholar of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803779115/-/DCSupplemental.
References
- 1.Bender CL, Alarcón-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–292. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katagiri F, Thilmony R, He SY. The Arabidopsis thaliana-Pseudomonas syringae interaction. Arabidopsis Book. 2002;1:e0039. doi: 10.1199/tab.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Geng X, Jin L, Shimada M, Kim MG, Mackey D. The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta. 2014;240:1149–1165. doi: 10.1007/s00425-014-2151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson A, Johnston J. Phaseolotoxin: Chlorosis, ornithine accumulation and inhibition of ornithine carbamoyltransferase in different plants. Physiol Plant Pathol. 1980;16:269–275. [Google Scholar]
- 5.Bachmann A, Matile P, Slusarenko A. Inhibition of ornithine decarboxylase activity by phaseolotoxin: Implications for symptom production in halo blight of French bean. Physiol Mol Plant Pathol. 1998;53:287–299. [Google Scholar]
- 6.Gulick AM. Nonribosomal peptide synthetase biosynthetic clusters of ESKAPE pathogens. Nat Prod Rep. 2017;34:981–1009. doi: 10.1039/c7np00029d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross H, Loper JE. Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep. 2009;26:1408–1446. doi: 10.1039/b817075b. [DOI] [PubMed] [Google Scholar]
- 8.Gimenez-Ibanez S, Chini A, Solano R. How microbes twist jasmonate signaling around their little fingers. Plants (Basel) 2016;5:9. doi: 10.3390/plants5010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toruño TY, Stergiopoulos I, Coaker G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol. 2016;54:419–441. doi: 10.1146/annurev-phyto-080615-100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Feng B, He P, Shan L. From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol. 2017;55:109–137. doi: 10.1146/annurev-phyto-080516-035649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fouts DE, et al. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc Natl Acad Sci USA. 2002;99:2275–2280. doi: 10.1073/pnas.032514099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mucyn TS, et al. Variable suites of non-effector genes are co-regulated in the type III secretion virulence regulon across the Pseudomonas syringae phylogeny. PLoS Pathog. 2014;10:e1003807. doi: 10.1371/journal.ppat.1003807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weingart H, Stubner S, Schenk A, Ullrich MS. Impact of temperature on in planta expression of genes involved in synthesis of the Pseudomonas syringae phytotoxin coronatine. Mol Plant Microbe Interact. 2004;17:1095–1102. doi: 10.1094/MPMI.2004.17.10.1095. [DOI] [PubMed] [Google Scholar]
- 14.Lan L, Deng X, Zhou J, Tang X. Genome-wide gene expression analysis of Pseudomonas syringae pv. tomato DC3000 reveals overlapping and distinct pathways regulated by hrpL and hrpRS. Mol Plant Microbe Interact. 2006;19:976–987. doi: 10.1094/MPMI-19-0976. [DOI] [PubMed] [Google Scholar]
- 15.Oh CS, Beer SV. Molecular genetics of Erwinia amylovora involved in the development of fire blight. FEMS Microbiol Lett. 2005;253:185–192. doi: 10.1016/j.femsle.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 16.Oh CS, Kim JF, Beer SV. The Hrp pathogenicity island of Erwinia amylovora and identification of three novel genes required for systemic infection. Mol Plant Pathol. 2005;6:125–138. doi: 10.1111/j.1364-3703.2005.00269.x. [DOI] [PubMed] [Google Scholar]
- 17.Kvitko BH, et al. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez-Gómez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999;18:277–284. doi: 10.1046/j.1365-313x.1999.00451.x. [DOI] [PubMed] [Google Scholar]
- 19.Bowles DJ. Defense-related proteins in higher plants. Annu Rev Biochem. 1990;59:873–907. doi: 10.1146/annurev.bi.59.070190.004301. [DOI] [PubMed] [Google Scholar]
- 20.Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci USA. 2003;100:8577–8582. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci USA. 2004;101:9927–9932. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Torres M, et al. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. Plant J. 2006;47:368–382. doi: 10.1111/j.1365-313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- 23.Nomura K, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 24.Underwood W, Zhang S, He SY. The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J. 2007;52:658–672. doi: 10.1111/j.1365-313X.2007.03262.x. [DOI] [PubMed] [Google Scholar]
- 25.Cunnac S, et al. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci USA. 2011;108:2975–2980. doi: 10.1073/pnas.1013031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silby MW, et al. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009;10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas WJ, Thireault CA, Kimbrel JA, Chang JH. Recombineering and stable integration of the Pseudomonas syringae pv. syringae 61 hrp/hrc cluster into the genome of the soil bacterium Pseudomonas fluorescens Pf0-1. Plant J. 2009;60:919–928. doi: 10.1111/j.1365-313X.2009.03998.x. [DOI] [PubMed] [Google Scholar]
- 28.Oh H-S, Park DH, Collmer A. Components of the Pseudomonas syringae type III secretion system can suppress and may elicit plant innate immunity. Mol Plant Microbe Interact. 2010;23:727–739. doi: 10.1094/MPMI-23-6-0727. [DOI] [PubMed] [Google Scholar]
- 29.He SY, Huang H-C, Collmer A. Pseudomonas syringae pv. syringae harpinPss: A protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 30.Charkowski AO, et al. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J Bacteriol. 1998;180:5211–5217. doi: 10.1128/jb.180.19.5211-5217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fawaz MV, Topper ME, Firestine SM. The ATP-grasp enzymes. Bioorg Chem. 2011;39:185–191. doi: 10.1016/j.bioorg.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael AJ, Furze JM, Rhodes MJ, Burtin D. Molecular cloning and functional identification of a plant ornithine decarboxylase cDNA. Biochem J. 1996;314:241–248. doi: 10.1042/bj3140241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamana K, Sakamoto A, Tachiyanagi S, Terauchi E. Polyamine profiles of some members of the gamma subclass of the class Proteobacteria: Polyamine analysis of twelve recently described genera. Microbiol Cult Collect. 2003;19:3–11. [Google Scholar]
- 34.Medema MH, Takano E, Breitling R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol Biol Evol. 2013;30:1218–1223. doi: 10.1093/molbev/mst025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baltrus DA, et al. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011;7:e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCann HC, et al. Genomic analysis of the Kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 2013;9:e1003503, and erratum (2013) 9:10.1371/annotation/af157ddc-200a-4105-b243-3f01251cc677. doi: 10.1371/journal.ppat.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L, et al. Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe. 2017;21:156–168. doi: 10.1016/j.chom.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell RE. Isolation and structure of a chlorosis-inducing toxin of Pseudomonas phaseolicola. Phytochemistry. 1976;15:1941–1947. [Google Scholar]
- 39.Hussain SS, Ali M, Ahmad M, Siddique KH. Polyamines: Natural and engineered abiotic and biotic stress tolerance in plants. Biotechnol Adv. 2011;29:300–311. doi: 10.1016/j.biotechadv.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Jiménez-Bremont JF, et al. Physiological and molecular implications of plant polyamine metabolism during biotic interactions. Front Plant Sci. 2014;5:95. doi: 10.3389/fpls.2014.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 42.Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunze G, et al. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell. 2004;16:3496–3507. doi: 10.1105/tpc.104.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoda H, et al. Polyamines as a common source of hydrogen peroxide in host- and nonhost hypersensitive response during pathogen infection. Plant Mol Biol. 2009;70:103–112. doi: 10.1007/s11103-009-9459-0. [DOI] [PubMed] [Google Scholar]
- 45.Ward JL, et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 2010;63:443–457. doi: 10.1111/j.1365-313X.2010.04254.x. [DOI] [PubMed] [Google Scholar]
- 46.Lou Y-R, Bor M, Yan J, Preuss AS, Jander G. Arabidopsis NATA1 acetylates putrescine and decreases defense-related hydrogen peroxide accumulation. Plant Physiol. 2016;171:1443–1455. doi: 10.1104/pp.16.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez ME, et al. Perturbation of spermine synthase gene expression and transcript profiling provide new insights on the role of the tetraamine spermine in Arabidopsis defense against Pseudomonas viridiflava. Plant Physiol. 2011;156:2266–2277. doi: 10.1104/pp.110.171413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knight H, Trewavas AJ, Knight MR. Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell. 1996;8:489–503. doi: 10.1105/tpc.8.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson JK, Frølund SU, Tikhonov DB, Kristensen AS, Strømgaard K. Synthesis and biological activity of argiotoxin 636 and analogues: Selective antagonists for ionotropic glutamate receptors. Angew Chem Int Ed Engl. 2009;48:3087–3091. doi: 10.1002/anie.200805426. [DOI] [PubMed] [Google Scholar]
- 50.Kromann H, et al. Solid-phase synthesis of polyamine toxin analogues: Potent and selective antagonists of Ca2+-permeable AMPA receptors. J Med Chem. 2002;45:5745–5754. doi: 10.1021/jm020314s. [DOI] [PubMed] [Google Scholar]
- 51.Ogasawara Y, Dairi T. Biosynthesis of oligopeptides using ATP‐grasp enzymes. Chemistry. 2017;23:10714–10724. doi: 10.1002/chem.201700674. [DOI] [PubMed] [Google Scholar]
- 52.Hollenhorst MA, Clardy J, Walsh CT. The ATP-dependent amide ligases DdaG and DdaF assemble the fumaramoyl-dipeptide scaffold of the dapdiamide antibiotics. Biochemistry. 2009;48:10467–10472. doi: 10.1021/bi9013165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arai T, Kino K. A novel L-amino acid ligase is encoded by a gene in the phaseolotoxin biosynthetic gene cluster from Pseudomonas syringae pv. phaseolicola 1448A. Biosci Biotechnol Biochem. 2008;72:3048–3050. doi: 10.1271/bbb.80439. [DOI] [PubMed] [Google Scholar]
- 54.Carrión VJ, Arrebola E, Cazorla FM, Murillo J, de Vicente A. The mbo operon is specific and essential for biosynthesis of mangotoxin in Pseudomonas syringae. PLoS One. 2012;7:e36709. doi: 10.1371/journal.pone.0036709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blasiak LC, Clardy J. Discovery of 3-formyl-tyrosine metabolites from Pseudoalteromonas tunicata through heterologous expression. J Am Chem Soc. 2010;132:926–927. doi: 10.1021/ja9097862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogasawara Y, et al. Exploring peptide ligase orthologs in Actinobacteria: Discovery of pseudopeptide natural products, ketomemicins. ACS Chem Biol. 2016;11:1686–1692. doi: 10.1021/acschembio.6b00046. [DOI] [PubMed] [Google Scholar]
- 57.Tornero P, Dangl JL. A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 2001;28:475–481. doi: 10.1046/j.1365-313x.2001.01136.x. [DOI] [PubMed] [Google Scholar]
- 58.Kim MG, et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–759. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 59.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carrión VJ, et al. The mangotoxin biosynthetic operon (mbo) is specifically distributed within Pseudomonas syringae genomospecies 1 and was acquired only once during evolution. Appl Environ Microbiol. 2013;79:756–767. doi: 10.1128/AEM.03007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.